ABSTRACT
Many pathogens express noncoding RNAs (ncRNAs) during infection processes. In the most extreme case, pathogenic ncRNAs alone (such as viroids) can infect eukaryotic organisms, leading to diseases. While a few pathogenic ncRNAs have been implicated in regulating gene expression, the functions of most pathogenic ncRNAs in host-pathogen interactions remain unclear. Here, we employ potato spindle tuber viroid (PSTVd) infecting tomato as a system to dissect host interactions with pathogenic ncRNAs, using comprehensive transcriptome analyses. We uncover various new activities in regulating gene expression during PSTVd infection, such as genome-wide alteration in alternative splicing of host protein-coding genes, enhanced guided-cleavage activities of a host microRNA, and induction of the trans-acting function of phased secondary small interfering RNAs. Furthermore, we reveal that PSTVd infection massively activates genes involved in plant immune responses, mainly those in the calcium-dependent protein kinase and mitogen-activated protein kinase cascades, as well as prominent genes involved in hypersensitive responses, cell wall fortification, and hormone signaling. Intriguingly, our data support a notion that plant immune systems can respond to pathogenic ncRNAs, which has broad implications for providing new opportunities for understanding the complexity of immune systems in differentiating “self” and “nonself,” as well as lay the foundation for resolving the long-standing question regarding the pathogenesis mechanisms of viroids and perhaps other infectious RNAs.
IMPORTANCE Numerous pathogens, including viruses, express pathogenic noncoding transcripts during infection. In the most extreme case, pathogenic noncoding RNAs alone (i.e., viroids) can cause disease in plants. While some work has demonstrated that pathogenic noncoding RNAs interact with host factors for function, the biological significance of pathogenic noncoding RNAs in host-pathogen interactions remains largely unclear. Here, we apply comprehensive genome-wide analyses of plant-viroid interactions and discover several novel molecular activities underlying nuclear-replicating viroid infection processes in plants, including effects on the expression and function of host noncoding transcripts, as well as the alternative splicing of host protein-coding genes. Importantly, we show that plant immunity is activated upon infection of a nuclear-replicating viroid, which is a new concept that helps to understand viroid-based pathogenesis. Our finding has broad implications for understanding the complexity of host immune systems and the diverse functions of noncoding RNAs.
KEYWORDS: Viroid, noncoding RNA, alternative splicing, inducible phasiRNA, immune responses
INTRODUCTION
Long noncoding RNAs (lncRNAs) are ubiquitous in living organisms (1). In general, they range from 200 to several thousand nucleotides (nt) in length. Many pathogens, including viruses, also express transcripts without protein-coding capacity (pathogenic ncRNAs) in host cells during infection. Intriguingly, some pathogenic ncRNAs, such as viroids, by themselves can cause diseases. However, in contrast to the explosive discovery of such noncoding sequences, the biological significance of most pathogenic ncRNAs during disease remains poorly understood.
Emerging evidence shows that some pathogenic ncRNAs can recruit host factors to exert their functions. EBER2, a pathogenic ncRNA encoded by Epstein-Barr virus, binds nascent viral RNA to drive host transcription factor paired box protein 5 (PAX5) to viral DNA for downstream viral gene expression (2). Potato spindle tuber viroid (PSTVd), a pathogenic ncRNA causing plant diseases, utilizes host transcription factors to mediate host DNA-dependent RNA polymerase II transcription using PSTVd RNA templates (3–5). Interestingly, a PSTVd relative, hop stunt viroid, can directly interact with a host histone deacetylase to alter the methylation states of rRNA genes and potentially other genes (6). Besides direct association with host proteins, pathogenic ncRNAs can serve as precursors to generate small interfering RNAs (siRNAs) for function. For example, two viroid-derived small RNAs (vdsRNAs) from peach latent mosaic viroid (PLMVd), which replicates in chloroplasts, guide the cleavage of target transcripts encoding a chloroplast heat shock protein. This regulation has been implicated in symptom development (7). Nevertheless, these pathogenic ncRNA-host factor combinations and/or vdsRNAs cannot fully explain the molecular mechanisms underpinning the pathogenic ncRNA-triggered symptoms.
In response to infectious RNAs (such as RNA viruses and viroids), RNA silencing is a widely employed defense mechanism in animals and plants (8, 9). Both animals and plants have Dicers or Dicer-like proteins (DCLs) to dice double-stranded viral or viroid RNAs into small RNAs, which are loaded onto Argonaute proteins (AGOs) to further inhibit infectious RNAs (8, 9). Therefore, Dicers and DCLs are proposed as pattern recognition receptors while AGOs are considered effectors in RNA-based immunity (8). In addition to the RNA-silencing pathway, animals have a broad arsenal of immune responses, such as type I interferon and double-stranded-RNA-activated protein kinases, to fight RNA viruses (10). These components work in concert as part of the immune system. Plants also have innate immunity machinery, such as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is triggered by recognizing PAMPs via the membrane-associated receptor-like protein kinases, which serve as the first layer of defense. Pathogens counter the host plant defense by “injecting” effectors into plant cells, which trigger ETI and downstream hypersensitive responses (HRs) (11–15). PTI and ETI are rarely used to describe plant defense against RNA viruses due to the lack of well-defined molecular patterns or effectors (11, 16, 17), despite the fact that some plant viruses can trigger gene-for-gene resistance and HRs. In such scenarios, the plant immune system employs various resistance genes to recognize certain viral proteins as avirulence determinants and consequently to confer gene-for-gene resistance. These avirulence determinants are critical for viral infection in compatible hosts and serve as virulence factors leading to pathogenesis (18). However, some of the pathogenic ncRNAs can directly cause diseases without introducing anything other than RNAs into plant cells. Whether this pathogenicity is attributable mainly to the activation of host RNA-silencing machinery or to the combined effects of the RNA-silencing machinery and other components (e.g., plant immunity) remains elusive. To better understand this, it is critical to dissect the biology of pathogenic ncRNAs apart from the complex host-pathogen interactions, where pathogens often have various functional components (e.g., toxins, proteins, and nucleic acids) that hinder the understanding of individual factors.
In this regard, viroids have distinct advantages to serve as a model. The genome of a viroid is a single-stranded circular noncoding RNA, ranging from 250 to 400 nt, that replicates and spreads in plants (19, 20). Its incredibly simple genome provides a clean system without the interference of other products from pathogens while retaining the natural infection status. Despite these advantages, however, the molecular basis of viroid diseases remains elusive. Global changes in gene expression (including multiple hormonal genes) certainly contribute to pathogenesis (19, 21–24), but these phenomena are more likely the consequences. vdsRNAs may contribute to posttranscriptional gene regulation (7, 25), but to date, these activities cannot fully explain viroid pathogenesis.
Here, we employ PSTVd infecting tomato as an experimental system and apply comprehensive genome-wide analyses to dissect the host responses to pathogenic ncRNAs. We reveal several novel activities triggered by PSTVd infection, such as affecting host lncRNA expression, genome-wide changes in alternative splicing of host protein-coding genes, alteration in host microRNA (miRNA)-guided cleavage of transcripts, and inducible trans-acting function of phased secondary siRNAs (phasiRNA). Our analyses also provide the first comprehensive functional assessment of the whole population of vdsRNAs. Importantly, we uncover the massive activation of genes involved in the signaling of basal defense, indicating the activation of plant immunity upon PSTVd infection. This concept helps elucidate global alteration in gene expression in the context of immune responses and facilitates the development of effective measurements against diseases caused by nuclear-replicating viroids. The discovery of plant immune responses to viroid infection has broad implications for understanding the complex host immune system and the diverse functions of ncRNAs.
RESULTS
Tomato transcriptome sequencing in response to a pathogenic noncoding RNA.
