ABSTRACT
Recent studies have shown that Fc-Fcγ receptor (FcγR) interactions are required for in vivo protection against influenza viruses by broadly reactive anti-hemagglutinin (HA) stem, but not virus strain-specific, anti-receptor binding site (RBS), antibodies (Abs). Since only a few Abs recognizing epitopes in the head region but outside the RBS have been tested against single-challenge virus strains, it remains unknown whether Fc-FcγR interactions are required for in vivo protection by Abs recognizing epitopes outside the RBS and whether the requirement is virus strain specific or epitope specific. In the present study, we therefore investigated the requirements for in vivo protection using two pan-H5 Abs, 65C6 and 100F4. We generated chimeric Abs, 65C6/IgG2a and 100F4/IgG2a, which preferentially engage activating FcγRs, and isogenic forms, 65C6/D265A and 100F4/D265A, which do not bind FcγR. Virus neutralizing activity, binding, antibody-dependent cellular cytotoxicity (ADCC), and in vivo protection of these Abs were compared using three H5 strains, A/Shenzhen/406H/2006 (SZ06), A/chicken/Shanxi/2/2006 (SX06), and A/chicken/Netherlands/14015526/2014 (NE14). We found that all four chimeric Abs bound and neutralized the SZ06 and NE14 strains but poorly inhibited the SX06 strain. 65C6/IgG2a and 100F4/IgG2a, but not 65C6/D265A and 100F4/D265A, mediated ADCC against target cells expressing HA derived from all three virus strains. Interestingly, both 65C6/IgG2a and 65C6/D265A demonstrated comparable protection against all three virus strains in vivo; however, 100F4/IgG2a, but not 100F4/D265A, showed in vivo protection. Thus, we conclude that Fc-FcγR interactions are required for in vivo protection by 100F4, but not by 65C6, and therefore, protection is not virus strain specific but epitope specific.
IMPORTANCE Abs play an important role in immune protection against influenza virus infection. Fc-FcγR interactions are required for in vivo protection by broadly neutralizing antistem, but not by virus strain-specific, anti-receptor binding site (RBS), Abs. Whether such interactions are necessary for protection by Abs that recognize epitopes outside RBS is not fully understood. In the present study, we investigated in vivo protection mechanisms against three H5 strains by two pan-H5 Abs, 65C6 and 100F4. We show that although these two Abs have similar neutralizing, binding, and ADCC activities against all three H5 strains in vitro, they have divergent requirements for Fc-FcγR interactions to protect against the three H5 strains in vivo. The Fc-FcγR interactions are required for in vivo protection by 100F4, but not by 65C6. Thus, we conclude that Fc-FcγR interactions for in vivo protection by pan-H5 Abs is not strain specific, but epitope specific.
KEYWORDS: influenza viruses, in vivo protection, monoclonal antibodies
INTRODUCTION
Human influenza epidemics cause 3 to 5 million cases of severe infection and up to half a million deaths per year worldwide (1). Zoonotic infections, in which humans have no preexisting immunity, could result in influenza pandemics and outbreaks, such as the emergence of the pandemic H1N1 virus in 2009 and the avian H5N1 and H7N9 viruses (2–4).
Influenza viruses are enveloped, negative-sense, single-strand RNA viruses with segmented genomes. Hemagglutinin (HA), neuraminidase (NA), and matrix 2 (M2) are three virion surface proteins. HA is composed of two major domains: the globular head (HA1) and the stem (HA2). These domains assemble into trimers of covalently linked HA1/HA2 heterodimers. HA1 mediates binding to sialic acid receptors, and HA2 mediates viral and endosomal membrane fusion (5). HA is also a major target of host antibody responses. It is well documented that anti-HA antibody responses elicited by vaccinations and passive administrations of anti-HA antibodies provide protection against influenza infection in humans (6).
In past years, quite a few antibodies (Abs) against the stem of HA have been isolated, and the epitopes for these Abs have been mapped. These Abs also provide various degrees of cross-protection (7–16). Epitopes of some of the Abs are (i) conserved within the HA subtypes of group 1 (7–12) or group 2 (13, 14), (ii) found in both groups 1 and 2 (15), or (iii) present even between influenza A and B viruses (16). In addition, Abs against the globular head with different degrees of cross-reactivity have also been isolated (17–30). Many of these Abs are virus strain specific and recognize epitopes located in the receptor binding site (RBS), but some Abs recognize conserved epitopes within or outside the RBS of diverse strains of different subtypes (17–19) or within a HA subtype (20–30). The antibody repertoire against epitopes located in the head is more diverse than those Abs targeting epitopes in the stem (31). This could be due to the occlusive (less accessible) nature of epitopes in the stem on virions. Few Abs with specific modes of action may be able to interact with these epitopes (32). As a result, antibody responses against the head of HA are more potent and dominant than those against the stem (31).
Recent studies have shown that interactions between the Fc portion of antibodies and family members of the Fcγ receptor (FcγR) are required for in vivo protection against influenza viruses by both broadly neutralizing or nonneutralizing, but not strain-specific, Abs (25, 33–35). For example, a study by DiLillo et al. (33) showed that broadly neutralizing antistem Abs require Fc-FcγR interactions to mediate antibody-dependent cellular cytotoxicity (ADCC) for in vivo protection, whereas strain-specific anti-RBS Abs do not. Another study by DiLillo et al. (34) tested the contribution of Fc-FcγR interactions to in vivo protection against the A/Netherlands/602/2009 (Neth09) H1N1 strain, using a panel of 13 anti-HA human Abs, which included 8 antihead Abs. They showed that broadly cross-reactive antibodies, regardless whether they are neutralizing and nonneutralizing or whether they are antihead or antistem Abs, depend upon the Fc-FcγR interactions for in vivo protection. However, since the epitopes recognizing the antihead Abs were not mapped, it is not clear whether these antibodies are directed to epitopes within or outside the RBS. Furthermore, Henry Dunand et al. (35) examined the contribution of Fc-FcγR interactions for in vivo protection against the A/Shanghai/1/2013 H7N9 strain using three nonneutralizing anti-H7 stem Abs. For the comparison, they also tested a neutralizing anti-RBS Ab 07-5G01. They found that two broadly cross-reactive antistem Abs, 07-5E01 and 41-5D06, but not a narrowly cross-reactive Ab, 24-4C01, or the anti-RBS Ab 07-5G01, are protective through Fc-mediated effector cell recruitment. In this study, antibodies directed to epitopes outside the RBS were not tested. Tan et al. (25) described four mouse Abs raised against the H7 HA. Of these Abs, only Ab 1H5 was used for testing the contribution of Fc-FcγR interactions to in vivo protection against the A/Shanghai/1/2013 H7N9 strain. Ab 1H5, a nonneutralizing antibody, reacts to H7 viruses from the North American lineage. Diminished binding of 1H5 was found in an HA mutant carrying the R65K mutation, suggesting that it binds to an epitope at the interface between the head and stem domains. The results of in vivo testing indicate that both Fc-FcγR interactions and Fc-complement interactions contribute to the protective effect of 1H5. More recently, it has been shown that optimal activation of Fc-mediated effector functions by anti-HA antibodies requires not only Fc-FcγR interactions but also interactions between HA and sialic acid receptor on effector cells (36–38). Since the requirement of the Fc-FcγR interactions for in vivo protection by the Abs in these studies was studied using a single-challenge virus strain and because among antihead Abs tested, 1H5 is the only Ab whose epitope was tentatively mapped to the head region outside the RBS, it remains to be determined whether the observed association between Fc-FcγR interactions and in vivo protection by these Abs is strain specific or epitope specific. It is also unknown whether Fc-FcγR interactions are necessary for in vivo protection by Abs that recognize epitopes in the head region but outside the RBS.
