ABSTRACT
Follicular helper CD4 T cells, TFH, residing in B-cell follicles within secondary lymphoid tissues, are readily infected by AIDS viruses and are a major source of persistent virus despite relative control of viral replication. This persistence is due at least in part to a relative exclusion of effective antiviral CD8 T cells from B-cell follicles. To determine whether CD8 T cells could be engineered to enter B-cell follicles, we genetically modified unselected CD8 T cells to express CXC chemokine receptor 5 (CXCR5), the chemokine receptor implicated in cellular entry into B-cell follicles. Engineered CD8 T cells expressing human CXCR5 (CD8hCXCR5) exhibited ligand-specific signaling and chemotaxis in vitro. Six infected rhesus macaques were infused with differentially fluorescent dye-labeled autologous CD8hCXCR5 and untransduced CD8 T cells and necropsied 48 h later. Flow cytometry of both spleen and lymph node samples revealed higher frequencies of CD8hCXCR5 than untransduced cells, consistent with preferential trafficking to B-cell follicle-containing tissues. Confocal fluorescence microscopy of thin-sectioned lymphoid tissues demonstrated strong preferential localization of CD8hCXCR5 T cells within B-cell follicles with only rare cells in extrafollicular locations. CD8hCXCR5 T cells were present throughout the follicles with some observed near infected TFH. In contrast, untransduced CD8 T cells were found in the extrafollicular T-cell zone. Our ability to direct localization of unselected CD8 T cells into B-cell follicles using CXCR5 expression provides a strategy to place highly effective virus-specific CD8 T cells into these AIDS virus sanctuaries and potentially suppress residual viral replication.
IMPORTANCE AIDS virus persistence in individuals under effective drug therapy or those who spontaneously control viremia remains an obstacle to definitive treatment. Infected follicular helper CD4 T cells, TFH, present inside B-cell follicles represent a major source of this residual virus. While effective CD8 T-cell responses can control viral replication in conjunction with drug therapy or in rare cases spontaneously, most antiviral CD8 T cells do not enter B-cell follicles, and those that do fail to robustly control viral replication in the TFH population. Thus, these sites are a sanctuary and a reservoir for replicating AIDS viruses. Here, we demonstrate that engineering unselected CD8 T cells to express CXCR5, a chemokine receptor on TFH associated with B-cell follicle localization, redirects them into B-cell follicles. These proof of principle results open a pathway for directing engineered antiviral T cells into these viral sanctuaries to help eliminate this source of persistent virus.
KEYWORDS: B-cell follicles, CD8 T cells, CXCR5, T-cell homing
INTRODUCTION
Recent observations and experiments have identified B-cell follicles in secondary lymphoid organs as significant reservoirs of residual human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) in “elite controllers,” i.e., individuals that manifest substantial spontaneous control of viremia or in those undergoing effective combination antiviral therapy (cART) (1–9). This viral persistence is due, at least in part, to the apparent absence of effective antiviral CD8 T-cell responses inside B-cell follicles (1–3, 10, 11). This deficiency is linked to a relative inability of CD8 T cells to effectively traffic to B-cell follicles within secondary lymphoid tissues, including lymph nodes, spleen, and tonsils, and suppress AIDS virus replication among the CD4 follicular helper T cells (TFH) (1, 3, 10).
Despite the paucity of CD8 effector T cells in follicles, this exclusion is not absolute with multiple reports of CD8 T cells inside B-cell follicles in human, mouse, and rhesus lymph nodes (10, 12–14). Indeed, lymph nodes in HIV-infected patients can have a low level of antiviral CD8 T cells (12). Recently, Li et al. and Miles et al. reported that most CD8 T cells found in B-cell follicles in SIV-infected rhesus macaques are Foxp3+ (15, 16), suggesting that these cells exert a regulatory T-cell (Treg) function that dampens antiviral responses. Nevertheless, CD8 T-cell depletion experiments in those same animals observed modest increases in SIV-infected TFH, suggesting some degree of CD8 T-cell-mediated control of virus replication in B-cell follicles (15). Despite the presence of some CD8 T cells inside B-cell follicles within secondary lymphoid tissues, these structures provide a relatively immune privileged sanctuary where HIV or SIV can replicate in TFH, shielded from antiviral effector CD8 cells which are largely excluded (1). Indeed, most of the residual virus in individuals who otherwise control viral replication comes from this reservoir (7, 8, 17–19). We and others have proposed improving the ability of antiviral CD8 T cells to traffic into B-cell follicles as an approach to reduce or eliminate this reservoir of infected TFH (11).
B-cell follicles are critical sites of antigen-driven, TFH-assisted B-cell activation and maturation by class switching and somatic mutation which yields high-affinity B cells and plasma cells (20). This takes place mostly in the interior of the follicles in germinal centers, histologically classified as the light zone, which are induced upon antigen exposure. CXC chemokine receptor 5-positive (CXCR5+) B cells enter the follicles directly by chemotaxis (21), following a gradient of CXC chemokine ligand 13 (CXCL13), the cognate ligand for the CXC chemokine receptor 5 (CXCR5) (22, 23). CXCL13 is produced by stromal and dendritic cells inside the follicles (24) and is required for B-cell entry and structural organization of the follicle (25). CXCR5+ B cells within the lymph node enter the follicle by crossing the interface between the T-cell zone and the B-cell follicle, i.e., the T-B border, which represents the junction of the outer edge of the follicles and the surrounding regions of secondary lymphoid tissues.
While the direct role of CXCR5 chemotaxis for follicular B-cell localization is clear, the exact role for CXCR5 in TFH entry into B-cell follicles is less so. Indeed, CXCR5 has been found to be insufficient for follicular entry of CD4 T cells in mice (26, 27). Additionally, circulating CXCR5+ CD4 T cells in peripheral blood, mostly memory T cells, do not appear to migrate to B-cell follicles in mice, consistent with CXCR5 expression alone being insufficient to target CD4 T cells to B-cell follicles (28–30). An extensive body of experiments in mice (reviewed in references 31, 32, and 33) has led to a current model for the genesis and trafficking of TFH that relies on follicular B cells forming antigen-stabilized complexes with their cognate naive T cells, contacts through which they induce TFH differentiation and actively bring the TFH into the B-cell follicles. Briefly, upon antigen stimulation in the T-cell zone, naive undifferentiated CD4 T cells start expressing moderate levels of CXCR5 and begin to downregulate CC chemokine receptor 7 (CCR7) (26, 34), resulting in their relocalization to the T-B border of the follicle (27, 35). There, they interact with follicular B cells brought to the T-B border by upregulation of CCR7 on their surface, a result of stimulation with soluble antigen in the follicle (36–38). At the border, follicular B cells form a complex with their naive CXCR5+ CD4 T-cell counterparts by antigen sharing, which induces further differentiation of the T cells into the full TFH phenotype with heightened levels of CXCR5. In the final step, the follicular B-cell/antigen/TFH complex crosses the border, apparently driven by B-cell chemotaxis to CXCL13. In addition to CXCR5, TFH also express inducible T-cell costimulator (ICOS), the expression of which on TFH appears to be required for entry (39). Thus, the physiologic entry of TFH into the follicle is considerably more involved than simple CXCR5 expression, requiring follicular B cells for coordinated TFH differentiation and antigen binding.
CD8 T cells are generally excluded from B-cell follicles; however, some CXCR5+ CD8 T cells are found in B-cell follicles (12–15). Indeed, adoptively transferred native CXCR5+ CD8 T cells from wild-type mice can migrate into B-cell follicles in CXCR5−/− knockout mutant mice (12, 13). However, it is unclear whether ordinary circulating CD8 T cells can be engineered to enter B-cell follicles. The ability to genetically engineer T cells with genes of interest, especially T-cell receptors (TCR) and chimeric antigen receptors, using retroviral and lentiviral vectors is a powerful technique, from the initial human gene therapy trial 25 years ago to its current use in cellular immunotherapeutics against cancer (40–42). In a pilot study, we engineered SIV-specific T cells with CCR7 and CD62L which preferentially localized to lymph nodes in a rhesus macaque adoptive transfer system (43). For this study, we tested whether circulating CD8 T cells can be engineered to enter B-cell follicles in rhesus macaques by ectopically expressing CXCR5 on unselected bulk peripheral blood-derived CD8 T cells, expanding the engineered T cells ex vivo, and infusing the autologous cells back into donor animals. We found that CXCR5 transductants, but not coinfused untransduced cells, could effectively traffic into B-cell follicles.
