ABSTRACT
Peptides derived from the C-terminal heptad repeat (CHR) region of the human immunodeficiency virus type 1 (HIV-1) fusogenic protein gp41 are potent viral entry inhibitors, and currently, enfuvirtide (T-20) is the only one approved for clinical use; however, emerging drug resistance largely limits its efficacy. In this study, we generated a novel lipopeptide inhibitor, named LP-19, by integrating multiple design strategies, including an N-terminal M-T hook structure, an HIV-2 sequence, intrahelical salt bridges, and a membrane-anchoring lipid tail. LP-19 showed stable binding affinity and highly potent, broad, and long-lasting antiviral activity. In in vitro studies, LP-19 efficiently inhibited HIV-1-, HIV-2-, and simian immunodeficiency virus (SIV)-mediated cell fusion, viral entry, and infection, and it was highly active against diverse subtypes of primary HIV-1 isolates and inhibitor-resistant mutants. Ex vivo studies demonstrated that LP-19 exhibited dramatically increased anti-HIV activity and an extended half-life in rhesus macaques. In short-term monotherapy, LP-19 reduced viral loads to undetectable levels in acutely and chronically simian-human immunodeficiency virus (SHIV)-infected monkeys. Therefore, this study offers an ideal HIV-1/2 fusion inhibitor for clinical development and emphasizes the importance of the viral fusion step as a drug target.
IMPORTANCE The peptide drug T-20 is the only viral fusion inhibitor in the clinic, which is used for combination therapy of HIV-1 infection; however, it requires a high dosage and easily induces drug resistance, calling for a new drug with significantly improved pharmaceutical profiles. Here, we have developed a short-lipopeptide-based fusion inhibitor, termed LP-19, which mainly targets the conserved gp41 pocket site and shows highly potent inhibitory activity against HIV-1, HIV-2, and even SIV isolates. LP-19 exhibits dramatically increased antiviral activity and an extended half-life in rhesus macaques, and it has potent therapeutic efficacy in SHIV-infected monkeys, highlighting its high potential as a new viral fusion inhibitor for clinical use.
KEYWORDS: HIV-1, HIV-2, fusion inhibitor, lipopeptide
INTRODUCTION
More than 36 million people are infected with human immunodeficiency virus (HIV) worldwide (www.unaids.org). Because of the lack of an effective vaccine, antiretroviral therapy (ART) has been highly appreciated as a vital strategy to control the HIV epidemic. Currently, there are six classes of approved anti-HIV drugs that target four steps of the viral life cycle, including entry, reverse transcription, integration, and virion maturation (www.fda.gov). Different from other drugs that act after infection occurs, HIV entry inhibitors intercept the virus before it invades target cells. There are two HIV entry inhibitors approved for clinical use (1–3). Maraviroc targets the cell coreceptor CCR5, thereby blocking viral binding, and enfuvirtide (T-20), a 36-mer peptide derived from the C-terminal heptad repeat (CHR) region of the HIV-1 transmembrane protein gp41, binds to the N-terminal heptad repeat (NHR) of gp41, thus preventing the formation of the viral six-helical bundle (6-HB) structure. T-20 is effective in combination therapy; however, it requires high dosages (90 mg twice daily) and easily induces drug resistance, calling for next-generation viral fusion inhibitors with improved pharmaceutical profiles.
Since its discovery, the deep pocket on the trimeric NHR helices of gp41 has been considered an ideal drug target because of its high degree of conservation and essential role in viral entry (4–7). However, small-molecule or -peptide inhibitors that specifically target the pocket site often lack high activity, mostly likely due to their weak binding. We recently discovered that the N-terminal motif of CHR-based peptides adopts a unique M-T hook structure, which can dramatically enhance the binding and inhibitory activities of inhibitors, especially for short peptides that are otherwise inactive (8–11). It was shown that inhibitors with a modified M-T hook structure also possessed potent activities in inhibiting drug-resistant mutants and conferring high genetic barriers to inducing resistance (12–14). Most recently, we successfully developed the helical short peptide 2P23 by introducing the M-T hook structure, HIV-2 sequences, and “salt bridge”-forming residues, which showed highly potent and broad inhibitory activity against HIV-1, HIV-2, and simian immunodeficiency virus (SIV) (15). In this study, we further generated a lipopeptide-based HIV-1/2 fusion inhibitor, termed LP-19, by adding a fatty acid group (palmitic acid [C16]) to the C terminus of 2P23. LP-19 exhibits high binding stability and extremely potent in vitro, ex vivo, and in vivo antiviral activity, providing an ideal candidate for clinical development. The new data also verify the viral fusion step as a pivotal drug target.
RESULTS
Generation of a highly stable lipopeptide-based HIV fusion inhibitor.
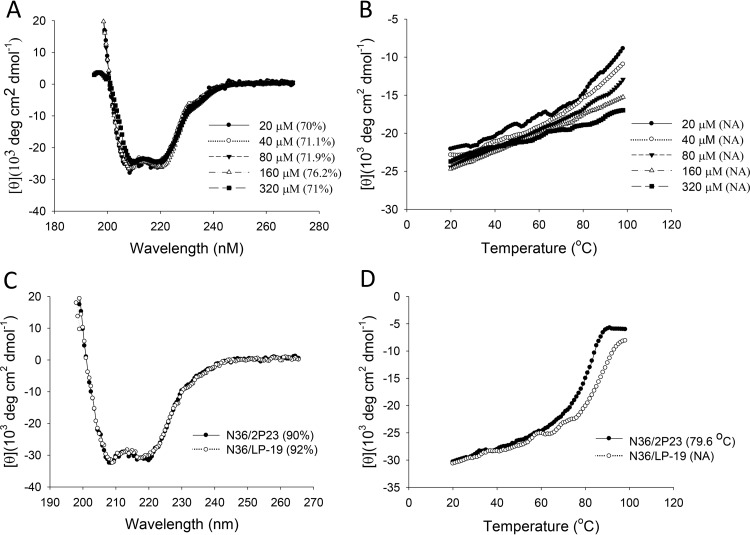
A peptide-based inhibitor usually has a short half-life in vivo, which critically limits its therapeutic efficacy. Emerging studies have suggested that lipid-conjugated peptides possess sharply increased antiviral potency and in vivo stability (16–19). In our case, we recently generated a panel of anti-HIV lipopeptides (LP-1 to LP-18) by using the short peptide HP23 as a template (16). Of these lipopeptides, LP-11 displayed potent and long-lasting antiviral activity. To develop a more effective HIV fusion inhibitor, here, we modified 2P23 with a C16 fatty acid. As illustrated in Fig. 1, the lipopeptide LP-19 was created by adding C16 to the C terminus of 2P23 through a flexible polyethylene glycol 8 (PEG8) linker. First, we performed circular dichroism (CD) experiments to examine the secondary structure and stability of LP-19 itself. As expected, LP-19 exhibited high α-helicity at different peptide concentrations (Fig. 2A), and its thermal unfolding transition (melting temperature [Tm]) could not be achieved (Fig. 2B), which indicated its high helical stability. We then compared LP-19 and its parent compound 2P23 for their α-helicity and thermostability in the presence of an NHR-derived peptide (N36). Typically, both inhibitors interacted with the target surrogate N36 to form 6-HB structures with high helical contents (Fig. 2C); however, while the N36/2P23-based 6-HB had a Tm value of 79.6°C, the N36/LP-19-based complex could not achieve its Tm owing to apparently incomplete unfolding (Fig. 2D). These results suggested that LP-19 is a highly stable helical lipopeptide with greatly increased binding stability.