Previously, we isolated two human Abs 65C6 and 100F4 from a highly pathogenic avian influenza (HPAI) H5N1-infected individual. These Abs potently neutralize all clades and subclades of HPAI H5N1 viruses except subclade 7.2. Furthermore, they protect against lethal challenge of both homologous and heterologous H5N1 strains (26, 27). More recently, we showed that 65C6 and 100F4 Abs also cross-neutralize newly emerging HPAI H5N6 and H5N8 reassortants. In mice, a single injection of 0.5 mg of 100F4 Ab/kg of body weight prophylactically or 10 mg/kg 100F4 therapeutically outperformed a 5-day course of 10-mg/kg/day oseltamivir treatment against lethal H5N6 or H5N8 challenge (39). Epitope mapping by cocrystal structures of Fab-HA complexes showed that the 65C6 epitope consists of 18 contact residues from the loop consisting of residues 121 to 129 and the β-strand consisting of residues 162 to 171 in the globular head region. The 100F4 epitope consists of 21 contact residues located right below the 65C6 epitope (40, 41). Among these 21 contact residues, single mutations of D77 or E119 almost completely abolish the neutralization activity of 100F4 (27, 40, 41). Thus, Abs 65C6 and 100F recognize two conformational epitopes in the head region outside the RBS. Mechanistically, in vitro neutralization mediated by Abs 65C6 and 100F4 differs from anti-RBS antibodies that block binding of virus to receptors and antistem antibodies that block HA-mediated viral-endosomal membrane fusion (27). Both 65C6 and 100F4 bind virus before and after virus attaches to target cells. Antibody-bound viruses are internalized into the perinuclear region. Antibody-bound HA on virion surfaces fails to mediate low-pH-triggered membrane fusion (27). Importantly, although Abs 65C6 and 100F4 neutralize A/Shenzhen/406H/2006 (SZ06) H5N1 and A/chicken/Netherlands/14015526/2014 (NE14) H5N8 strains but poorly neutralize or fail to neutralize the A/chicken/Shanxi/2/2006 (SX06) H5N1 strain, both Abs effectively protect against all three strains in vivo (26, 39). Thus, using Abs 65C6 and 100F4 would allow us to obtain definitive answers as to whether the requirement of Fc-FcγR interactions for in vivo protection is virus strain specific or epitope specific and whether Fc-FcγR interactions are necessary for in vivo protection by Abs that recognize epitopes in the head region but outside the RBS. To address these issues, we first compared in vitro neutralization and in vivo protection of Abs 65C6 and 100F4 against these three H5 viruses. We then constructed chimeric heavy chains by linking the Fv of the heavy chains of Abs 65C6 and 100F4 to the Fc of mouse IgG2a, which preferentially engages activating FcγRs, or a D265A mutant, which cannot bind FcγRs (40, 41). Chimeric Abs (cAbs) containing the chimeric heavy chains of 65C6 or 100F4 and corresponding light chains were produced and purified. The in vitro neutralizing, binding, and ADCC activity of these cAbs against the three HPAI H5 viruses were carefully studied and correlated with their ability to protect mice in challenge experiments with these viruses.
RESULTS
Abs 100F4 and 65C6 potently neutralize SZ06 and NE14 virus strains but have no or little neutralization against SX06 in vitro.
In this study, three representative HPAI H5 strains SZ06, SX06, and NE14 were selected for evaluation (Table 1). Previously, using pseudotype-based neutralization (PN) assays, it was found that Abs 65C6 and 100F4 potently neutralized SZ06 and NE14 pseudotypes, but poorly neutralized or did not neutralize a SX06 pseudotype (26, 39). To test whether the different neutralizing activities could be reproduced with replication-competent SZ06, NE14, and SX06 viruses, we performed microneutralization (MN) and plaque reduction assays.
TABLE 1.
Three HPAI H5 strains used in the present study
| Virus strain | Strain abbreviation | Subtype | Clade | TCID50/ml | PFU/ml | MLD50/ml |
|---|---|---|---|---|---|---|
| A/Shenzhen/406H/2006 | SZ06 | H5N1 | 2.3.4 | 1.5 × 108 | 1.1 × 108 | 1.0 × 108 |
| A/chicken/Shanxi/2006 | SX06 | H5N1 | 7.2 | 1.8 × 106 | 1.1 × 106 | 3.0 × 104 |
| A/chicken/Netherlands/14015526/2014 | NE14 | H5N8 | 2.3.4.4 | 2.9 × 107 | 1.9 × 107 | 6.2 × 107 |
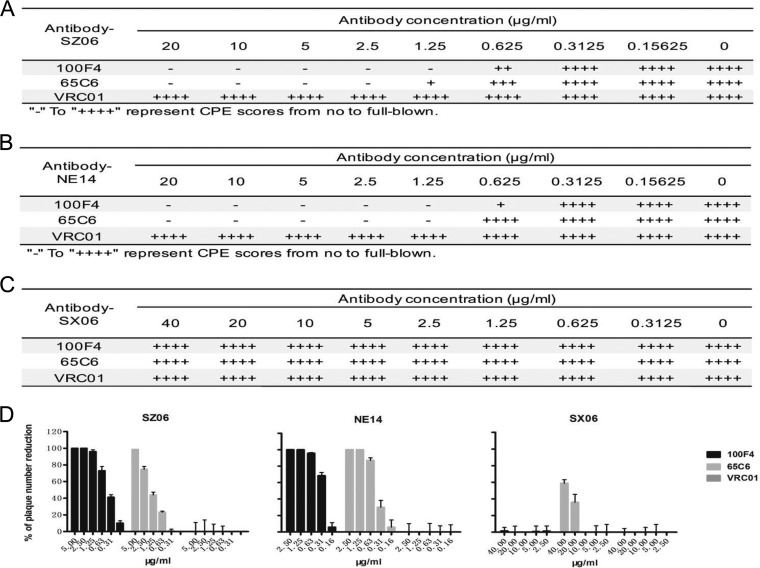
Figure 1A to C show the titrations of neutralizing activity of Abs 100F4 and 65C6 along with a control Ab VRC01 as measured by the microneutralization assay. Clearly, Ab 100F4 at concentrations of 1.25 μg/ml or higher or Ab 65C6 at concentrations of 2.5 μg/ml or higher completely neutralized SZ06 infection. In contrast, the control Ab, VRC01, even at a concentration of 20 μg/ml, did not have any neutralizing activity against this virus (Fig. 1A). Similarly, Abs 100F4 and 65C6 completely neutralized NE14 infection at concentrations of 1.25 μg/ml or higher (Fig. 1B). In contrast, neither 100F4 nor 65C6 neutralized SX06 infection, even at concentrations of 40 μg/ml (Fig. 1C).
FIG 1.
Assessment of neutralization by 100F4 and 65C6 antibodies and control antibody VRC01 using a microneutralization assay (A to C) or plaque reduction assay (D). Neutralization of the antibodies against virus strain SZ06 (A), NE14 (B), and SX06 (C) by a microneutralization assay (A to C) or plaque reduction assay (D) is shown. CPE, cytopathic effect.
Similar, but slightly different, results were also obtained when neutralization was measured by a plaque reduction assay (Fig. 1D). Ab 100F4 at a concentration of 1.25 μg/ml or Ab 65C6 at concentrations of 5 and 1.25 μg/ml completely blocked plaque formation of the SZ06 and NE14 strains, respectively. As expected, control Ab VRC01, even at a concentration of 20 μg/ml, did not inhibit these viruses (the left and middle panels in Fig. 1D). In contrast to those results, Ab 100F4 did not reduce plaque formation by the SX06 strain, even at a concentration of 40 μg/ml. However, a 60% reduction in plaque formation occurred with Ab 65C6 at the concentration of 40 μg/ml (the right panel in Fig. 1D). Thus, we conclude that similar to what was found using the PN assay (26, 39), Abs 100F4 and 65C6 potently neutralize both SZ06 and NE14 strains but have very little to no neutralizing activity against the SX06 strain in vitro, even though they both bind to the H5 HA of SX06 strain with much lower binding activity (see below).
Abs 65C6 and 100F4 completely or almost completely protect mice from lethal challenge in vivo.
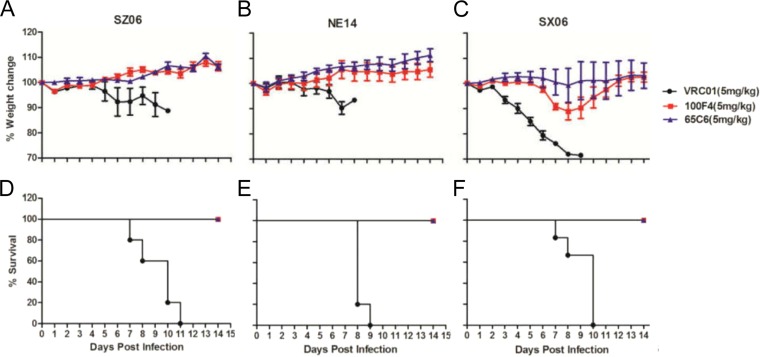
Figure 2 shows the prophylactic efficacies of 5 mg/kg of Abs 100F4 and 65C6 or control Ab VRC01 against 10 MLD50 (50% mouse lethal dose) lethal challenges with virus strains SZ06 (Fig. 2A and D), NE14 (Fig. 2B and E), and SX06 (Fig. 2C and F). Mice infused with control Ab VRC01 followed by lethal challenge with SZ06, NE14, or SX06 strain exhibited severe sickness on day 6 or 7 postchallenge, and all mice died. In contrast, all mice infused with Ab 100F4 followed by lethal challenge with SZ06 or NE14, or given Ab 65C6 followed by lethal challenge using SZ06, NE14, or SX06 exhibited no signs of illness, and all survived. While the mice given Ab 100F4 followed by lethal challenge with SX06 lost about 10% of their weight, they all survived the infection. Taking all these results together, we conclude that Abs 65C6 and 100F4 completely or almost completely protected mice from lethal challenges using virus strain SZ06, NE14, or SX06, even though they have no or very little neutralizing activity against SX06 strain in vitro (Fig. 1 and 3).
FIG 2.
In vivo efficacy of Abs 100F4 and 65C6 along with the VRC01 control Ab against virus strains SZ06, NE14, and SX06. (A and D) Time course of body weight changes (A) and survival rate of each group (D) against SZ06 strain. (B and E) Time course of body weight changes (B) and survival rate of each group (E) against strain NE14. (C and F) Time course of body weight changes (C) and survival rate of each group (F) against strain SX06. The survival rate was calculated as percent survival within each experimental group (five mice in each group).
FIG 3.
