ABSTRACT
An efficient adenovirus infection results in high-level accumulation of viral DNA and mRNAs in the infected cell population. However, the average viral DNA and mRNA content in a heterogeneous cell population does not necessarily reflect the same abundance in individual cells. Here, we describe a novel padlock probe-based rolling-circle amplification technique that enables simultaneous detection and analysis of human adenovirus type 5 (HAdV-5) genomic DNA and virus-encoded mRNAs in individual infected cells. We demonstrate that the method is applicable for detection and quantification of HAdV-5 DNA and mRNAs in short-term infections in human epithelial cells and in long-term infections in human B lymphocytes. Single-cell evaluation of these infections revealed high heterogeneity and unique cell subpopulations defined by differential viral DNA content and mRNA expression. Further, our single-cell analysis shows that the specific expression pattern of viral E1A 13S and 12S mRNA splice variants is linked to HAdV-5 DNA content in the individual cells. Furthermore, we show that expression of a mature form of the HAdV-5 histone-like protein VII affects virus genome detection in HAdV-5-infected cells. Collectively, padlock probes combined with rolling-circle amplification should be a welcome addition to the method repertoire for the characterization of the molecular details of the HAdV life cycle in individual infected cells.
IMPORTANCE Human adenoviruses (HAdVs) have been extensively used as model systems to study various aspects of eukaryotic gene expression and genome organization. The vast majority of the HAdV studies are based on standard experimental procedures carried out using heterogeneous cell populations, where data averaging often masks biological differences. As every cell is unique, characteristics and efficiency of an HAdV infection can vary from cell to cell. Therefore, the analysis of HAdV gene expression and genome organization would benefit from a method that permits analysis of individual infected cells in the heterogeneous cell population. Here, we show that the padlock probe-based rolling-circle amplification method can be used to study concurrent viral DNA accumulation and mRNA expression patterns in individual HAdV-5-infected cells. Hence, this versatile method can be applied to detect the extent of infection and virus gene expression changes in different HAdV-5 infections.
KEYWORDS: adenovirus, persistent infection, lytic infection, rolling-circle amplification, single-cell analysis
INTRODUCTION
Human adenoviruses (HAdV) are double-stranded DNA (dsDNA) viruses classified into seven distinct species, A to G, with over 60 species described so far (1). Typically, HAdV infection of epithelial cells causes an efficient cell lysis and release of infectious virus progeny within a few days postinfection (2). In addition, an HAdV infection can lead to long-term latent/persistent infections. In particular, the species C viruses (HAdV-1, HAdV-2, HAdV-5, and HAdV-6) have been detected in human adenoidal and tonsillar lymphocytes, where they are believed to establish long-term latent/persistent infections (3–5).
The HAdV infectious cycle is subdivided into an early and late phase based on the time of onset of virus DNA replication. The first viral gene to be expressed is the early region 1A (E1A) gene (6). The E1A pre-mRNA undergoes extensive alternative splicing, resulting in the translation of at least five different E1A protein isoforms (7, 8). The main role of the major E1A proteins, E1A(243R) and E1A(289R), is to force the host cell into S phase of the cell cycle via interaction with the pRb family of proteins. In addition, the E1A proteins activate expression of other early viral genes to initiate the viral DNA replication (9). Among the genes activated is the viral E2 transcription unit, coding for the viral DNA polymerase (Ad-Pol), the terminal protein (pTP), and the single-stranded DNA-binding protein (DBP) E2A-72K, all needed for the replication of the viral genome (10). Importantly, initiation of viral DNA replication triggers the accumulation of HAdV-2 late-phase transcripts (11). Essentially all HAdV late genes are expressed from a single transcription unit, the major late transcription unit (MLTU). Transcription from the MLTU gives rise to a 28,000-nucleotide-long major late pre-mRNA, which undergoes extensive posttranscriptional processing by alternative polyadenylation site usage and alternative splicing to produce five families of late mRNAs. All MLTU mRNAs receive through alternative splicing a common 5′ noncoding sequence, the so-called tripartite leader, which is created by joining the first three exons present at the 5′ end of the major late pre-mRNA (reviewed in reference 12). The proteins encoded by the MLTU are mainly structural proteins needed for virus particle formation. Notably, MLTU encodes an abundant histone-like protein, pVII, which is proteolytically processed into the mature protein VII during the final steps of virion maturation (13). The mature protein VII [also known as pVII(Δ24) (14)] organizes the viral genome into a nucleosome-like structure (15–17), protects viral DNA from the cellular DNA damage response (18), and also can act as a protein regulating transcription (19, 20).
An efficient HAdV infection results in high-level accumulation of viral DNA and early and late mRNAs in the infected cell population (12, 21). However, the average viral DNA and mRNA quantities in a heterogeneous cell population do not necessarily reflect the same abundance in the individual cells. For example, infection efficiency of HAdV-5 depends on the cell cycle status and expression of specific viral genes (22). Also, during long-term latent/persistent HAdV-5 infection, the amount of viral DNA does not necessarily correlate with viral gene transcription, as the viral genomes can be transcriptionally silenced in these infections (23). Hence, single-cell studies can be more informative for monitoring viral DNA accumulation and mRNA expression in individual HAdV-infected cells.
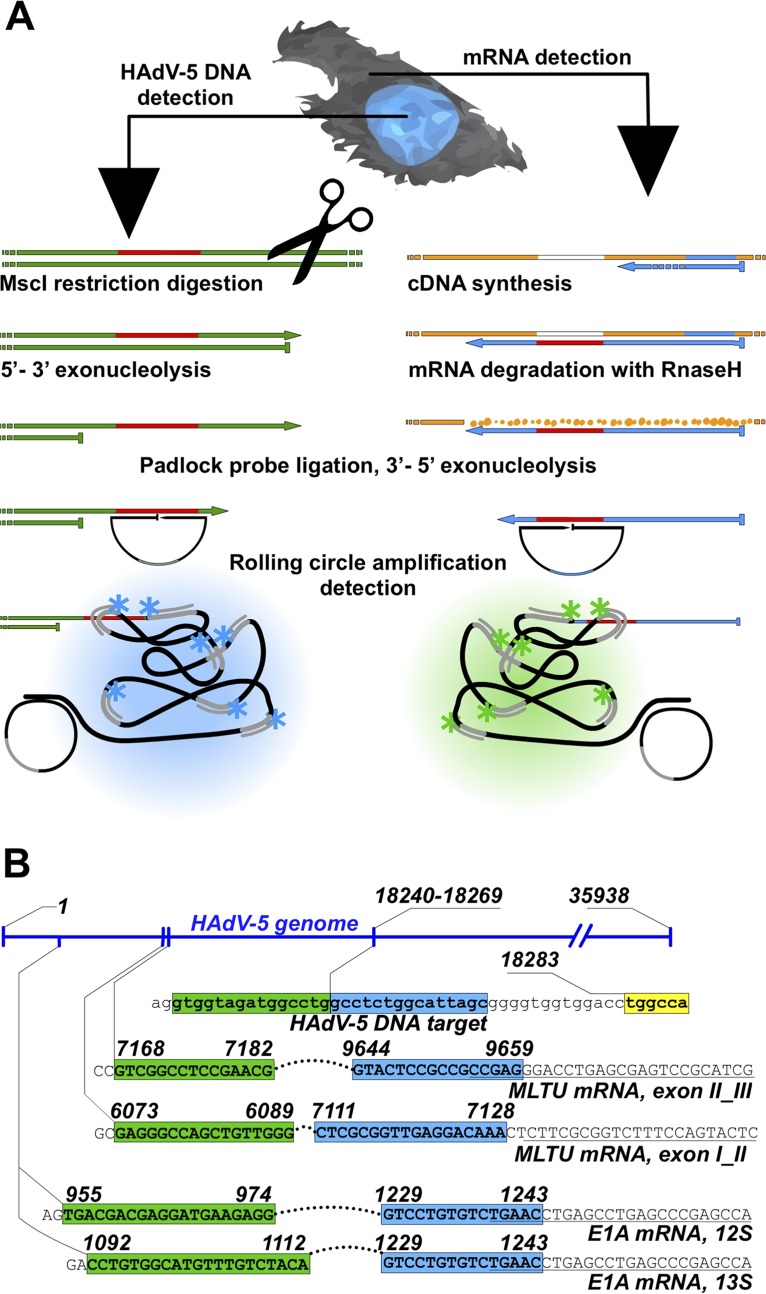
Rolling-circle amplification (RCA) of circularized padlock probes (PLP) is a sensitive method to detect and quantitate molecules in individual cells (Fig. 1A) (24). Indeed, a specific amplification of the PLPs by RCA has been successfully applied to detect genome-specific repeated sequences in human metaphase chromosomes (25), cellular genomic DNA (26), cDNA (24), and mRNA (27) in situ. PLP-based RCA methods have also been applied to detect HAdV-5 E1A alternatively spliced mRNAs ex situ (28) and porcine circovirus type 2 virus DNA in situ (29). Whereas the usage of PLPs for mRNA detection has received a standard molecular technique status already (30), simultaneous and multiplexed analyses of DNA and mRNAs across various biological specimens have not been reported.
FIG 1.