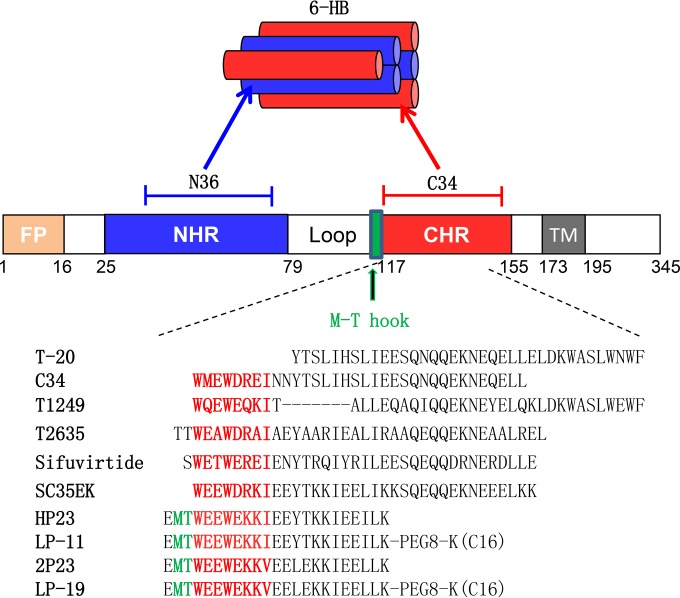
FIG 1.
Schematic illustration of the HIV-1 gp41 protein and its peptide-based inhibitors. The gp41 numbering for HIV-1HXB2 is used. FP, fusion peptide; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; TM, transmembrane domain. The sequences corresponding to the CHR pocket-binding domain (PBD) are marked in red. The position and sequence of the M-T hook structure are marked in green.
FIG 2.
Biophysical properties of LP-19 determined by CD spectroscopy. (A and B) The secondary structure (A) and thermostability (B) of LP-19 in isolation were measured at different concentrations in PBS. (C and D) The secondary structure (C) and thermostability (D) of 2P23- and LP-19-based 6-HBs were determined with the final concentration of each peptide in PBS at 10 μM. The α-helicity and Tm values are shown in parentheses. NA, not applicable for calculation. The experiments were repeated at least two times, and representative data are shown.
LP-19 shows potent inhibitory activity against HIV-1, HIV-2, and SIV.
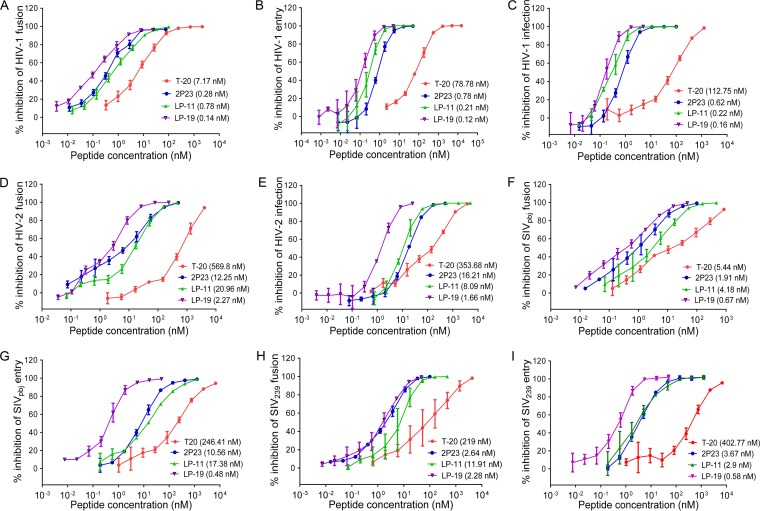
Initially, we compared the inhibitory activities of LP-19, 2P23, T-20, and LP-11 against HIV-1 isolate NL4-3-mediated cell fusion (Fig. 3A), viral entry (Fig. 3B), and infection (Fig. 3C), which validated that LP-19 was the most potent inhibitor. For inhibition of cell fusion, LP-19 showed a 50% inhibitory concentration (IC50) of 0.14 nM, whereas 2P23, T-20, and LP-11 showed IC50s of 0.28, 7.17, and 0.78 nM, respectively. For inhibition of pseudovirus entry, LP-19 showed an IC50 of 0.12 nM, whereas 2P23, T-20, and LP-11 showed IC50s of 0.78, 78.78, and 0.21 nM, respectively. For replicative virus, LP-19 gave an IC50 of 0.16 nM, whereas 2P23, T-20, and LP-11 exhibited IC50s of 0.62, 112.75, and 0.22 nM, respectively. Next, we determined the inhibitory potency against HIV-2 and SIV isolates. As shown in Fig. 3D and E, LP-19 possessed dramatically increased activity in inhibiting HIV-2ROD-mediated fusion and infection over T-20, 2P23, and LP-11. Specifically, LP-19 inhibited cell fusion with an IC50 of 2.27 nM, which indicated a 5.4-fold increase compared to that of 2P23 (12.25 nM), a 251.01-fold increase compared to that of T-20 (569.8 nM), and a 9.23-fold increase compared to that of LP-11 (20.96 nM). Similarly, LP-19 was the most potent inhibitor of SIV Env-mediated cell fusion and virus entry (Fig. 3F to I). In the fusion assay, LP-19 inhibited SIVpbj and SIV239 with IC50s of 0.67 and 2.28 nM, respectively, and 2P23, T-20, and LP-11 inhibited SIVpbj with IC50s of 1.91, 5.44, and 4.18 nM, respectively, and inhibited SIV239 with IC50s of 2.64, 219, and 11.91 nM, respectively. In the pseudovirus entry assay, LP-19 inhibited SIVpbj and SIV239 with IC50s of 0.48 and 0.58 nM, respectively, and 2P23, T-20, and LP-11 inhibited SIVpbj with IC50s of 10.56, 246.41, and 17.38 nM, respectively, and inhibited SIV239 with IC50s of 3.67, 402.77, and 2.9 nM, respectively. Taken together, these data demonstrated that LP-19 is a highly potent and broad fusion inhibitor against HIV-1, HIV-2, and SIV isolates and verified that lipid conjugation is an efficient strategy to improve the antiviral activity of CHR-derived peptide inhibitors.