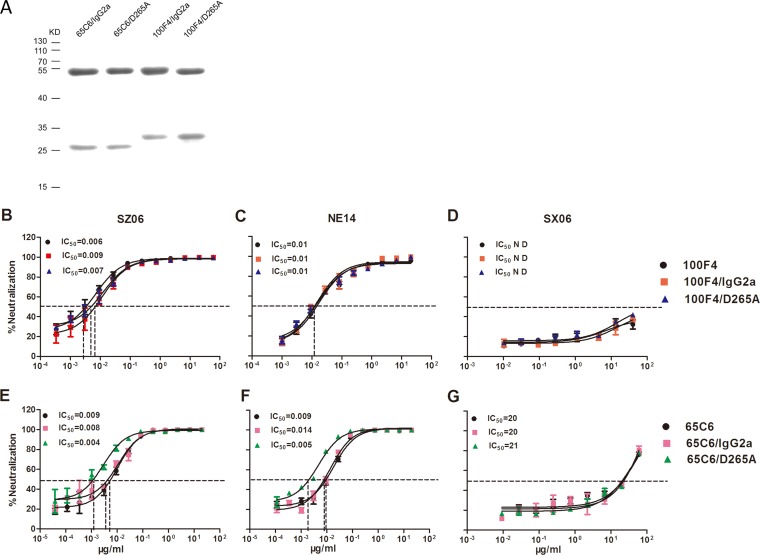
Detection and neutralizing activity of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A. (A) Purified 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A detected by SDS-PAGE followed by Coomassie blue staining. The top bands are chimeric heavy chains, and the bottom bands are kappa or lambda light chains. The positions of molecular mass markers (in kilodaltons [KD]) are shown to the left of the gel. (B to D) The titration and IC50s of neutralizing activity of 100F4/IgG2a and 100F4/D265A as well as human Ab 100F4 against SZ06 (B), NE14 (C), or SX06 (D) using a pseudotype-based neutralization (PN) assay. (E to G) The titration and IC50s of neutralizing activity of 65C6/IgG2a and 65C6/D265A as well as human Ab 65C6 against SZ06 (E), NE14 (F), or SX06 (G) using a PN assay. N D, not determined.
Chimeric Abs 65C6/IgG2a and 65C6/D265A or 100F4/IgG2a and 100F4/D265A have comparable neutralization activity in vitro.
Several recent studies have demonstrated the importance of Fc-FcγR interactions in the clearance of influenza virus infection in vivo (25, 33–35). To test whether Fc-FcγR interactions are critical for in vivo protection by Abs 100F4 and 65C6, we constructed four chimeric Abs (cAbs) by linking the Fv regions of Ab 100F4 or 65C6 heavy chain to mouse IgG2a or a D265A mutant. The latter mutation abrogates binding of mouse IgG2a to FcγR (40, 41). These fusion genes encoding chimeric heavy chains of 100F4 or 65C6, along with their corresponding human light chains (26), were cotransfected into Drosophila S2 cells. Stably transfected S2 clones were selected, grown in a BioWAVE reactor, and cAbs in the culture supernatants were purified. Figure 3A shows the Coomassie blue staining of the heavy- and light-chain bands of the four purified cAbs, 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A, separated using 12% SDS-PAGE.
To ensure that Fab functions of 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A were unaffected by altering the Fc region, we verified their binding (see below) and neutralizing activity using the PN assay. Figure 3B to D and Table 2 show that 100F4/IgG2a and 100F4/D265A had similar potent neutralizing activity against strain SZ06 (the 50% inhibitory concentrations [IC50s] of 100F4/IgG2a and 100F4/D265A were 0.009 and 0.007 μg/ml, respectively) and NE14 (the IC50s of both 100F4/IgG2a and 100F4/D265A were 0.01 μg/ml). In contrast, both 100F4/IgG2a and 100F4/D265A did not neutralize SX06. Similarly, 65C6/IgG2a and 65C6/D265A potently neutralized SZ06 (the IC50s of 65C6/IgG2a and 65C6/D265A were 0.008 and 0.004 μg/ml, respectively) and NE14 (the IC50s of 65C6/IgG2a and 65C6/D265A were 0.014 and 0.005 μg/ml, respectively) but had very little neutralizing activity against SX06 (the IC50s of 65C6/IgG2a and 65C6/D265A were 21 and 20 μg/ml, respectively) (Fig. 3E to G and Table 2). For comparison, we also tested the neutralizing activity of human Abs 100F4 and 65C6 against all three H5 pseudotypes and observed IC50s comparable to those of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A (Fig. 3B to G). Thus, taking these results together, we conclude that cAbs 65C6/IgG2a and 65C6/D265A as well as 100F4/IgG2a and 100F4/D265A had neutralizing activities against all three representative H5 strains that were comparable to the neutralizing activities of the parental human Abs 100F4 and 65C6.
TABLE 2.
Summary of the results of ELISA binding, neutralization, FACS analysis, ADCC, and in vivo testing of chimeric Abs 100F4/IgG2a, 100F4/D265A, 65C6/IgG2a, and 65C6/D265A
| Parameter and cAb | Value for parameter in virus straina: |
||
|---|---|---|---|
| SZ06 | NE14 | SX06 | |
| IC50 (μg/ml) | |||
| 100F4/IgG2a | 0.009 | 0.010 | ND |
| 100F4/D265A | 0.007 | 0.010 | ND |
| 65C6/IgG2a | 0.008 | 0.014 | 20 |
| 65C6/D265A | 0.004 | 0.005 | 21 |
| EC50 (ng/ml) | |||
| 100F4/IgG2a | 4.4 | 7.3 | 417.0 |
| 100F4/D265A | 13.1 | 27.0 | 340.0 |
| 65C6/IgG2a | 13.6 | 23.8 | 446.0 |
| 65C6/D265A | 8.1 | 8.8 | 381.0 |
| MFIb | |||
| 100F4/IgG2a | 254 | 155 | 137 |
| 100F4/D265A | 216 | 145 | 127 |
| 65C6/IgG2a | 290 | 214 | 128 |
| 65C6/D265A | 294 | 260 | 138 |
| ADCC (%) (mean ± SEM) | |||
| 100F4/IgG2a | 18.2 ± 2.6 | 21.2 ± 2.5 | 24.1 ± 6.6 |
| 100F4/D265A | 5.0 ± 3.1 | 0.3 ± 0.4 | 2.4 ± 3.2 |
| 65C6/IgG2a | 20.1 ± 2.7 | 13.9 ± 2.0 | 17.2 ± 2.8 |
| 65C6/D265A | 1.5 ± 3.4 | 5.5 ± 1.7 | −0.6 ± 5.5 |
| Survival rate (no. of mice that survived/total no. of mice) | |||
| 100F4/IgG2a | 4/5 | 5/5 | 5/5 |
| 100F4/D265A | 0/5 | 1/5 | 1/5 |
| 65C6/IgG2a | 4/5 | 5/5 | 5/5 |
| 65C6/D265A | 4/5 | 5/5 | 5/5 |
ND, not detected.
MFI, median fluorescence intensity of FACS staining by chimeric Abs.
cAbs 65C6/IgG2a and 65C6/D265A or 100F4/IgG2a and 100F4/D265A retain binding, but differ in their ADCC, capability.
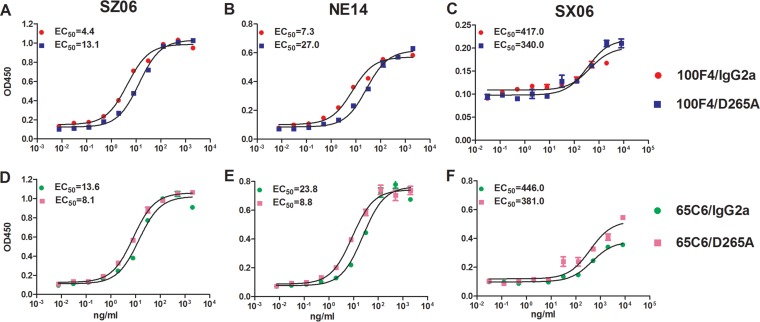
To test the binding activity of cAbs, we first performed enzyme-linked immunosorbent assays (ELISAs) against virus-like particles (VLPs) derived from SZ06, NE14, and SX06 strains (Fig. 4 and Table 2). Both 100F4/IgG2a and 100F4/D265A bound well against SZ06 and NE14 with 50% effective concentrations (EC50s) of 4.4 and 13.1 ng/ml to SZ06 and 7.3 and 27.0 ng/ml to NE14, respectively (Fig. 4A and B), but they bound poorly to SX06 with EC50s of 417 and 343 ng/ml, respectively (Fig. 4C). Similarly, both 65C6/IgG2a and 65C6/D265A bound well to SZ06 and NE14 with EC50s of 13.6 and 8.1 ng/ml to SZ06 and 23.8 and 8.8 ng/ml to NE14, respectively (Fig. 4D and E). However, 65C6/IgG2a and 65C6/D265A bound poorly to SX06 with EC50s of 446 and 381 ng/ml, respectively (Fig. 4F).
FIG 4.
Measurement of binding activity of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A using an ELISA. The titration and EC50s of binding activity of 100F4/IgG2a and 100F4/D265A against virus strains SZ06 (A), NE14 (B), and SX06 (C). The titration and EC50s of binding activity of 65C6/IgG2a and 65C6/D265A against SZ06 (D), NE14 (E), and SX06 (F).