RESULTS
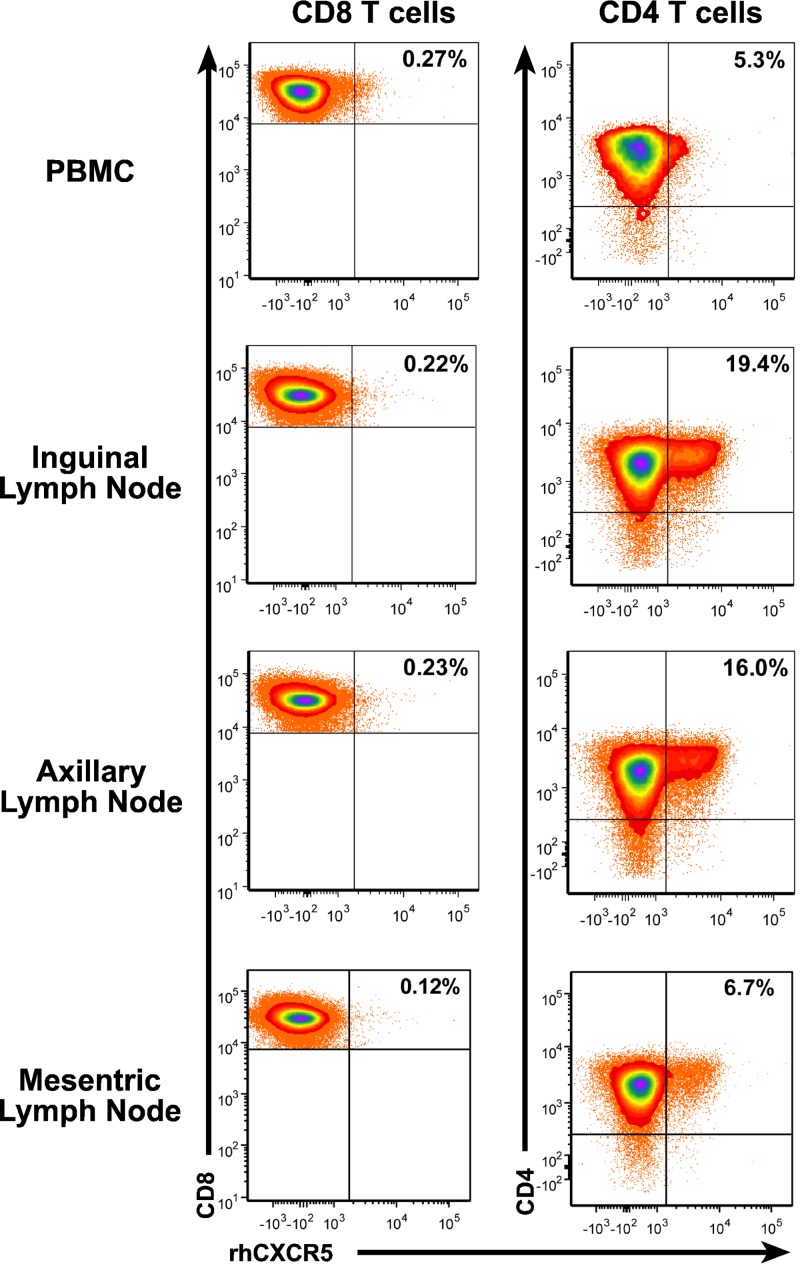
Several studies have reported a low but clearly present level of CXCR5+ CD8 T cells in peripheral blood and lymph nodes. To examine the presence of CXCR5+ in infected rhesus macaques, we analyzed necropsy samples of an animal infected with SIVmac239X, a molecularly tagged synthetic swarm of otherwise isogenic SIVmac239 (44). Flow cytometry of peripheral blood mononuclear cells (PBMC) detected only low frequencies of CD8 or CD4 T cells expressing low levels of CXCR5+ in circulation (Fig. 1). Analysis of CD8 T cells in lymph nodes showed a similar low-frequency, low-intensity staining for CXCR5 (Fig. 1). In contrast, CD4 T-cell analysis of the same samples detected a considerable population of bright CXCR5 staining cells, most likely TFH. These results are consistent with a general lack of high-level CXCR5 expression by CD8 T cells as a contributing factor in the functional exclusion of antiviral CD8 T cells from infected B-cell follicles.
FIG 1.
Low frequencies of CXCR5+ CD8 T cells in lymph nodes. Flow cytometry of PBMC and lymph node samples collected from an infected rhesus macaque is presented with CD8 or CD4 staining on the y axis and recombinant human CXCR5 (rhCXCR5) on the x axis. Frequencies of CXCR5+ T cells are provided in the upper right dot plot quadrant.
Engineering CXCR5 expression on CD8 T cells.
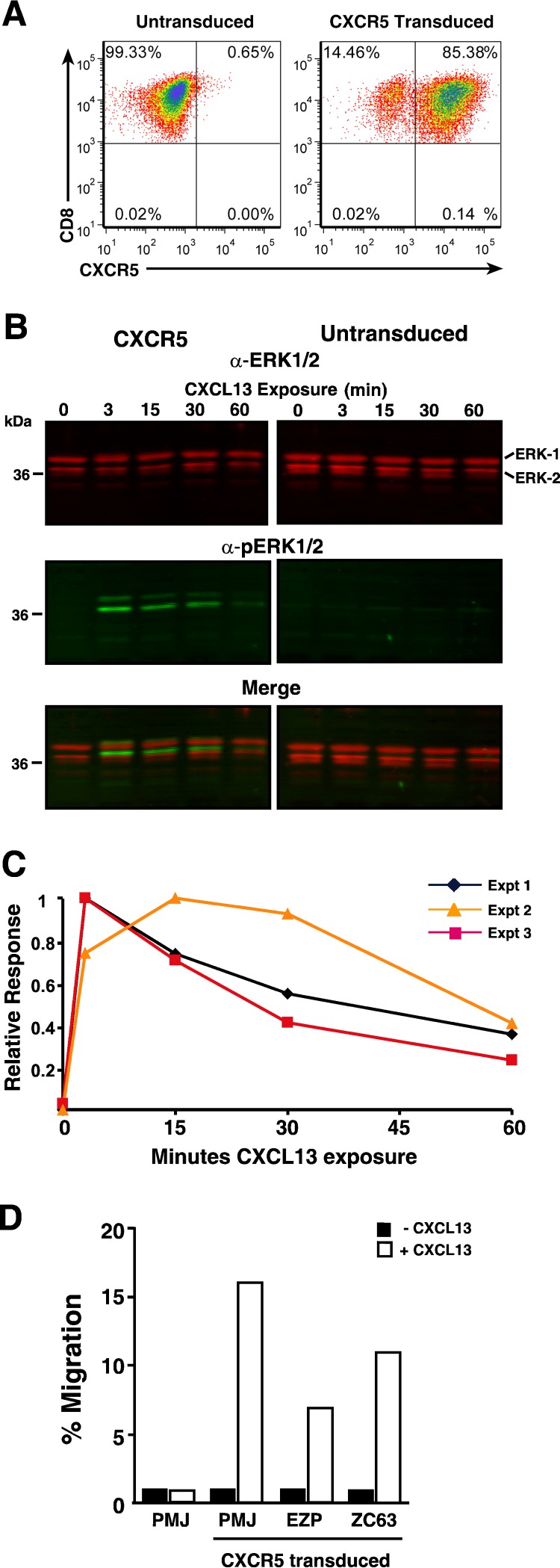
To redirect PBMC-derived CD8 T cells to B-cell follicles, we produced a human CXCR5 (hCXCR5) murine leukemia virus (MuLV)-based retroviral expression vector. The human gene was used due to its 97% protein sequence identity to rhesus macaque CXCR5. Also, by using a species-specific antibody that detects only human and not endogenous rhesus macaque CXCR5 protein, we could uniquely identify any engineered cells from the endogenous cells. Primary rhesus macaque CD8 T cells transduced with the hCXCR5 vector exhibited bright staining for hCXCR5 (Fig. 2A), demonstrating high-level expression of hCXCR5 by the vector.
FIG 2.
CXCR5 transduction of primary rhesus macaque T cells confers functional CXCL13-mediated signaling. Analyses of CXCR5-transduced CD8 T cells are presented. (A) Dot plot of CD8/CXCR5 flow cytometry. (B) Near-infrared LI-COR ERK1/2 and phosphorylated ERK1/2 (pERK1/2) immunoblots of cell lysates. The CXCL13 exposure time (in minutes) is indicated above each sample. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the blot, and the positions of bands are identified to the right of the blot. α-ERK1/2, ant-ERK1/2 antibody. (C) Graph of the kinetics of pERK1/2 induction. (D) Graph of cell counts from CXCL13-induced migration of transduced cells in a transwell assay.
In vitro functional evaluation of CD8 T cells transduced with hCXCR5.
To confirm the function of our hCXCR5 protein, we examined CXCL13-mediated signaling in hCXCR5-transduced CD8 cells by monitoring the induction of phosphorylation on extracellular signal-regulated kinase 1 (ERK1) and ERK2 protein kinases, a key point in the signaling cascade (45). Serum-starved hCXCR5 CD8 T-cell cultures were stimulated with CXCL13, and samples were analyzed by quantitative near-infrared immunoblot analyses. The results from three independent experiments showed rapid induction of phosphorylated ERK1 or ERK2 (phospho-ERK1/2) (pERK1/2) in the presence of CXCL13 which peaked at 3 min and declined with a half-life of 40 min as appropriate for CXCR5 signaling (46) (Fig. 2B and C). In contrast, the matching untransduced CD8 T cells failed to generate any detectable pERK1/2 in the presence of CXCL13 (Fig. 2B; data not shown), consistent with ligand-specific signaling in the hCXCR5 transductants.
To determine whether the hCXCR5 signaling in transduced cells resulted in chemotaxis, we examined the hCXCR5-transduced culture for specific migration toward CXCL13 in a transwell assay. The hCXCR5 transductants migrated into chambers containing CXCL13, but not into chambers without added chemokine (Fig. 1D). Furthermore, the matched untransduced cells failed to migrate in response to CXCL13. Taken together, the in vitro-specific CXCL13 signaling and chemotaxis conferred by hCXCR5 transduction indicate that our retroviral vector provides appropriate CXCR5 function to CD8 T cells.
Engineering CXCR5-transduced cells for infusion into rhesus macaques.
