ABSTRACT
The measles virus hemagglutinin (MeV-H) protein is the main target of protective neutralizing antibodies. Using a panel of monoclonal antibodies (MAbs) that recognize known major antigenic sites in MeV-H, we identified a D4 genotype variant that escapes neutralization by MAbs targeting the neutralizing epitope (NE) antigenic site. By site-directed mutagenesis, L249P was identified as the critical mutation disrupting the NE in this genotype D4 variant. Forty-two available D4 genotype gene sequences were subsequently analyzed and divided into 2 groups according to the presence or absence of the L249P MeV-H mutation. Further analysis of the MeV-N gene sequences of these 2 groups confirmed that they represent clearly definable, sequence-divergent D4 subgenotypes, which we named subgenotypes D4.1 and D4.2. The subgenotype D4.1 MeVs were isolated predominantly in Kenya and Ethiopia, whereas the MAb-resistant subgenotype D4.2 MeVs were isolated predominantly in France and Great Britain, countries with higher vaccine coverage rates. Interestingly, D4.2 subgenotype viruses showed a trend toward diminished susceptibility to neutralization by human sera pooled from approximately 60 to 80 North American donors. Escape from MAb neutralization may be a powerful epidemiological surveillance tool to monitor the evolution of new MeV subgenotypes.
IMPORTANCE Measles virus is a paradigmatic RNA virus, as the antigenic composition of the vaccination has not needed to be updated since its discovery. The vaccine confers protection by inducing neutralizing antibodies that interfere with the function of the hemagglutinin protein. Viral strains are indistinguishable serologically, although characteristic nucleotide sequences differentiate 24 genotypes. In this work, we describe a distant evolutionary branch within genotype D4. Designated subgenotype D4.2, this virus is distinguishable by neutralization with vaccine-induced monoclonal antibodies that target the neutralizing epitope (NE). The subgenotype D4.2 viruses have a higher predominance in countries with intermediary levels of vaccine coverage. Our studies demonstrate that subgenotype D4.2 lacks epitopes associated with half of the known antigenic sites, which significantly impacts our understanding of measles virus evolution.
KEYWORDS: virus evolution, antibody-mediated neutralization, antigenic variation, measles virus hemagglutinin, viral epitopes, measles virus genotypes, immune evasion
INTRODUCTION
Measles ranks as one of the most deadly diseases in the history of humankind (1, 2). The advent of an effective measles virus (MeV) vaccine 46 years ago dramatically reduced the number of measles deaths (3), making the MeV live-attenuated vaccine one of the most successful public health interventions ever undertaken (4, 5). Nonetheless, measles is still a leading cause of death globally for children younger than 5 years of age and was responsible for 134,200 deaths in 2015 (6). All 194 WHO member states have committed to reducing measles deaths, and the Global Measles and Rubella Strategic Plan set the goal of measles elimination by 2020 through increased vaccination coverage (7). In this regard, measles cases among vaccinated people have raised concerns about waning immunity in vaccinees and the occurrence of antigenic changes in currently circulating strains, raising concerns as to whether MeV elimination can be achieved (8–12).
MeV belongs to the family Paramyxoviridae and carries a nonsegmented negative-strand RNA genome tightly encapsidated by a helically arranged nucleocapsid (MeV-N) protein and packaged in a lipoprotein envelope (13). Two transmembrane glycoproteins are found in the virion, the MeV hemagglutinin (MeV-H) protein and the MeV F (MeV-F) protein. The former is responsible for receptor attachment and has a fusion support function when coexpressed with the latter (14, 15). Neutralizing antibodies to either of these antigens inhibit MeV infection by preventing the interaction of the MeV-H protein with its cellular receptor(s) and by blocking fusion activity (16, 17). Although both cellular and humoral immune responses are important during MeV infection, they have different effects. Antibodies to MeV-H and MeV-F are essential for protection, as recently demonstrated by the absence of protection in macaques with an MeV-specific T-cell response but without neutralizing antibodies (18).
Despite the fact that ex vivo studies confirm that the MeV polymerase mutation rate is high, similarly to other RNA viruses, MeV is considered antigenically stable and has only 1 serotype (19–21). Nevertheless, sequence analysis of the two most variable MeV genes, MeV-H and MeV-N, in naturally occurring field isolates has enabled MeV to be classified into 24 genotypes (22). During a recent genotype B3.1 measles outbreak, we observed a gradual nucleotide divergence in the MeV-H gene over a 6-month period and estimated a mutation rate of 2.66 × 10−3 substitutions per site per year (23), similar to the rates of antigenic drift in other RNA viruses (24, 25). In contrast, no changes were observed in the hypervariable carboxy end of the MeV-N gene or the gene encoding the other important surface antigen, MeV-F (23). This observation could open the question of immune-driven evolution into the major surface antigen MeV-H. Certainly, the introduction of the live-attenuated MeV vaccine not only has led to a dramatic decrease in MeV incidence but also has been accompanied by changes in the global distribution of measles virus genotypes (26, 27).
Currently, six MeV genotypes (genotypes B1, C1, D1, E, F, and G1) are considered extinct, and five other genotypes (genotypes D2, D3, D10, G2, and H2) have not been detected since 2006. Furthermore, all MeV isolates detected in the past 5 years belong to only seven genotypes and show some geographic restriction: genotypes B3, D4, D6, D8, D9, G3, and H1 (27–29). Because measles vaccination with a genetically restricted strain (genotype A) has been used throughout the world for 50 years, changes in genotype circulation patterns might reflect the immune selection of “fitter” viruses (30, 31). Antigenic differences of MeVs of various genotypes have been detected by using monoclonal antibodies (MAbs) and polyvalent antisera from vaccinees to identify differences in neutralization titers against certain wild-type viruses (32–36; M. A. Muñoz-Alía, J. Carabaña, A. Serrano-Pardo, R. Porras-Mansilla, C. Santiago, J. M. Casasnovas, M. L. Celma, and R. Fernández-Muñoz, presented at the X Spanish National Conference on Virology, Salamanca, Spain, 21 to 24 June 2009). Nevertheless, despite these subtle differences, all contemporary wild-type strains of MeV are readily neutralized in vitro with polyclonal serum from vaccinees (34, 37, 38).
During our studies on antigenic variation across MeV genotypes, we identified a difference in the neutralization sensitivities of two viruses belonging to the D4 genotype. Subsequent genetic and antigenic analyses of a larger number of genotype D4 viruses identified two definable D4 subgenotypes, which we named D4.1 and D4.2. In contrast to subgenotype D4.1 viruses, subgenotype D4.2 viruses are not neutralized by antibodies targeting the neutralizing epitope (NE), indicating that they lack three of the six known antigenic sites. Perhaps more significantly, the two subgenotypes differ in their susceptibilities to neutralization by pooled human sera from 60 to 80 North American donors.
RESULTS
Discrimination between genotype D4 viruses using antibodies targeting the NE.
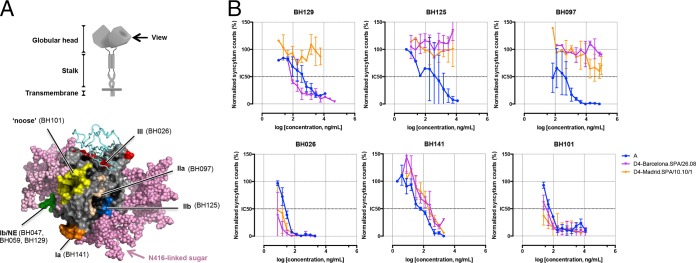
During our studies on antigenic variation between MeV genotypes, we noted that two MeV strains isolated in Spain and defined as belonging to genotype D4 showed distinct sensitivities to antibody-mediated neutralization. To ascertain a role of substitutions in MeV-H, we generated enhanced green fluorescent protein (EGFP)-expressing recombinant MeV in which the MeV-H gene was replaced with that of two Spanish genotype D4 isolates, Barcelona.SPA/26.08 and Madrid.SPA/10.10/1. We again tested their neutralization sensitivities with a panel of anti-MeV-H antibodies against known major antigenic sites in MeV-H (Fig. 1A). The results presented in Fig. 1B show that the BH026, BH141, and BH101 MAbs completely inhibited infection by both genotype D4 viruses and the vaccine strain (genotype A), used as a control for neutralization. In contrast, discrimination between the MeV vaccine strain and the genotype D4 viruses was observed when the BH125 and BH097 MAbs were tested instead. Surprisingly, the two genotype D4 viruses showed markedly different neutralization profiles when the NE-targeting BH129 MAb was used. Although the BH129 MAb completely blocked infection by the Barcelona.SPA/26.08 virus, it was ineffective against the genotype D4 Madrid.SPA/10.10/1 virus.