FIG 3.
Inhibitory activities of LP-19 and control peptides against HIV-1, HIV-2, and SIV isolates. Inhibition of HIV-1NL4-3-mediated cell fusion (A), entry (B), and infection (C); HIV-2ROD-mediated cell fusion (D) and infection (E); SIVpbj-mediated cell fusion (F) and entry (G); and SIV239-mediated cell fusion (H) and entry (I) by T-10, 2P23, LP-11, and LP-19 was determined. The experiments were performed in triplicate and repeated 3 times. Percent inhibition of the peptides and IC50s were calculated. Data are expressed as means ± standard deviations.
LP-19 is highly active against diverse primary HIV-1 isolates and resistant mutants.
To further validate the broad anti-HIV activity of LP-19, we determined its inhibition of diverse subtypes of HIV-1 isolates in comparison with T-20, 2P23, and LP-11. Two panels of 29 total HIV-1 pseudoviruses were prepared and used in single-cycle infection assays. The panel 1 Envs were routinely used in our laboratory; the panel 2 Envs, the so-called “global panel,” were recently selected on the basis of the genetic and antigenic variability of HIV-1 Envs representing the global AIDS epidemic (20). As shown in Table 1, LP-19 efficiently inhibited diverse subtypes of HIV-1 isolates with a mean IC50 of 0.5 nM (panel 1) or 0.41 nM (panel 2); in sharp contrast, 2P23, T-20, and LP-11 inhibited the panel 1 viruses with mean IC50s of 5.84, 39.43, and 1.29 nM, respectively, and inhibited the panel 2 viruses with mean IC50s of 4.35, 26.32, and 1.3 nM, respectively. These results demonstrated that LP-19 shows a large improvement over T-20, 2P23, and LP-11.
TABLE 1.
Inhibitory activity of fusion inhibitors against diverse primary HIV-1 isolates and drug-resistant mutantsa
| Pseudovirus | Subtype | Mean IC50 (nM) ± SD |
|||
|---|---|---|---|---|---|
| T-20 | 2P23 | LP-11 | LP-19 | ||
| Panel 1 primary viruses | |||||
| 92RW020 | A | 6.68 ± 1.15 | 1.96 ± 0.07 | 1.08 ± 0.18 | 0.39 ± 0.01 |
| 92UG037.8 | A | 9.88 ± 0.01 | 2.06 ± 0.43 | 1.04 ± 0.25 | 0.36 ± 0.03 |
| SF162 | B | 6.42 ± 0.13 | 17.52 ± 0.65 | 3.26 ± 0.77 | 0.69 ± 0.16 |
| JRFL | B | 29.3 ± 9.5 | 6.45 ± 1.1 | 2.7 ± 0.64 | 1.56 ± 0.27 |
| AC10.0.29 | B | 3.9 ± 1.08 | 1.36 ± 0.02 | 0.38 ± 0.07 | 0.17 ± 0.08 |
| SC422661.8 | B | 14.65 ± 2.35 | 2.21 ± 0.09 | 0.31 ± 0.08 | 0.1 ± 0.02 |
| B01 | B′ | 66.09 ± 9.23 | 6.2 ± 0.65 | 0.77 ± 0.19 | 0.22 ± 0.08 |
| B02 | B′ | 52.2 ± 10.91 | 8.57 ± 2.39 | 1.50 ± 0.44 | 0.49 ± 0.18 |
| B04 | B′ | 14.03 ± 4.77 | 4.75 ± 1.59 | 1.03 ± 0.12 | 0.31 ± 0.06 |
| CAP45.2.00.G3 | C | 176.24 ± 18.58 | 17.2 ± 3.6 | 1.61 ± 0.91 | 0.88 ± 0.25 |
| ZM109F.PB4 | C | 13.33 ± 4.9 | 1.07 ± 0.11 | 1 ± 0.28 | 0.37 ± 0.11 |
| ZM53M.PB12 | C | 33.68 ± 7.6 | 0.91 ± 0.04 | 0.69 ± 0.02 | 0.22 ± 0.08 |
| AE03 | A/E | 10.38 ± 7.47 | 4.74 ± 2.43 | 0.47 ± 0.15 | 0.32 ± 0.08 |
| AE04 | A/E | 18.32 ± 12.83 | 7.23 ± 2.21 | 0.98 ± 0.4 | 0.37 ± 0.18 |
| CH64.20 | B/C | 20.35 ± 1.88 | 0.54 ± 0.08 | 0.42 ± 0.17 | 0.14 ± 0.03 |
| CH070.1 | B/C | 159.45 ± 36.32 | 6.84 ± 1.04 | 3.18 ± 1.33 | 1.15 ± 0.13 |
| CH120.6 | B/C | 35.45 ± 9.03 | 9.69 ± 0.29 | 1.49 ± 0.54 | 0.83 ± 0.04 |