We inoculated tomato seedlings with PSTVd and verified the infection at 3 weeks postinfection via Northern blotting (Fig. 1A). We emphasize that this is a critical stage at which the PSTVd progeny start to accumulate in systemic leaves and symptoms begin to emerge (Fig. 1B). Using RNA from the same samples, we constructed and sequenced three sets of libraries: small RNA (sRNA), degradomal RNA (dRNA-Seq), and RNA-Seq for mock- and PSTVd-infected tomato leaves with three biological replicates. The data were analyzed in a coordinated manner to unravel the expression and functions of various RNA populations. After processing the sequencing data (see Table S1 in the supplemental material), for each sample we obtained 1 to 3 million high quality sRNA reads, 3 to 14 million dRNA-Seq reads, and 5 to 17 million RNA-Seq reads (except one PSTVd-infected library with 1.5 million reads). It is noteworthy that sequence abundance profiles of replicated samples showed high correlations (see Table S2 in the supplemental material), indicating the high quality of the data we generated. These data together captured the dynamics of various classes of regulatory RNAs and the protein-coding mRNAs.
FIG 1.

PSTVd-infected tomato plants. (A) Northern blots showing the presence of PSTVd in systemic leaves 3 weeks postinoculation. (B) Symptom development in PSTVd-infected plants over 5 weeks.
PSTVd infection impacts the expression of lncRNAs in tomato.
Previous studies on transcriptome changes during viroid infection focused on the expression dynamics of protein-coding genes (21–24). The extent to which the PSTVd infection affects host noncoding transcripts remains unexplored. Using the transcriptome data generated in this study, we identified 6,726 genomic loci that generated detectable lncRNAs (see Table S3 in the supplemental material). Interestingly, we also identified 217 previously unannotated protein-coding loci in the tomato genome (see Table S4 in the supplemental material).
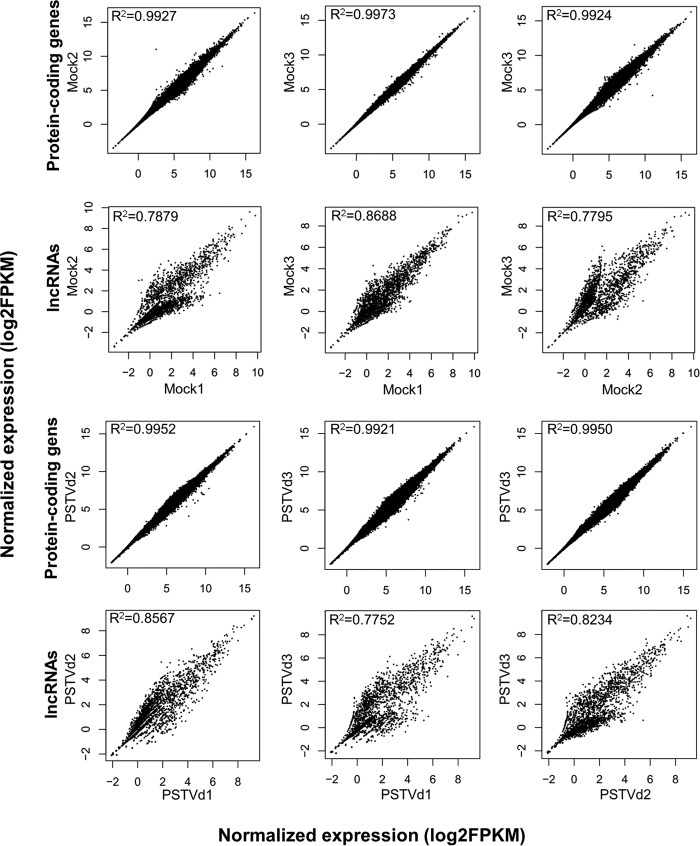
We further analyzed expression profiles of lncRNAs in response to viroid infection and found that tomato lncRNAs exhibited stochastic expression patterns even among the same treatments, which was in drastic contrast to the protein-coding transcripts (Fig. 2). To uncover lncRNAs that show specific expression patterns in response to PSTVd infection, we used stringent criteria (see Materials and Methods for details) and identified 44 lncRNAs that were PSTVd responsive (see Table S5 in the supplemental material).
FIG 2.
Normalized expression levels of protein-coding genes or lncRNAs among all biological replicates.
Many transcripts, including protein-coding as well as noncoding RNAs, can be processed by plant DCLs to generate sRNAs, such as pre-miRNAs and parental transcripts of phasiRNAs. Plant DCL1 generates ∼21- to 22-nt miRNAs, whereas DCL2, DCL3, and DCL4 generate siRNAs 22, 24, and 21 nt in length, respectively (26). To obtain a better understanding of the potential of host transcripts in generating sRNAs, we compared the rates of sRNA production from protein-coding transcripts versus those from lncRNAs and found that the average production of sRNAs from lncRNAs was about 10 times higher than that from protein-coding transcripts (Fig. 3A). Interestingly, while 24-nt size class sRNAs were predominantly mapped to protein-coding transcripts, similar amounts of sRNAs of 21-nt, 22-nt, and 24-nt size classes were mapped to lncRNAs (Fig. 3B). Thus, our observation implies that lncRNAs are more potent substrates of DCLs in generating 21- and 22-nt sRNAs than protein-coding transcripts. It is noteworthy that most of the 44 lncRNAs with significant expression changes upon PSTVd infection generated few or no sRNAs (see Table S5 in the supplemental material), which implies that these lncRNAs are unlikely to function via sRNA pathways.
FIG 3.
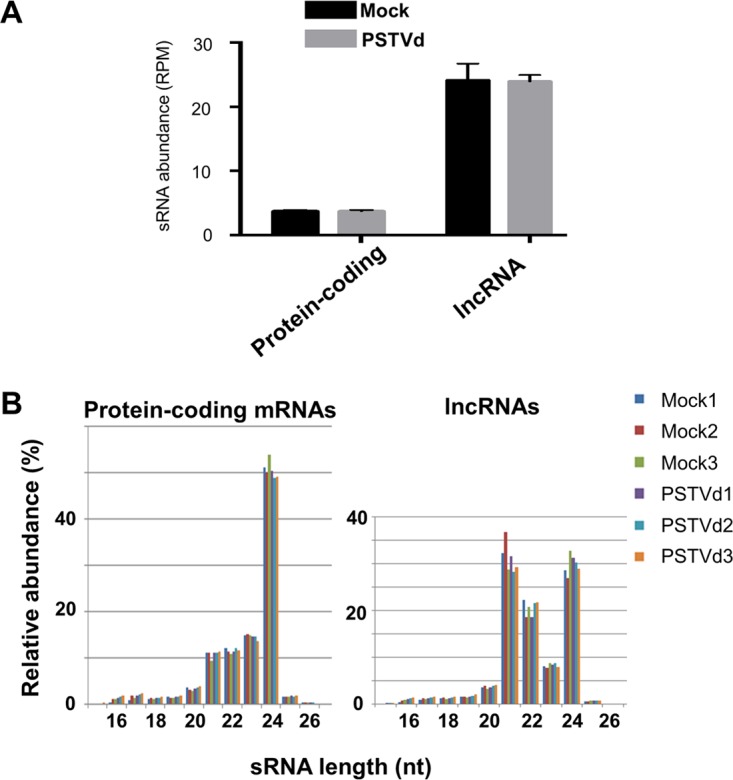
Features of sRNAs derived from tomato lncRNAs. (A) Average abundances of sRNAs generated from protein-coding transcripts and lncRNAs. (B) Size distribution of sRNAs mapped to protein-coding mRNAs (left) and lncRNAs (right). The error bars indicate standard deviations.