FIG 1.
Neutralization activity of a panel of anti-MeV-H MAbs against 2 genotype D4 viruses. (A, top) Diagram of the MeV-H homodimeric structure. (Bottom) Radiographic crystallographic structure of the MeV-H globular head showing the location of the 6 antigenic sites. Antigenic sites are color-coded (orange, Ia; green, Ib or NE; wheat, IIa; blue, IIb; red, III; yellow, noose), indicating a representative antibody target. The signaling lymphocytic activation molecule immunoglobulin V domain and modeled N-linked sugars are shown as cyan ribbons and pink spheres, respectively. A side view is shown, as indicated at the top. The N416-linked sugar present in genotype D4 is highlighted. (B) Virus neutralization assay. Recombinant measles virus expressing 2 different MeV-H genotype D4 sequences or genotype A (vaccine strain) was incubated in the absence or presence of the indicated neutralizing antibody for 1 h at 37°C. Two days later, the numbers of enhanced green fluorescent protein-expressing foci in the presence and absence of MAbs were counted and compared. Neutralization is plotted as a percentage of the control for residual infection (y axis) by MAb concentration (x axis). For a given MAb-virus pair, the data represent the geometric means of results from at least 2 independent experiments performed in quadruplicate. NE, neutralizing epitope.
Two sequence-divergent D4 subgenotypes defined within genotype D4.
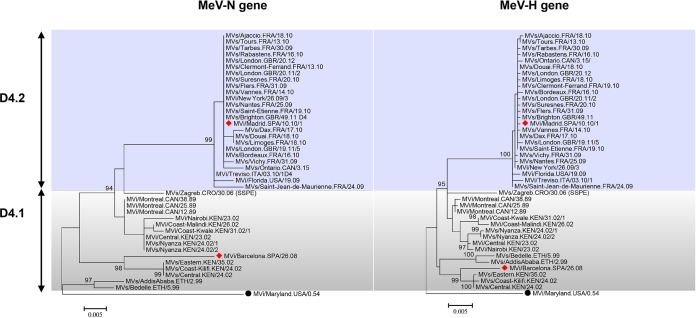
Having shown that the two genotype D4 Spanish isolates showed differing neutralization patterns by anti-MeV-H MAbs, we sought to determine the broader significance of this finding for other members of genotype D4. A new MeV genotype is typically defined by 2.5% and 2.0% nucleotide divergences in the MeV-N and MeV-H genes, respectively (85). We therefore retrieved both MeV-N and MeV-H gene sequences available for MeV isolates classified as belonging to genotype D4 from GenBank (n = 41) (Table 1). After removing redundant sequences, we reconstructed phylogenetic trees for both genes and rooted them on the reference strain for genotype A (MVi/Maryland.USA/0.54). Phylogenetic analysis showed that the two Spanish genotype D4 viruses were indeed representative of two definable subgenotypes, which were supported by high bootstrap values (Fig. 2). The average number of base substitutions per site for the MeV-N gene between the 2 subgenotypes was 4.0% (±0.8%), ranging from 2.5% to 6.5%. The nucleotide sequence divergence for the MeV-H gene was 2.3% (±0.2%) (range, 1.5% to 3.0%). These results show high intragenotypic variation for the sequences assigned to genotype D4 but do not support the designation of 2 different genotypes. Here, we distinguish subgenotype 1 (D4.1) and subgenotype 2 (D4.2).
TABLE 1.
List of MeV sequences used in the course of this work
| Genotype | WHO strain designation | Material | GenBank accession no. |
Reference | |
|---|---|---|---|---|---|
| MeV-N | MeV-H | ||||
| A | MVi/Maryland.USA/0.54 | Cell culture | U01987 | U03669 | 80 |
| D4 | MVi/Montreal.CAN/08.89 | Cell culture | U01976 | AF079554 | 80 |
| D4 | MVi/Montreal.CAN/12.89 | Cell culture | AF410990 | AF410975 | 81 |
| D4 | MVi/Montreal.CAN/25.89 | Cell culture | AF410991 | AF410976 | 81 |
| D4 | MVs/Brighton.GBR/49.11 | Cell culture | KT732227 | KT732227 | 82 |
| D4 | MVs/London.GBR/20.12 | Oral fluid | KT732229 | KT732229 | 82 |
| D4 | MVs/London.GBR/20.11/2 | Oral fluid | KT732226 | KT732226 | 82 |
| D4 | MVs/London.GBR/19.11/5 | Oral fluid | KT732225 | KT732225 | 82 |
| D4 | MVi/Treviso.ITA/03.10/1 | Cell culture (urine) | KC164757.1 | KC164757.1 | E. Franchin et al.a |
| D4 | MVi/New York/26.09/3 | Not specified | JN635402.1 | JN635402.1 | E. F. Kirkness et al.b |
| D4 | MVi/Florida.USA/19.09 | Not specified | JN635403.1 | JN635403.1 | E. F. Kirkness et al.b |
| D4 | MVi/Central.KEN/23.02 | Not specified | AY249250 | AY249260 | 46 |
| D4 | MVi/Nairobi.KEN/23.02 | Not specified | AY249251 | AY249261 | 46 |
| D4 | MVs/Coast-Kilifi.KEN/24.02 | Not specified | AY249252 | AY249262 | 46 |
| D4 | MVs/Central.KEN/24.02 | Not specified | AY249253 | AY249263 | 46 |
| D4 | MVs/Nyanza.KEN/24.02/1 | Not specified | AY249254 | AY249264 | 46 |
| D4 | MVs/Nyanza.KEN/24.02/2 | Not specified | AY249259 | AY249269 | 46 |
| D4 | MVi/Coast-Kwale.KEN/31.02/1 | Not specified | AY249255 | AY249265 | 46 |
| D4 | MVs/Eastern.KEN/35.02 | Not specified | AY249257 | AY249267 | 46 |
| D4 | MVi/Coast-Malindi.KEN/26.02 | Not specified | AY249258 | AY249268 | 46 |
| D4 | MVs/Zagreb.CRO/30.06 (SSPE) | Brain tissue | FJ475060.1 | JX126962.1 | 83 |
| D4 | MVs/Bedelle.ETH/5.99 | Oral fluid | AF280800 | AF280805 | 84 |
| D4 | MVs/AddisAbaba.ETH/2.99 | Oral fluid | AF280802 | AF280807 | 84 |
| D4 | MVs/Ontario.CAN/3.15/ | Urine | KU218405.1 | KU218406.1 | J. Hiebert |
| D4 | MVs/Saint-Jean-de-Maurienne.FRA/24.09 | Oral fluid | KJ183791.1 | KJ183852.1 | J. Dina |
| D4 | MVs/Suresnes.FRA/20.10 | Oral fluid | KJ183846.1 | KJ183910.1 | J. Dina |
| D4 | MVs/Saint-Etienne.FRA/19.10 | Oral fluid | KJ183840.1 | KJ183904.1 | J. Dina |
| D4 | MVs/Limoges.FRA/18.10 | Oral fluid | KJ183838.1 | KJ183902.1 | J. Dina |
| D4 | MVs/Douai.FRA/18.10 | Oral fluid | KJ183837.1 | KJ183901.1 | J. Dina |
| D4 | MVs/Ajaccio.FRA/18.10 | Oral fluid | KJ183836.1 | KJ183900.1 | J. Dina |
| D4 | MVs/Rabastens.FRA/16.10 | Oral fluid | KJ183833.1 | KJ183896.1 | J. Dina |
| D4 | MVs/Dax.FRA/17.10 | Oral fluid | KJ183834.1 | KJ183898.1 | J. Dina |
| D4 | MVs/Bordeaux.FRA/16.10 | Oral fluid | KJ183831.1 | KJ183894.1 | J. Dina |
| D4 | MVs/Tours.FRA/13.10 | Oral fluid | KJ183821.1 | KJ183882.1 | J. Dina |
| D4 | MVs/Nantes.FRA/25.09 | Oral fluid | KJ183795.1 | KJ183856.1 | J. Dina |
| D4 | MVs/Flers.FRA/31.09/ | Oral fluid | KJ183818.1 | KJ183879.1 | J. Dina |
| D4 | MVs/Vichy.FRA/31.09/ | Oral fluid | KJ183819.1 | KJ183880.1 | J. Dina |
| D4 | MVs/Clermont-Ferrand.FRA/13.10 | Oral Fluid | KJ183824.1 | KJ183885.1 | J. Dina |
| D4 | MVs/Tarbes.FRA/30.09 | Oral fluid | KJ183813.1 | KJ183874.1 | J. Dina |
| D4 | MVs/Vannes.FRA/14.10 | Oral fluid | KJ183826.1 | KJ183887.1 | J. Dina |
| D4 | MVi/Barcelona.SPA/26.08 | Cell culture | KY524301 | KY524302 | This work |
| D4 | MVi/Madrid.SPA/10.10/1 | Cell culture (urine) | KY524303 | KY524304 | This work |
E. Franchin, F. Dal Bello, M. Pacenti, R. Cusinato, E. Lavezzo, L. Barzon, G. Palu.
E. F. Kirkness, R. Halpin, J. Bera, N. Fedorova, L. Overton, T. Stockwell, P. Amedeo, B. Bishop, H. Chen, P. Edworthy, N. Gupta, D. Katzel, K. Li, S. Schobel, S. Shrivastava, V. Thovarai, S. Wang, B. Bankamp, L. Byrd, W. Bellini, P. Rota.