| Mean | 39.43 | 5.84 | 1.29 | 0.5 | |
| Panel 2 primary viruses | |||||
| 398-F1_F6_20 | A | 38.12 ± 4.62 | 1.43 ± 0.08 | 0.58 ± 0.22 | 0.11 ± 0.01 |
| TRO.11 | B | 14.24 ± 2.28 | 4.59 ± 1.08 | 3.07 ± 1.48 | 0.93 ± 0.08 |
| X2278_C2_B6 | B | 11.7 ± 1.99 | 1 ± 0.17 | 0.69 ± 0.02 | 0.13 ± 0.01 |
| CE703010217_B6 | C | 43.11 ± 9.81 | 5.20 ± 0.36 | 1.42 ± 0.19 | 0.4 ± 0.09 |
| CE1176_A3 | C | 10.9 ± 5.01 | 5.28 ± 0.31 | 1.46 ± 0.28 | 0.46 ± 0.1 |
| HIV_25710-2.43 | C | 20.12 ± 2.69 | 2.15 ± 0.33 | 0.7 ± 0.23 | 0.21 ± 0.1 |
| X1632-S2-B10 | G | 13.86 ± 2.57 | 4.64 ± 0.24 | 2.36 ± 0.61 | 0.99 ± 0.04 |
| 246_F3_C10_2 | A/C | 33.77 ± 7.54 | 4.13 ± 0.41 | 1.11 ± 0.26 | 0.33 ± 0.11 |
| CNE8 | A/E | 53.61 ± 6.38 | 11.94 ± 4.86 | 0.98 ± 0.16 | 0.38 ± 0.01 |
| CNE55 | A/E | 47.35 ± 23.37 | 3.25 ± 0.08 | 0.58 ± 0.18 | 0.25 ± 0.04 |
| CH119.10 | B/C | 6.82 ± 0.27 | 7.62 ± 0.6 | 1.35 ± 0.23 | 0.35 ± 0.1 |
| BJOX002000.03.2 | B/C | 22.21 ± 2.01 | 0.97 ± 0.14 | 1.24 ± 0.15 | 0.41 ± 0.1 |
| Mean | 26.32 | 4.35 | 1.3 | 0.41 | |
| T-20-resistant mutants | |||||
| NL4-3WT | B | 83.71 ± 9.56 | 0.72 ± 0.07 | 0.21 ± 0.01 | 0.12 ± 0.01 |
| NL4-3I37T | B | 559.02 ± 183.65 | 1.33 ± 0.12 | 0.52 ± 0.04 | 0.2 ± 0.07 |
| NL4-3V38A | B | 1,383.67 ± 137.65 | 0.81 ± 0.15 | 0.32 ± 0.09 | 0.15 ± 0.06 |
| NL4-3V38M | B | 774.8 ± 96.45 | 1.14 ± 0.19 | 0.42 ± 0.12 | 0.21 ± 0.01 |
| NL4-3Q40H | B | 1,910.34 ± 103.71 | 1.19 ± 0.12 | 0.28 ± 0.08 | 0.15 ± 0.07 |
| NL4-3N43K | B | 749.04 ± 157.25 | 1.19 ± 0.21 | 0.29 ± 0.07 | 0.15 ± 0.05 |
| NL4-3D36S/V38M | B | 423.45 ± 9.36 | 1.36 ± 0.33 | 0.63 ± 0.24 | 0.27 ± 0.06 |
| NL4-3I37T/N43K | B | 7,187 ± 4,941.26 | 1.28 ± 0.2 | 0.48 ± 0.14 | 0.22 ± 0.08 |
| NL4-3V38A/N42T | B | 3,167.67 ± 961.19 | 0.4 ± 0.06 | 0.14 ± 0.04 | 0.06 ± 0.02 |
| Mean | 1,804.3 | 1.05 | 0.37 | 0.17 | |
| HP23-resistant mutants | |||||
| NL4-3E49K | B | 176.7 ± 1.46 | 5.1 ± 0.51 | 0.44 ± 0.16 | 0.12 ± 0 |
| NL4-3L57R | B | 46.24 ± 11.17 | 39.62 ± 19.64 | 4.16 ± 1.05 | 0.43 ± 0.18 |
| NL4-3N126K | B | 209.87 ± 5.17 | 1.22 ± 0.61 | 0.67 ± 0.06 | 0.31 ± 0.03 |
| NL4-3E136G | B | 143.28 ± 77.63 | 4.68 ± 0.41 | 1.72 ± 0.84 | 0.62 ± 0.16 |
| NL4-3E49K/N126K | B | 190.03 ± 37.03 | 4.94 ± 0.48 | 2.7 ± 0.04 | 0.59 ± 0.04 |
| NL4-3L57R/E136G | B | 40.28 ± 12.6 | 168.49 ± 0.37 | 23.51 ± 3.57 | 2.76 ± 0.21 |
| Mean | 134.4 | 36.52 | 5.53 | 0.81 | |
The assay was performed in triplicate and repeated 3 times.
We also constructed two panels of inhibitor-resistant pseudoviruses using NL4-3 Env as a template. The panel 1 Envs were introduced with single or double mutations that rendered resistance to T-20, while the panel 2 Envs were generated to carry HP23-resistant mutations (21, 22). The data in Table 1 showed that T-20, 2P23, LP-11, and LP-19 inhibited T-20-resistant mutants with mean IC50s of 1,804.3, 1.05, 0.37, and 0.17 nM, respectively, while they inhibited HP23-resistant mutants with mean IC50s of 134.4, 36.52, 5.53, and 0.81 nM, respectively. Therefore, LP-19 has dramatically increased potency against T-20- and HP23-resistant viruses.
LP-19 has strong and long-acting ex vivo antiviral activity.