HAdV-5 DNA and mRNA detection by rolling-circle amplification (RCA). (A) Simplified schematic overview of concurrent viral DNA and mRNA detection. Viral mRNA is converted to cDNA by reverse transcription, followed by mRNA degradation by RNase H. HAdV-5 DNA endonucleolytic cleavage with MscI and λ-exonuclease digestion expose ssDNA target sequence for padlock probe (PLP) binding. Target annealed PLPs are circularized with DNA ligase and amplified by RCA. The generated rolling-circle product (RCP) is visualized upon binding of fluorophore-conjugated oligonucleotides under a fluorescence microscope. Arrows represent 3′ extremities, and flat ends represent 5′ extremities. (B) PLP target sites on HAdV-5 DNA and mRNAs. The E1A mRNA and MLTU mRNA PLP target sequences (uppercase letters) are presented on the corresponding HAdV-5 genome regions. 5′ and 3′ arms of the PLP target sequences are indicated in green and blue, respectively. The MscI restriction site adjacent to the HAdV-5 genomic DNA padlock probe sequence target is in yellow. RT primer binding sites adjacent to E1A and MLTU target sites (italic). HAdV-5 genome numbering is based on NCBI reference sequence AC_000008.1.
In the present study, we have developed a specific PLP-based RCA technique, which allows for concurrent detection of HAdV-5 DNA and virus-encoded, spliced mRNAs in individual infected cells. Importantly, we demonstrate that this versatile method can be used to monitor and characterize different HAdV-5 infections and to correlate virus DNA content with virus gene expression patterns in individual infected cells.
RESULTS
Padlock probe-based detection of HAdV-5 DNA and mRNAs in individual cells.
To study HAdV-5 DNA replication and mRNA accumulation in individual cells, we established a specific PLP-based nucleic acid detection method. The PLP is a linear DNA oligonucleotide containing terminal 5′ and 3′ arms needed for specific target recognition (Fig. 1) (25). Annealing and ligation of both PLP arms to a complementary target sequence generate a circularized DNA probe, which is amplified by RCA and detected using fluorophore-conjugated oligonucleotides specific to an embedded barcode sequence (Fig. 1A). The PLP-based target recognition mechanism differs when detecting DNA or mRNA. In the latter case, mRNAs are converted to cDNAs via reverse transcription, whereas genomic DNA is cleaved with the restriction endonuclease MscI and recessed by the λ-exonuclease (31). The λ-exonuclease treatment is important, as it generates single-stranded DNA (ssDNA) needed for specific PLP annealing to the viral DNA (Fig. 1A). Since the mRNA-specific PLP targets only the exon-exon junction in the spliced cDNA sequence, the cross-reactivity of those PLPs on viral genomic DNA is highly unlikely. To detect HAdV-5 genomic DNA, we designed a specific PLP annealing to the viral pVI gene sequence, since this region is adjacent to a suitable MscI cleavage site (Fig. 1B). To detect HAdV-5 E1A mRNAs, two specific PLPs were designed to discriminate between the abundant E1A 13S and 12S splice variants. Since we were also interested in detecting viral late mRNAs, the specific PLPs were designed to anneal to spliced exon I and II or exon II and III junctions in the HAdV-5 tripartite leader sequence present in all MLTU mRNAs (Fig. 1B).
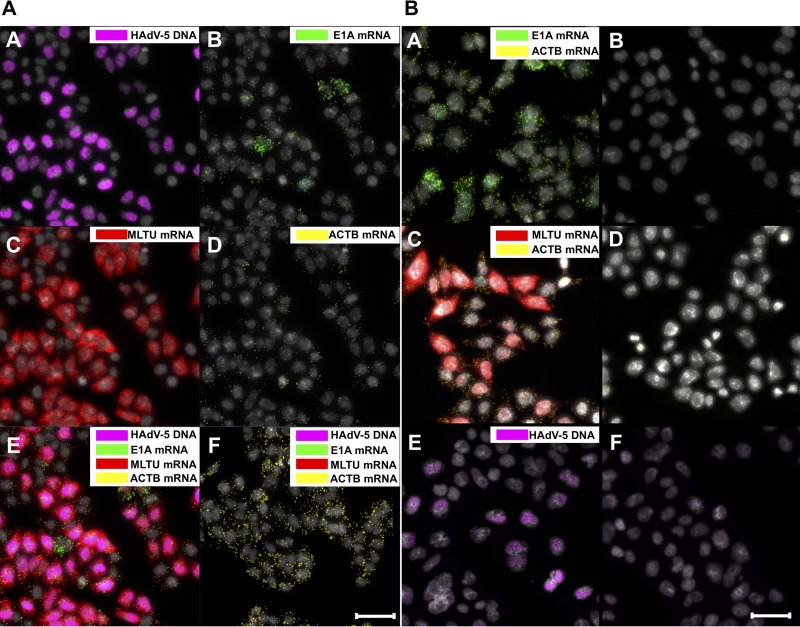
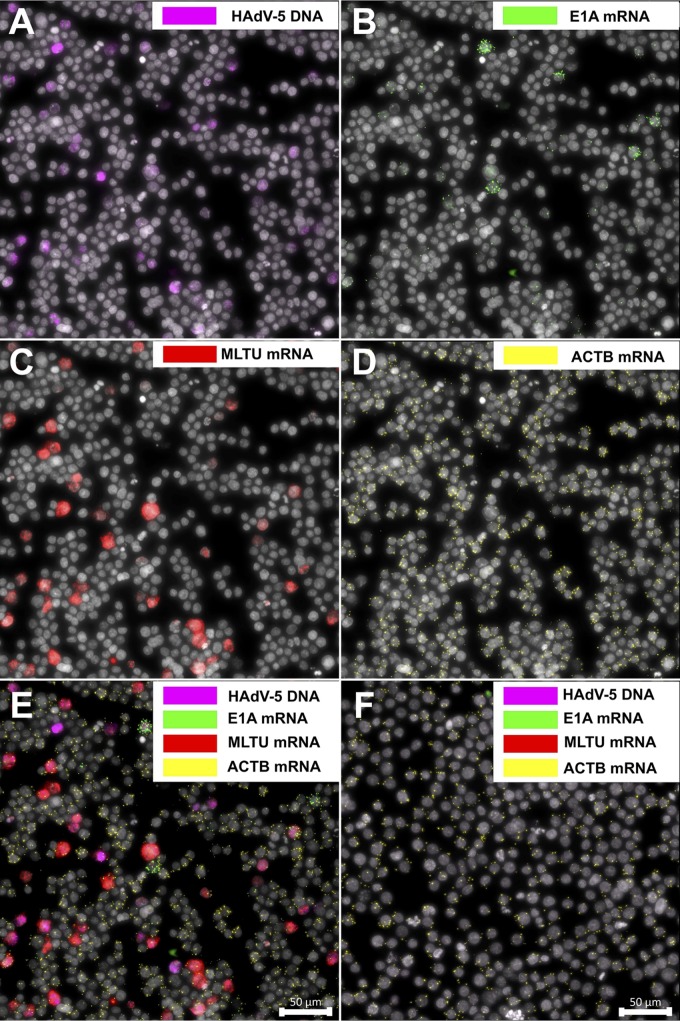
To test the specificity of the designed PLPs, HeLa cells were infected with HAdV-5 and the described PLPs applied to the fixed cells in a multiplexed manner at 25 h postinfection (hpi). Thereafter, the amplification of viral DNA and mRNA probes by RCA was visualized by fluorescence microscopy. As shown in Fig. 2A, the HAdV-5 DNA signal is clearly detectable within the nucleus boundaries (image A), demonstrating, as expected, an extensive HAdV-5 DNA replication at 25 hpi. Because our method allows for the simultaneous detection of viral DNA and mRNA, the same samples were also analyzed for viral early (here referred to as E1A) and late (here referred to as MLTU) mRNA expression. As expected, MLTU mRNAs, which constitute the major pool of viral mRNAs late in infection (32), were highly abundant at 25 hpi (Fig. 2A, image C). Similarly, the E1A transcripts were detectable (image B), although the signal intensity was lower than that of the MLTU mRNA signal. As a control for successful PLP amplification, the cellular beta-actin mRNA (here referred to as ACTB) was monitored in the same HAdV-5-infected cells (image D). As expected, viral DNA and mRNAs were undetectable in uninfected HeLa cells, confirming the specificity of the designed PLPs (image F). To further confirm that mRNA-specific PLPs anneal only to viral cDNA and not to the viral genomic DNA, RCA experiments were conducted in the absence of the reverse transcriptase treatment (Fig. 2B, images A to D). Similarly, the precision of viral genomic DNA-specific PLP was tested in the absence of MscI endonuclease and λ-exonuclease treatments (Fig. 2B, images E and F). No cross-reactivity with viral DNA was observed for all viral mRNA PLPs, and no viral DNA was detected when genomic DNA was not enzymatically preprocessed (Fig. 2B).
FIG 2.
Concurrent in situ detection of HAdV-5 DNA and mRNAs. (A) HeLa cells were infected with HAdV-5 (10 FFU/cell) and analyzed at 25 hpi. HAdV-5 genomic DNA is presented in magenta (image A), E1A mRNAs (13S and 12S) in green (image B), MLTU mRNAs (exon I_II and exon II_III) in red (image C), and ACTB mRNA in yellow (image D). A merged image (image E) and uninfected cells (image F) also are shown. (B) Detection of spliced E1A mRNAs (13S and 12S), MLTU mRNAs (exon I_II and exon II_III), and ACTB mRNA with PLPs in HeLa cells 25 hpi according to the standard protocol (images A and C). Reverse transcriptase was omitted from the cDNA synthesis reaction (images B and D). Detection of HAdV-5 DNA with PLP in HeLa cells 25 hpi was performed according to the standard protocol (image E). MscI endonuclease and λ-exonuclease treatments were omitted during HAdV-5 DNA preparation (image F). Scale bar, 50 μm.