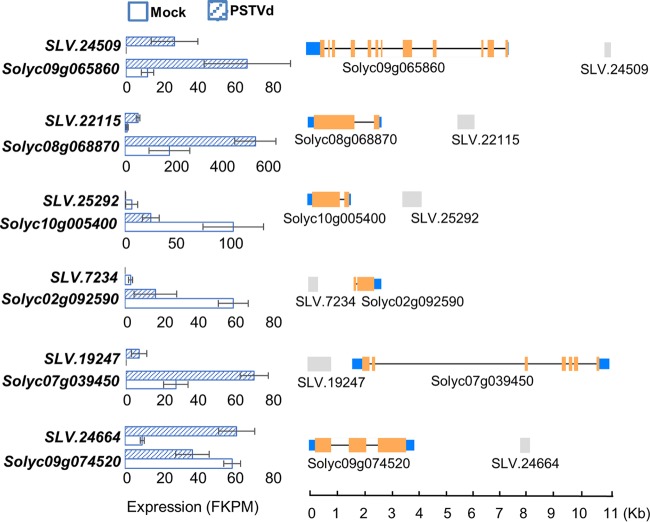
Accumulating evidence from recent reports supports the notion that various lncRNAs can serve as protein scaffolds for functions (27). Among them, some show specific coexpression patterns with their adjacent protein-coding genes, a phenomenon that has been proposed to be caused by (i) the conformational changes of local chromatin structure triggered by lncRNA expression or (ii) the induction or suppression caused by lncRNA-transcription factor complexes (28). In the present study, we found six lncRNAs showing tight coexpression patterns with their adjacent protein-coding genes upon PSTVd infection: three lncRNAs were coupregulated with their cognate adjacent protein-coding genes within 5 kb, two lncRNAs showed cosuppressed patterns with their cognate adjacent protein-coding genes within 5 kb, and the last showed opposite expression patterns (Fig. 4; see Table S6 in the supplemental material). These protein-coding genes are mostly involved in various metabolic and hormone signaling pathways (i.e., a uridine kinase, a peptidase A1, an inositol oxygenase, a density-regulated protein, an auxin F-box protein, and a conserved hypothetical protein) (see Table S6 in the supplemental material). Based on the working model of previously reported lncRNAs, we postulate that these lncRNAs possess regulatory functions to modulate the expression of adjacent genes.
FIG 4.
Tightly linked expression of lncRNAs and their nearby protein-coding genes. lncRNA–protein-coding transcript pairs showed significant changes in expression upon PSTVd infection. Shown are the expression dynamics (left) and genome coordinates (right) of lncRNAs and their cognate adjacent protein-coding transcripts. The orange and blue boxes depict the coding and untranslated regions, respectively. The gray boxes depict lncRNAs. The error bars indicate standard deviations.
PSTVd globally affects the expression and alternative splicing patterns of protein-coding transcripts.
Previous studies using macroarray and microarray analyses revealed that PSTVd infection in tomatoes triggers the expression changes of genes involved in stress response, cell wall structure, chloroplast function, protein metabolism, and hormonal pathways (21, 23). In this study, we employed RNA-Seq analysis to achieve higher resolution in dynamic changes of gene expression profiles in response to PSTVd infection in the Heinz 1706 cultivar. We identified 830 and 766 genes that were significantly up- and downregulated, respectively, upon PSTVd infection (Fig. 5A; see Table S7 in the supplemental material). The affected genes were predominantly metabolic genes and genes responsive to stimuli, which is in line with previous reports. Pathway analysis showed that 14 pathways were significantly elevated while 18 pathways were significantly suppressed upon PSTVd infection (P < 0.05) (see Table S8 in the supplemental material). The specific induction of the ascorbate glutathione cycle and l-ascorbate degradation V is of particular interest because both pathways are involved in the metabolism of reactive oxygen species (ROS) (29), which is commonly associated with plant responses to biotic and abiotic stresses.
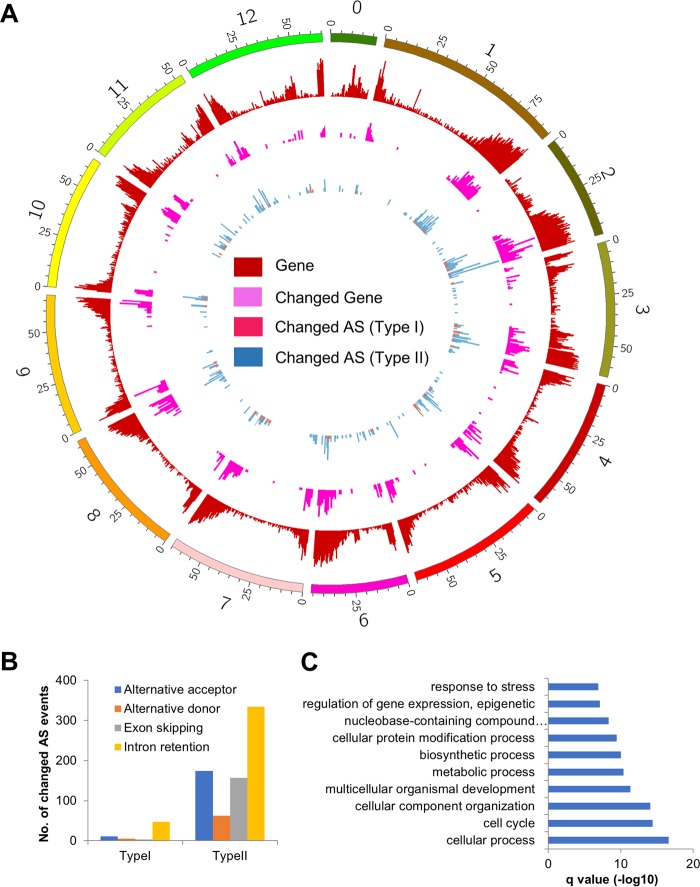
FIG 5.
Gene expression dynamics and global alterations in AS upon PSTVd infection. (A) Density plot of protein-coding genes, differentially expressed protein-coding genes, and protein-coding genes with changed AS (type I and type II) across the tomato genome. (B) Summary of different categories of changed AS events. (C) Top 10 GO terms significantly enriched in protein-coding genes with changed AS.
We further found that the differential expression of some protein-coding genes was associated with altered alternative splicing (AS) events. Two types of AS changes were observed: (i) splicing patterns were the same in mock-infected and infected samples, but only one of the splicing variants showed significant changes in expression, and (ii) splicing patterns changed directly between mock-infected and infected samples. We found that 57 loci had only one splicing variant selectively up- or downregulated (type I changes) (Fig. 5A; see Table S9 in the supplemental material). The type II changes included exon skipping, alternative 5′ donor sites, alternative 3′ acceptor sites, and intron retention. We identified 367 loci that showed distinct alternative splicing patterns between mock-infected and infected samples (Fig. 5A; see Table S10 in the supplemental material), among which intron retention was the most dominant AS event while exon skipping and alternative acceptors each accounted for one-fourth of the AS events (Fig. 5B). Gene ontology (GO) analysis showed that the genes with AS changes (types I and II) were predominantly involved in biosynthetic and metabolic processes, regulation of gene expression, and response to stress (Fig. 5C; see Table S11 in the supplemental material), indicating that PSTVd infection affects cellular processes by altering both the sequences and expression of regulatory and metabolic gene products.
PSTVd infection alters the functions of host sRNAs.
sRNAs, including miRNAs and siRNAs, are essential regulators involved in various biological processes. We tested if PSTVd, a pathogenic ncRNA, affected the expression and functions of host sRNAs. As shown in Fig. 6A (see Table S12 in the supplemental material), only miR171e and miR4376 among all known tomato miRNAs showed significant changes in expression upon PSTVd infection. This overall expression pattern is largely in agreement with previous reports that viroid infection has a limited effect on host miRNA expression (30).
FIG 6.
Expression and cleavage activity dynamics of tomato miRNAs upon PSTVd infection. (A) miRNA expression changes in PSTVd-infected samples compared with mock-infected samples. The average of miRNA counts in three replicates was used for plotting. **, P < 0.01. (B) PARE data showing alteration in miRNA-guided cleavage activities. (Left) miRNA and target pairings. The arrows indicate the guided cleavage positions. (Right) Abundances of degradome tags.
We then analyzed the expression of phasiRNAs, which are a unique class of plant siRNAs derived from truncated transcripts as products of miRNA-guided cleavages (31, 32) and display a head-to-tail phased pattern when mapped to parental transcripts. The phasiRNA pathway has an impact on plant innate immunity by regulating various nucleotide binding site–leucine-rich repeat (NBS-LRR) family genes (33–35). We identified 75 phasiRNA-generating loci (PHAS) and uncovered miRNA/sRNA triggers for 28 of them (see Table S13 in the supplemental material). None of the trigger miRNAs/siRNAs showed significant changes in their accumulation levels in response to PSTVd infection. However, the abundances of phasiRNAs generated from their parental PHAS loci varied significantly (2-fold changes in average sRNA production) in 17 loci (see Table S13 in the supplemental material). Some of these changes may be attributed to the differential accumulation of parental transcripts. For example, the reduction of phasiRNAs from the TIR1 (transport inhibitor response 1) gene (PHAS76; mapped to the tomato gene Solyc09g074520) in PSTV-infected samples was associated with less accumulation of TIR1 transcripts (see Table S13 in the supplemental material).