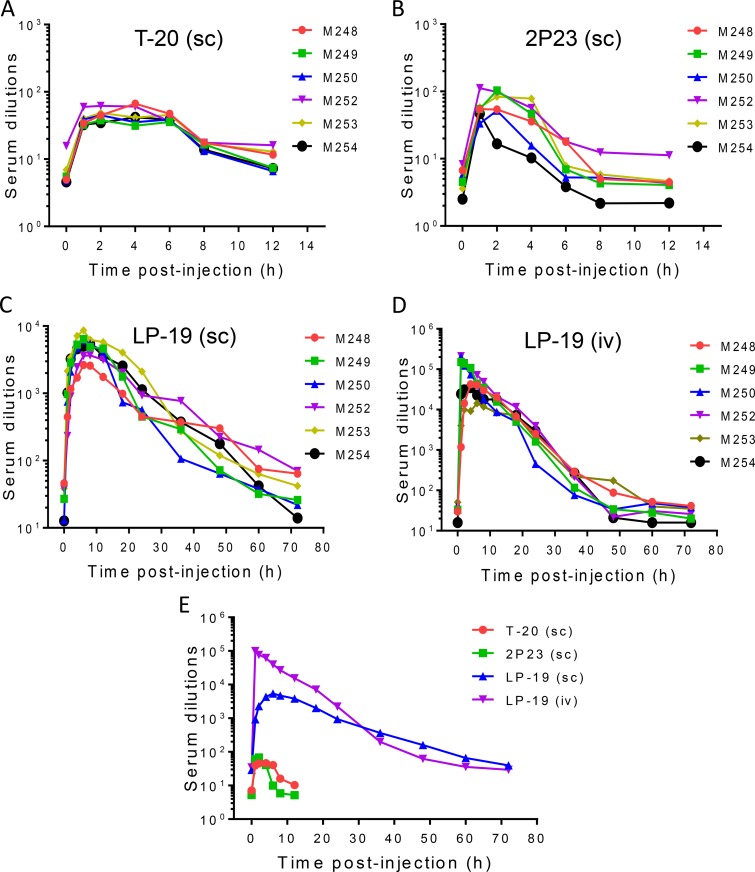
We previously reported the long-acting antiviral activity of LP-11 in a rat model (16). Obviously, LP-19 showed much better in vitro antiviral activity than did LP-11, making it an ideal candidate for further development. Thus, we decided to study the in vivo stability and antiviral activity of LP-19 in nonhuman primates. To this end, T-20 and 2P23 were also tested as controls. Each inhibitor was subcutaneously injected into six healthy rhesus monkeys, and the pharmacological kinetics in sera was measured pre- or postadministration. As shown in Fig. 4A, the drug T-20 exhibited a serum peak level during 1 to 4 h after administration, with dilutions that inhibited 50% of virus infection ranging from 40- to 46-fold. The unconjugated peptide 2P23 showed a serum peak level during 1 to 2 h with dilutions of 60- to 68-fold (Fig. 4B). For these two peptides, the anti-HIV activity of sera could not be detected after 6 h, implying their short half-lives in vivo. Strikingly, the lipopeptide LP-19 showed a serum peak level during 4 to 12 h with dilutions ranging from 3,845- to 4,329-fold (Fig. 4C). For example, LP-19 reached serum dilutions of 928-fold at 1 h and 2,282-fold at 2 h postinjection. Even after 60 or 72 h, LP-19 still exhibited potent inhibitory activity with a serum dilution of 66- or 40-fold, respectively, which was comparable to the peak levels of T-20 and 2P23. Furthermore, LP-19 was given to monkeys intravenously, and its pharmacological kinetics was evaluated similarly. It was found that LP-19 sharply reached a peak level with a serum dilution of 99,107-fold at 1 h postinjection and then maintained a high level of activity over 48 h (Fig. 4D). In comparison, lipid conjugation rendered the parental peptide 2P23 with dramatically improved pharmaceutical profiles (Fig. 4E). Therefore, we conclude that LP-19 is a highly potent, broad, and long-lasting lipopeptide HIV-1/2 fusion inhibitor.
FIG 4.
Ex vivo antiviral activities of LP-19 and control peptides. Each of the inhibitors was subcutaneously or intravenously injected into six rhesus monkeys at 3 mg/kg, and monkey sera were harvested at different time points before or after injection. (A to D) The antiviral activities of sera from monkeys administered T-20 (A), 2P23 (B), and LP-19 (C and D) were tested by an HIV-1NL4-3-based single-cycle infection assay, and serum dilutions required for 50% inhibition of virus infection were calculated. sc, subcutaneous injection; iv, intravenous injection. (E) Comparison of HIV-inhibitory activities of monkey sera. A Mann-Whitney U test was performed to judge the significance of the difference between treatment groups (P = 0.0026 for LP-19 versus 2P23, P = 0.0026 for LP-19 versus T-20, P = 0.62 for T-20 versus 2P23, P = 0.2382 for LP-19 [subcutaneous injection] versus LP-19 [intravenous injection], and P = 0.0433 for LP-19 [subcutaneous injection] versus LP-19 [intravenous injection] during 0 to 36 h).
Therapeutic efficacy of LP-19 in acute SHIV infection.