Temporal changes in HAdV-5 DNA replication and mRNA expression in infected HeLa cells.
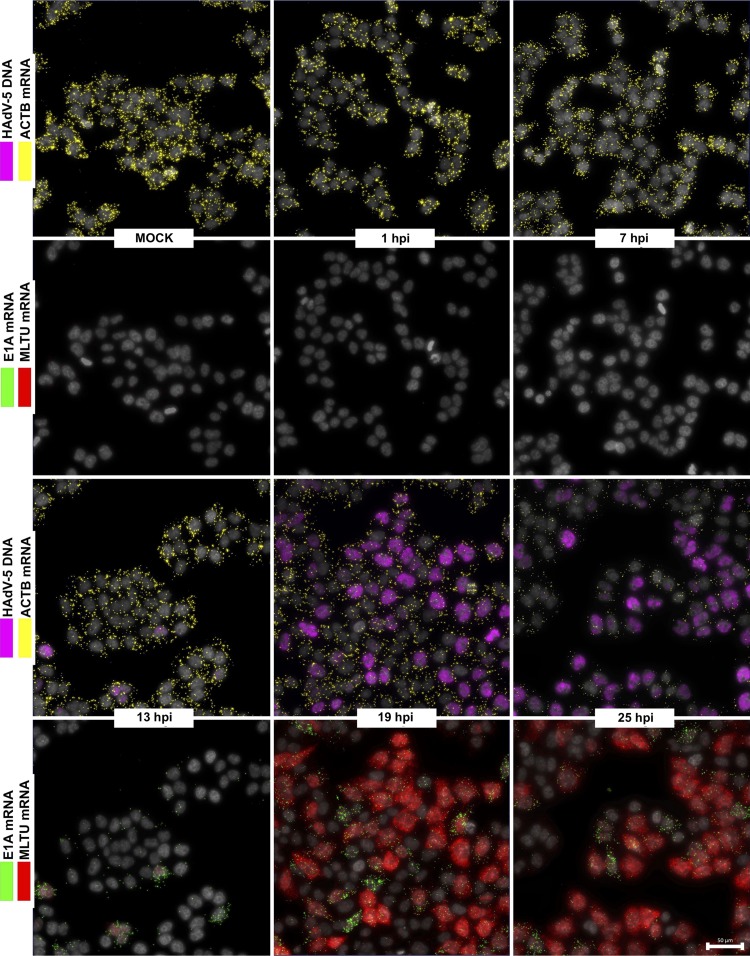
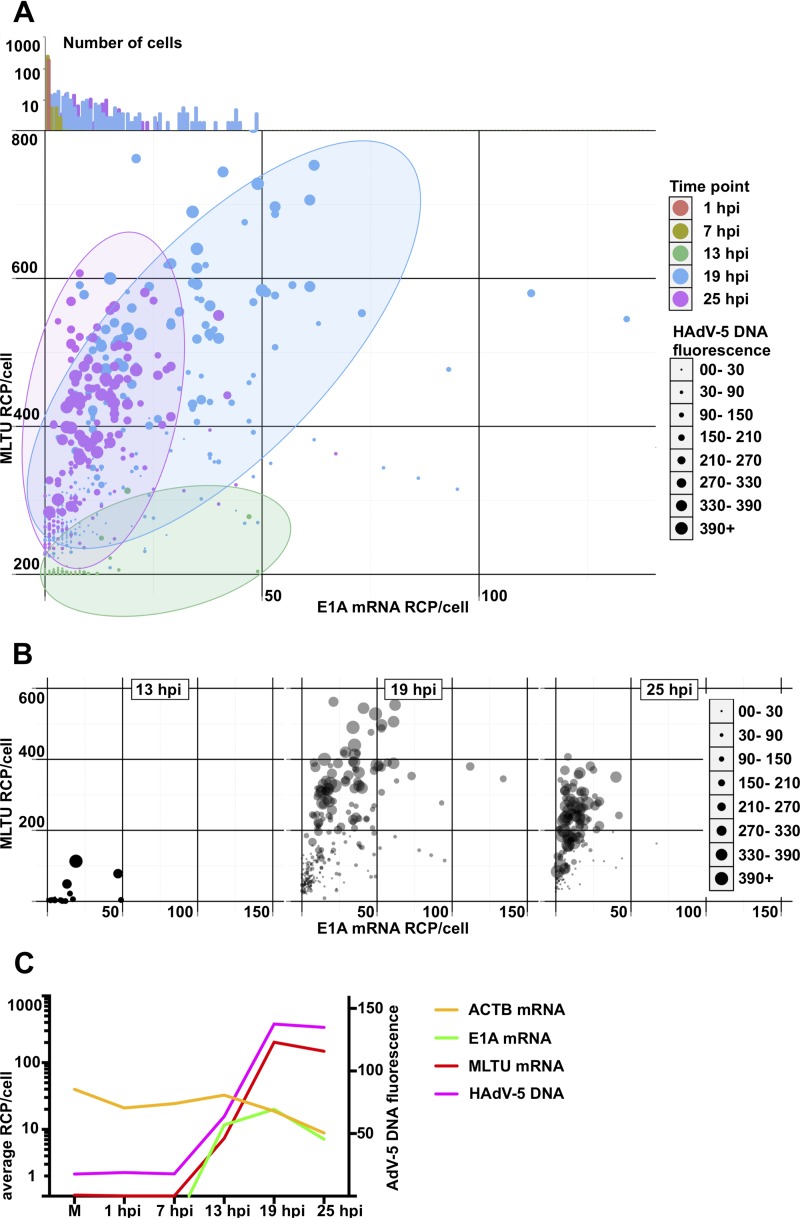
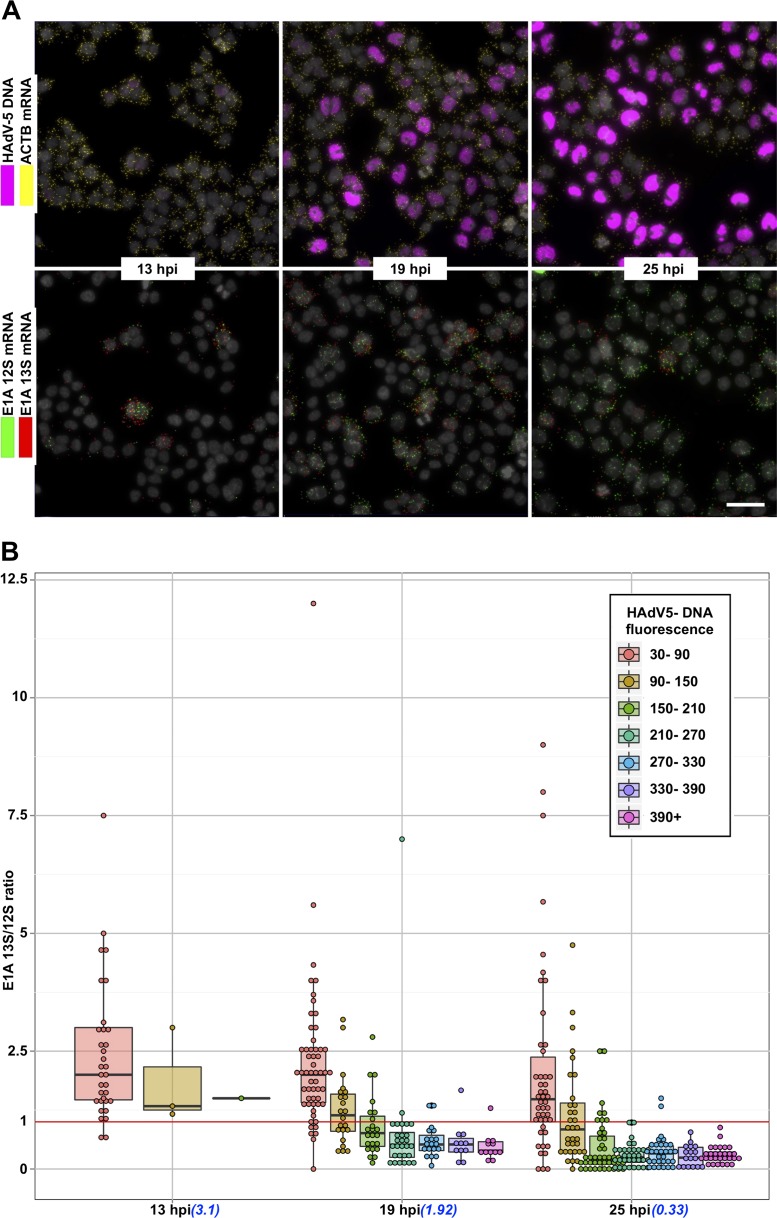
To gain more detailed information on how HAdV-5 infection proceeds in a temporal manner in individual cells, we monitored viral DNA replication and mRNA expression patterns during a 25-h infection period. HeLa cells were infected with HAdV-5, and the viral DNA and mRNA accumulation were simultaneously monitored at different times postinfection. As shown in Fig. 3, both HAdV-5 genomic DNA and viral early and late mRNAs were undetectable in uninfected or in early virus-infected cells (1 and 7 hpi). However, following entry into the late phase (13 hpi and onwards), the PLP amplification on the viral targets became clearly detectable. The HAdV-5 DNA and E1A (13S and 12S) and MLTU (exon I_II and exon II_III) mRNAs accumulated to high levels between 13 hpi and 25 hpi. Interestingly, E1A 13S and 12S mRNA signals were not detected at early times of infection (1 to 13 hpi), contradicting the expected, early expression pattern of this viral gene (Fig. 3) (6). A clear advantage of our method is that it allows us to correlate viral DNA amount to mRNA expression in the same cell. Therefore, we randomly picked >100 infected cells at different time points postinfection and quantified the fluorescence signals in the individual cells (Fig. 4). According to our single-cell analysis (Fig. 4A), cells sampled at 13-, 19-, and 25-hpi time points formed very characteristic, heterogeneous populations. For example, at 13 hpi almost all of the cells had a very low HAdV-5 fluorescence, consistent with very low E1A and MLTU mRNA accumulation (Fig. 4B). In contrast, cells sampled at 19 hpi exhibited noticeably high and heterogeneous viral mRNA expression and HAdV-5 DNA fluorescence compared to the 13-hpi sample (Fig. 4B, compare 13 hpi and 19 hpi). Interestingly, individual cells at 25 hpi showed reduced heterogeneity regarding the viral mRNA and DNA accumulation compared to the cells at the 19-hpi time point (Fig. 4B). In line with previous reports (11, 32), a quantitative analysis of viral DNA and mRNA signals in HeLa cell populations confirmed the interdependency of viral genome replication and MLTU mRNA accumulation (Fig. 4C).