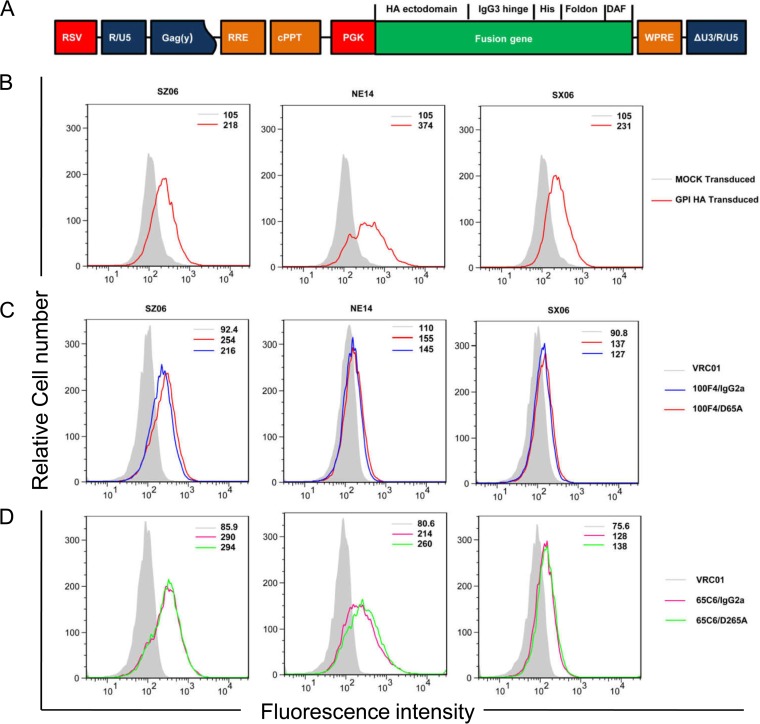
To further test binding activity of cAbs, we generated stably transduced CEM.NKR CCR5+ Luc+ (CEM.NKR) cells expressing trimeric forms of His-tagged glycosylphosphatidylinositol (GPI)-HA ectodomains from SZ06, NE14, or SX06 strain using recombinant lentiviral vectors (Fig. 5A). Figure 5B shows that the GPI-HA ectodomains of SZ06, NE14, or SX06 were expressed on the surfaces of stably transduced CEM.NKR cells, as measured by an anti-His tag Ab. Figure 5C and D and Table 2 show that 65C6/IgG2a and 65C6/D265A as well as 100F4/IgG2a and 100F4/D265A bound well to the GPI-HA ectodomain from SZ06. Furthermore, 65C6/IgG2a and 65C6/D265A also bound well to the GPI-HA ectodomains from strain NE14. Binding of 65C6/IgG2a and 65C6/D265A, as well as binding of 100F4/IgG2a and 100F4/D265A, to the GPI-HA ectodomain from SX06 was relatively low. Poor binding of 100F4/IgG2a and 100F4/D265A to the GPI-HA from NE14 was also observed. We speculate that the difference in the binding of 100F4/IgG2a and 100F4/D265A to NE14 as measured by fluorescence-activated cell sorting (FACS) analysis (Fig. 5C) and by ELISA (Fig. 4B) may be due to the different methods for displaying the HA antigens used in these two assays. For the ELISAs, VLPs expressing H5 HA were coated on the wells of the plates. However, for the FACS analysis, the ectodomains of H5 HAs were displayed on the surfaces of cells as a GPI-anchored form. Interestingly, although the levels of cAb binding differed among transduced cells expressing the different GPI-HA ectodomains, for each of the GPI-HA-modified cells, the levels of binding of 65C6/IgG2a and 65C6/D265A, as well as 100F4/IgG2a and 100F4/D265A, were always very similar, indicating that cAb pair 65C6/IgG2a and 65C6/D265A as well as cAb pair 100F4/IgG2a and 100F4/D265A had similar binding capabilities.
FIG 5.
Cell surface expression of GPI-HA ectodomains and binding activity of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A by FACS analysis. (A) Schematic diagram of the third-generation self-inactivating lentiviral vector pRRL-HA/hinge/his-tag/foldon/DAF. The fusion gene is made up of the ectodomain of H5 HA from SZ06, NE14, or SX06 strain (HA ectodomain) a human IgG3 hinge region (IgG3 hinge), a 6-histidine residue tag (His), foldon, a 27-residue trimerization domain at the C-terminal bacteriophage T4 fibritin, and the C-terminal 34 amino acid residues of the decay-accelerating factor (DAF). The fusion gene was driven by the phosphoglycerate kinase (PGK) promoter. RSV, RSV promoter; Gag(y), encapsidation signal of HIV-1 Gag; RRE, Rev-response element; cPPT, central polypurine track; Wpre, posttranscriptional regulatory element of woodchuck hepatitis virus. (B) FACS analysis of cell surface expression of GPI-HA ectodomain from strains SZ06, NE14, and SX06 detected by anti-His tag antibody. The light gray line shows mock-transduced CEM.NKR cells, and the red line shows GPI-HA-transduced CEM.NKR cells. (C) FACS analysis of cell surface expression of GPI-HA from SZ06, NE14, and SX06 detected by 100F4/IgG2a and 100F4/D265A along with VRC01 control. (D) FACS analysis of cell surface expression of GPI-HA from SZ06 (left), NE14 (middle), and SX06 (right) detected by 65C6/IgG2a and 65C6/D265A along with VRC01 control. The numbers within the graphs in panels B to D are the median values of fluorescent intensity.
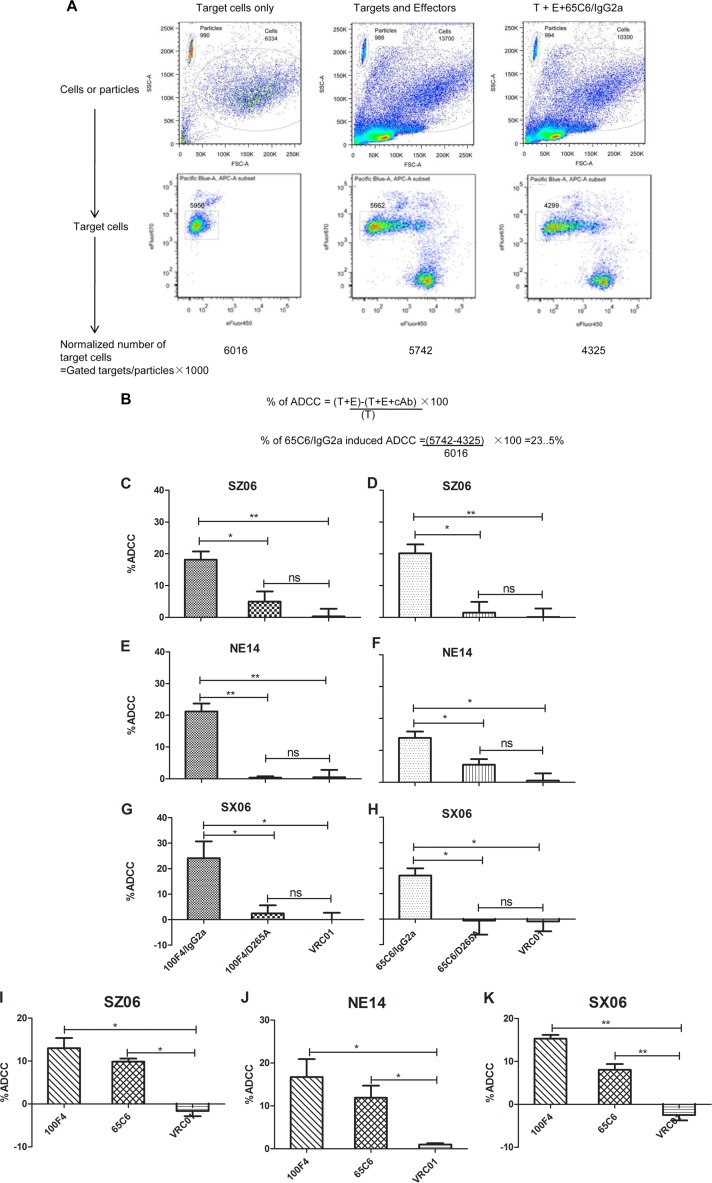
Richard et al. (42) recently developed an alternative flow cytometry-based assay to measure ADCC responses against target cells expressing HIV-1 envelope protein. By staining target and effector cells with different dyes, it allows precise gating and the calculation of the number of surviving target cells by normalization to the number of flow cytometry particles. To adopt this assay for measuring ADCC against target cells expressing influenza HA, we measured ADCC of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A along with control Ab VRC01 against target CEM.NKR cells stably transduced with lentiviral vectors expressing the GPI-HA ectodomains of SZ06, SX06, or NE14 strain (see above). Figure 6A and B show the gating strategy and calculation to determine the percentage of cells killed by ADCC from a representative experiment with 65C6/IgG2a at a concentration of 10 μg/ml.
FIG 6.