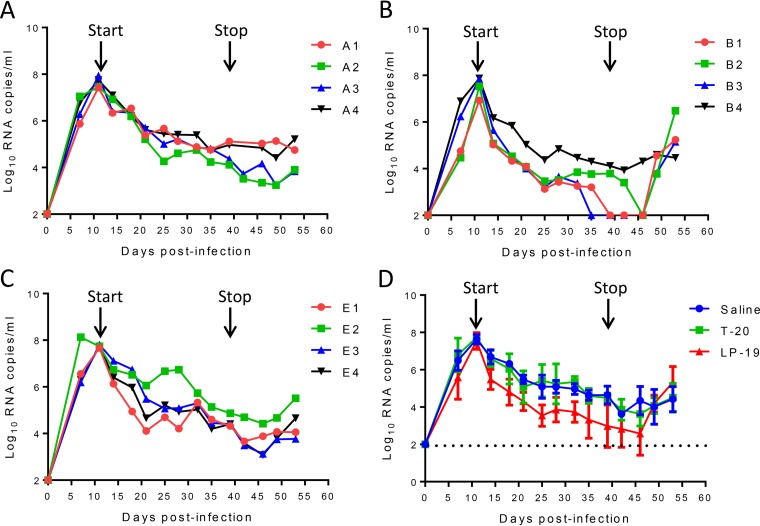
Highly encouraged by the in vitro and ex vivo inhibitory data, we sought to observe the in vivo therapeutic efficacy of LP-19. First, we initiated treatment of simian-human immunodeficiency virus SF162P3 (SHIVSF162P3)-infected rhesus monkeys on day 11 after virus inoculation, when acute infection had been established with the peak of virus replication. Twelve monkeys were randomly assigned to three groups and subcutaneously injected with normal saline (group A), LP-19 (group B), and T-20 (group E) once daily for 4 weeks. Virus replication reached the highest levels in all groups (average of 7.64 log10 RNA copies/ml) and then declined naturally (Fig. 5A to C); however, LP-19 treatment obviously increased the decline rate (Fig. 5D). As shown in Fig. 5B, the plasma viral loads in monkeys B3, B1, and B2 declined to undetectable levels (100 copies/ml) by days 24, 28, and 35, respectively, after the initiation of treatment. As expected, viral rebounds were observed in all three monkeys in which treatment was effective after LP-19 was withdrawn at 10 days. Unexpectedly, T-20 did not show appreciable therapeutic effects on SHIV-infected monkeys, as the viral kinetics in treated monkeys were similar to those in the group that received normal saline. Here, we speculate that T-20, due to a short half-life (∼3.8 h), might require more frequent injections to achieve efficacy, similar to its use in clinics (90 mg twice daily).
FIG 5.
Therapeutic efficacy of LP-19 in acutely SHIV-infected rhesus monkeys. (A to C) Starting from day 11 after virus inoculation, 12 SHIVSF162P3-infected monkeys were subcutaneously treated with normal saline (A), LP-19 (B), or T-20 (C) once daily for 4 weeks. The plasma viral loads of monkeys were measured at different time points by quantitative PCR. (D) Comparison of the viral load kinetics in monkeys treated with LP-19 and controls. A Mann-Whitney U test was performed to judge the significance of the difference during treatment (P = 0.0304 for LP-19 versus saline, P = 0.0315 for LP-19 versus T-20, and P = 1.000 for T-20 versus saline).
Therapeutic efficacy of LP-19 in chronic SHIV infection.
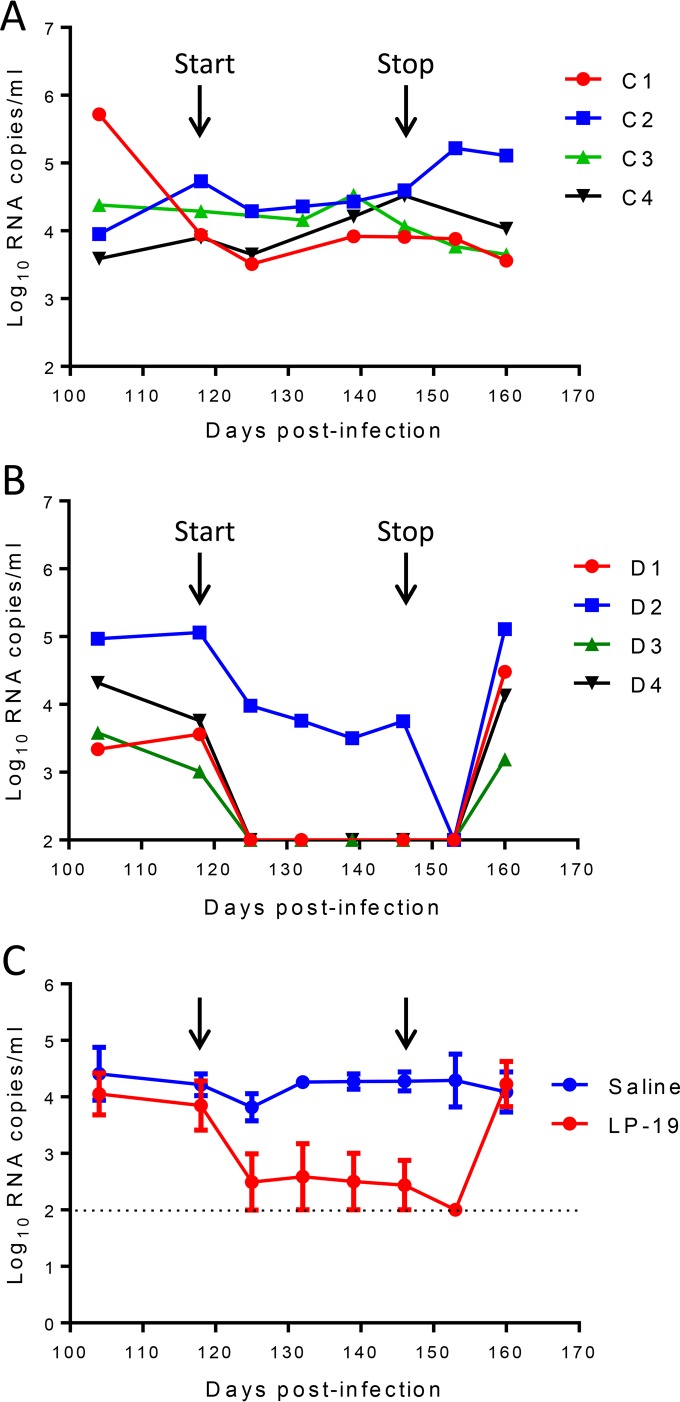
We were intrigued to know the therapeutic efficacy of LP-19 in chronic infection. For this, eight monkeys were intravenously infected with SHIVSF162P3 for 4 months, when chronic infection had been established with set point viral loads of 3.01 to 5.06 log10 RNA copies/ml. Group C monkeys (monkeys C1 to C4) were treated with normal saline, and group D monkeys (monkeys D1 to D4) were treated with LP-19 once daily for 4 weeks. As a control, treatment with saline had no effect on the viral loads in monkeys (Fig. 6A). Strikingly, we observed rapid and precipitous declines of plasma viral loads to undetectable levels by day 7 in 3 of 4 monkeys treated with LP-19 (Fig. 6B). Monkey D2, with the highest baseline viral load, also showed a fast decline during the first week of treatment, which gradually reached an undetectable level at week 5. As expected, the viral loads rebounded in all of the LP-19-treated monkeys after LP-19 was stopped at 2 weeks (Fig. 6C).