FIG 3.
Temporal expression pattern of HAdV-5 mRNAs in infected HeLa cells. HeLa cells infected with HAdV-5 (10 FFU/cell) and analyzed during different time points (1, 7, 13, 19, and 25 hpi). HAdV-5 genomic DNA (magenta) is presented along with ACTB mRNA (yellow). E1A mRNAs (13S and 12S, green) are presented along with MLTU mRNAs (exon I_II and exon II_III, red). Scale bar, 50 μm.
FIG 4.
Single-cell quantification of HAdV-5 DNA and viral mRNAs in HeLa cells at different time points postinfection. (A) All cells, color-coded depending on the infection time point, were distributed in the scatterplot depending on E1A (x axis) and MLTU mRNA expression (y axis). The diameter of the data point is proportional to HAdV-5 DNA content based on its fluorescence signal. Cells from 1 and 7 hpi were grouped at the plot origin, as no specific E1A/MLTU/virus DNA detection was observed in these cells. Since some data points are overlapping in the two-dimensional scatterplot, E1A mRNA expression histograms for all time points were plotted against the log10 cell number above the scatterplot. Dominant 1- and 7-hpi cell populations close the plot origin are clearly visible (orange- and olive-colored columns). (B) Individual scatterplots of 13, 19, and 25 hpi from experiments shown in panel A. The data points for 19 and 25 hpi are plotted with 40% transparency. (C) Geometric averages of HAdV-5 DNA and viral mRNAs in infected HeLa cells. Average RCP/cell (y axis) was calculated for MLTU mRNA (red), E1A mRNA (green), and ACTB (yellow) and plotted against time (x axis). HAdV-5 DNA arbitrary fluorescence is presented on the secondary vertical y axis.
Temporal expression pattern of the E1A 13S and 12S mRNAs in individual infected cells.
Since E1A mRNA PLPs were designed to detect the two most abundant E1A splice variants, 13S and 12S, we decided to monitor their expression in individual HAdV-5-infected HeLa cells. To achieve this, unique reporter sequence was embedded in each E1A probe, facilitating their discrimination in situ. Both E1A 13S and 12S mRNAs were detected in HAdV-5-infected HeLa cells at 13 hpi, although the detection level was relatively low. Interestingly, E1A 13S mRNA detection did not drastically improve during the infection, whereas E1A 12S mRNA showed accelerating accumulation as the infection proceeded (Fig. 5A). To further investigate this phenomenon, we binned cells into 7 groups based on their HAdV-5 DNA fluorescence and calculated the E1A 13S/12S mRNA expression ratio in individual infected cells (Fig. 5B). At all three studied time points (13 hpi, 19 hpi, and 25 hpi), we observed negative correlation between HAdV-5 DNA amount and E1A 13S/12S mRNA ratio. Curiously, in cells where HAdV-5 DNA was detected at a very low level (fluorescence between 30 and 90), E1A 13S mRNA expression was consistently higher than E1A 12S mRNA expression. In contrast, cells with higher HAdV-5 DNA levels (fluorescence between 90 and 150) showed only slightly higher E1A 13S mRNA levels than E1A 12S mRNA at 13 hpi and 19 hpi. Strikingly, the majority of infected cells analyzed at 19 hpi and 25 hpi with high HAdV-5 DNA content (DNA fluorescence of >150) clearly expressed more E1A 12S mRNA than E1A 13S mRNA (Fig. 5). Taken together, our single-cell analysis suggested the HAdV-5 DNA content is related to the differential accumulation of the spliced E1A 13S and 12S mRNA transcripts in the infected HeLa cells.
FIG 5.
Single-cell E1A 13S/12S splice variant expression ratio in HeLa cells at 13 hpi, 19 hpi, and 25 hpi. (A) E1A 13S mRNA (red) and 12S mRNA (green) detection in individual HAdV-5-infected (magenta) HeLa cells. Scale bar, 50 μm. (B) Cells at each time point were divided into groups and color coded based on their HAdV-5 DNA fluorescence. Each data point (which represents an individual cell) was distributed in the plot depending on the time point (x axis) and E1A 13S/12S splice variant expression (y axis). Finally, a box-and-whisker plot for each cell group was overlaid with data points. The lower and upper hinges of box plots correspond to the first and third quartiles (the 25th and 75th percentiles). Black line, median; upper whisker, highest value that is within 1.5× the interquartile range of the hinge; lower whisker, lowest value within 1.5× the interquartile range of the hinge; red line, E1A 13S/12S ratio of 1, showing equal expression of both splice variants. Calculated geometrical average E1A 13S/12S mRNA ratios at indicated time points are shown in parentheses below the x axes.
Detection of HAdV-5 DNA and viral mRNAs in long-term infections.
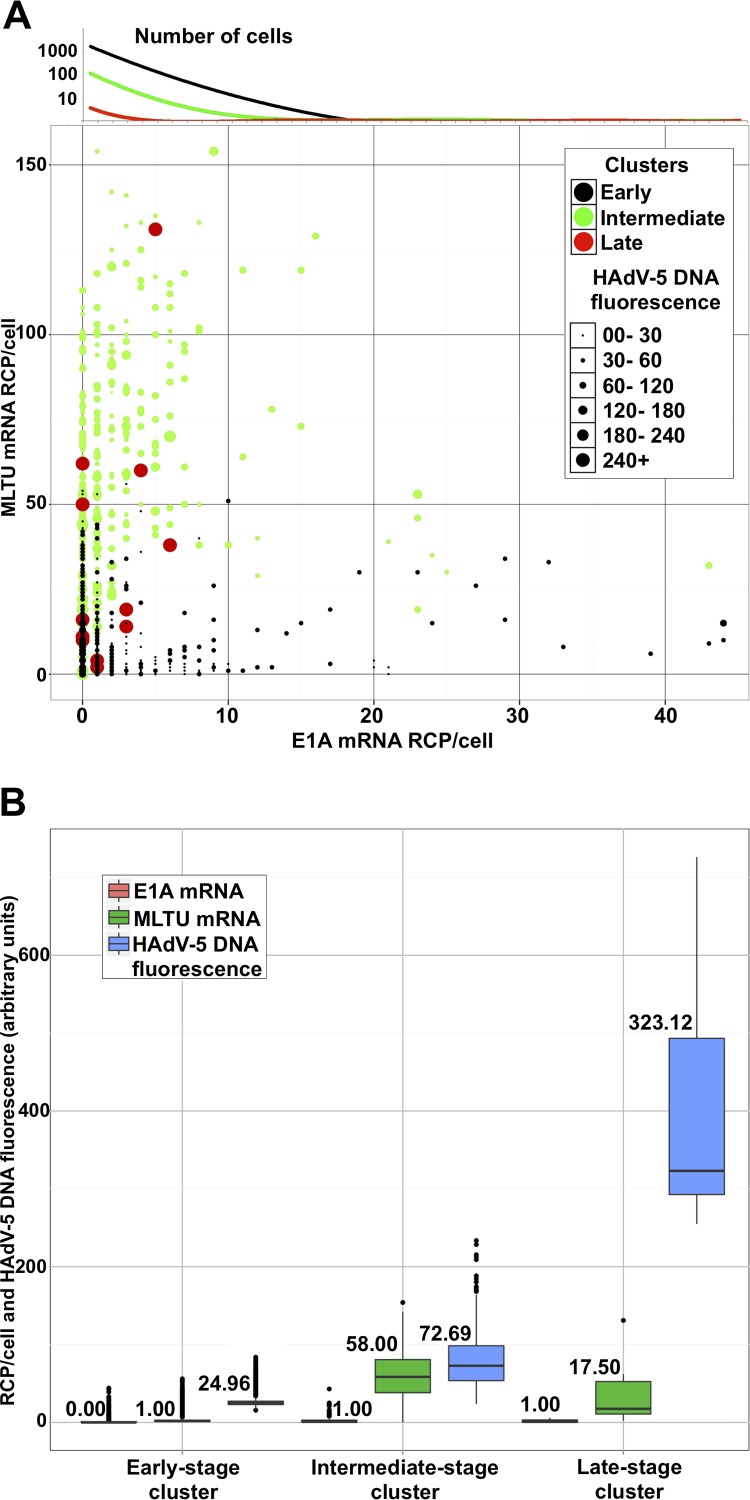
Recent studies have shown that HAdV-5 also can cause long-term latent/persistent infections in B- and T-lymphocyte cell lines (23, 33, 34). Therefore, it became of interest to test if the RCA method could define viral DNA accumulation and mRNA expression patterns in HAdV-5-infected B lymphocytes. For this purpose, the human Burkitt lymphoma cell line BJAB was infected with HAdV-5, followed by simultaneous detection of viral DNA and mRNA accumulation at 6 days postinfection (dpi). As shown in Fig. 6, HAdV-5 DNA was detectable with our PLP (Fig. 6A), thereby confirming the previous observation that HAdV-5 can infect and replicate in BJAB cells (23). Consequently, E1A (Fig. 6B, 13S and 12S) and MLTU (Fig. 6C, exon I_II and exon II_III) mRNAs were successfully detected in the BJAB cells. Finally, annealing of the PLPs was specific, as viral DNA or viral mRNAs were not detected in uninfected BJAB cells (Fig. 6F). To gain further quantitative information about the HAdV-5 infection, viral DNA and mRNA signals were quantified in >5,300 randomly picked BJAB cells. Out of these, 1,412 uninfected cells were removed from the data set. The remaining cells were plotted as individual data points on a scatterplot, where the size of the data point (an individual cell) was proportional to the viral DNA amount (Fig. 7A). Cells were grouped into three arbitrary infection stage clusters (early, intermediate, and late) based on HAdV-5 DNA fluorescence and viral mRNA expression. Cells (black; 3,584 cells) grouped into the early-infection-stage cluster were comprised of cells expressing some viral mRNAs without or with barely detectable viral DNA staining. The remaining BJAB cells were subdivided into intermediate (green; 358 cells)- and late-infection (red; 12 cells)-stage clusters. According to our calculations, median MLTU expression was highest in the intermediate cluster (58 rolling-circle products [RCP]/cell) and dropped in the late infection stage (17 RCP/cell). Interestingly, only a few cells grouped into the early-stage cluster showed signal for HAdV-5 DNA. Fluorescence from viral DNA was stably rising from 25, 72, and 323 arbitrary units in early-, intermediate-, and late-stage clusters, respectively (Fig. 7B).