ADCC activity of cAbs and human Abs. (A) Gating strategy and normalization of the numbers of viable target cells using a representative cAb 65C6/IgG2a as an example. (B) Calculation of percentage of ADCC using a representative cAb 65C6/IgG2a as an example. T, target cells; E, effector cells. (C to E) Percentage of ADCC by 100F4/IgG2a and 100F4/D265A along with VRC01 control against CEM.NKR cells transduced with GPI-HA from virus strains SZ06 (C), NE14 (D), and SX06 (E). (F to H) Percentage of ADCC by 65C6/IgG2a and 65C6/D265A along with VRC01 control against CEM.NKR cells transduced with GPI-HA from SZ06 (F), NE14 (G), and SX06 (H). (I to K) Percentage of ADCC by human Abs 100F4 and 65C6 along with VRC01 control against CEM.NKR cells transduced with GPI-HA from SZ06 (I), NE14 (J), and SX06 (K). *, P < 0.05; **, P < 0.01; ns, not significant.
Figure 6C to H and Table 2 summarize the relative levels of ADCC against CEM.NKR cells stably expressing GPI-HA ectodomains of strain SZ06, NE14, or SX06 and mediated by cAb pair 100F4/IgG2a and 100F4/D265A or by cAb pair 65C6/IgG2a and 65C6/D265A, compared to the VRC01 control Ab. At a concentration of 10 μg/ml, the VRC01 control Ab exhibited no ADCC activity; cAb 100F4/IgG2a had averages of 18.2%, 21.2%, and 24.1% ADCC against SZ06 (Fig. 6C), NE14 (Fig. 6D), and SX06 (Fig. 6E), respectively. In contrast, cAb 100F4/D265A had averages of only 5.0%, 0.4%, and 2.4% ADCC against SZ06 (Fig. 6C), NE14 (Fig. 6D), and SX06 (Fig. 6E), respectively. Statistically, the differences in ADCC against target cells expressing the GPI-HA ectodomains from all three H5 strains were significant (P < 0.05) or very significant (P < 0.01) between cAbs 100F4/IgG2a and 100F4/D265A.
Similar to cAb pair 100F4/IgG2a and 100F4/D265A, at a concentration of 10 μg/ml, cAb 65C6/IgG2a had averages of 20.1%, 13.9%, and 17.2% ADCC against strain SZ06 (Fig. 6F), NE14 (Fig. 6G), and SX06 (Fig. 6H), respectively. In contrast, 65C6/D265A had averages of only 1.5%, 5.5%, and 0% ADCC against SZ06 (Fig. 6F), NE14 (Fig. 6G), and SX06 (Fig. 6H). Statistically, the differences in ADCC against target cells expressing GPI-HA ectodomains from all three H5 strains were also significant (P < 0.05) between cAbs 65C6/IgG2a and 65C6/D265A. Thus, taking the results together, we conclude that although 65C6/IgG2a and 65C6/D265A, or 100F4/IgG2a and 100F4/D265A had similar HA binding capability, only 65C6/IgG2a and 100F4/IgG2a, but not the 65C6/D265A and 100F4/D265A mutants, mediated ADCC.
For the comparison, we also measured the relative level of ADCC against CEM.NKR cells stably expressing GPI-HA ectodomains of strains SZ06, NE14, and SX06 and mediated by the human antibodies 100F4 and 65C6, compared to the VRC01 control. The results show that human antibodies 100F4 and 65C6 also mediated ADCC activity, but the levels of ADCC were lower than those for cAbs 100F4/IgG2a and 65C6/IgG2a (Fig. 6I to K).
Divergent requirement of Fc-FcγR interactions by cAbs 65C6 and 100F4 for in vivo protection.
To measure the half-lives of cAbs in vivo, BALB/c mice (three mice in each group) were injected intravenously (i.v.) with 200 μg of purified 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, or 100F4/D265A and bled on days 1, 4, 7, 11 and 16 postinjection. The serum concentrations of 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A were measured by ELISA using H5 HA protein-coated plates. Table 3 shows that the average half-lives of 100F4/IgG2a and 100F4/D265A in blood were 3.5 and 5.3 days, respectively; the average half-lives in blood for 65C6/IgG2a and 65C6/D265A were 9.8 and 7.3 days, respectively.
TABLE 3.
Half-lives of chimeric Abs in blood
| Chimeric Ab | t1/2 (days) (mean ± SEM) |
|---|---|
| 100F4/IgG2a | 3.5 ± 1.9 |
| 100F4/D265A | 5.3 ± 1.1 |
| 65C6/IgG2a | 9.8 ± 1.0 |
| 65C6/D265A | 7.3 ± 1.9 |
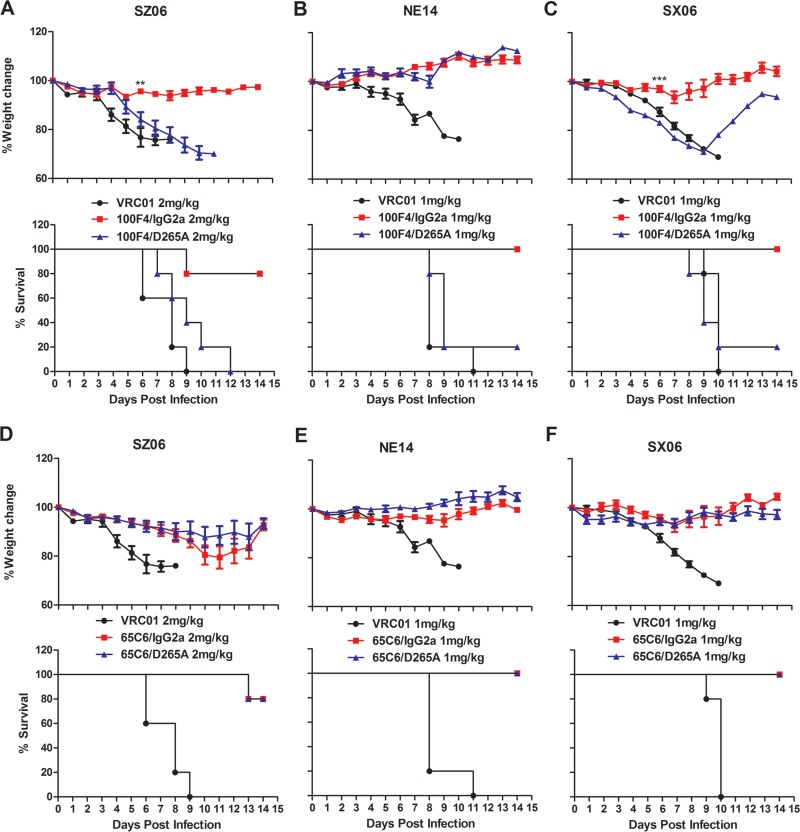
Bournazos et al. compared the in vivo activity of different cAbs of anti-HIV-1 gp120 antibody 3BNC117 and found that a mouse IgG2a variant exhibited significantly higher in vivo protective activity compared to D265A variants at a specific dose range (43). Therefore, to investigate whether the Fc-FcγR interactions were critical for in vivo protection by the anti-HA cAbs, we first determined the minimum doses of protection by 65C6/IgG2a or 100F4/IgG2a against SZ06, SX06, or NE14 by treating BALB/c mice with four different doses (0.5, 1, 2, and 4 mg/kg) of either 65C6/IgG2a or 100F4/IgG2a followed by intranasal (i.n.) challenge using 5 MLD50s of SZ06, SX06, or NE14 (S. Wang and P. Zhou, data not shown). We then used the minimum protective doses of 65C6/IgG2a or 100F4/IgG2a to compare the in vivo protective efficacy of cAb pair 65C6/IgG2a and 65C6/D265A or cAb pair 100F4/IgG2a and 100F4/D265A, as well as a control Ab VRC01. Figure 7 and Table 2 show that when challenged with the SZ06 strain, four out of five mice injected with 2 mg/kg of 100F4/IgG2a survived with slight weight loss. However, no mice injected with 2 mg/kg of 100F4/D265A or VRC01 control Ab survived (Fig. 7A). In contrast, four of five mice injected with 2 mg/kg of either 65C6/IgG2a or 65C6/D265A survived with less than 20% weight loss (Fig. 7D).
FIG 7.
In vivo efficacy of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A in minimum protective doses against the lethal challenge of virus strains SZ06, SX06, and NE14. (A to F) Time course of body weight changes (top panels) and survival rate of each group (bottom panels) against SZ06 (A and D), NE14 (B and E), and SX06 (C and F) by 100F4/IgG2a and 100F4/D265A cAbs (A to C) and by 65C6/IgG2a and 65C6/D265A cAbs (D to F) along with VRC01 control antibody. The survival rate was calculated as percent survival within each experimental group (five mice in each group).
When challenged with the NE14 strain, all five mice injected with 1 mg/kg of 100F4/IgG2a survived with slight weight loss, but only one mouse and no mice injected 1 mg/kg of 100F4/D265A or the VRC01 control Ab survived, respectively (Fig. 7B). In contrast, all five mice injected with 1 mg/kg of either 65C6/IgG2a or 65C6/D265A survived without weight loss (Fig. 7E). Similarly, when mice were challenged with the SX06 strain, all five mice injected with 1 mg/kg of 100F4/IgG2a survived with slight weight loss, but only one mouse and no mice injected with 1 mg/kg of 100F4/D265A or VRC01 control Ab survived, respectively (Fig. 7C). In contrast, all five mice injected with 1 mg/kg of either 65C6/IgG2a or 65C6/D265A survived without weight loss (Fig. 7F). Thus, the results from these challenge experiments using three H5 strains, SZ06, NE14, and SX06, clearly demonstrate that Fc-FcγR interactions are critical for in vivo protection by 100F4, but not by 65C6.