FIG 2.
Phylogenetic relationships of the MeV-N gene (left) and MeV-H gene (right) of measles virus genotype D4. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model. Measles virus genotype A (MVi/Maryland.USA/0.54) was used for rooting the tree. Evolutionary analyses were conducted with MEGA 6.06. Statistical support for grouping is shown as bootstrap values (1,000 replicated). Values of >80% are indicated at the deep node. The bar represents the genetic distance. The subgenotype classification is illustrated. Sequences were retrieved from the National Center for Biotechnology Information (NCBI) GenBank database and are named according to World Health Organization guidelines (Table 1).
Evolving residues in the MeV-H protein define a subgenotype classification.
MeV-neutralizing antibodies are directed mainly against the MeV-H protein (16). Because of our identification of two subgenotypes in MeV genotype D4, we further investigated the differences observed in the MeV-H gene at the level of the resulting protein.
The predicted amino acid differences between the MeV-H proteins of the two subgenotypes were 2.0% (±0.2%), ranging from 1.2% to 4.0%. Of note, both subgenotypes were characterized by 2 well-defined consensus sequences with variations at positions 212, 240, 249, 303, 316, 359, 412, and 481 (Fig. 3). These residues, together with residues 305 and 562, were also identified as directionally evolving sites through convergent evolution under a directional evolution of protein sequences test (Bayes factor, >105) (86). Likewise, 5 pairs of interactions were detected (P < 0.05): N238T←→P276L, R34K←→T307A, F476L←→S590T, M333L←→I564L, and S318N←→F571S. All sites except position 571 were exposed on the protein surface (not shown). However, none of them showed close proximity in the two crystallographic forms resolved so far for MeV-H (form I and form II [not shown]) (39).
FIG 3.
Sequence alignment of the MeV-H genes of genotype D4 viruses. The GenBank accession numbers for the MeV-H gene sequences used in the sequence alignment are shown in Table 1.
These results prompted us to analyze selection pressures on the MeV-H gene among genotype D4 viruses. To estimate positively and negatively selected sites, we used the following four methods: single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), internal fixed-effects likelihood (IFEL), and mixed-effect model of evolution (MEME). Four positively selected sites were estimated by using the MEME method (Table 2), which detected residues under both pervasive positive selection and episodic selection (40): E85Q, L172M, V562F/A, and Q574K. In addition, 15 sites were under negative selection, 5 of which were common to all four methods (Table 2): 3P, 113N, 256E, 342D, and 347D. Except for sites at positions 172 and 256, they were located on the protein surface (not shown). We also calculated the combined synonymous and nonsynonymous rate to be 0.22 (95% confidence interval [CI], 0.17 to 0.29) with the SLAC method. Overall, these results establish a basis for the distinction of D4 subgenotypes.
TABLE 2.
Selected sites in the MeV-H genea
| Amino acid | Detection of site by method |
|||
|---|---|---|---|---|
| SLAC | FEL | IFEL | MEME | |
| Positively selected sites | ||||
| Glu85Glnb | • | |||
| Leu172Metb | • | |||
| Val562Phe/Alab | • | |||
| Gln575Lysb | • | |||
| Negatively selected sites | ||||
| 3Pro | • | • | • | |
| 113Asn | • | • | • | |
| 173Glu | • | |||
| 174Ala | • | • | ||
| 256Glu | • | • | • | |
| 342Asp | • | • | • | |
| 347Asp | • | • | • | |
| 393Leu | • | |||
| 405Asn | • | |||
| 445Lys | • | |||
| 497Pro | • | • | ||
| 522Leu | • | |||
| 572Thr | • | • | ||
| 606Cys | • | |||
| 610Arg | • | • | ||
Abbreviations: FEL, fixed-effects likelihood; IFEL, internal fixed-effects likelihood; MeV-H, measles virus hemagglutinin; MEME, mixed-effects model of episodic diversifying selection; SLAC, single-likelihood ancestor counting.
P value of <0.10.
An L249P amino acid mutation in MeV-H is the main driver for subgenotype D4.2 escape from NE-specific antibodies.
Apart from the BH129 MAb, the BH047 and BH059 MAbs have also been shown to bind a sequential epitope encompassing amino acids 240 through 250 of MeV-H (41). To confirm that antigenic drift of the NE site defines D4 subgenotypes, we studied the neutralization capacity of the other NE-targeting MAbs. We additionally tested different recombinant MeVs encoding the genotype-specific MeV-H genes of genotypes B3.1, C2, D4, D6, D7, D8, D9, G3, and H1 (Fig. 4A). As expected, none of the NE-targeting MAbs (BH129, BH047, and BH059) were able to inhibit infection by subgenotype D4.2 viruses, whereas all of them inhibited viruses of subgenotype D4.1. Surprisingly, these antibodies did not inhibit infection by recombinant MeV possessing the MeV-H gene of genotype D6, although all other genotypes were neutralized.
FIG 4.
Leu249Pro amino acid change as the driver mutation for escape of MeV subgenotype D4.2. (A) Neutralization activity of the NE-targeting antibodies against MeV genotypes. The neutralization assay was performed and data are represented as indicated in the legend of Fig. 1. Of note, only genotype D6 and the so-called subgenotype D4.2 viruses escape neutralization by all 3 antibodies. (B) Sequence alignment (amino acids 235 to 255) of MeV-H proteins among the MeV genotypes used in this study. (C) Effect of amino acid mutations on NE-targeting antibody neutralization. (D) NE-targeting antibody binding to MeV-H proteins. MAb binding was determined by an enzyme-linked immunosorbent assay after the addition of a nonsaturating concentration of MAbs to microtiter plates coated with the FLAG-tagged MeV-H protein. The amount of anti-FLAG antibody binding was used as a loading control. Binding values were normalized to those obtained with MeV-H genotype A. Means and standard deviations for a representative experiment performed in duplicate are shown.
To understand in greater detail the neutralization selectivity of these NE-targeting antibodies, we aligned and compared the MeV-H amino acid sequences for all the viruses that we used (Fig. 4B). Whereas the NE antigenic site is one of the most variable regions of MeV-H, NE-targeting antibodies discriminated only the D4.2 and D6 genotypes. Subgenotype D4.2 viruses showed the single mutation L249P, whereas genotype D6 viruses also showed an S247P mutation. To further elucidate the role of these amino acid changes found within the NE antigenic site, we introduced each of the two single point mutations into MeV-H genotype A and rescued the corresponding viruses. Figure 4C illustrates that an L249P amino acid substitution completely changed the neutralization sensitivity of antibodies targeting the NE, yet the sensitivity of genotype A virus to antibodies remained unchanged with and without the single point mutation S247P. These observations demonstrate that a single point mutation, L249P, but not S247P, allows MeV to avoid neutralizing antibodies targeting the NE.
Viral escape is due to loss of MeV-H binding sites for NE-targeting antibodies.
Virus neutralization requires a certain level of electrostatic and shape complementarity between the viral antigen and neutralizing antibody (42). Since 2 different mutations were found in the NE antigenic site, we further investigated their effect on virus-antibody interactions. To this end, we first produced a soluble form of the MeV-H protein that displays a C-terminal FLAG epitope. This tag enables the control of the amount of protein coating the surface of the plastic well. As expected, a FLAG antibody equally recognized all FLAG-tagged MeV-H proteins (Fig. 4D). However, NE-targeting antibodies showed defects in binding to those MeV-H proteins whose viruses escaped neutralization. Overall, these results show that an L249P substitution, but not S247P, impairs the binding of neutralizing antibodies that recognize the NE antigenic site, thereby allowing virus neutralization escape.