FIG 6.
Concurrent detection of HAdV-5 DNA and mRNAs in long-term infected BJAB cells. BJAB cells were infected with HAdV-5 (100 FFU/cell) and analyzed 6 days postinfection (dpi). HAdV-5 DNA is presented in magenta (A), E1A mRNAs in green (B), MLTU mRNAs in red (C), and ACTB mRNA in yellow (D). (E) Merged image. (F) Uninfected cells stained with the aforementioned padlock probes. Scale bar, 50 μm.
FIG 7.
Quantification of HAdV-5 DNA and viral mRNAs in BJAB cells at 6 dpi. (A) Each data point (which represents an individual cell) was distributed in the scatterplot depending on E1A (13S and 12S; x axis) and MLTU expression (exon I_II and exon II_III; y axis). The diameter of the data point is proportional to HAdV-5 DNA content based on its fluorescence signal. Cells were grouped into 3 infection stage clusters (early [black], intermediate [green], and late [red]) using k-means clustering, based on HAdV-5 DNA abundance and viral mRNA expression. Since a two-dimensional scatterplot does not reflect cell abundance, the E1A mRNA expression trend line for all clusters is plotted above its corresponding axis against the log10 cell number. The dominant population of cells expressing low E1A mRNA levels is clearly indicated (black line). (B) BJAB cells were divided into clusters (x axis). The lower and upper hinges of box plots correspond to the first and third quartiles (the 25th and 75th percentiles). Black line, median (also the number above/below a box plot); upper whisker, highest value that is within 1.5× the interquartile range of the hinge; lower whisker, lowest value within 1.5× the interquartile range of the hinge.
The mature protein VII affects HAdV-5 DNA recognition by RCA.
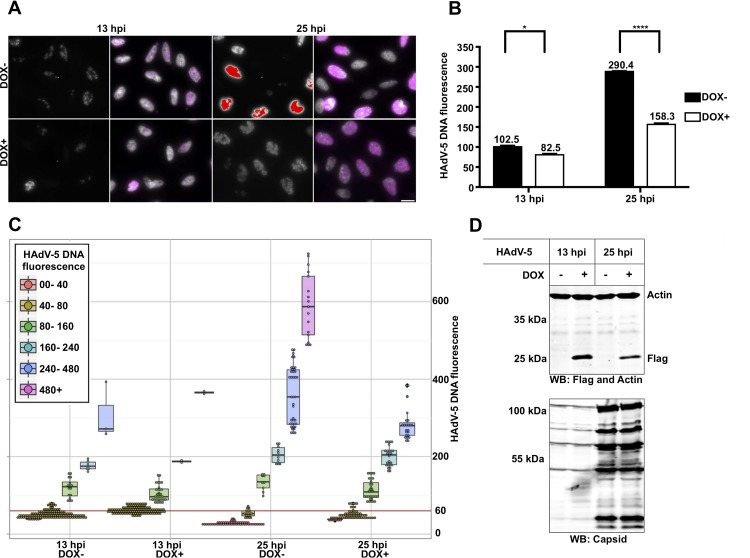
A unique feature of HAdVs is that the viral DNA is associated with the virus-encoded histone-like protein VII [here designated pVII(Δ24)]. A curious finding was that the incoming HAdV-5 DNA was difficult to detect with the RCA method at the early time of infection (Fig. 3 and 4). Therefore, we hypothesized that association of the pVII(Δ24) protein with virus genome affected the accessibility of the endo/exonucleases and/or the PLP to the virus DNA. To test this hypothesis, we took advantage of a HeLa stable cell line where expression of the pVII(Δ24)Flag protein was induced with doxycycline (Dox) (Fig. 8). HeLa-pVII(Δ24)Flag cells were treated with Dox for 24 h and infected with HAdV-5 in the presence of Dox, and viral DNA was detected with RCA at 13 hpi and 25 hpi (Fig. 8A). Notably, quantitative analysis of the HAdV-5 DNA signals in the Dox-treated and the untreated cells indicated that the presence of the pVII(Δ24)Flag protein significantly reduced detection of the HAdV-5 DNA by the PLP at 25 hpi (P ≤ 0.0005) and 13 hpi (P ≤ 0.05) (Fig. 8B and C).
FIG 8.
Expression of the pVII(Δ24) protein reduces HAdV-5 DNA detection with RCA. (A) HeLa-pVII(Δ24)Flag cells were treated with doxycycline (Dox+) or were left untreated (Dox−) for 24 h, followed by HAdV-5 infection in the presence or absence of Dox. RCA with PLP recognizing HAdV-5 genomic DNA (magenta or raw black-and-white exposure) at 13 hpi and 25 hpi is shown. The scale bar is 20 μm. (B) Geometric averages and standard deviations of HAdV-5 DNA signal. Average HAdV-5 DNA fluorescence was plotted against infection time (x axis). ****, P ≤ 0.0005; *, P ≤ 0.05. (C) Single-cell HAdV-5 DNA fluorescence in HeLa cells 13 hpi and 25 hpi. Cells were divided into groups and color coded based on their HAdV-5 DNA fluorescence. Each data point (which represents an individual cell) was distributed in the plot depending on the time point, Dox treatment (x axis), and HAdV-5 DNA fluorescence (y axis). A box-and-whisker plot for each cell group was overlaid with data points. The red line indicates an HAdV-5 DNA fluorescence of 60, showing the fluorescence cutoff used to exclude uninfected cells to generate the accurate and unbiased averages and standard deviations shown in panel B. (D) Detection of the pVII(Δ24)Flag protein with anti-Flag antibody by Western blotting (WB).
DISCUSSION
HAdVs have been extensively used as model systems to elucidate various aspects of eukaryotic gene expression and genome organization (12, 35–37). However, the vast majority of those studies are based on standard experimental procedures carried out in a heterogeneous cell population. Since the infection efficiency of HAdV-5 varies with the cell cycle and the expression profile of specific viral genes (22), the analysis of HAdV-5 gene expression and genome organization would benefit from a method that permits single-cell analysis in the heterogeneous cell population. As we show here, the PLP-based RCA method can be used to simultaneously study viral genomic DNA accumulation and mRNA expression in individual HAdV-infected cells.
To validate the sensitivity and specificity of the RCA method, we analyzed the temporal gene expression profiles in relation to the viral DNA content in different HAdV-5 infections. Our results from short-term infections showed that HAdV-5 DNA and viral early (E1A) and late (MLTU) mRNAs are simultaneously well detected in individual infected HeLa cells from 13 hpi onwards (Fig. 3). This observation is in line with previous reports showing that HAdV DNA replication and accumulation of the full-length MLTU mRNAs starts between 8 and 12 hpi in HeLa cells (20, 32, 38). Our results also show that the HAdV-5 DNA and E1A mRNAs are not detectable at early times (≤7 hpi) of infection. This observation indicates that the PLPs have a detection limit toward HAdV-5 DNA and E1A mRNAs during the early phase of infection, when the viral DNA copy number and viral mRNA expression are low. Truly, infection of HeLa cells with a 10× higher (100 FFU/cell) virus amount improved HAdV-5 DNA and E1A mRNA detection during early time points (7 hpi and 13 hpi) (data not shown). Hence, this observation indicates that the RCA method has some limited sensitivity under our experimental conditions and that further improvement of the protocol sensitivity is needed to study low-copy-number molecules.