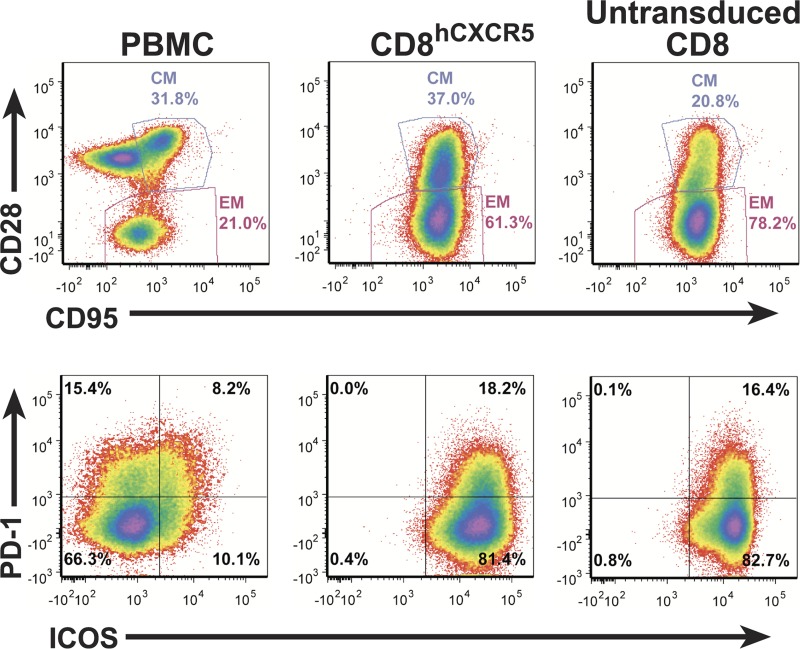
To test the ability of our hCXCR5 vector to direct CD8 T cells into B-cell follicles, we isolated unselected bulk CD8 T cells from PBMC collected from six SIVmac239-infected animals and produced autologous untransduced CD8 and transduced and sorted hCXCR5-expressing CD8 (CD8hCXCR5) cell lines which were then expanded ex vivo to provide large numbers of cells for infusion. Due to the considerable logistical demands of these experiments, including coordinating transductions, T-cell expansion, animal manipulations, and postnecropsy analyses, two groups with three animals in each group was used in this study. The first group, animals 1 to 3, was infused and analyzed 2 weeks prior to the second group, animals 4 to 6, resulting in the latter ex vivo expansion cultures receiving an additional round of stimulation. The T-cell lines for all animals were analyzed 1 week before their infusion by flow cytometry to confirm similar phenotypes (Fig. 3). The analyses showed the presence of considerable frequencies of cells with a central memory phenotype (CD95+ CD28+) in both the untransduced CD8 and CD8hCXCR5 T-cell cultures. For example, for animal 1, the untransduced T-cell cultures had 23% of the cells with a central memory phenotype versus 37% for the CD8hCXCR5 T cells with the balance being effector memory cells (CD95+ CD28−) (Fig. 3). As expected for anti-CD3-expanded T cells, there were no cells with a naive phenotype (CD95− CD28+) in either culture, compared to a typical rhesus macaque PMBC sample (Fig. 3). Additionally, two markers associated with TFH, ICOS and programmed cell death protein 1 (PD-1), were present to the same extent in both cultures, at nearly 100% and 17% frequencies, respectively.
FIG 3.
Expanded CD8hCXCR5 and untransduced CD8 T-cell cultures have similar phenotypic profiles. Flow cytometry analysis for memory differentiation marker expression (CD95+/CD28+) and PD-1 and ICOS expression on CD8hCXCR5 and untransduced CD8 T-cell cultures expanded ex vivo from animal 1 1 week before infusion and a fresh PBMC sample from a similar infected rhesus macaque outside the study group are presented. Samples are identified above each column with central memory and effector memory gates denoted by CM and EM labels, respectively.
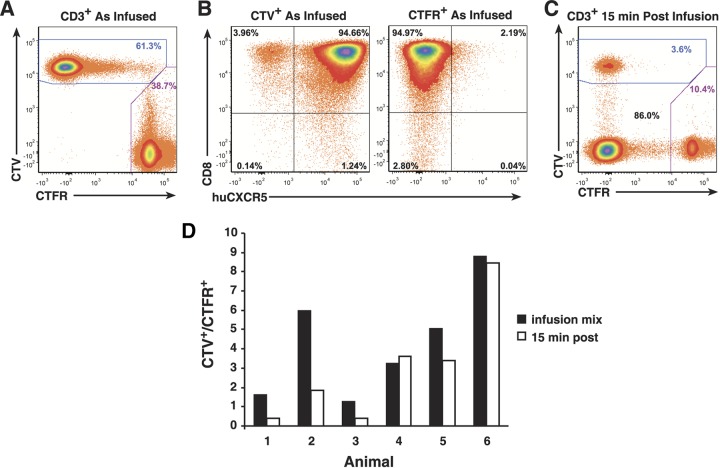
Immediately prior to infusion, the cells in the cultures were collected, washed, and concentrated. To differentiate between the two cultures, the CD8hCXCR5 cells were stained with CellTrace violet (CTV) and the untransduced cells were stained with CellTrace far red (CTFR), then the numbers of cells for each culture were counted, and the cultures were combined into the infusion preparation. The total numbers of cells and the ratios of CD8hCXCR5 to untransduced CD8 T cells varied in the animals due to differences in the initial numbers of cells transduced and their subsequent cell expansion rates in the untransduced and CD8hCXCR5 cultures. In all cases, there were more CD8hCXCR5 T cells than untransduced CD8 T cells in the infusion mixtures. Flow cytometry of the infused preparation confirmed bright differential CTV/CTFR labeling of the cells (representative data for animal 1 shown in Fig. 4A). Gating on both the CTV+ and CTFR+ populations for CD8 signal in the animal 1 infusion mixture revealed that 95% of both cultures were CD8 T cells with minor levels of non-CD8 T cells, 1% and 3%, respectively (Fig. 4B). Additionally, a minor portion, 4%, of the CTV+ cells infused were hCXCR5−, apparently nontransductant carryover from the CXCR5 sorting. Because the antibody used for CXCR5 detection specifically detects the human receptor and not the rhesus receptor, the minor (2%) low-intensity CXCR5 staining population in the untransduced culture is likely background signal. Similar results were obtained for the other animals.
FIG 4.
Analysis of T-cell infusion mixtures and initial postinfusion PMBC samples. Representative flow cytometry analysis results are shown for animal 1. (A) Preinfusion analysis of differential dye labeling of transduced CD8hCXCR5 CTV-labeled CD8 T cells, and untransduced CTFR-labeled CD8 T cells. (B) CD8- and hCXCR5-specific preinfusion analyses gated on CTV and CTFR (labeled) above their respective plots. (C) 15-min postinfusion analysis of differential dye labeling of CD8hCXCR5 T cells. (D) Graphs of the CTV+-to-CTFR+ ratios in the infusion and the 15-min postinfusion analyses.
Initial analysis of infused cells in PBMC.
Each fluorescently labeled cell mixture was infused into its autologous donor animal via the femoral vein. Flow cytometry for CTV+ and CTFR+ cells within the total CD3+ T-cell population in PBMC from 15-min postinfusion blood samples showed that the infused cells were present in between 2 and 23% of the CD3+ T cells (n = 6; x̄ = 12%; standard deviation [SD] = 7%) (Fig. 4C). For the group 1 animals, the ratios of the transductant-to-nontransductant (CTV+/CTFR+) frequencies in the 15-min postinfusion samples were reversed from those of the infusion preparations with proportionally more untransduced cells present than initially infused, consistent with the CD8hCXCR5 T cells leaving the circulatory system more rapidly than the CD8 T cells did. However, the results for group 2, while variable among the animals, did not exhibit a similar extent of ratio reversal as found in group 1, with the infused preparation and 15-min ratios being nearly the same or, at most only slightly reversed (Fig. 4C). Considering that the group 2 cells were in culture longer with an extra stimulation, the difference in behavior of the two groups might be due to this factor, apparently though not due to an obvious difference in central memory/effector memory or PD-1/ICOS phenotypes (Fig. 3).
Preferential localization of CXCR5-transduced CD8 cells to secondary lymphoid tissues.
The study animals were euthanized and necropsied 48 h postinfusion, and PMBC and cells isolated from tissue samples were analyzed by flow cytometry for the levels of CTV+ and CTFR+ cells within the total CD3+ T-cell population to determine the relative distributions of the infused cells. The frequencies of the infused T cells in PBMC from necropsy samples were decreased approximately 10-fold from those of the 15-min postinfusion samples, ranging from 0.3 to 1% (n = 6; x̄ = 0.7%; SD = 0.25%), suggesting movement of the infused cells out of the peripheral circulatory system. One common observation in adoptive transfer studies is that bronchoalveolar lavage sampling of animals contain high frequencies of infused cells, presumed to be “trapped” in the lung (43, 47, 48). Flow cytometry of CD3+ cells isolated from the prenecropsy bronchoalveolar lavage samples showed a high frequency of infused cells relative to the endogenous T cells, from 7 to 76% (n = 6; x̄ = 54%; SD = 24%) consistent with retention of some of the infused cells in the lung.