Subgenotype D4.2 shows diminished sensitivity to neutralization by pooled human sera.
The data presented in Fig. 1 show that a naturally occurring MeV strain can escape neutralization by MAbs targeting 3 of 6 antigenic sites of the MeV-H protein. Because 90% of the antibodies of MeV patients are directed against the MeV-H protein (17), we next assessed whether the mutation of the NE antigenic site in the subgenotype D4.2 MeV-H protein translates into a different serum neutralization pattern than those for subgenotype D4.1 and vaccine genotype A viruses.
To obtain a representative sample for population immunity against measles, we used pooled human serum collected from approximately 60 to 80 blood type AB donors in North America. To rule out potential interference by MeV-F-specific antibodies, we first absorbed MeV-F-specific antibodies from pooled human sera using MeV-F of genotype A, the F protein expressed by all of our recombinant MeVs. This depletion was performed by incubation with MeL-JuSo cells expressing the MeV-F protein. Following this depletion procedure, the remaining measles neutralization activity of the depleted serum is mediated exclusively by anti-MeV-H antibodies. Figure 5A shows that incubation with untransfected MeL-JuSo cells results in no depletion of MeV-H/F-specific antibodies, whereas incubation with MeL-JuSo/MeV-F cells results in the elimination of MeV-F-specific antibodies while maintaining the MeV-H-specific antibodies. As an additional control for the presence of residual MeV-F-specific antibodies due to the limit of detection of the fluorescence-activated cell sorter (FACS) analysis, we used an envelope-chimeric MeV/canine distemper virus (CDV) (M. A. Muñoz-Alía and Stephen J. Russell, unpublished data). This envelope-chimeric virus expresses the MeV-F protein in combination with the Onderstepoort strain CDV-H protein possessing a Y537D point mutation (43). This envelope-chimeric MeV-F/CDV-H showed complete resistance to neutralization by MeV-F-depleted pooled human sera (Fig. 5B). Neutralization assays using recombinant MeVs expressing the two D4 subgenotype-specific MeV-H proteins showed a diminished sensitivity for subgenotype D4.2 (50% inhibitory concentration [IC50] = 33.00 ± 3.50 milli-international units [mIU]/ml) compared to subgenotype D4.1 (IC50 = 30.93 ± 4.31) or the genotype A vaccine strain (IC50 = 30.05 ± 3.35) (P = 0.21, according to a paired t test). Therefore, the absence of the NE in subgenotype D4.2 MeV-H was associated with a definite reduction (albeit nonsignificant) in neutralization sensitivity compared to those of subgenotype D4.1 and vaccine genotype A viruses.
FIG 5.
Neutralization of MeV by anti-MeV-H antibodies from pooled human sera. (A) Reactivity of pooled human sera after 0 days (untreated) or 4 days (MeV-F absorbed) of incubation at 37°C with cells expressing MeV-F. Binding reactivity was measured by fluorescence-activated cell sorter analysis using MeL-JuSo cells expressing or not expressing the MeV glycoproteins (MeV-H and MeV-F). Anti-human immunoglobulin G(H+L) Alexa Fluor-488 was used for staining. (B) Neutralization of MeV by anti-H pooled human sera. The IC50s and standard deviations (StDev) were calculated from 3 experiments performed independently in quadruplicate. Values were calculated after nonlinear regression of the corresponding neutralization curves with GraphPad Prism software. CDV, canine distemper virus; NA, not applicable.
DISCUSSION
RNA virus populations are well known for their high levels of genetic diversity and high mutation rates, which provide a continuous challenge for investigators who must develop and constantly update vaccines and antiviral drugs (44). However, MeV stands apart from many other RNA viruses because of its limited antigenic diversity. In the present study, we focused on intragenotype variation among genetically and antigenically closely related viruses belonging to genotype D4. Viruses belonging to genotype D4 represent about 20% of the MeV sequences reported in databases (http://www.who-measles.org/), reflecting the high current prevalence of this genotype. The abundance of available genotype D4 sequences has allowed us to analyze a considerable number of MeV-N and MeV-H nucleotide sequences to probe their genetic diversity. MeV-H phylogeny analysis was congruent with MeV-N gene phylogeny, in support of the current standard for measles virus genotype identification. It allowed us to distinguish two subgenotypes, D4.1 and D4.2, that were paired with different neutralization sensitivities by anti-MeV-H antibodies targeting the NE antigenic site.
Distinct unrelated subgroups for genotype D4 viruses were previously proposed exclusively on the basis of phylogenic analysis of the MeV-N gene (45–47). We are aware that the minimum genetic distance observed in the MeV-H genes between the proposed D4 subgenotypes (1.5%) does not match WHO criteria for assigning a new genotype (>2%), and the MeV-N minimum genetic distance (2.5%) is at best borderline (>2.5%). Nevertheless, a similar subgenotype classification was proposed for subgenotypes B3.1 and B3.2 without fulfilling the divergence criteria (48, 49). Regardless of the definition for measles virus lineages, the subgenotype B3.1 and B3.2 viruses and the subgenotype D4.1 and D4.2 viruses are antigenically distinguishable within their respective genotypic clades.
Rapid genetic evolution drives the emergence of viral variants escaping immune surveillance. Mutations of the influenza virus hemagglutinin (HA) protein, the respiratory syncytial virus F protein, and the E2 protein of hepatitis C virus are known to underlie their escape from neutralization by polyclonal antisera (50–53). The MeV-H protein is the primary target of protective anti-measles virus antibodies (16, 17). We observed that subgenotype D4.2 viruses are resistant to monoclonal antibodies targeting three of the six known MeV-H protein antigenic sites (BH129, BH047, BH059, BH125, and BH097). These viruses were nevertheless efficiently neutralized by pooled human sera. The presence of other antigenic sites might mask such an effect.
The NE antigenic site is thought to encompass a linear epitope consisting of the region spanning amino acids 244 to 250, together with the region spanning amino acids 233 to 240, recognized by MAb BH1 (41). Although the region spanning amino acids 240 to 245 has not been visualized in the MeV-H crystal structures because of low electron density (39, 54, 55), it is predicted to display an α-helix where E245L-QL249 would be located on the protein surface (56). As a consequence, S247 would not be accessible, explaining the lack of an effect on both virus neutralization and antibody binding for the MeV-H S247P mutant. Conversely, a mimotope containing the S247P mutation associates with the NE-targeting BH129 MAb up to 135 times more efficiently (56). Discrepancies between antipeptide serum binding to the virus and virus neutralization activity have been reported for mimotopes (57, 58), which emphasizes the utility of testing linear epitope-based vaccines in the context of the mutant viruses. The NE antigenic site is immunogenic because MAb BH047 inhibits the binding of 20% of human serum antibodies in measles patients (59). In this regard, the MeV genotype C2 mutant variants S246L and S247L present in 2 of 5 measles isolates sequenced during a Madrid, Spain, outbreak may reflect immune selection (60).
To date, 24 MeV genotypes are recognized with preferential geographic circulation. A Bayesian skyline plot analysis suggested that prevalent genotypes such as genotype D4 are more adaptive to humans (61). The fact that most of the subgenotype D4.1 sequences that we retrieved from GenBank were derived from Kenya and Ethiopia, whereas the MAb-resistant subgenotype D4.2 MeV sequences were derived predominantly from France and Great Britain, might suggest that an intermediary level of vaccine coverage provides an environment more prone to adaptive mutations. Our inability to discern antigenic differences between the 2 subgenotypes by human serum neutralization could be intrinsic to the heterogeneous composition of the human sera used. Their high neutralization potency is indicative of natural MeV infection; therefore, some of the human patients whose sera were included in the pool could have even been infected with a genotype D4 virus strain, potentially masking antigenic differences. The use of vaccine samples with lower neutralization titers could potentially help in discerning this problem.
Nevertheless, the 2 D4 subgenotypes circulated in Spain at the same point, even in the same geographic area. Thus, although our subgenotype D4.1 virus was isolated in the Barcelona region of Catalonia (northeastern Spain) (MVi/Barcelona.SPA/26.08) and our subgenotype D4.2 virus was isolated in the autonomous region of Madrid (central Spain) (MVi/Madrid.SPA/10.10/1), a sporadic case of a subgenotype D4.1 virus circulating in Madrid could be identified (MVs/Madrid.SPA/50.05; GenBank accession no. EU085473). Interestingly, the sequence is similar to the sequence circulating in the United Kingdom at that time, which was epidemiologically linked to Somalia (45). However, MeV endemicity was established 2 years later in the United Kingdom with a subgenotype D4.2 virus (MVs/Enfield.UNK/14.07), and subgenotype D4.2 viruses have become predominant in Europe since then (47, 62). In 2011, these subgenotype D4.2 viruses were imported from France to the United States in 2011, causing the highest number of measles cases since it was declared eliminated (63).