DISCUSSION
In the present study, we investigated whether Fc-FcγR interactions were necessary for two pan-H5 Abs to confer in vivo protection against three H5 strains, SZ06, SX06, and NE14. The two pan-H5 Abs that we studied, 65C6 and 100F4, recognize two conformational epitopes in the globular head region but outside the RBS (26, 27, 30). Since we previously showed that SZ06, SX06, and NE14 pseudotypes exhibit different neutralization sensitivities to 65C6 and 100F4 in vitro (26, 39), testing Abs 65C6 and 100F4 with these H5 strains would allow us to determine whether Fc-FcγR interactions are required for in vivo protection against different individual virus strains.
Here, we demonstrated that both 100F4/IgG2a and 65C6/IgG2a, but not 100F4/D265A and 65C6/D265A, mediate ADCC to a similar extent against target cells expressing GPI-anchored HA ectodomains from all three H5 strains, SZ06, NE14, and SX06 (Fig. 6 and Table 2). This was unexpected because the cAbs had very little or no neutralizing activity and a 1- to 2-log-unit reduction in binding activity against the SX06 strain (Fig. 3 to 5 and Table 2). The data indicate that different neutralizing and binding activities of these cAbs against different H5 strains have little influence on their ADCC activity. These paradoxical results could be explained by the recent finding that optimal activation of Fc-mediated effector functions by anti-HA antibodies requires not only Fc-FcγR interactions but also interactions between HA and sialic acid receptor on effector cells (36–38). Since in our previous study we found that both 65C6 and 100F4 bind and neutralize H5 viruses before and after viruses bind to target cells (27), we speculate that binding of 65C6 and 100F4 to HA probably does not interfere with the interaction between HA and the sialic acid receptor. Because of this, Fc-FcγR interactions mediated by low binding activity between 65C6 or 100F4 and the SX06 HA, plus the interaction between the SX06 HA and sialic acid receptor on effector cells, may be sufficient to mediate ADCC.
In the present study, we demonstrated that a single injection of 1 or 2 mg/kg (minimum protective doses) of 65C6/IgG2a, 65C6/D265A, or 100F4/IgG2a, but not 100F4/D265A, completely or almost completely protected mice from lethal challenge of all three H5 challenge virus strains, SZ06, NE14, and SX06 (Fig. 7 and Table 2). This occurred despite the fact that the cAb pair 65C6/IgG2a and 65C6/D265A and cAb pair 100F4/IgG2a and 100F4/D265A have comparable and decent neutralizing and binding activities against SZ06 and NE14 strains but very little to no neutralizing activity as well as 1- to 2-log-unit reduction in binding activity against the SX06 strain (Fig. 3 to 5 and Table 2). Furthermore, only 65C6/IgG2a and 100F4/IgG2a, but not 65C6/D265A or 100F4/D265A, mediated ADCC activity against all three strains (Fig. 6 and Table 2). Thus, for cAbs 65C6 and 100F4, there is a divergent association between Fc-FcγR interactions and in vivo protection. The Fc-FcγR interactions are required for in vivo protection against all three H5 strains by 100F4, but not by 65C6. These results strongly suggest that individual strains with different binding and neutralization sensitivity in vitro do not influence whether Fc-FcγR interactions are necessary for Abs to mediate in vivo protection. Instead epitope specificity and location recognized by a given Ab could strongly influence such a requirement. The 100F4 epitope is located away from the RBS, while the 65C6 epitope is close in proximity to it. Also, binding of 65C6 to its epitope might slightly hinder the engagement of HA to sialic acid (SA) receptors (30). This may explain why Ab 65C6 exhibits low but measureable inhibition of virus attachment, but Ab 100F4 does not (27). This also indicates that the absence of a requirement for ADCC (or Fc-FcγR interactions) to mediate in vivo protection by cAb 65C6 is unlikely due to a potential occlusive interaction between HA and SA induced by engagement of cAb 65C6 with its HA epitope. Our data show that 65C6/IgG2a can mediate ADCC (Fig. 6), and after incubation with Ab 65C6, more than 70% of viruses still bind to SA and induce receptor-mediated endocytosis (27).
In summary, in the present study, we investigated the requirement of Fc-FcγR interactions for in vivo protection against three representative H5 strains by pan-H5 Abs 65C6 and 100F4 that recognize two epitopes outside the RBS. We found that although these two Abs have similar neutralizing, binding, and ADCC activities against all three H5 strains in vitro, they have divergent associations between Fc-FcγR interactions and in vivo protection against all three strains. The Fc-FcγR interactions are required for in vivo protection by 100F4, but not by 65C6. Thus, the dependence of Fc-FcγR interactions for protection against H5 strains in vivo is not virus strain specific, but epitope specific.
MATERIALS AND METHODS
Ethics statement.
The experimental protocol (CULATR-3064-13) was approved by the Animal Use Committee and by the Safety Committee on BSL-3 Facility and Infectious Agents at the Li Ka-Shing Faculty of Medicine, The University of Hong Kong. All infection experiments were conducted at the biosafety level 3 (BSL-3) facilities and were in compliance with the Ethics Committee regulations of The University of Hong Kong in accordance with EC directive 86/609/CEE.
Cells.
The human embryonic kidney cell line 293T was purchased from Invitrogen Life Technologies (Waltham, MA, USA) and maintained in complete Dulbecco modified Eagle medium (DMEM) (high-glucose DMEM supplemented with 10% fetal bovine serum [FBS], 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]; Corning). The Madin-Darby canine kidney (MDCK) cell line was maintained in complete DMEM. The CEM.NKR CCR5+ Luc+ (CEM.NKR) cell line (44, 45) was obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP) (Germantown, MD) and maintained in complete RPMI 1640 medium (RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 μg/ml]). Drosophila S2 cells were maintained in complete Express Five SFM medium (i.e., Express Five SFM medium supplemented with 10% FBS, 100 U of penicillin/ml, 100 U of streptomycin/ml, and 100 mg of l-glutamine/liter) at 28°C without CO2. The cells were split at the density of 106 cells per ml every 3 or 4 days.
Viruses.
Wild-type virus A/Shenzhen/406H/2006 (SZ06) (H5N1 subclade 2.3.4) and a reassortant A/chicken/Netherlands/14015526/2014 (NE14) (RG6+2; H5N8 subclade 2.3.4.4) were described before (26). A reassortant A/chicken/Shanxi/2/2006 (SX06) (RG6+2, H5N1 subclade 7.2) virus was generated by cotransfecting a mixture of 293T and MDCK cells with gene segments encoding HA and neuraminidase (NA) proteins derived from corresponding wild-type strains and the remaining six gene segments encoding nucleoprotein (NP), polymerase acidic (PA), PB1, PB2, matrix (M), and nonstructural protein (NS) from A/WSN/1933 strains as described by Hoffmann et al. (46). The wild-type virus and reassortants were propagated on MDCK cells using standard viral culturing techniques. The 50% tissue culture infectious dose (TCID50), number of PFU, and 50% mouse lethal dose (MLD50) were determined by serial titration of viruses in MDCK cells and in BALB/c mice, respectively (Table 1).
Generation of chimeric Abs.
For the generation of chimeric Abs (cAbs), the Fv of the heavy chain of 100F4 or 65C6 was PCR amplified and cloned into S2 cell expression vectors containing the Fc of the mouse IgG2a or D265A mutant. The D265A mutant was generated using specific primers and the QuikChange site-directed mutagenesis kit II (Agilent Technologies) and validated by direct sequencing. The expression vectors containing genes encoding the chimeric 100F4 or 65C6 heavy chains along with the expression vectors containing genes encoding the 100F4 or 65C6 light chain, respectively, were cotransfected into Drosophila S2 cells, and stably transfected S2 cell clones were selected as described before (47). The cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A were produced by the stably transfected Drosophila S2 clones and purified by affinity chromatography using protein A agarose (Pierce; Thermo Fisher Scientific) and quantified by bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) according to the manufacturer's instructions.
Generation of HA and NA pseudotypes.
The methods used to produce H5N1 (SZ06), H5N1 (SX06), and H5N8 (NE14) pseudotypes were as described before (48). The relative luciferase activity (RLA) of the pseudotypes was determined on MDCK cells as described before (48).
Microneutralization (MN) assay.
Neutralizing activity of Abs 100F4 and 65C6 against the SZ06, SX06, and NE14 strains was analyzed in an MN assay based on the methods of the WHO Global Influenza Program (49). Briefly, Abs were serially twofold diluted (starting at 20 μg/ml for SZ06 and NE14 strains and 40 μg/ml for SX06 strain) prior to mixing with 100 TCID50 of virus for 1 h at 37°C, and the mixture was added to the MDCK monolayer. Cytopathic effects (CPE) were recorded and scored after 72 h. The MN assay was performed in triplicate.
Plaque reduction assay.