To evaluate the functions of host sRNAs in tomato responses to PSTVd infection, we performed parallel analysis of RNA ends (PARE) of our dRNA data to uncover sRNA-guided cleavages of mRNAs. PARE has been widely applied to validate sRNA-guided transcript cleavage experimentally at a genome-wide level (36, 37). Statistical analysis with the CleaveLand suite (36) identified sRNA-guided transcript cleavages and provided a calculated P value, as well as a category score ranking from 0 (most promising) to 5 (least promising). In general, categories 0 and 1 can be considered strong support of sRNA-guided cleavage (36). Using our deep-sequencing data on sRNAs (21- and 22-nt populations) and dRNAs, we uncovered 1,073 positive events of sRNA-guided transcript cleavage satisfying both the criteria of a P value of <0.05 in at least one sample and a category score of 1 or 0 in at least one sample (see Table S14 in the supplemental material).
Most of the miRNA activities remained largely unaffected based on the category scores and the accumulation levels of their target transcripts. For example, miR396 regulation of Solyc12g096070 (encoding growth-regulating factor 5 [GRF5]) did not affect the accumulation of Solyc12g096070, and the category scores were 0 in all mock-infected and PSTVd-infected samples (see Table S15 in the supplemental material). These serve as positive controls supporting the quality of the data. Interestingly, we found that activities were significantly enhanced for miR167-guided cleavage of Solyc02g037530 (encoding auxin response factor 8 [ARF8]) in PSTVd-infected samples. While all PSTVd-infected samples had category 0 for this regulation, two samples had category 2 and one sample had category 0 in mock-infected samples. This enhanced activity was correlated with a significant (P < 0.008), more than 2-fold reduction in Solyc02g037530 accumulation (Fig. 6B; see Table S15 in the supplemental material). It is noteworthy that changed activities appear to be restricted for specific targets. For instance, activities remained unaffected for miR167-guided cleavage of Solyc03g031970 (ARF8-1) (see Table S15 in the supplemental material). We also observed an interesting mode in which the activities of miRNA-guided cleavages were reduced but were associated with less target transcript accumulation. For instance, miR393-guided cleavage of Solyc09g074520 (encoding TIR1) had category 0 in two mock-infected samples and category 2 in the other mock-infected sample but could be detected in only one PSTVd-infected sample with category 4 (Fig. 6B; see Table S15 in the supplemental material). This phenomenon may be caused by the reduction of target transcripts via other regulatory means, resulting in less target for the miRNA activities. This is further supported by the observation that the secondary siRNAs derived from miR393-guided cleaved TIR1 transcripts (PHAS76) were also significantly reduced in PSTVd-infected samples (see Table S13 in the supplemental material).
Furthermore, we identified a phasiRNA (phasiRNAX [5′-UCAGCUGUUGAGCUACGGUGU-3′]) that acquired the trans-acting activity in mediating cleavage of transcripts from Solyc09g042380 (encoding an unknown protein) only in PSTVd-infected samples (Fig. 7). Similar inducible behavior was observed in another phasiRNA (phasiRNAY [5′-UGUUGAUCGCGCUUCAGUUGU-3′]), which showed highly increased efficiency in guiding the cleavage of Solyc07g052760 (encoding an unknown protein) specifically upon PSTVd infection (Fig. 7).
FIG 7.
Inducible trans-acting activities of phasiRNAs by PSTVd. (Left) Pairings of phasiRNA targets. The arrows indicate the cleavage sites. (Right) Abundances of degradome tags.
Plant silencing machinery responsive to the infection of PSTVd.
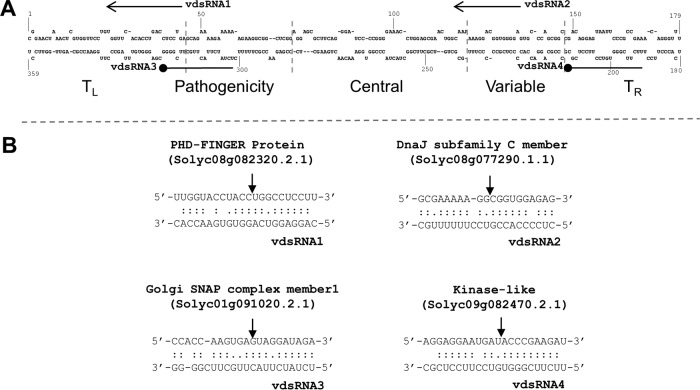
In plants, RNA silencing has been proposed to play a major role in defending the infection of RNA viruses and viroids (38). Viroids and their derived double-stranded RNAs are substrates for DCLs to generate vdsRNAs, which are loaded into host Argonaute proteins for function (39–41). Taking advantage of our dRNA data, we found that several vdsRNAs showed the capacity to guide the cleavage of host transcripts, supported by PARE analysis, in all three replicated samples (Fig. 8; see Table S14 in the supplemental material). The transcripts that were targeted by vdsRNAs included those encoding a PHD-Finger protein (Solyc08g082320), a DnaJ homolog protein (Solyc08g077290), a kinase-like protein (SLV.24849.1), and a Golgi SNAP receptor complex member (Solyc01g091020), all of which can be described as genes responsive to stimuli. We also observed that additional vdsRNAs could potentially guide cleavage of host transcripts, supported by a category score of 0 or 1 in the PARE analysis (see Table S14 in the supplemental material). However, the guided-cleavage function for this set of vdsRNAs was validated in only one out of the three replicates. This observation implies that some vdsRNAs guide the cleavage of target transcripts in a stochastic fashion, which might lack biological significance as individual events. Most of the above-mentioned vdsRNAs were present at relatively low levels (<50 transcripts per million [TPM]), except vdsRNA1 (∼380 TPM), in our sRNA deep-sequencing data (see Table S12 in the supplemental material). It is noteworthy that none of the vdsRNA targets showed significant expression changes in our RNA-Seq data sets, raising the possibility that vdsRNAs could regulate these host target genes through translational inhibition or other means.
FIG 8.
Cleavage of host transcripts guided by vdsRNAs of PSTVd. (A) Genomic locations of four vdsRNAs with potential cleavage activities. Arrows indicate the complementary orientations, while lines with a black circle indicate the sense orientations. The five domains on the PSTVd genome are labeled. (B) Four high-confidence vdsRNA-target pairs. The arrows indicate the guided-cleavage positions confirmed by PARE data.
PSTVd infection triggers plant immune responses.
Viroid infection generally leads to disease symptoms, including stunting, leaf epinasty, and chlorosis (42). How viroids cause plant diseases is an outstanding question. Through gene ontology analyses, we noticed the significant and massive induction of genes involved in immune responses upon PSTVd infection, as well as ROS production and responses (Fig. 9A; see Table S16 in the supplemental material).
FIG 9.
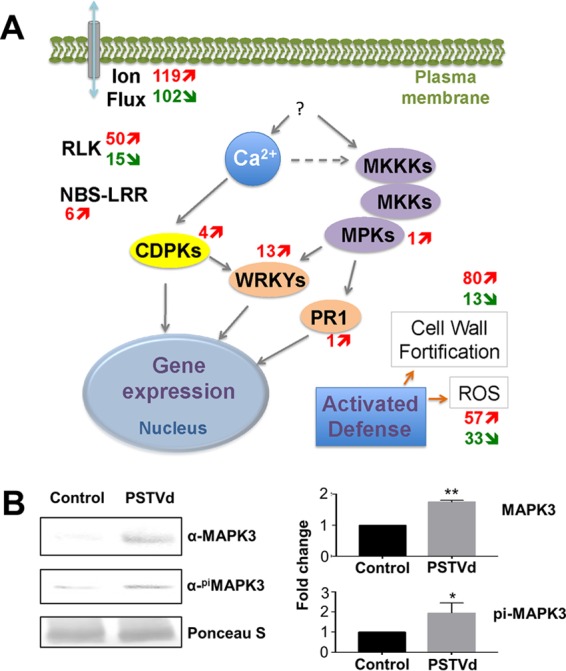
Activation of plant immune responses upon PSTVd infection. (A) Simplified view of plant immune response. The numbers of genes upregulated (red arrows) or downregulated (green arrows) by PSTVd infection are shown next to each gene family/category. (B) Elevated accumulation of the MAPK3 protein upon PSTVd infection. (Left) Western blots with Ponceau S staining as the loading control. (B) Statistical analysis of the upregulation of the MAPK3 protein. *, P < 0.05; **, P < 0.01. pi, phosphorylated. The error bars indicate standard deviations.