Flow cytometric analysis of cell preparations from lymphoid tissue samples for the infused fluorescently labeled cells within the total CD3+ T-cell population detected, with the spleen containing the highest frequency of infused cells, ranging between 2% and 5% (n = 6; x̄ = 3%; SD = 1%) in all of the animals with lymph node samples having lower frequencies (0.02% to 0.09% [n = 6; x̄ = 0.04%; SD = 0.03%]).
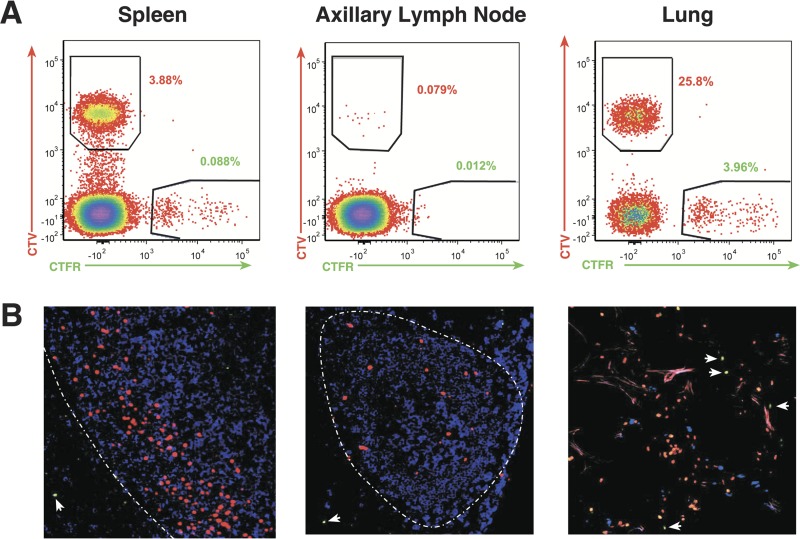
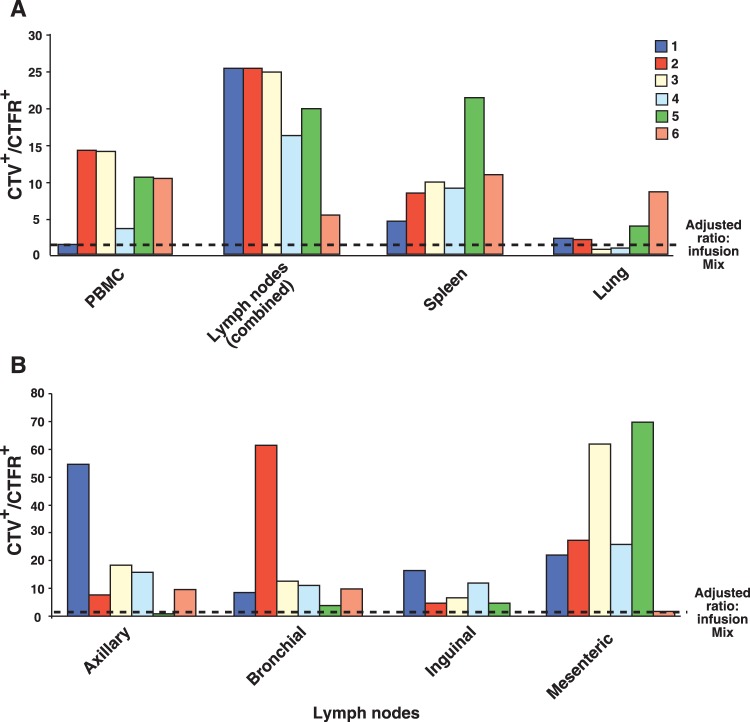
To evaluate potential preferential localization of infused CD8hCXCR5 T cells in lymphoid tissues, the CD3+ CD8+ cells were gated into CTV+ and CTFR+ populations to determine the ratio of CD8hCXCR5 T cells to untransduced CD8 T cells. As expected, analysis of spleen and lymph node cells confirmed that the CTV+ cells present in these tissues also expressed high levels of hCXCR5+ T cells (data not shown). Figure 5A presents an example of representative flow cytometry results from animal 4. Analyses of the CD8 T cells from spleen showed a 40-fold predominance of CTV+ CD8hCXCR5 T cells compared to the CTFR+ untransduced control cells. For all of the spleen samples from the animals, the number of CXCR5 transductants was greater than that of the untransduced cells. Normalized for the greater numbers of CD8hCXCR5 T cells in the infusion preparation for animal 4 (Table 1), the number of CD8hCXCR5 T cells was 20-fold greater than that of the nontransduced CD8 T cells, consistent with preferential trafficking. Similarly, analysis of spleen samples from the other animals also detected considerably more CD8hCXCR5 T cells than untransduced CD8 T cells (Fig. 6A).
FIG 5.
Detection of infused cells in lymphoid tissues. (A) Representative flow cytometry analysis for CTV and CTFR staining of T cells isolated from of 48-h tissue samples from animal 4 with samples identified above each plot. (B) Thin-section confocal merged micrographs of tissue samples from animal 4 with CTV+ cells pseudocolored red (CD8CXCR5 T cells), CTFR+ cells pseudocolored green (CD8 T cells), and anti-CD20+ cells pseudocolored blue (B cells). The T-cell zone/B-cell follicle interface is indicated by a white broken line. Untransduced CTFR+ cells are indicated with white arrows.
TABLE 1.
Cell infusion parameters
| Animal | Wt (kg) | CXCR5+ cell counta | Untransduced cell counta |
|---|---|---|---|
| 1 | 8.2 | 4.25 × 109 | 2.41 × 109 |
| 2 | 6.4 | 2.77 × 109 | 0.66 × 109 |
| 3 | 9.6 | 1.34 × 109 | 0.88 × 109 |
| 4 | 6.4 | 4.37 × 109 | 2.16 × 109 |
| 5 | 5.9 | 4.72 × 109 | 1.43 × 109 |
| 6 | 5.7 | 4.22 × 109 | 1.81 × 109 |
Cell count of each culture used for the infusion mixture.
FIG 6.
Summary of flow cytometry data detecting the relative presence of CD8hCXCR5 T cells versus untransduced CD8 T cells. A graph of the ratios of CTV+-to-CTFR+ staining cells observed by flow cytometry is presented. (A) Results from lymphoid and nonlymphoid tissues from the six animals with the ratios for axillary, inguinal, mesenteric, and bronchial lymph nodes averaged. (B) Results from the anatomically distinct individual lymph nodes. All values were normalized for the ratios of the cell counts in the infusion mixture.
Even though fewer infused cells were observed in lymph nodes, there was a correspondingly greater frequency of CD8hCXCR5 T cells compared to untransduced CD8 T cells in these tissues as well. For example, flow cytometry of samples from an axillary lymph node from animal 4 detected frequencies of 0.08% CTV+ transductants versus 0.01% for the CTFR+ untransduced cells, corresponding to an approximately fivefold greater presence of CD8hCXCR5 T cells after normalization (Fig. 5A and Table 1). Preferential accumulation of CD8hCXCR5 T cells was observed in the four anatomically distinct lymph nodes sampled, axillary, inguinal, bronchial, and mesenteric lymph nodes (Fig. 6B). While mesenteric lymph nodes overall had high ratios of CD8CXCR5-to-untransduced CD8 T cells, different lymph nodes from some animals had the highest ratios, axillary lymph nodes in animal 1 and bronchial lymph nodes in animal 2.
Analysis of CD3+ cells isolated from lung tissue samples, a nonlymphoid organ, showed that, while the lung tissue samples also contained a higher frequency of CTV+ T cells than CTFR+ T cells for nearly all of the animals (representative data from animal 4 in Fig. 5), the normalized ratios in lung were considerably lower than those of the lymphoid tissues (Fig. 6), indicating preferential accumulation of CD8hCXCR5 T cells in secondary lymphoid versus nonlymphoid tissues (Fig. 6). Taken together, these flow cytometry data are consistent with preferential targeting of CD8hCXCR5 T cells to secondary lymphoid tissues containing B-cell follicles.
CXCR5 targets CD8 T cells into B-cell follicles.
While our results demonstrate a preferential localization of the infused CD8hCXCR5 cells to secondary lymphoid tissues, flow cytometry itself cannot determine their localization within the tissue architecture. To determine whether the CD8hCXCR5 T cells entered the B-cell follicles, the anatomical localization of the CTV+ stained transduced cells in tissues was examined using fluorescence confocal microscopy of frozen lymph node and spleen thin sections. In both spleen and lymph node samples, the vast majority of the CTV+ cells observed were present within B-cell follicles (representative data from animal 4 in Fig. 5B). Consistent with the flow cytometry data, there were more CTV+ CD8hCXCR5 cells present in splenic B-cell follicles than in lymph nodes in all of the animals (Fig. 5B). In all lymphoid samples, the few CTFR+ cells present, representing the untransduced CD8 T cells, were observed almost exclusively in the T-cell zone and not inside the adjacent follicles (Fig. 5B). In contrast, while analyses of lung tissue samples, a nonlymphoid source, showed more CTV+ cells than CTFR+ cells, there was no distinct localization of either cell type: the cells appear to be distributed randomly (Fig. 5B).