Finally, we studied the adaptive significance of amino acid substitutions in MeV-H by different methods. We detected 5 pairs of amino acidic interactions. Whether they have functional significance warrants reverse-genetics systems and virus fitness experiments. Of the estimated positively and negatively selected sites in the gene, 5 negatively selected sites were detected by FEL, IFEL, and SLAC methods (3P, 113N, 256E, 342D, and 347D). Of these sites, only 3P has been detected previously by analyzing genotypes D3, D5, D9, and H1 (64).
In summary, we genetically identified 2 different lineages, or subgenotypes, in MeV genotype D4 strains that are antigenically distinguishable by virtue of their neutralization by antibodies targeting the NE antigenic site. This close genetic and antigenic relatedness might indicate selective pressure. Although subtle differences in human sera of MeV patients were detected, the absence of half of the known antigenic sites present in MeV-H warrants close antigenic monitoring in virological surveillance for measles.
MATERIALS AND METHODS
Cells and media.
B95a cells (an adherent marmoset B-cell line) were maintained in Roswell Park Memorial Institute medium (RPMI 1640; Corning) supplemented with 10% (vol/vol) heat-inactivated (56°C for 30 min) fetal bovine serum (FBS) (Gibco). African green monkey kidney (Vero) cells stably expressing human SLAM (65) were maintained in Dulbecco's modified minimal essential medium (DMEM) (HyClone; GE Healthcare Life Science) containing 5% FBS and 0.5 mg of Geneticin ml−1 (G418; Corning). The human melanoma cell line Mel-JuSo/wt, transfected or not with the Edmonston MeV-H (Mel-JuSo/MeV-H) or MeV-F (Mel-JuSo/MeV-F) protein, was described previously (16, 66) and cultured in RPMI 1640. In antibody depletion assays, the medium was replaced with human serum diluted 1:10 in RPMI medium, and cells were cultured for 4 days at 37°C in a humidified atmosphere of 5% carbon dioxide (17). All cell culture media were additionally supplemented with 100 IU penicillin ml−1, 100 μg streptomycin ml−1, and 10 mM HEPES (Gibco). All cells routinely tested negative for mycoplasma contamination.
Viruses.
Growth of the Edmonston strain (67) and the MeV primary isolates was performed as described previously (60). The wild-type MeVs used in this work were MVi/Madrid.SPA/15.06/3 (subgenotype B3.1) (23), FV (genotype C2) (60), MVi/Barcelona.SPA/26.08 (genotype D4), MVi/Madrid.SPA.10.10/1 (genotype D4), BCL (genotype D6) (60), MVi/Madrid.SPA/25.03 (genotype D7), MVi/Alicante.SPA/22/03 (genotype D8), MVi/Granada.SPA/32.08 (genotype D9), 65045397 (genotype G3) (68), and MVi/Madrid.SPA/50.10 (genotype H1). Viral stocks were prepared by repeated cycles of freezing and thawing and were frozen in aliquots at −80°C. The MeV titer was determined as PFU by using Vero/hSLAM cells.
Antibodies and virus neutralization assay.
Monoclonal antibodies and human sera were heat inactivated and serially diluted in Opti-MEM (Thermo Fisher Scientific). Equal volumes of the respective virus at 30 PFU per well were mixed with the respective antibodies at various 2-fold serial dilutions in 96-well plates (Costar Corp.), incubated at 37°C for 1 h, and inoculated onto 80% to 90% confluent Vero/hSLAM cells. The number of EGFP-expressing syncytia per well was counted under a fluorescence microscope after 2 days of culture and converted to a percentage in relation to infection in the absence of antibodies (100%). Each sample was run in 4 wells, and each experiment was repeated at least twice on different days.
The following anti-MeV-H protein MAbs used for neutralization were previously described: BH129, BH047, BH059 (69), BH101, BH026 (70), BH141 (71), and BH097 (72). Protein G affinity-purified MAbs were used, except for BH141 and BH059. BH141 and BH059 antibody concentrations were determined with a mouse isotype-specific immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA) quantitation set (Bethyl Laboratories).
Human serum used in this study was commercially purchased (product no. HP1022, lot no. C80553; Valley Biomedical). This serum represents a pool of 60 to 80 blood type AB healthy donors (communication with the supplier). The antibody concentration that causes a 50% reduction in plaque numbers (IC50) was determined by nonlinear regression using software (Prism version 7.00 for Mac; GraphPad Software). The titers were transformed to international units per milliliter by using National Institute for Biological Standards and Control (NIBSC) reference serum (WHO/BS/06.2031), according to procedures described previously (73).
Viral RNA isolation and generation of recombinant viruses.
RNA was extracted from infected B95a cells by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. A hypervariable region of the MeV-N gene and the entire region of the MeV-H gene were reverse transcribed by using 200 U of reverse transcriptase (SuperScript II; Invitrogen) and primers (Integrated DNA Technologies) MVN(+)958 (TTGGACTGCATGAATTTGCTGG) and V181 (TTAATTAAAACTTAGGGTGCAAGATCATCGATAATG), respectively. The resulting cDNA was amplified by PCR (HotStarTaq DNA polymerase; Qiagen) with primer pairs V181/H1931 (CACTAGTGGGTATGCCTGATGT) and MVN(+)958/MVP(−)3355 (GGTTGGCAGGTAAGTTGAGCT). Amplified DNA fragments were analyzed by electrophoresis in a 0.7% agarose gel and purified from the gel for sequencing (ABI Prism 3730xl DNA analyzer; Perkin-Elmer Applied Biosystems) by using a gel extraction kit (QIAquick; Qiagen). The entire MeV-H gene flanked with SpeI/PacI restriction sites (underlined in the primer sequences) was cloned into the equally restricted pCG vector (New England BioLabs Inc.) (74). At least 4 clones were sequenced, and the direct sequences of the PCR products were used to define the consensus sequences. Only those clones with the consensus sequence were transferred into an infectious MeV genome expressing EGFP upstream of N [MVvac2(GFP)N], using PacI and SpeI restriction sites. The MVvac2(GFP)N coding capacity is identical to those of the Moraten/Schwartz vaccine strains (75). Recombinant viruses were rescued by using standard protocols with modifications (76, 77). The integrity of the insert was verified by sequencing, and the corresponding virus was renamed according to the genotype of the MeV-H gene. The sequence of the MeV-N gene was determined directly from the PCR products.
Cloning and expression of the MeV-H protein.
The soluble MeV-H protein was produced by amplification of the MeV-H ectodomain (residues 61 to 617) preceded by the murine IgG κ-chain leader and a Strep-tag II epitope. This construct was cloned in frame with the C-terminal FLAG tag present into the MluI- and EcoRV-restricted pCMV6-AC-IRES-GFP vector by using an InFusion HD kit (Clontech Laboratories Inc.). Expi293F cells (Gibco) were transiently transfected and grown in suspension for 6 days. The culture supernatant was harvested every 3 days, filtered through a 0.45-μm-pore-size filter, concentrated by using a centrifugal filter device (Amicon Ultra 50K; EMD Millipore Corp.), and buffer exchanged with HEPES buffer (10 mM HEPES, 150 mM sodium chloride [pH 7.5]).
FACS analysis.
MeV-H- and MeV-F-specific IgG antibody levels were determined as described previously (16, 17, 66), with the following modifications: RPMI 1600 medium supplemented with 2% FBS and 2-mM EDTA (catalogue no. 155575; Thermo Fisher Scientific) was used as a FACS buffer, and cells were stained with human IgG(H+L) Alexa Fluor-488.
Enzyme-linked immunosorbent assay.
Coated microplates (Strep-Tactin XT; IBA GmbH) were incubated at 37°C for 1 h with 0.1 ml of soluble MeV-H protein, previously diluted to give a final optical density of 1 under the conditions described above. The plates with the MeV-H protein absorbed through its amino-terminal Strep-tag II sequence were washed 3 times with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBS-Tween), and consequently, 0.1 ml of antibody in PBS (5 μg/ml) was added. In parallel, mouse anti-FLAG M2 antibody (Sigma-Aldrich) was included as a control for the absorbed protein. The plates were incubated for 1 h at 37°C and washed with PBS-Tween as described above. The mixtures were then allowed to react with 0.1 ml of goat anti-mouse IgG(H+L)-horseradish peroxidase (HRP) (catalogue no. 62-6520; Thermo Fisher Scientific) in PBS-Tween at a dilution of 1:2,000 to 1:4,000 for 1 h at room temperature. The peroxidase bound to the wells was detected by incubation with 1-Step Ultra TMB (3,3′,5,5′-tetramentylbenzidine; Thermo Fisher Scientific) according to the manufacturer's protocol. The Epstein-Barr virus VCA IgG titer was assessed by using a commercial ELISA as recommended by the manufacturer (catalogue no. 57351; IBL International GmbH).