Most of the assays used to study HAdV DNA content and RNA expression rely on whole-cell populations, where the sample heterogeneity is not considered. While population average (Fig. 4C) can give valuable insight into general infection pattern, single-cell observations can reveal the subtle subpopulations, which display characteristics far different from the average. For example, our single-cell analysis identified cells without detectable viral DNA signals but with considerable expression of viral E1A and/or MLTU mRNAs (Fig. 3 and 4B, 19 hpi). However, at present it remains enigmatic why this type of cell exists and what role they play in the HAdV-5 life cycle. Another advantage of the single-cell analysis is that it allows detailed understanding of how transitions between different infection phases may occur. For example, cell population-based analysis (Fig. 4C) showed reduced accumulation of HAdV-5 DNA and mRNAs from 19 hpi to 25 hpi. Analysis of the same samples at single-cell resolution revealed that the cells at 19 hpi showed more heterogeneity in viral DNA and mRNA content than the cells at 25 hpi (Fig. 4B). It is likely that the observed heterogeneity (19 hpi) reflects the asynchronous nature of the initial HAdV-5 infection, which is reduced as the infection time course proceeds (25 hpi). Collectively, these results show the advantage of single-cell analysis, revealing discrete cell subpopulations in HAdV-5 infection, which would be missed by population averaging.
A specific organization of the HAdV-5 genome during the early phase of infection (37) might reduce sensitivity of the method by blocking the PLP accessibility to the condensed viral genomic DNA. In line with this hypothesis, we found that overexpression of the pVII(Δ24) protein reduces HAdV-5 DNA detection (Fig. 8). During the early phase of infection, the pVII(Δ24) protein is tethered to the HAdV-5 DNA to protect the viral genome from the cellular DNA damage response (18, 39). As infection proceeds, the pVII(Δ24) protein is gradually removed from the viral DNA to allow proper HAdV-5 DNA replication and gene expression (20, 40). Quantification of the viral DNA content in the individual pVII(Δ24)-overexpressing cells showed reduced HAdV-5 DNA fluorescence signal at 13 hpi and 25 hpi (Fig. 8B and C). Interestingly, overexpression of the pVII(Δ24) protein reduced detection of the HAdV-5 DNA in cells with high DNA content (Fig. 8C, 25 hpi, DNA fluorescence of >240), whereas detection of the HAdV-5 DNA in cells with lower viral DNA content (DNA fluorescence of <240, both time points) was not affected to the same extent by pVII(Δ24) protein expression. Hence, we imply that association of the native pVII(Δ24) protein with the virus genome blocks viral DNA detection by RCA up to 7 hpi. Considering that the incoming viral DNA is embedded with the native pVII(Δ24) protein until the onset of virus DNA replication (20), it is possible that the Dox-induced pVII(Δ24) protein exerts only a minor effect on HAdV-5 DNA detection at 13 hpi. Conversely, extended overexpression of the pVII(Δ24) protein can interact with newly replicated viral genomes and block virus DNA detection with a greater effect at 25 hpi. Collectively, our data suggest that the presence of the pVII(Δ24) protein alters HAdV-5 genome organization, which in turn reduces the PLP binding to the viral DNA under the experimental conditions used.
Our study revealed, for the first time, a different accumulation pattern of the E1A 13S and 12S mRNAs depending on the HAdV-5 DNA amount in the individual cells. This is exemplified by the single-cell quantification, where the high level of E1A 12S mRNA correlated with high viral DNA content in individual infected cells (Fig. 5, 19 hpi and 25 hpi). Surprisingly, individual cells with low viral DNA content showed preferential accumulation of the E1A 13S mRNA. This observation was valid at all three analyzed infection time points (13 hpi, 19 hpi, and 25 hpi), suggesting that E1A 13S mRNA is expressed when the HAdV-5 viral replication rate or DNA abundance is low. Considering that E1A 13S expression enhances HAdV-5 replication better than E1A 12S expression (41), we speculate that the initial accumulation of E1A 13S-encoded protein E1A(289R) enhances viral DNA replication, which in turn leads to preferred splicing and accretion of the E1A 12S mRNA. The E1A 12S-encoded E1A(243R) protein in turn will be responsible for alteration of host gene expression to promote productive virus growth (42). Our interpretation is further supported by a previous observation whereby E1A 13S expression has a positive effect on viral E2A-72K (DBP) protein expression (41), which is an essential factor for viral DNA replication (10). A recent study has analyzed E1A 13S and 12S mRNA levels during the course of infection in HAdV-5 (dl309)-infected human fibroblast (41). Although in this study the E1A 13S and 12S mRNA accumulation pattern was established in the whole-cell population, the differences in E1A 13S and 12S mRNA accumulation were clearly detectable. In line with our single-cell quantification (Fig. 5B, 19 hpi and 25 hpi), the E1A 12S mRNA levels were reported to be continuously higher than the E1A 13S mRNA levels during a 96-h infection time course (41). When geometrical averages were calculated for the E1A 13S/12S ratio from the bulk HeLa cell population, the ratio was steadily declining from 3.1 (13 hpi) to 0.33 (25 hpi) (Fig. 5B). Thus, the observation that ∼3× less E1A 13S was detected at 25 hpi is also in line with a previous report (41). In conclusion, our method and analysis provide a useful tool to visualize and quantitate the heterogeneity of mRNA splice variants in individual cells.
In contrast to the well-characterized HAdV lytic infections, much less is known about the virus behavior in long-term latent/persistent infections in B and T lymphocytes (23). Therefore, we applied our method to study an HAdV-5 infection in the B lymphocyte cell line, BJAB. In contrast to HeLa cells at 25 hpi (Fig. 2), only a minor proportion (<20%) of the BJAB cells were positively stained for viral DNA at 6 dpi (Fig. 6). To analyze individual infected BJAB cells, we first excluded uninfected cells from the whole-cell population using viral RNA and DNA expression as the selection criteria. We next grouped the cells based on their DNA and RNA expression patterns into three clusters, named by us the early, intermediate, and late infection stages (Fig. 7A). A big fraction of cells in the early stage showed a tendency to express high levels of E1A and had little or no HAdV-5 DNA signal. Thus, contradicting lytic infection in HeLa cells (Fig. 4), the majority of the BJAB cells with high viral mRNA expression showed no or very weak viral DNA signal. Therefore, we speculate that the early-infection-stage BJAB cells represent cells having detectable viral mRNAs but containing specific viral DNA modifications, rendering viral genomes resistant to the PLP probing. Consequently, the vast majority of cells grouped into the intermediate-infection-stage cluster exhibit a high MLTU mRNA expression and HAdV-5 DNA content compared to the early infection cluster. Notably, a very small population of 12 cells, out of more than 5,300 imaged, were grouped into the late-infection-stage cluster and showed considerably higher HAdV-5 DNA fluorescence than all other imaged cells. Hence, cells with high viral DNA content but low E1A and MLTU mRNA expression (e.g., cells in the late infection stage) indicate that a subpopulation of infected cells have silenced their early and late gene expression. Those cells exhibit certain similarities, with a small subpopulation of HeLa cells infected and collected at the 25-hpi time point (Fig. 4B). Theoretically, low viral RNA and high DNA content in HeLa cells at 25 hpi may mark the commencement of virus release from the cell, whereas a similar scenario in BJAB cells at 6 dpi might specify the cells where HAdV-5 establishes persistent infections. Notably, Zhang and colleagues (23) have reported that HAdV-5-infected BJAB cells show a drastic reduction in viral hexon protein expression while maintaining relatively high viral DNA amounts during long-term infection.
The most commonly used quantitative methods to quantitate viral infections are quantitative PCR and quantitative reverse transcription-PCR. These methods consequently are blind to population heterogeneity and can only deliver averaged expression data. Our method provides a tool to quantitatively monitor individual infected cells with respect to their viral DNA content and early/late gene expression. Bearing in mind that the proposed procedure does not comprise known protein-degrading agents, our method has the potential to be applied on clinical specimens, optionally stained for immune response markers, to link infected cells and their expression profiles with the host cell immune response. In fact, simultaneous mRNA and protein detection with PLPs has been reported recently (43).
In summary, our results show that the RCA method should be a welcome addition to the method repertoire to further characterize the molecular details of the HAdV life cycle in cell culture experiments and potentially in patient tissue samples.
MATERIALS AND METHODS
Padlock probe design.