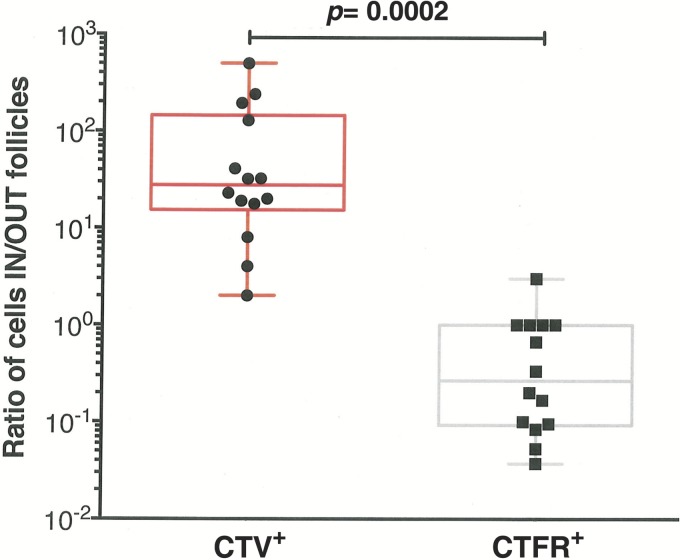
To quantitatively evaluate the preferential localization of CD8hCXCR5 in B-cell follicles, the distributions of fluorescently labeled cells were counted in 850-mm2 total area of both spleen and lymph node tissue sections, and the ratios of cells localized within and outside B-cell follicles were calculated for both the CTV+ and CTFR+ labeled cells. For most samples, there were high ratios of CD8hCXCR5 inside the B-cell follicles compared to extrafollicular sites with an average of approximately 120-fold-more cells inside B-cell follicles (Fig. 7). Thus, CD8hCXCR5 T cells preferentially localized within the B-cell follicles, being relatively absent from the surrounding T-cell zone. In contrast, the rare CTFR+ untransduced cells observed were situated almost exclusively outside the B-cell follicles with a mean ratio of 0.15, or approximately sevenfold-more untransduced CD8 T cells found outside the follicles than inside (Fig. 7). Taken together, our flow cytometry and tissue analyses demonstrate that CD8hCXCR5 T cells preferentially localized into spleen and lymph nodes with nearly all of those cells found within B-cell follicles.
FIG 7.
Preferential localization of CD8hCXCR5 T cells to B-cell follicles. A graph of the ratios for CTV+ and CTFR+ cells inside versus outside B-cell follicles in 850 mm2 of total area for each animal observed by thin-section fluorescence confocal microscopy analyses is presented. The tissues analyzed were mesenteric lymph nodes (animals 1, 3, 5, and 6), axillary lymph nodes (animals 1, 5, and 6), an inguinal lymph node (animal 4), bronchial lymph nodes (animals 1, 4, and 5), and spleen (animals 1, 4, and 5).
Phenotype of CD8hCXCR5 T cells inside lymph nodes.
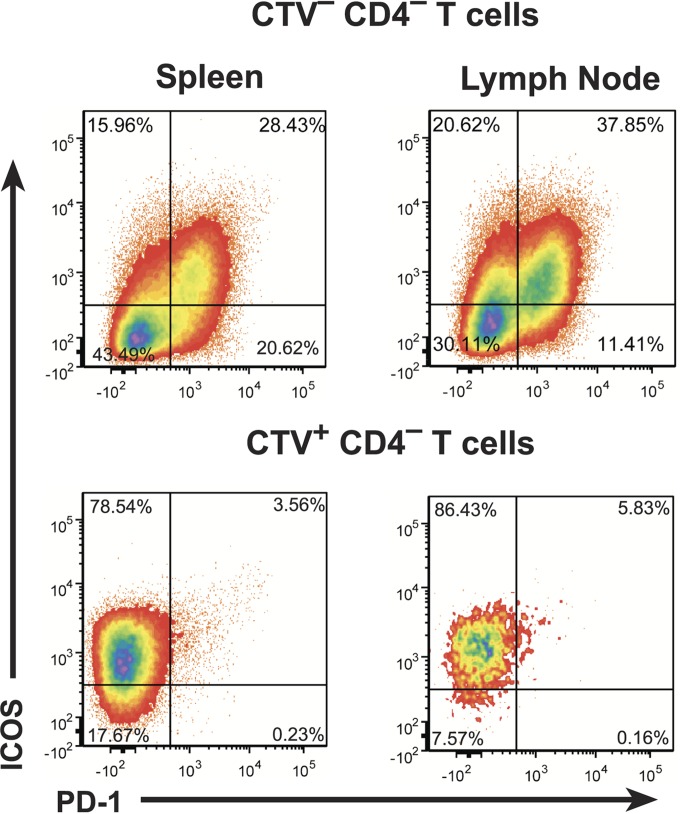
While our results show that CXCR5 expression is necessary for CD8 T-cell trafficking into B-cell follicles, its sole sufficiency for entry is not clear. Given the intricate nature of the physiologic development of TFH and their entry into B-cell follicles, the follicular targeting of CD8hCXCR5 T cells derived from bulk CD8 T cells could require expression of other proteins acting as cofactors. Two proteins associated with post-CXCR5 expression of TFH entry into and maintenance within B-cell follicles are PD-1, which is tightly correlated with both TFH and CXCR5+ CD8 T-cell trafficking (27, 32), and ICOS which has been proposed as a key molecule for entry (39). As presented above in Fig. 3, both the CD8hCXCR5 and untransduced CD8 T-cell cultures contained mostly ICOS+ cells with 16% of the cells being PD-1+/ICOS+. However, flow cytometry analysis of both spleen and lymph node necropsy tissue samples showed that the majority of the CXCR5+ transductants present were ICOS+ and PD-1− (Fig. 8, representative data for animal 4). The frequencies of CD8hCXCR5 T cells with a memory phenotype (CD28+/CD95+) were similar (data not shown) to those of the preinfusion cultures (Fig. 3). The absence of PD-1 in the infused CD8hCXCR5 cells that localized to follicles suggests that this protein was either downregulated or selected against on the CD8hCXCR5 T cells that successfully entered the follicles. In contrast, the maintenance of ICOS in the CD8hCXCR5 cells trafficking to B-cell follicles is consistent with the previously described role of ICOS as a necessary cofactor for normal TFH entry into B-cell follicles (39) and suggests that this cofactor could also play a yet undefined role in the CXCR5-forced targeting of CD8 T cells into follicles.
FIG 8.
Maintenance of ICOS expression in CD8hCXCR5 T cells. Representative flow cytometry analysis plots for ICOS and PD-1 expression in CD3+ T cells isolated from spleen and lymph node samples from animal 4 are presented. Gating for endogenous CTV− CD4− (top) and CTV+ CD8hCXCR5 T cells (bottom) cells are denoted above each plot.
CXCR5 transductants migrate to follicular regions containing SIV-infected cells.
Our ultimate goal in CXCR5-mediated targeting of CD8 T cells to B-cell follicles is to direct antiviral cytotoxic CD8 T cells to these sites to mediate effective viral suppression and/or clearance of infection in this sanctuary site. B-cell follicles have subregions, particularly light and dark zones, through which TFH specifically traffic in association with follicular B cells (49). To determine whether the CD8hCXCR5 T cells have access to the same follicular regions as SIV-infected TFH, we used a duplex amplified in situ hybridization (ISH) technique to visualize the presence of both vector-transduced and SIV-infected TFH cells in the follicle (9, 50). Thin tissue sections of lymph nodes were hybridized to two sets of double-Z probes, one specific for SIV RNA (shown in red) and another specific for the MuLV-derived sequences present in our CXCR5 retroviral vector (shown in yellow) (Fig. 9). The micrographs revealed the presence of punctuate red staining in the interstitial spaces around the cells, primarily in the light region/germinal center (Fig. 9A). Inspection at a higher magnification reveals that this viral RNA staining is made up of distinct small dots, each resulting from individual SIV virions present in the follicle (Fig. 9B) as detected by our highly sensitive hybridization technique (9). The vector-expressing cells were distributed throughout the B-cell follicle, including the light and dark zones (some highlighted with yellow arrows [Fig. 9]). ISH of tissues from different sections of the gut (ileum, cecum, and transverse colon) detected somewhat less CXCR5 transductants as well as infected cells in Peyer's patches and lymphoid aggregates from animals 1 and 6 (data not shown). Overall, the localization of the CXCR5 transductants was not obviously limited to any subregion of the follicle. Importantly, although we infused bulk CD8 cells that were not selected for SIV-specific cells, in several fields, some CD8hCXCR5 T cells colocalized in close proximity to SIV-infected cells, demonstrating that they can access the regions of the follicle that contain infected TFH.
FIG 9.