Sequence analysis.
Sequence data available for MeV-N and MeV-H of genotype D4 MeV strains were retrieved from GenBank and analyzed by using BioEdit version 7.0.9.0. Multisequence alignments were performed with ClustalX (http://www.genome.jp/tools/clustalw/), and identical sequences were removed from the data set. Phylogenetic reconstructions were inferred by the maximum likelihood method on the basis of the general time-reversible model available in MEGA version 6.06 (78). To estimate the robustness of the phylogenetic inference, we performed 1,000 bootstrap analyses. Estimation of the evolutionary divergence among sequences was conducted by using the Kimura 2-parameter model in MEGA version 6. The codon positions included were the first plus second plus third plus noncoding positions.
The selective pressure acting on the MeV-H gene was investigated by using 4 different methods: SLAC, FEL, IFEL, and MEME (all available in the HYPHY package on a Web-based interface [DataMonkey]) (79).
Structural modeling.
An MeV-H model was generated from the crystallographic structure of the purified protein alone (PDB accession no. 2ZB5) and in complex with SLAM (PDB accession no. 3ALZ). A complex-type N-glycan model was constructed manually by using a complex penta-antennary glycan whose coordinates were obtained online (see http://www.glycosciences.de/). Crystallographic structures were analyzed and modeled by using PyMOL software (http://pymol.org/).
Accession number(s).
The nucleotide sequences determined in this work were deposited in the GenBank database under accession no. KY524301 to KY524304. A list of nucleotide sequences used in the course of this work can be found in Table 1.
ACKNOWLEDGMENTS
We are indebted to Rafael Fernandez-Munoz and Maria Luisa Celma (formerly of the Spanish National Reference Laboratory for Measles Virus) for providing us with the MeV isolates. We are grateful to Rik de Swart (Erasmus University Medical Center, The Netherlands) for kindly allowing us to use the MeL-JuSo cell lines and to Roberto Cattaneo (Mayo Clinic) for recombinant MVvac2(GFP)N. We sincerely thank Takao Hashiguchi for sharing his PDB files and expertise, which aided in our glycan modeling onto MeV-H.
There is no conflict of interest.
This work was funded by Al and Mary Agnes McQuinn and the Mayo Clinic. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Torrey EF, Yolken RH. 3 April 2005. Their bugs are worse than their bite, p B01 Washington Post, Washington, DC. [Google Scholar]
- 2.Dumer AM. 2009. Swine flu: what you need to know. Brownstone Books, Brooklyn, NY. [Google Scholar]
- 3.World Health Organization. 2015. Global routine vaccination coverage, 2014. Wkly Epidemiol Rec 90:617–632. [PubMed] [Google Scholar]
- 4.Mina MJ, Metcalf CJ, de Swart RL, Osterhaus AD, Grenfell BT. 2015. Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Science 348:694–699. doi: 10.1126/science.aaa3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries RD, McQuaid S, van Amerongen G, Yuksel S, Verburgh RJ, Osterhaus AD, Duprex WP, de Swart RL. 2012. Measles immune suppression: lessons from the macaque model. PLoS Pathog 8:e1002885. doi: 10.1371/journal.ppat.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. 2016. Measles, fact sheet. WHO, Geneva, Switzerland. http://who.int/mediacentre/factsheets/fs286/en/ Accessed 13 January 2017.
- 7.World Health Organization. 2012. Global measles and rubella strategic plan 2012-2020. WHO, Geneva, Switzerland. [Google Scholar]
- 8.Shi J, Zheng J, Huang H, Hu Y, Bian J, Xu D, Li F. 2011. Measles incidence rate and a phylogenetic study of contemporary genotype H1 measles strains in China: is an improved measles vaccine needed? Virus Genes 43:319–326. doi: 10.1007/s11262-011-0638-0. [DOI] [PubMed] [Google Scholar]
- 9.Rosen JB, Rota JS, Hickman CJ, Sowers SB, Mercader S, Rota PA, Bellini WJ, Huang AJ, Doll MK, Zucker JR, Zimmerman CM. 2014. Outbreak of measles among persons with prior evidence of immunity, New York City, 2011. Clin Infect Dis 58:1205–1210. doi: 10.1093/cid/ciu105. [DOI] [PubMed] [Google Scholar]
- 10.Vardas E, Kreis S. 1999. Isolation of measles virus from a naturally-immune, asymptomatically re-infected individual. J Clin Virol 13:173–179. doi: 10.1016/S1386-6532(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 11.Rota JS, Hickman CJ, Sowers SB, Rota PA, Mercader S, Bellini WJ. 2011. Two case studies of modified measles in vaccinated physicians exposed to primary measles cases: high risk of infection but low risk of transmission. J Infect Dis 204(Suppl 1):S559–S563. doi: 10.1093/infdis/jir098. [DOI] [PubMed] [Google Scholar]
- 12.Risco-Risco C, Masa-Calles J, Lopez-Perea N, Echevarria JE, Rodriguez-Caravaca G. 1 June 2016. Epidemiology of measles in vaccinated people, Spain 2003-2014. Enferm Infecc Microbiol Clin. (In Spanish.) doi: 10.1016/j.eimc.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Griffin DE. 2013. Measles, p 1042–2069. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 14.Wild TF, Malvoisin E, Buckland R. 1991. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J Gen Virol 72:439–442. doi: 10.1099/0022-1317-72-2-439. [DOI] [PubMed] [Google Scholar]
- 15.Navaratnarajah CK, Leonard VHJ, Cattaneo R. 2009. Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr Top Microbiol Immunol 329:59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Swart RL, Yüksel S, Osterhaus ADME. 2005. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J Virol 79:11547–11551. doi: 10.1128/JVI.79.17.11547-11551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Swart RL, Yüksel S, Langerjs CN, Muller CP, Osterhaus ADME. 2009. Depletion of measles virus glycoprotein-specific antibodies from human sera reveals genotype-specific neutralising antibodies. J Gen Virol 90:2982–2989. doi: 10.1099/vir.0.014944-0. [DOI] [PubMed] [Google Scholar]
- 18.Lin WH, Pan CH, Adams RJ, Laube BL, Griffin DE. 2014. Vaccine-induced measles virus-specific T cells do not prevent infection or disease but facilitate subsequent clearance of viral RNA. mBio 5:e01047-14. doi: 10.1128/mBio.01047-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Rennick LJ, Duprex WP, Rima BK. 19 December 2012. Determination of spontaneous mutation frequencies in measles virus under nonselective conditions. J Virol doi: 10.1128/JVI.02146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrag SJ, Rota PA, Bellini WJ. 1999. Spontaneous mutation rate of measles virus: direct estimation based on mutations conferring monoclonal antibody resistance. J Virol 73:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anonymous. 2012. Measles virus nomenclature update: 2012. Wkly Epidemiol Rec 87:73–81. [PubMed] [Google Scholar]
- 23.Munoz-Alia MA, Fernandez-Munoz R, Casasnovas JM, Porras-Mansilla R, Serrano-Pardo A, Pagan I, Ordobas M, Ramirez R, Celma ML. 2015. Measles virus genetic evolution throughout an imported epidemic outbreak in a highly vaccinated population. Virus Res 196:122–127. doi: 10.1016/j.virusres.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins GM, Rambaut A, Pybus OG, Holmes EC. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol 54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Davis CT, Christman MC, Rivailler P, Zhong H, Donis RO, Lu G. 2012. Evolutionary history and phylodynamics of influenza A and B neuraminidase (NA) genes inferred from large-scale sequence analyses. PLoS One 7:e38665. doi: 10.1371/journal.pone.0038665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riddell MA, Rota JS, Rota PA. 2005. Review of the temporal and geographical distribution of measles virus genotypes in the prevaccine and postvaccine eras. Virol J 2:87. doi: 10.1186/1743-422X-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodson JL, Seward JF. 2015. Measles 50 years after use of measles vaccine. Infect Dis Clin North Am 29:725–743. doi: 10.1016/j.idc.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Mulders MN, Rota PA, Icenogle JP, Brown KE, Takeda M, Rey GJ, Ben Mamou MCB, Patamadilok S, Zhang Y, Gacic-Dobo M, Strebel PM, Goodson JL. 2016. Global measles and rubella laboratory network support for elimination goals, 2010-2015. Wkly Epidemiol Rec 91:240–246.27156255 [Google Scholar]