FIG 6.
Therapeutic efficacy of LP-19 in chronically SHIV-infected rhesus monkeys. (A and B) After virus inoculation for 4 months, eight SHIVSF162P3-infected monkeys were subcutaneously treated once daily for 4 weeks with normal saline (A) or LP-19 (B). The plasma viral loads of monkeys were measured at different time points by quantitative PCR. (C) Comparison of the viral load kinetics in monkeys treated with saline and LP-19. A Mann-Whitney U test was performed to judge the significance of the difference after treatment (P = 0.007 for LP-19 versus saline).
DISCUSSION
Highly active antiretroviral therapy (HAART) has been very successful in treating HIV-1 infection. However, the persistence of HIV-1 infection requires lifelong therapy that can result in drug resistance. Thus, there is a need to develop antiretroviral drugs that work earlier in the virus life cycle, such as HIV-1 entry inhibitors targeting its membrane fusion step. In this study, we generated LP-19 as a helical lipopeptide-based viral fusion inhibitor. Ideally, it showed high binding stability and potent inhibitory activity against HIV-1, HIV-2, and even SIV isolates. We also demonstrated that LP-19 exhibited broad activity in inhibiting diverse subtypes of HIV-1 isolates and T-20- and HP23-resistant mutants. Ex vivo experiments confirmed that LP-19 was an extremely potent and long-lasting fusion inhibitor in rhesus macaques. Furthermore, LP-19 had strong therapeutic efficacy in both acutely and chronically SHIV-infected monkeys. Therefore, we believe that LP-19 is a promising candidate for clinical use.
In the last decade, we dedicated our efforts to exploring the mechanism of HIV-1 fusion and to developing new viral fusion inhibitors. We previously found that the intra- and interhelical salt bridges of the gp41 6-HB structure play important roles for HIV-1 entry and its inhibition (23–25). We discovered that the upstream motif of the gp41 CHR critically determines the binding affinity and inhibitory activity of CHR-derived inhibitors (26, 27). By determining the crystal structures of newly designed peptides containing the upstream CHR sequence, an M-T hook structure was surprisingly identified, which was formed by two residues (Met115 and Thr116) immediately preceding the pocket-binding domain (PBD) of peptides (9, 10, 28). The M-T hook structure dramatically enhances the binding and inhibitory activities of diverse CHR-derived inhibitors and confers a high genetic barrier to inducing resistance (11, 13, 14). A potent short-peptide inhibitor specifically targeting the gp41 pocket site was designed by incorporating two M-T hook residues (8, 12). To overcome the problem of a peptide-based inhibitor that usually has a short in vivo half-life, we generated lipopeptides by conjugating different lipids that can anchor the inhibitors to the cell membranes, thus elevating the local drug concentration at the site where viral fusion occurs (16, 17, 29–31). Apart from its membrane-tropic feature, a lipopeptide can bind to serum proteins that can largely extend its in vivo half-life (32–34). To improve the inhibitory function of an inhibitor against HIV-2 isolates, we recently generated a short helical peptide (2P23) by incorporating an HIV-2 sequence (15). Actually, the present inhibitor, LP-19, was created by integrating all these strategies, including an M-T hook structure at its N terminus, multiple intrahelical salt bridges at its i and i+3 or i+4 positions, and a lipid anchor at its C terminus. It is conceivable that LP-19 mainly targets the highly conserved pocket region of gp41 on the targeting cell membrane where fusion occurs, and a relatively longer linker between the peptide sequence and the fatty acid chain would render its functional flexibility.
The most prominent result for LP-19 was its robust therapeutic efficacy in SHIV-infected rhesus monkeys. Short-term LP-19 monotherapy rapidly reduced viral loads to undetectable levels in both acute and chronic SHIV infections. To our knowledge, there has been no viral fusion inhibitor tested in a nonhuman primate model. The in vivo potency of LP-19 should be highly appreciated if we review previous short-term monotherapy with other anti-HIV drugs in clinics (2, 35–39), such as T-20 (−1.96 log10 RNA copies/ml); the second-generation fusion inhibitor T-1249 (−1.26 log10 RNA copies/ml); the CCR5 antagonist maraviroc (−1.6 log10 RNA copies/ml); the reverse transcriptase inhibitors zidovudine (−0.52 log10 RNA copies/ml), lamivudine (−1.19 log10 RNA copies/ml), and efavirenz (−1.68 log10 RNA copies/ml); as well as the protease inhibitor ritonavir (−1.38 log10 RNA copies/ml). Ten-day monotherapy with a high dose (5 g per day) of VIR-576, a naturally occurring HIV-1 fusion inhibitor targeting the fusion peptide of gp41, reduced the viral load by 1.23 orders of magnitude (40, 41). It is highly expected that the potency of LP-19 in monkeys could be translated to the clinical setting.
HIV-2 isolates have already spread to different regions worldwide, and currently, about 1 million to 2 million people have been infected, mostly in West Africa. HIV-1 and HIV-2 have different evolutionary histories and share only 50% genetic similarity (42, 43). However, all currently available antiretroviral drugs were specifically developed to inhibit HIV-1 entry and replication, and consequently, some drugs in clinical use have limited or no anti-HIV-2 activity, including all nonnucleoside reverse transcriptase inhibitors, some protease inhibitors, and the fusion inhibitor T-20 (44–47). It is also expected that LP-19 would provide a powerful tool for treating HIV-2 infection. Additionally, as a strong and stable HIV-1/2 entry inhibitor, LP-19 would serve as a prophylactic agent such as a topical microbicide. Taken together, these features make LP-19 a highly potent, broad, and long-lasting fusion inhibitor, and thus, it has high potential for clinical development. The lipopeptide-based strategy may also be applicable for the development of fusion inhibitors against other enveloped viruses, such as influenza virus and respiratory syncytial virus (RSV).
MATERIALS AND METHODS
Cells and reagents.