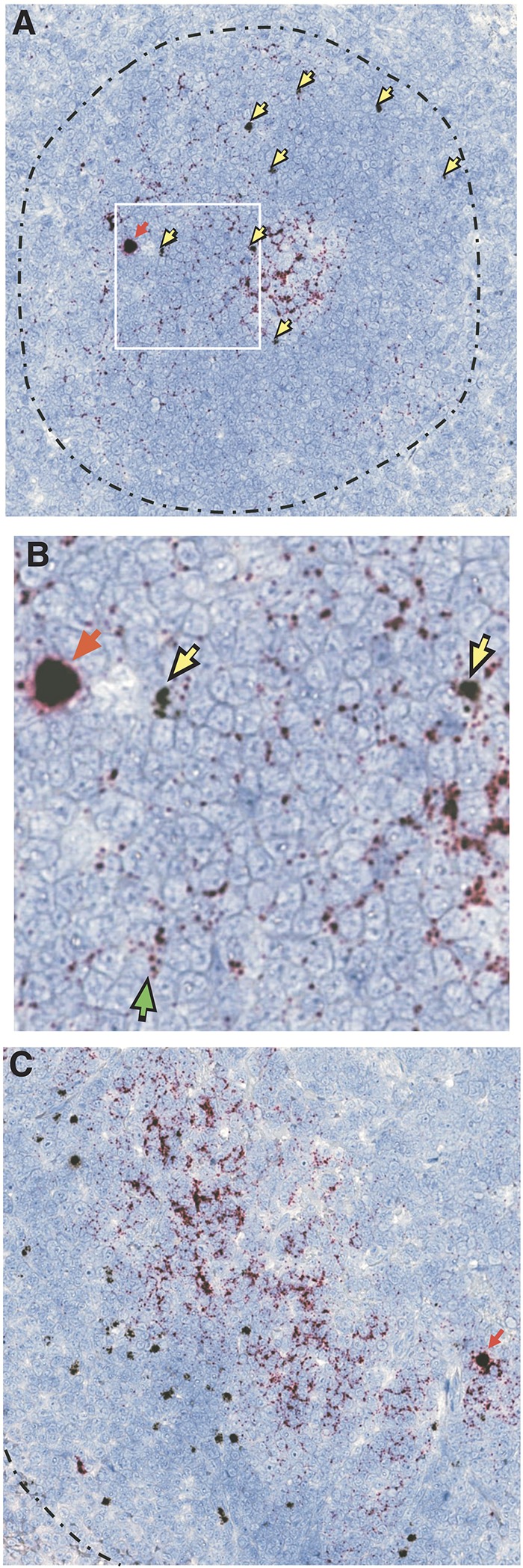
Duplex ISH detection of CD8hCXCR5 T-cell and SIV-infected cells in B-cell follicles. Representative duplex ISH analysis of thin tissue sections with retroviral vector RNA probe-hybridizing cells in black (examples highlighted with yellow arrows) and SIV hybridization in red (examples of red-staining cells highlighted with red arrows), with the T-cell zone/B-cell follicle border indicated by a broken black line. (A) Lymph node sample with an example of close proximity between infected and CD8hCXCR5 T cells boxed in white. (B) Magnification of the boxed lymph node area with a green arrow indicating an example of the presence of individual virions. (C) Spleen sample.
DISCUSSION
Here we show that engineering unselected PBMC-derived CD8 T cells to express CXCR5 results in their preferential localization to B-cell follicles in both spleen and lymph nodes, with approximately 120-fold-more CD8hCXCR5 T cells localizing to B-cell follicles than to the surrounding T-cell zone in secondary lymphoid tissues. It is unlikely that our engineered CD8hCXCR5 T cells enter B-cell follicles in the same complex antigen-specific differentiation-linked mechanism used by native TFH, where CXCR5+ B cells tow their cognate TFH into the follicles. Indeed, even though our experimental design used unselected CD8 T cells, the infused CD8hCXCR5 T cells, being derived from infected animals, included low, but clearly present frequencies of SIV-specific T cells detected by flow cytometry with a tetramer specific for the immunodominant MamuA*01-restricted SIVGagCM9 epitope (approximately 0.5 to 1% SIVGag CM9+ [data not shown]). However, these SIV-specific cells, which were a minor fraction of the infused cells, were not detected in flow cytometry analyses of either spleen or lymph node necropsy samples (data not shown) and therefore inconsistent with antigen-driven localization. Instead, the trafficking was more likely due to direct entry into B-cell follicles by CXCL13-driven chemotaxis, similar to B-cell entry (31–33).
While our results demonstrate that CXCR5 is necessary for preferential localization of unselected CD8 T cells, we cannot conclude that CXCR5 is sufficient for entry of the cells into the follicles. Indeed, the maintenance of ICOS on nearly all of the transduced cells is consistent with its proposed role in the de novo TFH differentiation/entry pathway, suggesting that ICOS coexpression might also be required for effective CXCR5-mediated targeting of CD8 T cells to B-cell follicles. In contrast, the CD8hCXCR5 T cells localized to B-cell follicles were uniformly PD-1−, suggesting that this factor was dispensable for trafficking.
Other adoptive transfer studies relying on longer, more extensive in vitro selection and expansion of naturally occurring SIV-specific T-cell clones in culture rather than the limited ex vivo expansion made possible by engineering T cells presented here have observed no detectable persistence in either blood or tissues other than the lung (47, 48). Recently, we applied T-cell engineering techniques to express CCR7/CD62L on T cells and observed limited, but unequivocal trafficking to lymph nodes and avoidance of the lung, demonstrating targeting of infused T cells to lymphoid tissues in a nonhuman primate system (43). Here, we extend this concept with cells that had less extensive expansion ex vivo than used in our prior CCR7/CD62L study, and we observed greater frequencies of infused cells in the secondary lymphoid tissues. Our ongoing efforts are focused on increasing the numbers of localizing CD8hCXCR5 T cells and their persistence potential by optimizing CXCR5 transduction procedures and cell culture conditions with the goal of minimizing the ex vivo culture/expansion manipulations.
The ability to directly place CD8 T cells into B-cell follicles theoretically allows for the localization of antiviral CD8 T cells into this otherwise immune privileged site (11). Other groups have observed in vivo trafficking and antiviral activity of native CXCR5 CD8 T cells in mice. Recently, Leong et al. isolated CXCR5+ CD8 T cells from mice expressing a transgenic TCR specific for lymphocytic choriomeningitis virus (LCMV) which entered B-cell follicles when infused into chronically infected CXCR5−/− knockout mice and reduced LCMV levels in TFH twofold (12). In the same report, adoptive transfer of endogenously derived CXCR5+ CD8 T cells engineered with a TCR specific for the B-cell-tropic murid herpesvirus-4 (MuHV-4) also homed to follicles and reduced the levels of MuHV-4 by fivefold. Furthermore, He et al. observed that anti-LCMV CXCR5+ CD8 T cells were present in chronically infected mice, but not in acutely infected mice. Transfer experiments demonstrated a requirement for CXCR5 expression for CD8 T-cell entry and a reduction in the LCMV viral load of 3 orders of magnitude (13). These adoptive transfer experiments in mice highlight the potential for placing effective AIDS virus-specific CD8 cytotoxic T lymphocytes (CTLs) into B-cell follicles to reduce infection in these viral sanctuaries.
One obstacle to this approach is the relatively low frequency of naturally occurring antiviral CD8 T cells, which require extensive expansion in culture to generate sufficient numbers of cells for infusion with its attendant poor persistence of adoptively transferred cells in lymphoid tissues (47, 48). To overcome this limitation, we have developed an approach for transferring TCRs isolated from highly effective virus-suppressing T-cell clones by retroviral transduction (51–53). Large-scale transductions with these vectors can rapidly transfer well-characterized, functional antiviral TCR specificities into large numbers of T cells, thereby generating large and effective CD8 T-cell responses for adoptive transfer experiments (43, 51, 53). Recently, we observed an antiviral effect by infusing engineered SIV-specific T cells in conjunction with SIV inoculation, manifested in a reduction of the number of transmitted/founder viruses in an acute infection model (43). Preliminary experiments combining this TCR engineering approach with the CXCR5-mediated targeting demonstrated here are testing the feasibility of designing CD8 T cells with both effective antiviral TCR specificities and CXCR5-mediated B-cell follicle targeting to place CD8 T cells with effective antiviral properties into these follicular AIDS virus sanctuaries. Importantly, success in these SIV/rhesus macaque studies could be translatable to a clinical immunotherapeutic aimed at reducing persistent virus present in cART-treated patients (54).
MATERIALS AND METHODS
Rhesus macaques.
Six MamuA*01 Indian rhesus macaques (Macaca mulatta), chronically infected with SIVmac239, were cared for and housed at the National Cancer Institute (NCI) in Bethesda, MD. All animal care and procedures, including infusions, blood draws, tissue biopsy specimens, and euthanasia/necropsy were carried out under protocols approved by the NCI Institutional Animal Care and Use Committee. Macaques were caged individually to reduce the risk of transmission of SIV or other viruses. All clinical procedures, including administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Steps were taken to ensure the welfare of the animals and minimize discomfort of all animals used in this study. Animals were closely monitored daily for any signs of illness, and appropriate medical care was provided as needed. NCI and Frederick National Laboratory are accredited by AAALAC International and follow the Public Health Service Policy for the Care and Use of Laboratory Animals (55). Animal care was provided in accordance with the procedures outlined in the Guide for Care and Use of Laboratory Animals (56).
Cell culture.
Primary rhesus T cells were expanded and maintained as previously described (47, 57, 58) in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum (Gemini Bio Products), 10 mM HEPES buffer, 2 mM glutamine (Life Technologies), interleukin-2 (IL-2) (50 IU/ml; NIH AIDS reagent repository), penicillin (100 μg/ml; Life Technologies), and streptomycin (100 μg/ml; Life Technologies). Cells were expanded through biweekly stimulation with anti-CD3 (30 ng/ml; clone SP34-2; BD Biosciences), IL-2 (50 IU/ml), irradiated human peripheral blood mononuclear cells (PBMC). and an Epstein-Barr virus transformed B-cell line (TM B-LCL) (47, 57, 58).
CXCR5 vector construction and production.