The mitogen-activated protein kinase (MAPK) cascade is the central player in signal transduction of PTI and ETI pathways to activate symptom-associated genes, such as the PR1 and WRKY transcription factors. Our data showed high elevation of genes encoding the MAPK3 (Solyc06g005170), as well as the PR1 (Solyc01g106620), and 13 WRKY transcription factors (see Table S7 in the supplemental material). Meanwhile, calcium-based signal transduction cascades, through calcium-dependent protein kinases (CDPKs), also play a critical role in sensing and responding to pathogen infection. Our analysis uncovered four upregulated CDPKs (see Table S7 in the supplemental material). Receptor-like kinases (RLKs) are membrane-localized proteins that sense PAMPs in the intracellular space. Although viroids have no reported PAMPs, PSTVd infection activated and repressed the expression of 50 and 15 RLKs, respectively (see Table S7 in the supplemental material). A dominant group of genes involved in innate immunity in plants is the NBS-LRR genes, which often recognize pathogen avirulence determinants. Viroids do not encode any proteins and have no products except their RNA genome and derivatives, but PSTVd could induce the expression of six NBS-LRR genes (see Table S7 in the supplemental material).
Upon pathogen attack, the activation of plant innate immunity is often associated with ion exchange, generation of ROS, cell wall fortification, and induction of hormone pathways. A functional module underlying the HRs to viral infection relies on the interaction of EDS1 (enhanced disease susceptibility 1), PAD4 (phytoalexin deficient 4), and SAG101 (senescence-associated gene 101) (43–46). All three of these components were activated in PSTVd-infected samples (see Table S7 in the supplemental material). Gene ontology analysis showed significant expression changes of 93 genes involved in cell wall biogenesis, 221 genes involved in ion transport, 90 genes involved in ROS metabolism, and 107 genes involved in ROS responses upon PSTVd infection (see Table S16 in the supplemental material). It has been shown that PSTVd infection greatly impacts hormone signaling (21). In this study, we uncovered significant changes in expression of (i) 93 and 146 genes involved in salicylic acid biogenesis and response, respectively; (ii) 83 genes involved in auxin responses; (iii) 40 and 98 genes involved in ethylene biogenesis and response, respectively; and (iv) 135 genes involved in response to abscisic acid (see Table S16 in the supplemental material).
To validate our observations, we analyzed the induction of MAPK3 via Western blotting. Using specific antibodies against plant MAPK3 and against phosphorylated MAPK3, we observed the induction of MAPK3 and phosphorylated MAPK3 in PSTVd-infected samples (Fig. 9B). This is in line with our RNA-Seq data and indicates the activation of MAPK3, and possibly its signaling cascades, upon PSTVd infection.
DISCUSSION
A pathogenic ncRNA has global effects on host alternative splicing and lncRNA expression.
Previous studies using macroarray and microarray analyses showed that PSTVd infection affects the expression of tomato genes, mainly those involved in stress responses, cell wall structure, chloroplast function, protein metabolism, and hormonal pathways (21, 23). Here, our high-resolution genome-wide analyses reveal the transcriptome changes upon PSTVd infection that are not only in agreement with previous observations, but also uncover a novel regulatory mode of PSTVd in affecting host alternative splicing. Two types of AS changes were observed: (i) mRNA splicing gave rise to the same patterns, but only one splicing variant showed significant changes in expression, and (ii) splicing patterns changed between mock-infected and PSTVd-infected samples. Each mode had dozens to hundreds of examples, supporting the global effects on alternative splicing upon PSTVd infection. It is not clear how PSTVd manipulates alternative splicing; however, it is intuitive to hypothesize that PSTVd likely interacts with host factors to achieve genome-wide changes in alternative splicing. Interestingly, a plant lncRNA has been shown to bind a splicing factor(s) that leads to the alteration of alternative splicing (47). Thus, a future search for splicing regulators that interact with PSTVd will likely provide mechanistic insights into pathogenic ncRNA-triggered changes in alternative splicing.
Alternative splicing has pivotal implications in host-microbe interactions. For example, a specific splicing form of the SYNTAXIN132 gene is critical for the symbiosis between legumes and rhizobia (48). In another example, a land plant-specific splicing form of eukaryotic general transcription factor IIIA is critical for aiding Pol II transcription using a PSTVd RNA template (3). It is emerging as a new layer of gene regulation as global changes in alternative splicing of host transcripts elicited by viruses have started to be observed (49). Hence, the altered alternative splicing patterns of a host may impact the replication and infectivity of PSTVd and other pathogens. Interestingly, the dominant groups affected are proteins involved in various catalytic reactions. Due to the changes in protein sequence caused by alternative splicing, their catalytic and regulatory functions are likely affected. Thus, our analysis uncovered a novel layer of gene regulation underpinning host-viroid interactions.
Several recent reports have revealed the role of lncRNAs in host immunity (50–52); however, it is unclear if pathogenic ncRNAs alone can alter the expression of host lncRNAs under native infection conditions. Our analysis demonstrated that pathogenic ncRNAs globally affect the expression of host lncRNAs. With the accumulating evidence supporting the regulatory role of lncRNAs in gene expression, this discovery reveals a new direction to explore the functions of pathogenic RNAs during infection. Interestingly, we showed that tomato lncRNAs are more potent in sRNA production, as well as serving as substrates for DCLs generating 21- and 22-nt size class sRNAs, than protein-coding transcripts, which might be attributed to the lack of protection from ribosome binding. Therefore, our analysis, together with the recent advances from other systems, provides a solid foundation for future exploration of lncRNA functions in host responses to pathogens.
PSTVd infection impacts the host miRNA-guided cleavage activity.
miRNAs are essential regulators of gene expression underlying various biological processes, including host responses to pathogen infection. A battery of miRNAs in Arabidopsis, including miR158a, miR160a, miR167, miR169, miR391, miR393, and miR396, showed elevated expression and enhanced capacity to guide the cleavage of target transcripts upon treatment with flg22 (53). These miRNAs regulate plant innate immunity in response to pathogen infection. Regarding viral infections in plants, it has been shown that miR168 is elicited by the activity of viral P19 protein to suppress the expression of host AGO1, which represents a viral counterdefense mechanism (54).
In our analyses, we found that the activity of miR167-guided cleavage of ARF8 was enhanced upon PSTVd infection, leading to the suppression of ARF8 expression. This likely impacts the auxin signaling pathways. It is noteworthy that miR167 regulation of ARF8 is also affected during plant defense against bacteria and fungi (53, 55), representing a common strategy in plant defense against a broad spectrum of pathogens. Besides the enhancement of miRNA activities, we observed another mode of action in which less accumulation of target transcripts (caused by other pathways) leads to less miRNA-guided cleavage. For example, miR393-guided cleavage of TIR1 transcripts was disturbed (see Table S15 in the supplemental material), likely due to changes in the accumulation of target transcripts. It is intriguing that miR393 is elevated during plant defense against bacterial infections to suppress the expression of TIR1 (53). Here, we observed a convergent pathway resulting in similarly reduced accumulation of TIR1. In summary, it appears that miRNA-based regulation of genes involved in auxin signaling is a general activity during plant-pathogen interactions.