- 29.Rima BK, Earle JAP, Yeo RP, Herlihy L, Baczko K, ter Meulen V, Carabaña J, Caballero M, Celma ML, Fernández-Muñoz R. 1995. Temporal and geographical distribution of measles virus genotypes. J Gen Virol 76:1173–1180. doi: 10.1099/0022-1317-76-5-1173. [DOI] [PubMed] [Google Scholar]
- 30.Santibañez S, Tischer A, Heider A, Siedler A, Hengel H. 2002. Rapid replacement of endemic measles virus genotypes. J Gen Virol 83:2699–2708. doi: 10.1099/0022-1317-83-11-2699. [DOI] [PubMed] [Google Scholar]
- 31.Woelk CH, Jin L, Holmes EC, Brown DWG. 2001. Immune and artificial selection in the haemagglutinin (H) glycoprotein of measles virus. J Gen Virol 82:2463–2474. doi: 10.1099/0022-1317-82-10-2463. [DOI] [PubMed] [Google Scholar]
- 32.Tamin A, Rota PA, Wang ZD, Heath JL, Anderson LJ, Bellini WJ. 1994. Antigenic analysis of current wild type and vaccine strains of measles virus. J Infect Dis 170:795–801. doi: 10.1093/infdis/170.4.795. [DOI] [PubMed] [Google Scholar]
- 33.Klingele M, Hartter HK, Adu F, Ammerlaan W, Ikusika W, Muller CP. 2000. Resistance of recent measles virus wild-type isolates to antibody-mediated neutralization by vaccinees with antibody. J Med Virol 62:91–98. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Finsterbusch T, Wolbert A, Deitemeier I, Meyer K, Mosquera MM, Mankertz A, Santibañez S. 2009. Measles virus of genotype H1 evade recognition by vaccine-induced neutralizing antibodies targeting the linear haemagglutinin noose epitope. J Gen Virol 90:2739–2745. doi: 10.1099/vir.0.013524-0. [DOI] [PubMed] [Google Scholar]
- 35.Tahara M, Ito Y, Brindley MA, Ma X, He J, Xu S, Fukuhara H, Sakai K, Komase K, Rota PA, Plemper RK, Maenaka K, Takeda M. 2013. Functional and structural characterization of neutralizing epitopes of measles virus hemagglutinin protein. J Virol 87:666–675. doi: 10.1128/JVI.02033-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birrer MJ, Udem SA, Nathenson S, Bloom BR. 1981. Antigenic variants of measles virus. Nature 293:67–69. doi: 10.1038/293067a0. [DOI] [PubMed] [Google Scholar]
- 37.Santibañez S, Niewiesk S, Heider A, Schneider-Schaulies J, Berbers GA, Zimmermann A, Halenius A, Wolbert A, Deitemeier I, Tischer A, Hengel H. 2005. Probing neutralizing-antibody responses against emerging measles viruses (MVs): immune selection of MV by H protein-specific antibodies? J Gen Virol 86:365–374. doi: 10.1099/vir.0.80467-0. [DOI] [PubMed] [Google Scholar]
- 38.Tahara M, Burckert JP, Kanou K, Maenaka K, Muller CP, Takeda M. 2016. Measles virus hemagglutinin protein epitopes: the basis of antigenic stability. Viruses 8:E216. doi: 10.3390/v8080216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol 18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 40.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fournier P, Brons NH, Berbers GA, Wiesmüller KH, Fleckenstein BT, Schneider F, Jung G, Muller CP. 1997. Antibodies to a new linear site at the topographical or functional interface between the haemagglutinin and fusion proteins protect against measles encephalitis. J Gen Virol 78:1295–1302. doi: 10.1099/0022-1317-78-6-1295. [DOI] [PubMed] [Google Scholar]
- 42.Epa VC, Colman PM. 2001. Shape and electrostatic complementarity at viral antigen-antibody complexes. Curr Top Microbiol Immunol 260:45–53. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Wallace OL, Domi A, Wright KJ, Driscoll J, Anzala O, Sanders EJ, Kamali A, Karita E, Allen S, Fast P, Gilmour J, Price MA, Parks CL. 2015. Canine distemper virus neutralization activity is low in human serum and it is sensitive to an amino acid substitution in the hemagglutinin protein. Virology 482:218–224. doi: 10.1016/j.virol.2015.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansky LM, Cunningham FS. 2000. Virus mutators and antimutators: roles in evolution, pathogenesis and emergence. Trends Genet 16:512–517. doi: 10.1016/S0168-9525(00)02125-9. [DOI] [PubMed] [Google Scholar]
- 45.Kremer JR, Brown KE, Jin L, Santibanez S, Shulga SV, Aboudy Y, Demchyshyna IV, Djemileva S, Echevarria JE, Featherstone DF, Hukic M, Johansen K, Litwinska B, Lopareva E, Lupulescu E, Mentis A, Mihneva Z, Mosquera MM, Muscat M, Naumova MA, Nedeljkovic J, Nekrasova LS, Magurano F, Fortuna C, de Andrade HR, Richard JL, Robo A, Rota PA, Samoilovich EO, Sarv I, Semeiko GV, Shugayev N, Utegenova ES, van Binnendijk R, Vinner L, Waku-Kouomou D, Wild TF, Brown DW, Mankertz A, Muller CP, Mulders MN. 2008. High genetic diversity of measles virus, World Health Organization European Region, 2005-2006. Emerg Infect Dis 14:107–114. doi: 10.3201/eid1401.070778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mbugua FM, Okoth FA, Gray M, Kamau T, Kalu A, Eggers R, Borus P, Kombich J, Langat A, Maritim P, Lesiamon J, Tipples GA. 2003. Molecular epidemiology of measles virus in Kenya. J Med Virol 71:599–604. doi: 10.1002/jmv.10515. [DOI] [PubMed] [Google Scholar]
- 47.Necula G, Lazar M, Stanescu A, Pistol A, Santibanez S, Mankertz A, Lupulescu E. 2013. Transmission and molecular characterisation of wild measles virus in Romania, 2008 to 2012. Euro Surveill 18:20658. doi: 10.2807/1560-7917.ES2013.18.50.20658. [DOI] [PubMed] [Google Scholar]
- 48.El Mubarak HS, van de Bildt MW, Mustafa OA, Vos HW, Mukhtar MM, Ibrahim SA, Andeweg AC, El Hassan AM, Osterhaus AD, de Swart RL. 2002. Genetic characterization of wild-type measles viruses circulating in suburban Khartoum, 1997-2000. J Gen Virol 83:1437–1443. doi: 10.1099/0022-1317-83-6-1437. [DOI] [PubMed] [Google Scholar]
- 49.Hanses F, Truong AT, Ammerlaan W, Ikusika O, Adu F, Oyefolu AO, Omilabu SA, Muller CP. 1999. Molecular epidemiology of Nigerian and Ghanaian measles virus isolates reveals a genotype circulating widely in western and central Africa. J Gen Virol 80(Part 4):871–877. [DOI] [PubMed] [Google Scholar]
- 50.Tome L, Frabasile S, Candia C, Pittini A, Farina N, Melero JA, Arbiza J. 2012. Selection and characterization of human respiratory syncytial virus escape mutants resistant to a polyclonal antiserum raised against the F protein. Arch Virol 157:1071–1080. doi: 10.1007/s00705-012-1274-2. [DOI] [PubMed] [Google Scholar]
- 51.Melero JA, Moore ML. 2013. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity. Curr Top Microbiol Immunol 372:59–82. doi: 10.1007/978-3-642-38919-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahid A, Dubuisson J. 2013. Virus-neutralizing antibodies to hepatitis C virus. J Viral Hepat 20:369–376. doi: 10.1111/jvh.12094. [DOI] [PubMed] [Google Scholar]
- 53.Knossow M, Skehel JJ. 2006. Variation and infectivity neutralization in influenza. Immunology 119:1–7. doi: 10.1111/j.1365-2567.2006.02421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. 2 December 2012. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat Struct Mol Biol doi: 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 55.Santiago C, Celma ML, Stehle T, Casasnovas JM. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 56.Deroo S, El Kasmi KC, Fournier P, Theisen D, Brons NH, Herrmann M, Desmet J, Muller CP. 1998. Enhanced antigenicity of a four-contact-residue epitope of the measles virus hemagglutinin protein by phage display libraries: evidence of a helical structure in the putative active site. Mol Immunol 35:435–443. doi: 10.1016/S0161-5890(98)00057-1. [DOI] [PubMed] [Google Scholar]