The human CXCR5 gene was cloned from cDNA prepared from RNA isolated from anonymous donor PBMC. The CXCR5 gene was sequenced and found to match the human coding sequence previously published in GenBank (accession no. NM_001716.4) except for a nucleotide difference at codon 334 (TCG-TTG) in the unprocessed protein resulting in a serine-to-leucine difference with the reference sequence. The CXCR5 vector DNA construct was produced by inserting the CXCR5 gene into a modified MSGV1 vector at PacI (the NcoI at nucleotide 1813 of MSGV1 removed and replaced with a PacI site) and EcoRI (59). Retroviral vectors were generated by transfecting vector constructs into Phoenix-RD114 packaging cells (60).
Flow cytometry.
The antibodies used in flow cytometry were as follows: CD3 (SP34-2), CD45 (HI30), CD4 (OKT4), CD8 (SK1), CCR7 (3D12), CD28 (CD28.2), CD95 (DX2), and CXCR4 (12g5) [all from BD Biosciences]; human CXCR5 (J252D4), PD-1 (EH12.h7), and ICOS (C398.4a) from BioLegend; and rhesus macaque CXCR5 (MU5UBEE) from eBioscience. More than 100,000 events were acquired on a BD LSRII or Fortessa flow cytometer (BD Biosciences). Complete blood cell counts were monitored using BD TruCount tubes (BD Biosciences) on a BD FACSVerse analyzer (BD Biosciences). Data analysis was performed using FCS Express 4 (De Novo Software). For flow cytometry analysis, lung tissue was prepared by a preliminary collagenase digestion, followed by mechanical disruption to isolate mononuclear cells.
CXCR5 transduction.
Transductions of CD8 T cells with CXCR5 retroviral vector supernatants were carried out as previously described (61). Supernatants containing retroviral vector were loaded onto retronectin (TaKaRa/Clontech, Inc.)-coated six-well non-tissue-coated plates (treated with 20 μg/ml) and centrifuged at 2,000 × g for 2 h at 32°C. Half of the vector supernatant was removed, and 1 × 106 stimulated T cells were added per well. The cells were then centrifuged at 2,000 × g for 15 min at 32°C followed by incubation at 37°C for 48 h before flow cytometric analysis for CXCR5 expression to determine transduction efficiency. After staining with either CD8 or CXCR5 phycoerythrin (PE)-conjugated antibody, cells were sorted with anti-PE paramagnetic beads and LS columns (Miltenyi Biotech, Inc.).
ERK1/2 phosphorylation analysis.
CXCR5-mediated ERK1/2 signaling was examined by immunoblotting as previously described (61). Briefly, the levels of ERK1/2 and phosphorylated ERK1/2 (pERK1/2) were measured in lysates produced from 2 × 106 CD8 T cells using phosphorylation-specific antibodies following stimulation with 2 μg/ml of human CXCL13 (ProSpec) for 3, 15, 30, and 60 min in serum-free medium. The cells were harvested by centrifugation at 10,000 × g for 1 min. The cell pellets were lysed with Pierce immunoprecipitation (IP) lysis buffer containing 1× Halt protease and phosphatase inhibitor cocktail (ThermoScientific) on ice for 30 min. The nuclear fraction was separated from the cytosolic lysate by centrifugation for 10 min at 10,000 × g, and the supernatant was transferred to a new tube. Prior to SDS fractionation, Laemmli sample loading buffer (62) was added to the lysates at a final concentration of 2×, and samples were boiled for 5 min. Samples were analyzed by quantitative near-infrared immunoblotting (63) with modifications using phospho-ERK1/2 (pERK1/2)-specific rabbit antiserum (ab4819; Abcam; at a 1:1,000 dilution) and an ERK1/2-specific mouse monoclonal antibody (66192-1-Ig; Proteintech Group, Inc.; at a 1:5,000 dilution) followed, after washing, by two-color detection with anti-mouse IRDye 680LT and anti-rabbit IRDye 800CW fluorescently labeled donkey secondary antibodies (LI-COR) and then analyzed after washing with an Odyssey infrared imaging system (LI-COR) using a laser intensity of between 1 and 5. The signal densities of bands were measured by the Odyssey 3.0 application software.
In vitro chemotaxis assay.
Chemotaxis was measured using transwell inserts (5-μm pore size) on a 24-well plate (Costar). The upper and lower chambers were filled with serum-free media. A total of 200,000 cells were added to the upper chamber, and the cells were allowed to settle for 1 h. Then 1 μg/ml CXCL13 (PROSPEC) was added to the lower chamber, and cell migration was measured by counting the cells in the lower chamber 3 h after the addition of CXCL13. Specific cell migration was calculated by subtracting the number of cells migrating in the absence of chemokine from the number of cells migrating in the presence of chemokine and then calculating the ratio of cells in the lower chamber relative to the number of input cells.
Adoptive T-cell transfer.
CD8 T cells from each animal to be infused were labeled with either 5 μM CellTrace violet (CTV) (Life Technologies) for the CXCR5 transductants or 5 μM CellTrace far red (CTFR) for the matching untransduced cells, resuspended in 50 ml saline solution and 2% (vol/vol) autologous serum, and infused into the femoral vein (flow rate of 2 ml/min) as previously described (47, 48). To monitor the transfer of cells, a 15-min postinfusion sample was collected from the femoral vein. Macaques were administered IL-2 subcutaneously (104 international units [IU]/kg of body weight) on the day of infusion and the day following infusion. The numbers of infused cells and weights of animals are provided in Table 1.
Confocal microscopic tissue analysis.
Briefly, Tissue-Tek OCT frozen tissue sections were placed in 4% paraformaldehyde-water (vol/vol) for 15 min at 4°C, followed by a brief submersion in ethanol and air drying. To visualize B-cell follicles, fixed sections were incubated in a 1:500 dilution of anti-CD20 (catalog no. 3807; Dako, Aligent Pathology Solutions) in Tris-buffered saline (Boston Biosciences) with 0.05% Tween 20 (vol/vol) and 0.25% casein (wt/vol) for 1 h at room temperature (RT) and then incubated in a 1:500 dilution of Alexa Fluor 594-conjugated donkey anti-rabbit secondary antibody for 15 min at RT. Slides were rinsed and mounted with Gold Seal cover glass with a thickness of 1.5 (Electron Microscopy Services) using Prolong Gold mounting medium (Invitrogen). Regions of interest were acquired using an Olympus Fluoview FV10i system at a magnitude of 600. The transduced (CTV+) and untransduced (CTFR+) cells present in 850 mm2 of combined fields were counted. Preferential localization of either CTV+ or CTFR+ cells is expressed as the ratio of the cell counts within follicles to those without follicles. Cumulative fields with no cells present for a particular category are assigned our limit of detection, ≤1 cell. A total of 3 spleen and 11 lymph node samples taken from five animals were examined.
Duplex SIV RNA and MuLV vector in situ hybridization.
Simultaneous visualization of both SIV RNA and the MuLV regions of the expressed vector RNAs in all animals, lymph nodes, and spleen tissues was performed as previously described (9). SIV RNA was visualized by hybridization to an alkaline phosphatase-conjugated RNA probe and MuLV vector RNA with a horseradish peroxidase-conjugated probe. Bright-field images were acquired and extracted using the APERIO AT2 digital scanning system (Leica Biosystems) after scanning the slides at high-resolution and ×400 magnification settings.
ACKNOWLEDGMENTS
We thank Vicky Coalter, Don Johnson, Jacob Kiser, Adam Wiles, and Rodney Wiles for processing tissues. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: human recombinant IL-2 (rIL-2) from Maurice Gately, Hoffmann-La Roche Inc.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E.