Inducible trans-acting function of phasiRNAs in response to PSTVd infection.
phasiRNAs are generated from miRNA-guided cleaved transcripts. If a given phasiRNA possesses guided-cleavage function for RNA transcripts other than its parental transcripts, it is termed a trans-acting siRNA. Members of the miR482/2118 miRNA superfamily, as well as several other miRNAs, such as miR6022 and miR6023, control the expression of various NBS-LRR genes by triggering the generation of phasiRNAs in many plant species (33–35). This regulation is proposed to balance the plant's normal growth and responses to pathogen infection (56). A recent kingdom-wide survey of phasiRNAs revealed the dynamic association of phasiRNA expression during viral infection (31). However, the biological functions of most phasiRNAs were not analyzed or validated.
Besides observation of the expression dynamics of phasiRNAs, our analysis also revealed the trans-acting functions of several phasiRNAs in guiding target cleavage of protein-coding genes. Intriguingly, we found that the trans-acting activities of two phasiRNAs were specifically induced by PSTVd infection, which showed tight regulation of the trans-acting activity of phasiRNAs. Whether this tight-regulation mechanism is also involved in various other biological processes, such as development and responses to biotic and abiotic stresses, deserves future exploration.
PSTVd infection triggers plant immune responses.
Viroids are plant pathogens that cause symptoms such as stunting, epinasty, and leaf rugosity (42). The molecular basis of viroid pathogenicity is poorly understood. PSTVd infection alters the host transcriptome (21, 23, 24); however, these observations were descriptive and lacked in-depth mechanistic insights into plant-viroid interactions.
Plant innate immunity, such as PTI and ETI, is conserved in defending against pathogens, such as bacteria, fungi, and parasitic plants (11, 57). Plants perceive the presence of bacterial and fungal pathogens using various membrane-associated receptor-like kinases and consequently activate PTI. Some pathogens have evolved effectors to successfully interfere with PTI response as a counterdefense mechanism. Plants also utilize cellular NBS-LRR proteins to recognize effectors and activate ETI response. A successful immune response results in the activation of MAPK cascades, as well as other related activities, such as induction of the PR1 protein, generation of ROS, and an increase in callose deposition (11, 14, 17). Although viruses do not have identified effectors, some of them can trigger a hypersensitive response via the activation of defense genes (45).
Our analysis clearly showed the activation of a plant immune response to PSTVd infection. We unraveled the massive activation of genes in various signaling pathways (e.g., MAPK3 and CDPKs), as well as prominent marker genes involved in the associated activities (e.g., PR1, 1,3-beta-glucanase, and ROS biogenesis genes) (Fig. 9A). It is noteworthy that the induction of MAPK3 was verified at both transcriptional and translational levels in this study (Fig. 9). Interestingly, a recent report showed that the ROS level was elevated in the metabolic profile in PSTVd-infected tomato plants (58). Thus, it was clear that HRs were present in PSTVd-infected plants. PSTVd does not encode any proteins, so it is unlikely to possess any known effectors to trigger immune responses. Nevertheless, our data demonstrated that the tomato likely has effective perception of the presence of this “nonself” RNA through direct recognition of the infectious RNA and/or detection of the cellular environmental changes that result from PSTVd replication.
Whether PSTVd activates a plant double-stranded-RNA-binding protein kinase to trigger immune responses is an outstanding question. No double-stranded-RNA-activated protein kinase has been identified in plants to date; however, previous reports have suggested that a protein kinase viroid-induced (PKV) gene (Solyc12g017390) is related to symptom development upon PSTVd infection (59, 60). Surprisingly, this PKV gene was not expressed in any of our samples (mock infected or infected). This discrepancy might be attributed to a cultivar-specific response, because we used the Heinz 1706 tomato cultivar, while previous studies used Rutgers. Nevertheless, it suggests a yet-to-be-identified player that connects the sensing of PSTVd and the activation of plant immune responses, which might be one or a few of the elevated protein kinases in our data set (see Table S7 in the supplemental material).
A molecular framework underlying PSTVd-triggered disease.
Associated with the activation of plant immune responses, plant cells often undergo significant changes in cell wall dynamics, alteration in hormone signaling pathways, and generation of ROS. Previous studies have reported some of the metabolic changes in viroid-infected plants (21, 58, 61, 62), but none has explicitly addressed whether host immune responses are activated upon viroid infection. Our data clearly showed that PSTVd infection activated the plant basal defense, as evidenced by the massive activation of marker genes. We propose a new model in which the RNA-silencing pathway and plant basal defense cooperate to combat infection by PSTVd and possibly other nuclear-replicating viroids. RNA-silencing machinery mainly functions to eliminate or inhibit the viroid genomic RNAs and their derivatives by producing vdsRNAs. A few PSTVd vdsRNAs stochastically target some host transcripts, but they hardly affect the accumulation of the host transcripts. Meanwhile, the presence of PSTVd activates the plant basal defense and leads to HRs inducing symptom development. Global changes in host gene expression and the metabolic profiles reflect the activation of plant immunity and the HRs. The plant alternative splicing machinery is manipulated upon PSTVd infection to further regulate gene expression controlling various metabolic reactions, as well as stress responses. Endogenous sRNAs contribute to the regulation of essential genes in signaling pathways, such as an auxin-responsive transcription factor gene. This new model will serve as a foundation to further elucidate plant-viroid interactions, as well as to contribute to the development of preventive measures against viroid diseases.
MATERIALS AND METHODS
Plant materials.
Tomato plants (cv. Heinz 1706) were grown in a greenhouse at 25°C with a 16-h/8-h light/dark cycle. Seedlings with the first two true leaves just emerging were inoculated with water or water containing 150 ng of PSTVdInt RNA (39). Three weeks postinoculation, the leaf samples were collected and the PSTVd infection was verified by Northern blotting.
Total RNA isolation and enrichment for sRNAs.
Total RNA from leaves was isolated and fractioned into >200-nt and <200-nt populations using RNAzolRT reagent (Sigma-Aldrich, St. Louis, MO). The sRNA species were further purified using the mirVana miRNA isolation kit (Thermo Fisher Scientific, Grand Island, NY) following the manufacturer's instructions. The mRNA populations were further purified using the Magnetic mRNA isolation kit (NEB, Ipswich, MA).
Northern and Western blots.
Total RNA was run on 5% (wt/vol) polyacrylamide-8 M urea gels, transferred to Hybond-XL nylon membranes using a vacuum blotting system (Amersham Biosciences, Little Chalfont, United Kingdom), and then immobilized by UV cross-linking. After overnight hybridization to [α-32P]UTP-labeled riboprobes at 70°C in Ultrahyb reagent (Thermo Fisher Scientific), the membranes were washed four times and exposed to a storage phosphor screen (Kodak, Rochester, NY). To detect PSTVd, probes were obtained by transcribing in vitro HindIII-linearized pRZ6-2 template using a T7 Maxiscript kit (Thermo Fisher Scientific).
For Western blotting, total proteins were extracted from leaves using RIPA buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich). The protein samples were then subjected to SDS-PAGE and semidry membrane transfer following the standard protocol. The following antibodies were diluted for the detection of MAPK3: primary antibody against Pi-MAPK3 (Novus Biologicals, Littleton, CO) at 1:1,000, primary antibody against MAPK3 (Sigma-Aldrich) at 1:1,000, and horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (Sigma-Aldrich) at 1:4,000. After supplying HRP substrates (Amersham Biosciences), the fluorescence signals were captured and analyzed using MyECL (Thermo Fisher Scientific).
Library construction and sequencing.
sRNA libraries were constructed following the established protocol (63). Briefly, urea-PAGE gel-purified 18- to 30-nt sRNA populations were ligated with 3′ and 5′ adapters. The sRNA populations with adapters were reverse transcribed, PCR amplified, and then purified from the 3% agarose gel. dRNA libraries were generated following the established protocol (36) with minor modifications. Briefly, a 5′ RNA adapter containing an Ecop15I site (5′-CAGAGUUCUACAGUCCGACGAUCCAGCAG-3′) was ligated to the 5′ end of sRNA-guided cleaved mRNA species (dRNAs). An oligo(dT) primer containing an Ecop15I site (5′-CTGATCTAGAGGTACCGGATCCCAGCAGT-3′) was used to reverse transcribe the dRNAs. The products were then subjected to Ecop15I digestion (New England BioLabs), urea-PAGE gel purification of 97-mers, 3′ DNA adapter ligation, and PCR amplification. Strand-specific RNA-Seq libraries were constructed using the protocol described previously (64). All the library constructs were analyzed and quantified with a bioanalyzer and sequenced on an Illumina HiSeq 2500 system.