To detect HAdV-5 (NCBI reference sequence AC_000008.1) genomic DNA PLP hybridization site between nucleotides 18240 and 18269 was selected. This position was specifically chosen, as it is adjacent to an MscI restriction site in the longest MscI restriction fragment in the HAdV-5 genome. Such an experimental design was previously reported to be the most efficient in detecting mitochondrial DNA in situ by RCA (31). To ensure detection of only spliced E1A mRNAs, PLP targets were selected to detect E1A 13S splice variants (nucleotides 1092 to 1112 and 1229 to 1243) and E1A 12S splice variants (nucleotides 955 to 974 and 1229 to 1243) (Fig. 1B). The ligation site was positioned on the junction between the E1A splice variant-specific 5′ exon and a common E1A last 3′ exon. The PLP arm hybridizing to a cDNA at the 5′ end was intentionally extended (if a conserved 3′ exon arm would be extended instead, PLP for another splice variant could occupy the cDNA, making ligation and detection impossible). Consequently, the melting points (Tm) of the arms toward 3′ and 5′ exons were set to ∼45°C and ∼55°C, respectively. To enable differentiation between E1A 13S and 12S splice variants, unique reporter sequence was embedded in the PLP backbones. To detect viral late mRNAs, two PLPs were designed to anneal to the HAdV-5 spliced tripartite leader-containing mRNAs. We designed two PLPs targeting junctions between MLTU tripartite exons I and II (exon I_II, nucleotides 6073 to 6089 and 7111 to 7128) and II and III (exon II_III, nucleotides 7168 to 7182 and 9644 to 9659). For each MLTU PLP, an LNA primer was designed to facilitate efficient synthesis of MLTU cDNAs. Similarly, a single LNA primer was designed for E1A mRNA that was compatible with both E1A PLPs. Detection of cellular beta-actin mRNA (ACTB; GenBank accession no. NM_001101) has been reported previously (43). All oligonucleotides were purchased from Integrated DNA Technologies (IDT), and the exact sequences are available on request from the authors.
Cell culture, virus infections, drug treatment, and transient transfections.
HeLa cells (ATCC CCL-2) were propagated in growth medium consisting of Dulbecco's modified Eagle medium (DMEM; Invitrogen), 10% fetal calf serum (FCS; PAA), and 1% penicillin-streptomycin mix (PEST; Gibco). BJAB cells (generously provided by Göran Akusjärvi) were grown as suspension cultures using Roswell Park Memorial Institute 1640 medium (RPMI 1640; Sigma), 10% FCS, 10 mM l-glutamine (Gibco), and 1% PEST. Both cell lines were grown in a humidified cell incubator at 37°C in the presence of 7% CO2. The stable HeLa cell line expressing the HAdV-5 pVII(Δ24)Flag protein was generated by integrating a codon-optimized pVII(Δ24)Flag cDNA (14) into the HeLa-Flp-In T-Rex cell line (generously provided by Stephen Taylor [44]). pVII(Δ24)Flag protein expression with doxycycline treatment and detection in whole-cell lysates were performed as reported previously (14).
The HAdV-5 (dl309) virus (45) was used to infect HeLa, HeLa-pVII(Δ24), and BJAB cells. HeLa and HeLa-pVII(Δ24) cells were infected at a multiplicity of infection (MOI) of 10 fluorescence-forming units (FFU) per cell in DMEM plus 2% FCS for 1 h at 37°C. Thereafter, cells were washed with growth medium, and finally, culture plates were filled with fresh growth medium. BJAB cells were infected at 100 FFU/cell. The infection was performed in 2 ml of RPMI 1640 medium containing 2 × 107 cells for 3 h at 37°C. Following the adsorption phase, cells were washed and finally diluted to 0.2 × 106 cells/ml with RPMI 1640 growth medium (including all supplements).
Cell fixation.
BJAB cells (1.5 × 106 cells/ml) were collected by centrifugation, washed with phosphate-buffered saline (PBS), and finally resuspended in PBS. Cells were spun down onto Superfrost Plus microscope slides with a Cytospin cytocentrifuge at 800 rpm for 5 min. HeLa and HeLa-pVII(Δ24) cells were grown in 100-mm cell culture dishes with submerged Superfrost Plus microscope slides. Slides with the attached cells were fixed in freshly prepared diethyl pyrocarbonate (DEPC)-treated PBS containing 3.4% formaldehyde for 20 min on ice. Thereafter, slides were washed twice with DEPC-PBS, dehydrated in an ethanol gradient (70%, 85%, and 99%; 5 min each), air dried, and stored at −80°C. Prior to the experiment, cells on the microscope slides were isolated by covering with an 8-mm-diameter and 50-μl-volume Secure Seal chamber (Invitrogen). Cells were rehydrated with DEPC–TBS-T (TBS with 0.05% Tween 20) buffer and permeabilized with 0.1 M HCl for 5 min at room temperature. Each incubation step was followed by two DEPC–TBS-T washes. All incubations were performed in a humid chamber to avoid evaporation of the reaction mixture.
Target RNA/cDNA and genomic DNA processing.
Target mRNAs were reverse transcribed at 45°C for 2 h as reported previously (46). Thereafter, samples were fixed with 3.7% formaldehyde in DEPC-PBS for 10 min at 25°C. HAdV-5 DNA was first fragmented in 1× CutSmart buffer (NEB) supplemented with 0.2 μg/μl bovine serum albumin (BSA) and 0.5 U/μl MscI endonuclease (NEB) for 60 min at 37°C. Finally, dsDNA fragments were converted to single-stranded DNA (ssDNA) in 1× λ-exonuclease buffer (NEB) with 10% glycerol, 0.2 μg/μl BSA, and 0.2 U/μl λ-exonuclease (NEB) for 30 min at 37°C. The PLP ligation reaction was performed as reported previously (47). Circularized PLPs were amplified in RCA reactions, and RCPs (rolling circle products) were detected as reported previously (46).
Image acquisition and analysis.
Stacks of images were acquired for each region of interest with an Axioplan II epifluorescence microscope (Zeiss) equipped with an Orca Flash 4.0 v2 camera (Hamamatsu) with a total magnification of ×400 using ZEN software. To image RCPs from all focal planes, stacks were superimposed using maximum intensity projection (MIP) in ZEN and finally exported as original black-and-white (BW) pictures. All individual BW images were processed with ImageJ and Cell Profiler software (48). Briefly, each cell nucleus was identified and separated based on shape features using a custom-made ImageJ script. Images were loaded into Cell Profiler, and cell area and cytoplasm were defined by expanding nucleus seed outward by 200 pixels for HeLa and HeLa-pVII(Δ24)Flag cells and 50 pixels for BJAB cells. Each signal-specific image (E1A mRNAs, MLTU mRNA, and ACTB mRNA) was preprocessed using automated top-hat filtering. Objects were identified using manually adjusted thresholding and separated based on object intensity. High signal abundance for the HAdV-5 DNA probe in heavily infected cells prevented accurate quantification of RCPs. To assess the extent of infection in a single cell, defined nuclei were used as masks, and the average fluorescence intensity within the mask was calculated from the image specific for the HAdV-5 DNA probe signals. The average fluorescence intensity was multiplied by 1,000× to generate values between 0 and 700. Finally, E1A, MLTU, and ACTB mRNA signals were related to whole-cell area. Data for each cell were exported as a .csv file and processed in MS Excel (2016) or RStudio (0.98.1091).
Data processing and statistics.
To visualize the heterogeneity and emphasize the cell-to-cell differences, the Cell Profiler output data were analyzed in RStudio. In HeLa cells, geometrical means and geometrical standard deviations were calculated for viral RNAs and DNA, as they are more informative and accurate than arithmetic equivalents for log-normally distributed features (49). Imaging and assessing mRNA and HAdV-5 DNA expression in single cells allowed us to accurately measure average mRNA and HAdV-5 DNA expression values. Uninfected HeLa cells (showing no viral mRNA signal and nuclear fluorescence in the HAdV-5 channel above the background level) were excluded from calculations. The HAdV-5 fluorescence cutoff was user-defined individually for each experiment, since background in the fluorescein isothiocyanate channel fluctuated slightly from experiment to experiment. A cutoff of 30 was used in the HeLa time course experiment (Fig. 3 and 4C). A total of 178 of 207 imaged HeLa cells were analyzed at 25 hpi, 153/262 at 18 hpi, and 12/202 at 13 hpi. No cells were excluded at 1 and 7 hpi due to no HAdV-5 DNA fluorescence being recorded. The same cutoff was applied in 12S E1A and 13S E1A splice variant analyses (Fig. 5B), where 233/359 cells were included at 25 hpi, 170/442 at 19 hpi, and 37/605 at 13 hpi. A cutoff of 60 was used for HeLa-pVII(Δ24) (Fig. 8B and C), where 154/323 (13 hpi) and 173/300 (25 hpi) cells were included for analyses. To check if viral DNA was distributed log normally between different groups in HeLa-pVII(Δ24) cells, HAdV-5 DNA fluorescence values were log transformed for each cell to approximate a normal distribution followed by Shapiro-Wilk normality testing. Since virus genomic DNA was not log-normally distributed in cells, the Wilcoxon signed-rank test was applied to compare Dox+ and Dox− samples. P values lower than 0.05 were considered statistically significant. In total, 3,956 BJAB cells were included for analysis (Fig. 7). A total of 1,412 noninfected BJAB cells were removed from the analysis using the following rejection criteria: E1A mRNA, 0; MLTU mRNA, 0; HAdV-DNA fluorescence, <30. Remaining cells were grouped into 3 individual clusters using k-means unsupervised clustering according to their E1A (13S and 12S) mRNA and MLTU (exon I_II and exon II_III) mRNA expression and HAdV-5 DNA fluorescence (Fig. 7). Expression data in BJAB cells was presented as a single-cell, two-dimensional scatterplot (Fig. 7A), or median expression for each feature was shown as a box-and-whisker plot (Fig. 7B). BJAB cells additionally were grouped into 6 bins according to their HAdV-5 DNA fluorescence in the nucleus. A similar binning strategy was applied to HeLa cells, where the E1A 12S/13S mRNA splice variant expression ratio was measured, and in the time course experiment (7 bins were set, covering a broader fluorescence range).