The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M Jr, Estes JD, Lifson JD, Picker LJ. 2015. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Folkvord JM, Armon C, Connick E. 2005. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res Hum Retroviruses 21:363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- 3.Connick E, Mattila T, Folkvord JM, Schlichtemeier R, Meditz AL, Ray MG, McCarter MD, Mawhinney S, Hage A, White C, Skinner PJ. 2007. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol 178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- 4.Streeck H. 2015. AIDS virus seeks refuge in B cell follicles. Nat Med 21:111–112. doi: 10.1038/nm.3795. [DOI] [PubMed] [Google Scholar]
- 5.Brenchley JM, Vinton C, Tabb B, Hao XP, Connick E, Paiardini M, Lifson JD, Silvestri G, Estes JD. 2012. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood 120:4172–4181. doi: 10.1182/blood-2012-06-437608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD III, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M Jr, Lifson JD, Sekaly RP, Picker LJ. 2012. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med 18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. 2016. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22:754–761. doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 9.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M Jr, Bess J, Anderson JL, Perkey KE, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD. 2016. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun 1:68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, Santiago ML, Schmitt K, Stephens EB, Kim HO, Wagstaff R, Li S, Abdelaal HM, Kemp N, Watkins DI, MaWhinney S, Skinner PJ. 2014. Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol 193:5613–5625. doi: 10.4049/jimmunol.1401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skinner PJ, Connick E. 2014. Overcoming the immune privilege of B cell follicles to cure HIV-1 infection. J Hum Virol Retrovirol 1:00001. [Google Scholar]
- 12.Leong YA, Chen Y, Ong HS, Wu D, Man K, Deleage C, Minnich M, Meckiff BJ, Wei Y, Hou Z, Zotos D, Fenix KA, Atnerkar A, Preston S, Chipman JG, Beilman GJ, Allison CC, Sun L, Wang P, Xu J, Toe JG, Lu HK, Tao Y, Palendira U, Dent AL, Landay AL, Pellegrini M, Comerford I, McColl SR, Schacker TW, Long HM, Estes JD, Busslinger M, Belz GT, Lewin SR, Kallies A, Yu D. 2016. CXCR5+ follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 13.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L. 2016. Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537:412–416. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 14.Quigley MF, Gonzalez VD, Granath A, Andersson J, Sandberg JK. 2007. CXCR5+ CCR7− CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur J Immunol 37:3352–3362. doi: 10.1002/eji.200636746. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Folkvord JM, Rakasz EG, Abdelaal HM, Wagstaff RK, Kovacs KJ, Kim HO, Sawahata R, MaWhinney S, Masopust D, Connick E, Skinner PJ. 2016. Simian immunodeficiency virus-producing cells in follicles are partially suppressed by CD8+ cells in vivo. J Virol 90:11168–11180. doi: 10.1128/JVI.01332-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles B, Miller SM, Folkvord JM, Levy DN, Rakasz EG, Skinner PJ, Connick E. 2016. Follicular regulatory CD8 T cells impair the germinal center response in SIV and ex vivo HIV infection. PLoS Pathog 12:e1005924. doi: 10.1371/journal.ppat.1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. 2012. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest 122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pissani F, Streeck H. 2014. Emerging concepts on T follicular helper cell dynamics in HIV infection. Trends Immunol 35:278–286. doi: 10.1016/j.it.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. 2012. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest 122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma CS, Deenick EK, Batten M, Tangye SG. 2012. The origins, function, and regulation of T follicular helper cells. J Exp Med 209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. 1998. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature 391:799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 22.Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. 1996. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell 87:1037–1047. doi: 10.1016/S0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 23.Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG. 2000. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406:309–334. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 24.Cyster JG, Ansel KM, Reif K, Ekland EH, Hyman PL, Tang HL, Luther SA, Ngo VN. 2000. Follicular stromal cells and lymphocyte homing to follicles. Immunol Rev 176:181–193. doi: 10.1034/j.1600-065X.2000.00618.x. [DOI] [PubMed] [Google Scholar]
- 25.Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol 5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- 26.Ansel KM, McHeyzer-Williams LJ, Ngo VN, McHeyzer-Williams MG, Cyster JG. 1999. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J Exp Med 190:1123–1234. doi: 10.1084/jem.190.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol 179:5099–1508. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 28.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med 192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. 2001. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med 193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med 192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 32.Qi H, Chen X, Chu C, Lu P, Xu H, Yan J. 2014. Follicular T-helper cells: controlled localization and cellular interactions. Immunol Cell Biol 92:28–33. doi: 10.1038/icb.2013.59. [DOI] [PubMed] [Google Scholar]
- 33.Vinuesa CG, Cyster JG. 2011. How T cells earn the follicular rite of passage. Immunity 35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. 2008. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature 455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardtke S, Ohl L, Forster R. 2005. Balanced expression of CXCR5 and CCR7 on follicular T helper cells determines their transient positioning to lymph node follicles and is essential for efficient B-cell help. Blood 106:1924–1931. doi: 10.1182/blood-2004-11-4494. [DOI] [PubMed] [Google Scholar]
- 36.Reif K, Ekland EH, Ohl L, Nakano H, Lipp M, Forster R, Cyster JG. 2002. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature 416:94–99. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, O'Garra A, Cahalan MD, Cyster JG. 2005. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol 3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi H, Egen JG, Huang AY, Germain RN. 2006. Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 312:1672–1666. doi: 10.1126/science.1125703. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, Zhang L, Qi H. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 40.Culver KW, Osborne WR, Miller AD, Fleisher TA, Berger M, Anderson WF, Blaese RM. 1991. Correction of ADA deficiency in human T lymphocytes using retroviral-mediated gene transfer. Transplant Proc 23:170–171. [PubMed] [Google Scholar]
- 41.Barrett DM, Grupp SA, June CH. 2015. Chimeric antigen receptor- and TCR-modified T cells enter Main Street and Wall Street. J Immunol 195:755–761. doi: 10.4049/jimmunol.1500751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Morgan RA. 2012. Genetic engineering with T cell receptors. Adv Drug Deliv Rev 64:756–762. doi: 10.1016/j.addr.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayala VI, Trivett MT, Barsov EV, Jain S, Piatak M Jr, Trubey CM, Alvord WG, Chertova E, Roser JD, Smedley J, Komin A, Keele BF, Ohlen C, Ott DE. 2016. Adoptive transfer of engineered rhesus simian immunodeficiency virus-specific CD8+ T cells reduces the number of transmitted/founder viruses established in rhesus macaques. J Virol 90:9942–9952. doi: 10.1128/JVI.01522-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Prete GQ, Park H, Fennessey CM, Reid C, Lipkey L, Newman L, Oswald K, Kahl C, Piatak M Jr, Quinones OA, Alvord WG, Smedley J, Estes JD, Lifson JD, Picker LJ, Keele BF. 2014. Molecularly tagged simian immunodeficiency virus SIVmac239 synthetic swarm for tracking independent infection events. J Virol 88:8077–8090. doi: 10.1128/JVI.01026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Haibi CP, Singh R, Sharma PK, Singh S, Lillard JW Jr. 2011. CXCL13 mediates prostate cancer cell proliferation through JNK signalling and invasion through ERK activation. Cell Prolif 44:311–319. doi: 10.1111/j.1365-2184.2011.00757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi GX, Harrison K, Wilson GL, Moratz C, Kehrl JH. 2002. RGS13 regulates germinal center B lymphocytes responsiveness to CXC chemokine ligand (CXCL)12 and CXCL13. J Immunol 169:2507–2515. doi: 10.4049/jimmunol.169.5.2507. [DOI] [PubMed] [Google Scholar]
- 47.Minang JT, Trivett MT, Bolton DL, Trubey CM, Estes JD, Li Y, Smedley J, Pung R, Rosati M, Jalah R, Pavlakis GN, Felber BK, Piatak M Jr, Roederer M, Lifson JD, Ott DE, Ohlen C. 2010. Distribution, persistence, and efficacy of adoptively transferred central and effector memory-derived autologous simian immunodeficiency virus-specific CD8+ T cell clones in rhesus macaques during acute infection. J Immunol 184:315–326. doi: 10.4049/jimmunol.0902410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolton DL, Minang JT, Trivett MT, Song K, Tuscher JJ, Li Y, Piatak M Jr, O'Connor D, Lifson JD, Roederer M, Ohlen C. 2010. Trafficking, persistence, and activation state of adoptively transferred allogeneic and autologous simian immunodeficiency virus-specific CD8(+) T cell clones during acute and chronic infection of rhesus macaques. J Immunol 184:303–314. doi: 10.4049/jimmunol.0902413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamel KM, Liarski VM, Clark MR. 2012. Germinal center B-cells. Autoimmunity 45:333–347. doi: 10.3109/08916934.2012.665524. [DOI] [PubMed] [Google Scholar]
- 50.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barsov EV, Trivett MT, Minang JT, Sun H, Ohlen C, Ott DE. 2011. Transduction of SIV-specific TCR genes into rhesus macaque CD8+ T cells conveys the ability to suppress SIV replication. PLoS One 6:e23703. doi: 10.1371/journal.pone.0023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coren LV, Jain S, Trivett MT, Ohlen C, Ott DE. 2015. Production of retroviral constructs for effective transfer and expression of T-cell receptor genes using Golden Gate cloning. Biotechniques 58:135–139. doi: 10.2144/000114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain S, Trivett MT, Ayala VI, Ohlen C, Ott DE. 2015. African green monkey TRIM5alpha restriction in simian immunodeficiency virus-specific rhesus macaque effector CD4 T cells enhances their survival and antiviral function. J Virol 89:4449–4456. doi: 10.1128/JVI.03598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Institutes of Health. 2002. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD. [Google Scholar]
- 56.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 57.Berger C, Huang ML, Gough M, Greenberg PD, Riddell SR, Kiem HP. 2001. Nonmyeloablative immunosuppressive regimen prolongs in vivo persistence of gene-modified autologous T cells in a nonhuman primate model. J Virol 75:799–808. doi: 10.1128/JVI.75.2.799-808.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riddell SR, Greenberg PD. 1990. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods 128:189–201. doi: 10.1016/0022-1759(90)90210-M. [DOI] [PubMed] [Google Scholar]
- 59.Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA, Morgan RA. 2005. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther 16:457–472. doi: 10.1089/hum.2005.16.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neff T, Peterson LJ, Morris JC, Thompson J, Zhang X, Horn PA, Thomasson BM, Kiem HP. 2004. Efficient gene transfer to hematopoietic repopulating cells using concentrated RD114-pseudotype vectors produced by human packaging cells. Mol Ther 9:157–159. doi: 10.1016/j.ymthe.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Ayala VI, Trivett MT, Coren LV, Jain S, Bohn PS, Wiseman RW, O'Connor DH, Ohlen C, Ott DE. 2016. A novel SIV gag-specific CD4(+)T-cell clone suppresses SIV replication in CD4(+)T cells revealing the interplay between antiviral effector cells and their infected targets. Virology 493:100–112. doi: 10.1016/j.virol.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 63.Coren LV, Trivett MT, Jain S, Ayala VI, Del Prete GQ, Ohlen C, Ott DE. 2015. Potent restriction of HIV-1 and SIVmac239 replication by African green monkey TRIM5α. Retrovirology 12:11. doi: 10.1186/s12977-015-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]