RNA-Seq read processing, transcript assembly, and differential expression.
Paired-end RNA-Seq reads were processed to remove adapters, as well as low-quality bases, using Trimmomatic (65), and trimmed reads shorter than 40 bp were discarded. The remaining high-quality reads were subjected to rRNA sequence removal by aligning to an rRNA database (66) using Bowtie (67), allowing up to three mismatches. The resulting read pairs were aligned to the tomato genome (83) using Tophat2 (68), allowing up to two mismatches. Only the aligned read pairs with no mismatch were assembled into transcripts using Cufflinks (69). The expression of the transcripts was measured and normalized to the number of fragments per kilobase of exon per million mapped fragments (FPKM), based on all mapped read pairs, using Cuffnorm, which is included in the Cufflinks package. Differential expression analysis was performed using Cuffdiff, also included in the Cufflinks package. Protein-coding genes with adjusted P values of <0.05 and no less than twofold changes were considered differentially expressed.
Functional annotation and coding potential assessment of assembled transcripts.
The assembled transcripts were compared to the Arabidopsis thaliana protein (70) and the UniProt (TrEMBL and Swiss-Prot) databases (71) using the BLASTX program (72) with an E value cutoff of 1e−4. The coding potential calculator (CPC) (73) was used to assess the coding potential of the transcripts.
Identification of lncRNAs and AS events.
Assembled transcripts derived from the tomato gene models or shorter than 200 bp were discarded. The remaining transcripts were digitally translated into proteins in 3 forward frames, and the longest amino acid sequences were used to determine the open reading frame (ORF) length. Only transcripts with CPC scores of <0 and ORF lengths of <100 were determined to be lncRNAs. The identified lncRNAs were classified into different categories according to their anatomical properties (68). Basically, lncRNAs with no intersection with any other transcripts and located 500 bp from any tomato gene loci were annotated as large intergenic ncRNAs (lincRNAs). LncRNAs that overlapped the minus strand of any region (intronic or exonic) of an annotated transcript model were categorized as antisense lncRNAs. lncRNAs that overlapped, in the same orientation, intronic regions of an annotated transcript model were categorized as intronic lncRNAs. Differentially expressed lncRNAs were identified using the same approach as for protein-coding genes described above, except that we required lncRNAs to be induced or repressed in all three replicated samples due to their stochastic expression.
AS events were identified from the expressed isoforms using AStalavista (74). Different types of AS events were extracted and counted as previously described (75), using an in-house Perl script.
sRNA sequence processing.
sRNA reads were processed to remove adaptors, low-quality bases, and short reads (less than 15 nt). The resulting sRNA reads were further cleaned by removing those that matched the sequences of tRNAs, snoRNAs, snRNAs (collected from GenBank), or rRNAs (66) using Bowtie (67). Raw counts for each unique sRNA were derived and normalized into TPM. sRNAs that were expressed at >5 TPM in at least one sample were processed using DESeq (76) to identify sRNAs differentially expressed between mock-infected and PSTVd-infected tomatoes. sRNAs with adjusted P values of <0.05 and >2-fold changes were considered differentially expressed sRNAs.
Identification of miRNAs.
We mainly followed previously described methods (77) to identify miRNAs from cleaned sRNA reads. sRNAs with >10 TPM in at least one sample were mapped to the tomato genome (version SL2.40) using Bowtie (67), allowing no mismatches. sRNAs mapped to more than 20 loci in the genome were discarded. The mapped loci and 200-bp flanking sequences on each side were extracted and then folded in silico using RNAfold (78). The resulting folded structures were checked with miRcheck (79) to identify candidate miRNAs, which were further compared with miRBase (80) to identify conserved miRNAs.
Identification of candidate PHAS loci.
We used previously described methods (31) to identify the candidate PHAS loci. In short, the cleaned sRNA sequences were mapped to the tomato reference sequences (genome or transcriptome) using Bowtie (67), allowing no mismatches and no more than six hits. The reference sequences were then scanned with a sliding window of 189 bp (nine 21-nt phase registers). A positive window was considered to contain no less than 10 unique sRNAs, with more than half of the unique sRNAs being 21 nt in length, and with no less than three 21-nt unique sRNAs falling into the phase registers. Windows were combined if they shared the same phase registers and fell into the same gene loci. P values and phasing scores for positive windows were calculated following methods described previously (81, 82).
dRNA read processing and identification of cleavage sites.
dRNA data were processed to remove adapters, low-quality bases, and short reads (<15 nt), as well as those mapped to the rRNA database. The cleaned dRNA reads were aligned with the assembled transcripts to generate a degradome density file, and the cleavage sites of miRNAs and siRNAs were identified using CleaveLand v4.3 (36).
Accession number(s).
The raw sequences of sRNAs, dRNA, and RNA-Seq have been deposited in the NCBI SRA with accession number SRP093503.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gregory Martin and Asuka Itaya for critical readings of the manuscript.
This work was supported by grants from the U.S. National Science Foundation (IOS-1354636 to B.D.; IOS-1025642 to Z.F.) and an Agriculture and Food Research Initiative Competitive Grant from the USDA National Institute of Food and Agriculture to B.D. and Z.F. (2014-67013-21550).
We declare that we have no conflicts of interest.
Y.Z., Y.W., B.D., and Z.F. designed the research; Y.Z. and Y.W. performed the research; Y.Z., Y.W., B.D., and Z.F. analyzed the data; Y.Z., Y.W., and Z.F. wrote the article.
This article is dedicated to Biao Ding, a caring mentor and exceptional colleague, who provided oversight of the investigation since its inception.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00247-17.