- 57.El Kasmi KC, Theisen D, Brons NH, Ammerlaan W, Klingele M, Truong AT, Muller CP. 1999. A hemagglutinin-derived peptide-vaccine ignored by virus-neutralizing passive antibodies, protects against murine measles encephalitis. Vaccine 17:2436–2445. doi: 10.1016/S0264-410X(99)00008-0. [DOI] [PubMed] [Google Scholar]
- 58.Obeid OE, Partidos CD, Howard CR, Steward MW. 1995. Protection against morbillivirus-induced encephalitis by immunization with a rationally designed synthetic peptide vaccine containing B- and T-cell epitopes from the fusion protein of measles virus. J Virol 69:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ertl OT, Wenz DC, Bouche FB, Berbers GAM, Muller CP. 2003. Immunodominant domains of the measles virus hemagglutinin protein eliciting a neutralizing human B cell response. Arch Virol 148:2195–2206. doi: 10.1007/s00705-003-0159-9. [DOI] [PubMed] [Google Scholar]
- 60.Rima BK, Earle JAP, Baczko K, ter Meulen V, Liebert UG, Carstens C, Carabaña J, Caballero M, Celma ML, Fernández-Muñoz R. 1997. Sequence divergence of measles virus haemagglutinin during natural evolution and adaptation to cell culture. J Gen Virol 78:97–106. doi: 10.1099/0022-1317-78-1-97. [DOI] [PubMed] [Google Scholar]
- 61.Kimura H, Saitoh M, Kobayashi M, Ishii H, Saraya T, Kurai D, Tsukagoshi H, Shirabe K, Nishina A, Kozawa K, Kuroda M, Takeuchi F, Sekizuka T, Minakami H, Ryo A, Takeda M. 2015. Molecular evolution of haemagglutinin (H) gene in measles virus. Sci Rep 5:11648. doi: 10.1038/srep11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santibanez S, Hubschen JM, Muller CP, Freymuth F, Mosquera MM, Mamou MB, Mulders MN, Brown KE, Myers R, Mankertz A. 2015. Long-term transmission of measles virus in Central and continental Western Europe. Virus Genes 50:2–11. doi: 10.1007/s11262-015-1173-1. [DOI] [PubMed] [Google Scholar]
- 63.Centers for Disease Control and Prevention. 2011. Measles: United States, January-May 20, 2011. MMWR Morb Mortal Wkly Rep 60:666–668. [PubMed] [Google Scholar]
- 64.Saitoh M, Takeda M, Gotoh K, Takeuchi F, Sekizuka T, Kuroda M, Mizuta K, Ryo A, Tanaka R, Ishii H, Takada H, Kozawa K, Yoshida A, Noda M, Okabe N, Kimura H. 2012. Molecular evolution of hemagglutinin (H) gene in measles virus genotypes D3, D5, D9, and H1. PLoS One 7:e50660. doi: 10.1371/journal.pone.0050660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75:4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Swart RL, Vos HW, UytdeHaag FGCM, Osterhaus ADME, van Binnendijk RS. 1998. Measles virus fusion protein- and hemagglutinin-transfected cell lines are a sensitive tool for the detection of specific antibodies by a FACS-measured immunofluorescence assay. J Virol Methods 71:35–44. doi: 10.1016/S0166-0934(97)00188-2. [DOI] [PubMed] [Google Scholar]
- 67.Alkatib G, Briedis DJ. 1986. The predicted primary structure of the measles virus hemagglutinin. Virology 150:479–490. doi: 10.1016/0042-6822(86)90312-0. [DOI] [PubMed] [Google Scholar]
- 68.Cilla G, Montes M, Artieda J, Pineiro L, Arriola L, Perez-Trallero E. 2011. Measles genotypes D4 and G3 reintroduced by multiple foci after 15 years without measles virus circulation, Gipuzkoa, the Basque Country, Spain, March to June 2011. Euro Surveill 16(43):pii=19997 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19997. [PubMed] [Google Scholar]
- 69.Ziegler D, Fournier P, Berbers GA, Steuer H, Wiesmuller KH, Fleckenstein B, Schneider F, Jung G, King CC, Muller CP. 1996. Protection against measles virus encephalitis by monoclonal antibodies binding to a cystine loop domain of the H protein mimicked by peptides which are not recognized by maternal antibodies. J Gen Virol 77(Part 10):2479–2489. [DOI] [PubMed] [Google Scholar]
- 70.Bouche FB, Ertl OT, Muller CP. 2002. Neutralizing B cell response in measles. Viral Immunol 15:451–471. doi: 10.1089/088282402760312331. [DOI] [PubMed] [Google Scholar]
- 71.Lech PJ, Tobin GJ, Bushnell R, Gutschenritter E, Pham LD, Nace R, Verhoeyen E, Cosset FL, Muller CP, Russell SJ, Nara PL. 2013. Epitope dampening monotypic measles virus hemagglutinin glycoprotein results in resistance to cocktail of monoclonal antibodies. PLoS One 8:e52306. doi: 10.1371/journal.pone.0052306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lech PJ, Pappoe R, Nakamura T, Tobin GJ, Nara PL, Russell SJ. 2014. Antibody neutralization of retargeted measles viruses. Virology 454–455:237–246. doi: 10.1016/j.virol.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Binnendijk RS, Poelen MC, van Amerongen G, de Vries P, Osterhaus AD. 1997. Protective immunity in macaques vaccinated with live attenuated, recombinant, and subunit measles vaccines in the presence of passively acquired antibodies. J Infect Dis 175:524–532. doi: 10.1093/infdis/175.3.524. [DOI] [PubMed] [Google Scholar]
- 74.Cathomen T, Buchholz CJ, Spielhofer P, Cattaneo R. 1995. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 75.del Valle JR, Devaux P, Hodge G, Wegner NJ, McChesney MB, Cattaneo R. 2007. A vectored measles virus induces hepatitis B surface antigen antibodies while protecting macaques against measles virus challenge. J Virol 81:10597–10605. doi: 10.1128/JVI.00923-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parks CL, Lerch RA, Walpita P, Sidhu MS, Udem SA. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J Virol 73:3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 80.Rota PA, Bloom AE, Vanchiere JA, Bellini WJ. 1994. Evolution of the nucleoprotein and matrix genes of wild-type strains of measles virus isolated from recent epidemics. Virology 198:724–730. doi: 10.1006/viro.1994.1086. [DOI] [PubMed] [Google Scholar]
- 81.Tipples GA, Gray M, Garbutt M, Rota PA, Canadian Measles Surveillance Program. 2004. Genotyping of measles virus in Canada: 1979-2002. J Infect Dis 189(Suppl 1):S171–S176. doi: 10.1086/377716. [DOI] [PubMed] [Google Scholar]
- 82.Penedos AR, Myers R, Hadef B, Aladin F, Brown KE. 2015. Assessment of the utility of whole genome sequencing of measles virus in the characterisation of outbreaks. PLoS One 10:e0143081. doi: 10.1371/journal.pone.0143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ivancic-Jelecki J, Baricevic M, Santak M, Harcet M, Tesovic G, Marusic Della Marina B, Forcic D. 2013. The first genetic characterization of a D4 measles virus strain derived from a patient with subacute sclerosing panencephalitis. Infect Genet Evol 17:71–78. doi: 10.1016/j.meegid.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 84.Nigatu W, Jin L, Cohen BJ, Nokes DJ, Etana M, Cutts FT, Brown DW. 2001. Measles virus strains circulating in Ethiopia in 1998-1999: molecular characterisation using oral fluid samples and identification of a new genotype. J Med Virol 65:373–380. doi: 10.1002/jmv.2044. [DOI] [PubMed] [Google Scholar]
- 85.WHO 2001. Nomenclature for describing the genetic characteristics of wild-type measles viruses (update). Part I. Wkly Epidemiol Rec 76:242–247. [PubMed] [Google Scholar]
- 86.Kosakovsky Pond SL, Poon AF, Leigh Brown AJ, Frost SD. 2008. A maximum likelihood method for detecting directional evolution in protein sequences and its application to influenza A virus. Mol Biol Evol 25:1809–1824. doi: 10.1093/molbev/msn123. [DOI] [PMC free article] [PubMed] [Google Scholar]