ACKNOWLEDGMENTS
We thank Raviteja Inturi and Stephen Taylor for kindly providing reagents, Anette Carlsson for excellent technical assistance, and Catharina Svensson for initiating the project and for giving valuable comments. We are grateful to Göran Akusjärvi for fruitful discussions, moral support throughout the study, and critical reading of the manuscript.
This work was supported by the Swedish Cancer Society (CAN 2013/350), Åke Wibergs Foundation (M14-0155), Marcus Borgströms Foundation, and Swedish Research Council through a grant to the Uppsala RNA Research Centre (2006-5038-36531-16). Research in M.N.'s group has been supported by FORMAS project Biobridges, the Swedish Foundation for Strategic Research project FLU-ID, and the Swedish Research Council.
REFERENCES
- 1.Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tollefson AE, Ryerse JS, Scaria A, Hermiston TW, Wold WS. 1996. The E3-11.6-kDa adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152–162. doi: 10.1006/viro.1996.0295. [DOI] [PubMed] [Google Scholar]
- 3.Assadian F, Sandstrom K, Bondeson K, Laurell G, Lidian A, Svensson C, Akusjarvi G, Bergqvist A, Punga T. 2016. Distribution and molecular characterization of human adenovirus and Epstein-Barr virus infections in tonsillar lymphocytes isolated from patients diagnosed with tonsillar diseases. PLoS One 11:e0154814. doi: 10.1371/journal.pone.0154814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnett CT, Erdman D, Xu W, Gooding LR. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol 76:10608–10616. doi: 10.1128/JVI.76.21.10608-10616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. 2009. Latent species C adenoviruses in human tonsil tissues. J Virol 83:2417–2428. doi: 10.1128/JVI.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nevins JR, Ginsberg HS, Blanchard JM, Wilson MC, Darnell JE Jr. 1979. Regulation of the primary expression of the early adenovirus transcription units. J Virol 32:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulfendahl PJ, Linder S, Kreivi JP, Nordqvist K, Sevensson C, Hultberg H, Akusjarvi G. 1987. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J 6:2037–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow LT, Broker TR, Lewis JB. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol 134:265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- 9.Frisch SM, Mymryk JS. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol 3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan S, Thimmapaya B. 1995. Regulation of adenovirus E2 transcription unit. Curr Top Microbiol Immunol 199(Part 3):177–194. [DOI] [PubMed] [Google Scholar]
- 11.Thomas GP, Mathews MB. 1980. DNA replication and the early to late transition in adenovirus infection. Cell 22:523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- 12.Akusjarvi G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front Biosci 13:5006–5015. [DOI] [PubMed] [Google Scholar]
- 13.Blainey PC, Graziano V, Perez-Berna AJ, McGrath WJ, Flint SJ, San Martin C, Xie XS, Mangel WF. 2013. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space. IV. Viral proteinase slides along DNA to locate and process its substrates. J Biol Chem 288:2092–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inturi R, Thaduri S, Punga T. 2013. Adenovirus precursor pVII protein stability is regulated by its propeptide sequence. PLoS One 8:e80617. doi: 10.1371/journal.pone.0080617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corden J, Engelking HM, Pearson GD. 1976. Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci U S A 73:401–404. doi: 10.1073/pnas.73.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirza MA, Weber J. 1982. Structure of adenovirus chromatin. Biochim Biophys Acta 696:76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- 17.Sung MT, Cao TM, Coleman RT, Budelier KA. 1983. Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc Natl Acad Sci U S A 80:2902–2906. doi: 10.1073/pnas.80.10.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karen KA, Hearing P. 2011. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol 85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JS, Osheim YN, Xue Y, Emanuel MR, Lewis PW, Bankovich A, Beyer AL, Engel DA. 2004. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J Virol 78:6459–6468. doi: 10.1128/JVI.78.12.6459-6468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu T, Haruki H, Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res 39:889–901. doi: 10.1093/nar/gkq783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong RN, van der Vliet PC, Brenkman AB. 2003. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr Top Microbiol Immunol 272:187–211. [DOI] [PubMed] [Google Scholar]
- 22.Goodrum FD, Ornelles DA. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol 71:548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Huang W, Ornelles DA, Gooding LR. 2010. Modeling adenovirus latency in human lymphocyte cell lines. J Virol 84:8799–8810. doi: 10.1128/JVI.00562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson C, Grundberg I, Soderberg O, Nilsson M. 2010. In situ detection and genotyping of individual mRNA molecules. Nat Methods 7:395–397. doi: 10.1038/nmeth.1448. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. 1994. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 26.Henriksson S, Shaposhnikov S, Nilsson M, Collins A. 2011. Study of gene-specific DNA repair in the comet assay with padlock probes and rolling circle amplification. Toxicol Lett 202:142–147. doi: 10.1016/j.toxlet.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Lagunavicius A, Merkiene E, Kiveryte Z, Savaneviciute A, Zimbaite-Ruskuliene V, Radzvilavicius T, Janulaitis A. 2009. Novel application of Phi29 DNA polymerase: RNA detection and analysis in vitro and in situ by target RNA-primed RCA. RNA 15:765–771. doi: 10.1261/rna.1279909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conze T, Goransson J, Razzaghian HR, Ericsson O, Oberg D, Akusjarvi G, Landegren U, Nilsson M. 2010. Single molecule analysis of combinatorial splicing. Nucleic Acids Res 38:e163. doi: 10.1093/nar/gkq581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henriksson S, Blomstrom AL, Fuxler L, Fossum C, Berg M, Nilsson M. 2011. Development of an in situ assay for simultaneous detection of the genomic and replicative form of PCV2 using padlock probes and rolling circle amplification. Virol J 8:37. doi: 10.1186/1743-422X-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaspar I, Ephrussi A. 2015. Strength in numbers: quantitative single-molecule RNA detection assays. Wiley Interdiscip Rev Dev Biol 4:135–150. doi: 10.1002/wdev.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M. 2004. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat Methods 1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 32.Shaw AR, Ziff EB. 1980. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3′ coterminal mRNAs and five late families. Cell 22:905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- 33.Murali VK, Ornelles DA, Gooding LR, Wilms HT, Huang W, Tollefson AE, Wold WS, Garnett-Benson C. 2014. Adenovirus death protein (ADP) is required for lytic infection of human lymphocytes. J Virol 88:903–912. doi: 10.1128/JVI.01675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furuse Y, Ornelles DA, Cullen BR. 2013. Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA. Virology 447:140–145. doi: 10.1016/j.virol.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuesta R, Xi Q, Schneider RJ. 2001. Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harbor Symp Quant Biol 66:259–267. doi: 10.1101/sqb.2001.66.259. [DOI] [PubMed] [Google Scholar]
- 36.Flint SJ, Gonzalez RA. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr Top Microbiol Immunol 272:287–330. [DOI] [PubMed] [Google Scholar]
- 37.Giberson AN, Davidson AR, Parks RJ. 2012. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu T, Nagata K. 2012. Replication-uncoupled histone deposition during adenovirus DNA replication. J Virol 86:6701–6711. doi: 10.1128/JVI.00380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang IH, Suomalainen M, Andriasyan V, Kilcher S, Mercer J, Neef A, Luedtke NW, Greber UF. 2013. Tracking viral genomes in host cells at single-molecule resolution. Cell Host Microbe 14:468–480. doi: 10.1016/j.chom.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Morral N, Engel DA. 2007. Transcription releases protein VII from adenovirus chromatin. Virology 369:411–422. doi: 10.1016/j.virol.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Radko S, Jung R, Olanubi O, Pelka P. 2015. Effects of adenovirus type 5 E1A isoforms on viral replication in arrested human cells. PLoS One 10:e0140124. doi: 10.1371/journal.pone.0140124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, Berk AJ. 2014. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weibrecht I, Lundin E, Kiflemariam S, Mignardi M, Grundberg I, Larsson C, Koos B, Nilsson M, Soderberg O. 2013. In situ detection of individual mRNA molecules and protein complexes or post-translational modifications using padlock probes combined with the in situ proximity ligation assay. Nat Protoc 8:355–372. doi: 10.1038/nprot.2013.006. [DOI] [PubMed] [Google Scholar]
- 44.Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. 2010. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol 190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bett AJ, Krougliak V, Graham FL. 1995. DNA sequence of the deletion/insertion in early region 3 of Ad5 dl309. Virus Res 39:75–82. doi: 10.1016/S0168-1702(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 46.Krzywkowski T, Hauling T, Nilsson M. 2017. In situ single-molecule RNA genotyping using padlock probes and rolling circle amplification. Methods Mol Biol 1492:59–76. doi: 10.1007/978-1-4939-6442-0_4. [DOI] [PubMed] [Google Scholar]
- 47.Grundberg I, Kiflemariam S, Mignardi M, Imgenberg-Kreuz J, Edlund K, Micke P, Sundstrom M, Sjoblom T, Botling J, Nilsson M. 2013. In situ mutation detection and visualization of intratumor heterogeneity for cancer research and diagnostics. Oncotarget 4:2407–2418. doi: 10.18632/oncotarget.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. 2005. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res 15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]