REFERENCES
- 1.Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Lee N, Moss WN, Yario TA, Steitz JA. 2015. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Qu J, Ji S, Wallace AJ, Wu J, Li Y, Gopalan V, Ding B. 2016. A land plant-specific transcription factor directly enhances transcription of a pathogenic noncoding RNA template by DNA-dependent RNA polymerase II. Plant Cell 28:1094–1107. doi: 10.1105/tpc.16.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlbach HP, Sanger HL. 1979. Viroid replication is inhibited by alpha-amanitin. Nature 278:185–188. doi: 10.1038/278185a0. [DOI] [PubMed] [Google Scholar]
- 5.Rackwitz HR, Rohde W, Sanger HL. 1981. DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 291:297–301. doi: 10.1038/291297a0. [DOI] [PubMed] [Google Scholar]
- 6.Castellano M, Pallas V, Gomez G. 2016. A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host histone deacetylase 6 activity. New Phytol 211:1311–1322. doi: 10.1111/nph.14001. [DOI] [PubMed] [Google Scholar]
- 7.Navarro B, Gisel A, Rodio ME, Delgado S, Flores R, Di Serio F. 2012. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J 70:991–1003. doi: 10.1111/j.1365-313X.2012.04940.x. [DOI] [PubMed] [Google Scholar]
- 8.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Lu J, Han Y, Fan X, Ding SW. 2013. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gantier MP, Williams BR. 2007. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev 18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 12.Schwessinger B, Ronald PC. 2012. Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol 63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 13.Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 14.Bent AF, Mackey D. 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 15.Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 16.Mandadi KK, Scholthof KB. 2013. Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25:1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spoel SH, Dong X. 2012. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 18.Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP. 2005. Mechanisms of plant resistance to viruses. Nat Rev Microbiol 3:789–798. doi: 10.1038/nrmicro1239. [DOI] [PubMed] [Google Scholar]
- 19.Ding B. 2009. The biology of viroid-host interactions. Annu Rev Phytopathol 47:105–131. doi: 10.1146/annurev-phyto-080508-081927. [DOI] [PubMed] [Google Scholar]
- 20.Flores R, Gago-Zachert S, Serra P, Sanjuan R, Elena SF. 2014. Viroids: survivors from the RNA world? Annu Rev Microbiol 68:395–414. doi: 10.1146/annurev-micro-091313-103416. [DOI] [PubMed] [Google Scholar]
- 21.Owens RA, Tech KB, Shao JY, Sano T, Baker CJ. 2012. Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol Plant Microbe Interact 25:582–598. doi: 10.1094/MPMI-09-11-0258. [DOI] [PubMed] [Google Scholar]
- 22.Rizza S, Conesa A, Juarez J, Catara A, Navarro L, Duran-Vila N, Ancillo G. 2012. Microarray analysis of Etrog citron (Citrus medica L.) reveals changes in chloroplast, cell wall, peroxidase and symporter activities in response to viroid infection. Mol Plant Pathol 13:852–864. doi: 10.1111/j.1364-3703.2012.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itaya A, Matsuda Y, Gonzales RA, Nelson RS, Ding B. 2002. Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol Plant Microbe Interact 15:990–999. doi: 10.1094/MPMI.2002.15.10.990. [DOI] [PubMed] [Google Scholar]
- 24.Katsarou K, Wu Y, Zhang R, Bonar N, Morris J, Hedley PE, Bryan GJ, Kalantidis K, Hornyik C. 2016. Insight on genes affecting tuber development in potato upon Potato spindle tuber viroid (PSTVd) infection. PLoS One 11:e0150711. doi: 10.1371/journal.pone.0150711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adkar-Purushothama CR, Brosseau C, Giguere T, Sano T, Moffett P, Perreault JP. 2015. Small RNA derived from the virulence modulating region of the Potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell 27:2178–2194. doi: 10.1105/tpc.15.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 27.Mercer TR, Mattick JS. 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 28.Clark BS, Blackshaw S. 2014. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet 5:164. doi: 10.3389/fgene.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martín R, Arenas C, Daròs J, Covarrubias A, Reyes J, Chua N. 2007. Characterization of small RNAs derived from Citrus exocortis viroid (CEVd) in infected tomato plants. Virology 367:135–146. doi: 10.1016/j.virol.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, Wang Y, Wu J, Ding B, Fei Z. 2015. A dynamic evolutionary and functional landscape of plant phased small interfering RNAs. BMC Biol 13:32. doi: 10.1186/s12915-015-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. 2012. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A 109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. 2012. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Addo-Quaye C, Miller W, Axtell MJ. 2009. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ. 2008. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 38.Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itaya A, Zhong X, Bundschuh R, Qi Y, Wang Y, Takeda R, Harris A, Molina C, Nelson R, Ding B. 2007. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J Virol 81:2980–2994. doi: 10.1128/JVI.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minoia S, Carbonell A, Di Serio F, Gisel A, Carrington JC, Navarro B, Flores R. 2014. Specific argonautes selectively bind small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J Virol 88:11933–11945. doi: 10.1128/JVI.01404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsarou K, Mavrothalassiti E, Dermauw W, Van Leeuwen T, Kalantidis K. 2016. Combined activity of DCL2 and DCL3 is crucial in the defense against Potato spindle tuber viroid. PLoS Pathog 12:e1005936. doi: 10.1371/journal.ppat.1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovalskaya N, Hammond RW. 2014. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci 228:48–60. doi: 10.1016/j.plantsci.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429. doi: 10.1046/j.1365-313X.2002.01297.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. 2011. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog 7:e1002318. doi: 10.1371/journal.ppat.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marathe R, Anandalakshmi R, Liu Y, Dinesh-Kumar SP. 2002. The tobacco mosaic virus resistance gene, N. Mol Plant Pathol 3:167–172. doi: 10.1046/j.1364-3703.2002.00110.x. [DOI] [PubMed] [Google Scholar]
- 46.Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE. 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 14:619–630. doi: 10.1016/j.chom.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JW, Crespi M. 2014. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30:166–176. doi: 10.1016/j.devcel.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 48.Pan H, Oztas O, Zhang X, Wu X, Stonoha C, Wang E, Wang B, Wang D. 2016. A symbiotic SNARE protein generated by alternative termination of transcription. Nat Plants 2:15197. doi: 10.1038/nplants.2015.197. [DOI] [PubMed] [Google Scholar]
- 49.Mandadi KK, Scholthof KB. 2015. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 27:71–85. doi: 10.1105/tpc.114.133991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satpathy AT, Chang HY. 2015. Long noncoding RNA in hematopoiesis and immunity. Immunity 42:792–804. doi: 10.1016/j.immuni.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Cao X. 2016. Long noncoding RNAs in innate immunity. Cell Mol Immunol 13:138–147. doi: 10.1038/cmi.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Au PC, Zhu QH, Dennis ES, Wang MB. 2011. Long non-coding RNA-mediated mechanisms independent of the RNAi pathway in animals and plants. RNA Biol 8:404–414. doi: 10.4161/rna.8.3.14382. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. 2010. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varallyay E, Valoczi A, Agyi A, Burgyan J, Havelda Z. 2010. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, Zhang HY, Xu PZ, Zhou JM, Wu XJ, Wang PR, Wang WM. 2014. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164:1077–1092. doi: 10.1104/pp.113.230052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fei Q, Zhang Y, Xia R, Meyers BC. 2016. Small RNAs add zing to the zig-zag-zig model of plant defenses. Mol Plant Microbe Interact 29:165–169. doi: 10.1094/MPMI-09-15-0212-FI. [DOI] [PubMed] [Google Scholar]
- 57.Hegenauer V, Furst U, Kaiser B, Smoker M, Zipfel C, Felix G, Stahl M, Albert M. 2016. Detection of the plant parasite Cuscuta reflexa by a tomato cell surface receptor. Science 353:478–481. doi: 10.1126/science.aaf3919. [DOI] [PubMed] [Google Scholar]
- 58.Bagherian SA, Hamzehzarghani H, Izadpanah K, Djavaheri M. 2016. Effects of potato spindle tuber viroid infection on tomato metabolic profile. J Plant Physiol 201:42–53. doi: 10.1016/j.jplph.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Hammond RW, Zhao Y. 2000. Characterization of a tomato protein kinase gene induced by infection by Potato spindle tuber viroid. Mol Plant Microbe Interact 13:903–910. doi: 10.1094/MPMI.2000.13.9.903. [DOI] [PubMed] [Google Scholar]
- 60.Hammond RW, Zhao Y. 2009. Modification of tobacco plant development by sense and antisense expression of the tomato viroid-induced AGC VIIIa protein kinase PKV suggests involvement in gibberellin signaling. BMC Plant Biol 9:108. doi: 10.1186/1471-2229-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marton L, Duran-Vila N, Lin JJ, Semancik JS. 1982. Properties of cell cultures containing the citrus exocortis viroid. Virology 122:229–238. doi: 10.1016/0042-6822(82)90223-9. [DOI] [PubMed] [Google Scholar]
- 62.Lison P, Tarraga S, Lopez-Gresa P, Sauri A, Torres C, Campos L, Belles JM, Conejero V, Rodrigo I. 2013. A noncoding plant pathogen provokes both transcriptional and posttranscriptional alterations in tomato. Proteomics 13:833–844. doi: 10.1002/pmic.201200286. [DOI] [PubMed] [Google Scholar]
- 63.Chen YR, Zheng Y, Liu B, Zhong S, Giovannoni J, Fei Z. 2012. A cost-effective method for Illumina small RNA-Seq library preparation using T4 RNA ligase 1 adenylated adapters. Plant Methods 8:41. doi: 10.1186/1746-4811-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 65.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 73.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foissac S, Sammeth M. 2007. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35:W297–W299. doi: 10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sammeth M, Foissac S, Guigo R. 2008. A general definition and nomenclature for alternative splicing events. PLoS Comput Biol 4:e1000147. doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fei Z, Joung JG, Tang X, Zheng Y, Huang M, Lee JM, McQuinn R, Tieman DM, Alba R, Klee HJ, Giovannoni JJ. 2011. Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res 39:D1156–D1163. doi: 10.1093/nar/gkq991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 80.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z. 2013. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Paoli E, Dorantes-Acosta A, Zhai J, Accerbi M, Jeong DH, Park S, Meyers BC, Jorgensen RA, Green PJ. 2009. Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA 15:1965–1970. doi: 10.1261/rna.1706109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomato Genome Consortium. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.