MDCK cells (5 × 105 cells per well) in complete DMEM were seeded into six-well plates. When the cells formed monolayers, the medium was removed, and the cells were washed twice with phosphate-buffered saline (PBS). Seven hundred microliters of serum-free DMEM was then added to each well. The SZ06, SX06, and NE14 strains (60 PFU per well) were incubated with twofold dilutions of antibody in a final volume of 100 μl at 37°C for 1 h. The mixtures were then added onto the MDCK monolayers. After incubation, 0.8% low-melting-point agarose in MEM (3 ml per well) was added. Following a 72-h culture period, the agarose overlay was discarded, and the cells were stained with 0.5% (wt/vol) crystal violet for 1 h. Crystal violet was then removed, and the cells were washed with H2O. The number of plaques in each well was counted. The plaque reduction assay was performed in triplicate.
HA and NA pseudotype-based neutralization (PN) assay.
The PN assay was described before (48). Titration curves were generated using sigmoid dose-response of nonlinear fit from GraphPad, and 50% inhibitory concentrations (IC50s) were determined as the concentration of a given antibody that resulted in 50% reduction of RLA.
ELISAs.
To compare the binding activity of cAbs, 96-well ELISA plates (Costar) were coated overnight at 4°C with virus-like particles (VLPs) derived from the SZ06, SX06, and NE14 strains. VLP production was described previously (50). The plates were blocked with PBS containing 5% bovine serum albumin (BSA) for 2 h at 37°C. They were then washed five times with PBST buffer (PBS containing 0.05% Tween 20). Fourfold-diluted Abs in assay diluent (PBS containing 10% BSA and 0.5% Triton X-100) were added to wells of the VLP-coated 96-well plates for 2 h at 37°C. The plates were then washed five times with PBST buffer. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Chemicon) at a 1:5,000 dilution was added. Colorimetric analysis was performed using a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate kit (Pierce), and the absorbance was read at 450 nm with a spectrophotometer (Thermo Scientific). Titration curves were generated using sigmoid dose-response of nonlinear fit from GraphPad, and 50% effective concentrations (EC50s) were determined as the concentrations of antibodies that generated 50% of maximal optical density at 450 nm (OD450) values.
Generation of stably transduced CEM.NKR cells.
Previously, we developed trimeric glycosylphosphatidylinositol (GPI)-HCDR3s and demonstrated that trimeric GPI-HCDR3 (PG16) are not only highly expressed on transduced cells but also dramatically improve anti-HIV-1 neutralization (51). Therefore, in the present study, we generated stably transduced CEM.NKR cells expressing trimeric GPI-anchored ectodomain HA and used these cells to measure binding and ADCC activities of the cAbs (see below).
To construct fusion genes encoding various GPI-anchored HA ectodomains, codon-optimized sequences encoding HA ectodomains of the SZ06, SX06, and NE14 strains, the IgG3 hinge region, foldon (a 27-residue trimerization domain at the C-terminal bacteriophage T4 fibritin), and a histidine tag were genetically linked to the sequence encoding a GPI attachment signal (a C-terminal 34 amino acid residues of delay-accelerating factor). The fusion genes were inserted into a third-generation lentiviral transfer vector pRRLsin-18.PPT.hPGK.Wpre (52). The resulting transfer constructs were designated pRRL-GPI-HAs.
Recombinant lentiviruses were generated as described previously (53). Briefly, to generate each recombinant virus, 4 × 106 293T cells were seeded into a P-100 dish in 10 ml complete DMEM. After culturing overnight, cells were cotransfected with 14 μg of a pRRL-GPI-HA transfer construct, 14 μg packaging construct encoding HIV-1 Gag/Pol (CMVRΔ8.2), and 2 μg of plasmid encoding the vesicular stomatitis virus (VSV) G protein envelope (pLP/VSV-G), using a calcium phosphate precipitation method. Sixteen hours later, culture supernatants were removed and replaced with fresh complete DMEM plus 1 mM sodium butyrate (Sigma). Eight hours later, supernatants were again removed and replaced with fresh DMEM plus 4% FBS. After another 20 h, the culture supernatants were harvested and concentrated by ultracentrifugation as described previously (53). The vector pellets were resuspended in a small volume of DMEM and stored in aliquots in a −80°C freezer. Vector titers were determined as previously described (53).
To transduce CEM.NKR cells, 1 × 105 cells per well were seeded into the wells of a 24-well plate. Cells were transduced with lentiviral vectors expressing GPI-HA at a multiplicity of infection (MOI) of 20 in the presence of 8 μg/ml of Polybrene. Twenty-four hours later, the cells were washed with fresh complete RPMI 1640 medium and cultured in complete RPMI 1640 medium. The stability of GPI-HA transgene expression in transduced CEM.NKR cells was checked periodically using anti-His tag Ab followed by FACS analysis (see below).
FACS analysis.
To analyze cell surface expression of H5 HA, CEM.NKR cells stably transduced with GPI-HA of SZ06, SX06, or NE14 strain were incubated with a mouse anti-His tag Ab (Sigma) followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG Ab (H&L) (Life Technologies). The cells were washed twice with FACS buffer (PBS containing 1% BSA and 0.02% NaN3) and fixed with 2% formaldehyde in 0.3 ml of FACS buffer. FACS analysis was performed on a LSR II flow cytometer (Becton Dickinson, Mountain View, CA).
To compare binding activity of cAbs, CEM.NKR cells stably transduced with GPI-HA of virus strains SZ06, SX06, and NE14 were incubated with 10 μg of 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, or 100F4/D265A for 45 min on ice. The cells were washed twice with FACS buffer. For cells stained with 65C6/IgG2a or 65C6/D265A as a primary Ab, 1:100 diluted FITC-conjugated mouse anti-human kappa Ab (Southern Biotech) was used as a secondary Ab. For cells stained with 100F4/IgG2a or 100F4/D265A as a primary Ab, 1:100 diluted FITC-conjugated anti-human lambda Ab (Life Technologies) was used as a secondary Ab. The cells were then washed twice with FACS buffer and fixed with 2% formaldehyde in 0.3 ml of FACS buffer. FACS analysis was performed on a LSR II flow cytometer.
ADCC assay.
A rapid fluorometric antibody-dependent cell-mediated cytotoxicity (ADCC) assay was performed as described by Richard et al. (42) with some modifications. Briefly, 10,000 CEM.NKR cells stably transduced with GPI-HA of virus strain SZ06, NE14, or SX06 (target cells) were labeled with 5 μM eFluor 670 (eBioscience). Labeled target cells were resuspended in RPMI 1640 medium containing 10% FBS. Splenocytes (effector cells) from naive mice were labeled with 5 μM eFluor 450 (eBioscience). Effector cells and target cells at a 10:1 effector cell/target cell (E:T) ratio were mixed in a 96-well V-bottom plate (Corning, Corning, NY) and incubated for 10 min at room temperature. The cAb 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, or 100F4/D265A, human Ab 65C6 or 100F4, and control antibody VRC01 were added to a final concentration of 10 μg/ml and incubated for 15 min at room temperature. The plates were centrifuged at 300 × g for 1 min to promote cell-cell interactions and then incubated for 6 h at 37°C in 5% CO2. The cells were washed once with PBS and then fixed in PBS containing 2% paraformaldehyde and 5 × 104/ml beads for flow cytometry. A total of 1,000 beads per well in triplicate wells were acquired within 18 h using a BD Fortessa flow cytometer. Data were analyzed using FlowJo (Tree Star, Inc.). The percent ADCC killing was determined by gating on the eFluor 670 populations of target cells. The ADCC assay was performed in triplicate.
Measurement of the in vivo half-life of each cAb.
To measure the half-life of each cAb in vivo, 6- to 8-week-old female BALB/c mice (three mice in each group) were injected intravenously (i.v.) with 200 μg of purified 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, or 100F4/D265A. Mice were bled at days 1, 4, 7, 11, and 16 postinjection. The serum concentrations of 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, and 100F4/D265A were measured in batches by ELISA using H5 HA protein-coated plates.
Animal experiments.
To test the in vivo efficacy of Abs 65C6 and 100F4, female BALB/c mice at ages of 6 to 8 weeks were randomly divided into groups (five mice in each group). Each group of mice was intraperitoneally (i.p.) injected with 5 mg of the 100F4 or 65C6 Ab or control antibody VRC01 per kg of body weight. Four hours later, mice were intranasally (i.n.) challenged with 5 MLD50 of virus strain SZ06, SX06, or NE14. After the inoculation, mice were monitored and any sign of illness was recorded daily for more than 14 days. Mice that were challenged with SZ06 and SX06 and lost 30% or more of their initial body weight were euthanized and counted as dead. Mice challenged with NE14 died without losing much weight and were not euthanized.
To determine minimum protective doses of cAbs 65C6/IgG2a and 100F4/IgG2a, female BALB/c mice that were 6 to 8 weeks old were randomly divided into groups (five mice in each group). The groups of mice were injected i.p. with 0.5, 1, 2, and 4 mg/kg of 65C6/IgG2a or 100F4/IgG2a or control antibody VRC01. Four hours later, the mice were challenged i.n. with 5 MLD50s of SZ06, SX06, or NE14. Following the challenge, the mice were monitored as described above. The minimum protective doses of 65C6/IgG2a or 100F4/IgG2a against a given H5 virus were determined by the lowest doses that had significant in vivo protection.
To test the in vivo efficacy of cAbs, female BALB/c mice that were 6 to 8 weeks old were randomly divided into groups (five mice in each group). Mice in groups were injected i.p. with the indicated doses of cAbs 65C6/IgG2a, 65C6/D265A, 100F4/IgG2a, 100F4/D265A, or control antibody VRC01. Four hours later, the mice were challenged i.n. with 5 MLD50s of SZ06, SX06, or NE14. After the virus challenge, mice were monitored as described above.
Statistical analysis.
The data from PN, plaque reduction, and ADCC assays and ELISAs were collected from three independent experiments. Titration curves of PN and ELISA data were generated using sigmoid dose-response of nonlinear fit from GraphPad, and IC50 and EC50 were determined by the best-fit values. The mean ± standard deviation of the plaque reduction and ADCC assay data were determined by using Microsoft Excel. The response of each mouse was counted as an individual data point for statistical analysis. The data obtained from animal studies were analyzed with GraphPad using two-way analysis of variance.
ACKNOWLEDGMENTS
We thank Jason T. Kimata at the Baylor College of Medicine for critically reviewing the manuscript, L. Naldini at the University Torino Medical School, Torino, Italy, for providing the lentiviral transfer vector, and other members of the Unit of Anti-Viral Immunity and Genetic Therapy, Institute Pasteur of Shanghai, Chinese Academy of Sciences, for helpful discussions during the course of this study. CEM.NKR CCR5+ Luc+ cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Germantown, MD. The cell line was originally developed and contributed by John Moore and Catherine Spenlehauer.
This work was supported by the 12-5 Mega Project (2013ZX10004003003003) and the 863 Project (2012AA02A404) from the Ministry of Science and Technology in China and by a grant from the Li Ka-Shing Foundation in Hong Kong.
REFERENCES
- 1.Grant EJ, Quinones-Parra SM, Clemens EB, Kedzierska K. 2016. Corrigendum to ‘Human influenza viruses and CD8+ T cell responses’ [Curr Opin Virol 16 (2016) 132-142]. Curr Opin Virol 19:99. doi: 10.1016/j.coviro.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Cowling BJ, Feng L, Lau EH, Liao Q, Tsang TK, Peng Z, Wu P, Liu F, Fang VJ, Zhang H, Li M, Zeng L, Xu Z, Li Z, Luo H, Li Q, Feng Z, Cao B, Yang W, Wu JT, Wang Y, Leung GM. 2013. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet 382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang TT, Palese P. 2011. Biochemistry. Catching a moving target. Science 333:834–835. doi: 10.1126/science.1210724. [DOI] [PubMed] [Google Scholar]
- 6.Ren H, Zhou P. 2016. Epitope-focused vaccine design against influenza A and B viruses. Curr Opin Immunol 42:83–90. doi: 10.1016/j.coi.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67:2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfus C, Ekiert DC, Wilson IA. 2013. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 87:7149–7154. doi: 10.1128/JVI.02975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyrzucki A, Dreyfus C, Kohler I, Steck M, Wilson IA, Hangartner L. 2014. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J Virol 88:7083–7092. doi: 10.1128/JVI.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, Korse HJ, Ophorst C, Brinkman-van der Linden EC, Throsby M, Kwakkenbos MJ, Bakker AQ, Beaumont T, Spits H, Kwaks T, Vogels R, Ward AB, Goudsmit J, Wilson IA. 2014. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A 111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 16.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. 2012. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A 109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota-Koketsu R, Mizuta H, Oshita M, Ideno S, Yunoki M, Kuhara M, Yamamoto N, Okuno Y, Ikuta K. 2009. Broad neutralizing human monoclonal antibodies against influenza virus from vaccinated healthy donors. Biochem Biophys Res Commun 387:180–185. doi: 10.1016/j.bbrc.2009.06.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt AG, Xu H, Khan AR, O'Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, Settembre EC, Dormitzer PR, Kepler TB, Zhang R, Moody MA, Haynes BF, Liao HX, Shaw DE, Harrison SC. 2013. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, Krammer F. 2016. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog 12:e1005578. doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, Calder L, Gamblin SJ, Zhang L, Deubel V, Zhou B, Skehel JJ, Zhou P. 2012. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol 86:2978–2989. doi: 10.1128/JVI.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian M, Hu H, Zuo T, Wang G, Zhang L, Zhou P. 2013. Unraveling of a neutralization mechanism by two human antibodies against conserved epitopes in the globular head of H5 hemagglutinin. J Virol 87:3571–3577. doi: 10.1128/JVI.01292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, Qin CF, Che XY, Wilson IA. 2013. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, Kurosawa Y, Yokoyama S. 2014. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol 88:7130–7144. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, Shi X, Liu X, Fan S, Ren H, Hu H, Sun L, Zhou B, Liang M, Zhou P, Wang X, Zhang L. 2015. Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun 6:8855. doi: 10.1038/ncomms9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. 2015. Viral receptor-binding site antibodies with diverse germline origins. Cell 161:1026–1034. doi: 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S. 2013. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci U S A 110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiLillo DJ, Palese P, Wilson PC, Ravetch JV. 2016. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox F, Kwaks T, Brandenburg B, Koldijk MH, Klaren V, Smal B, Korse HJWM, Geelen E, Tettero L, Zuijdgeest D, Stoop EJM, Saeland E, Vogels R, Friesen RHE, Koudstaal W, Goudsmit J. 2016. HA antibody-mediated FcγRIIIa activity is both dependent on FcR engagement and interactions between HA and sialic acids. Front Immunol 7:399. doi: 10.3389/fimmu.2016.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leon PE, He W, Mullarkey CE, Bailey MJ, Miller MS, Krammer F, Palese P, Tan GS. 2016. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci U S A 113:E5944–E5951. doi: 10.1073/pnas.1613225113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He W, Tan GS, Mullarkey CE, Lee AJ, Lam MM, Krammer F, Henry C, Wilson PC, Ashkar AA, Palese P, Miller MS. 2016. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci U S A 113:11931–11936. doi: 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren H, Wang G, Wang S, Chen H, Chen Z, Hu H, Cheng G, Zhou P. 2016. Cross-protection of newly emerging HPAI H5 viruses by neutralizing human monoclonal antibodies: a viable alternative to oseltamivir. MAbs 8:1156–1166. doi: 10.1080/19420862.2016.1183083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimmerjahn F, Ravetch JV. 2005. Divergent immunoglobulin G subclass activity through selective Fc receptor binding. Science 310:1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 41.Baudino L, Shinohara Y, Nimmerjahn F, Furukawa J, Nakata M, Martinez-Soria E, Petry F, Ravetch JV, Nishimura S, Izui S. 2008. Crucial role of aspartic acid at position 265 in the CH2 domain for murine IgG2a and IgG2b Fc-associated effector functions. J Immunol 181:6664–6669. doi: 10.4049/jimmunol.181.9.6664. [DOI] [PubMed] [Google Scholar]
- 42.Richard J, Veillette M, Batraville LA, Coutu M, Chapleau JP, Bonsignori M, Bernard N, Tremblay C, Roger M, Kaufmann DE, Finzi A. 2014. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J Virol Methods 208:107–114. doi: 10.1016/j.jviromet.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. 2014. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol 73:8966–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spenlehauer C, Gordon CA, Trkola A, Moore JP. 2001. A luciferase-reporter gene-expressing T-cell line facilitates neutralization and drug-sensitivity assays that use either R5 or X4 strains of human immunodeficiency virus type 1. Virology 280:292–300. doi: 10.1006/viro.2000.0780. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Hu H, Yang J, Wang F, Kaisermayer C, Zhou P. 2012. High yield of human monoclonal antibody produced by stably transfected Drosophila Schneider 2 cells in perfusion culture using Wave bioreactor. Mol Biotechnol 52:170–179. doi: 10.1007/s12033-011-9484-5. [DOI] [PubMed] [Google Scholar]
- 48.Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, Zhou B, Wang S, Lu S, Buchy P, Deubel V, Vogel FR, Zhou P. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790. doi: 10.1016/j.vaccine.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.World Health Organization. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. WHO Global Influenza Surveillance Network, World Health Organization, Geneva, Switzerland. [Google Scholar]
- 50.Ding H, Tsai C, Zhou F, Buchy P, Deubel V, Zhou P. 2011. Heterosubtypic antibody response elicited with seasonal influenza vaccine correlates partial protection against highly pathogenic H5N1 virus. PLoS One 6:e17821. doi: 10.1371/journal.pone.0017821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Wen M, Zhu Q, Kimata JT, Zhou P. 2016. Glycosyl phosphatidylinositol-anchored C34 peptide derived from human immunodeficiency virus type 1 gp41 is a potent entry inhibitor. J Neuroimmune Pharmacol 11:601–610. doi: 10.1007/s11481-016-9681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet 25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 53.Wen M, Arora R, Wang H, Liu L, Kimata JT, Zhou P. 2010. GPI-anchored single chain Fv–an effective way to capture transiently-exposed neutralization epitopes on HIV-1 envelope spike. Retrovirology 7:79. doi: 10.1186/1742-4690-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]