HEK293T cells were purchased from the American Type Culture Collection (ATCC) (Rockville, MD). U87 CD4+ CXCR4+ cells stably expressing CD4 and CXCR4; TZM-bl indicator cells stably expressing CD4 and CCR5 along with endogenously expressed CXCR4; and plasmids for HIV-1 Env panels (subtypes A, B, B′, C, G, A/C, A/E, and B/C), a molecular clone of HIV-1 reference strain NL4-3 (pNL4-), and SHIVSF162P3 were obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. A molecular clone of HIV-2 strain ROD (pROD) was kindly provided by Nuno Taveira at the Faculty of Pharmacy, University of Lisbon, Lisbon, Portugal. Two plasmids encoding SIV Env (pSIVpbj-Env and pSIV239-Env) were kindly provided by Jianqing Xu at the Shanghai Public Health Clinical Center and Institutes of Biomedical Sciences, Fudan University, Shanghai, China. Cells were cultured in complete growth medium that consisted of Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin-streptomycin, 2 mM l-glutamine, 1 mM sodium pyruvate, and 1× MEM nonessential amino acids (Gibco/Invitrogen, USA) and were maintained at 37°C in 5% CO2.
Peptide synthesis.
Peptides were synthesized on rink amide 4-methylbenzhydrylamine (MBHA) resin using a standard solid-phase 9-flurorenylmethoxycarbonyl (FMOC) method as described previously (16). All peptides were acetylated at the N terminus and amidated at the C terminus. For lipopeptides, the template peptides contain a lysine residue at the C terminus with a 1-(4,4-dimethyl-2,6-dioxocyclohexylidene)ethyl side-chain-protecting group, enabling the conjugation of a fatty acid that requires a special deprotection step in a solution of 2% hydrazinehydrate–N,N-dimethylformamide (DMF). Peptides were purified by reverse-phase high-performance liquid chromatography (HPLC) to more than 95% homogeneity and were identified by mass spectrometry. Concentrations of the peptides were determined by UV absorbance and a theoretically calculated molar extinction coefficient based on the tryptophan and tyrosine residues.
CD spectroscopy.
CD spectroscopy was performed according to protocols that we described previously (10). Briefly, a CHR inhibitor was diluted in phosphate-buffered saline (PBS) (pH 7.2) and incubated at 37°C for 30 min in the presence or absence of an equal molar concentration of the NHR peptide N36. CD spectra were acquired on a Jasco spectropolarimeter (model J-815) using a 1-nm bandwidth with a 1-nm step resolution from 195 to 270 nm at room temperature. Spectra were corrected by the subtraction of a solvent blank. The α-helical content was calculated from the CD signal by dividing the mean residue ellipticity ([θ]) at 222 nm by the value expected for 100% helix formation (−33,000° · cm2 · dmol−1). Thermal denaturation was performed by monitoring the ellipticity change at 222 nm from 20°C to 98°C at a rate of 2°C/min, and the Tm was defined as the midpoint of the thermal unfolding transition.
Cell fusion assay.
A dual-split-protein (DSP)-based assay was performed to determine HIV or SIV Env-mediated cell-cell fusion as described previously (48, 49). Briefly, a total of 1.5 × 104 HEK293T cells (effector cells) were seeded onto a 96-well plate, and a total of 8 × 104 U87 CD+ CXCR4+ cells (target cells) were seeded onto a 24-well plate. On the following day, effector cells were transfected with a mixture of an Env-expressing plasmid and a DSP1–7 plasmid, and target cells were transfected with a DSP8–11 plasmid. At 48 h posttransfection, the target cells were resuspended in 300 μl prewarmed culture medium, and 0.05 μl EnduRen live-cell substrate (Promega, Madison, WI) was added to each well. Aliquots of 75 μl of the target cell suspension were then transferred over each well of the effector cells in the presence or absence of serially 3-fold-diluted fusion inhibitors. The cells were then spun down to maximize cell-cell contact and incubated for 1 h at 37°C. Luciferase activity was measured by using luciferase assay regents and a luminescence counter (Promega). The percent inhibition of cell fusion and IC50 values were calculated by using GraphPad Prism software (GraphPad Software Inc., San Diego, CA).
Single-cycle infection assay.
A single-cycle infection assay was performed as described previously (50). Briefly, HIV-1 or SIV pseudoviruses were generated via the cotransfection of HEK293T cells with an Env-expressing plasmid and a backbone plasmid, pSG3Δenv, that encodes an Env-defective, luciferase-expressing HIV-1 genome. Culture supernatants were harvested 48 h after transfection, and 50% tissue culture infectious doses (TCID50) were determined in TZM-bl cells. To measure the antiviral activity of inhibitors, peptides were prepared in 3-fold dilutions, mixed with 100 TCID50 of viruses, and then incubated for 1 h at room temperature. The mixture was added to TZM-bl cells (104 cells/well), and the cells were incubated for 48 h at 37°C. Luciferase activity was measured by using a luminescence counter (Promega), and IC50s were calculated as described above.
Inhibition of infectious HIV-1NL4-3 and HIV-2ROD isolates.
The antiviral activity of fusion inhibitors was assessed by using molecular clones of replication- and infection-competent HIV-1NL4-3 and HIV-2ROD as two indicator viruses. Briefly, viral stocks were prepared by transfecting a plasmid (pNL4-3 or pROD) into HEK293T cells. Culture supernatants were harvested at 48 h posttransfection, and the TCID50 in TZM-bl cells was quantified. Viruses were used (100 TCID50) to infect TZM-bl cells in the presence or absence of serially 3-fold-diluted inhibitors. Cells were harvested at 2 days postinfection and lysed in reporter lysis buffer, and luciferase activity was measured as described above.
Ex vivo anti-HIV activity of inhibitors.
We previously developed a simple and sensitive system for evaluating the ex vivo antiviral activity of an inhibitor without animal infection facilities (16). Here, we adapted this method to a nonhuman primate model. Six Chinese rhesus macaques (Macaca mulatta) (3 males and 3 females aged 3 to 4 years and weighing 3.4 to 4.7 kg) were used to assess the ex vivo anti-HIV activity of LP-19, its parent peptide 2P23, as well as T-20. Briefly, an inhibitor was subcutaneously administered to macaques at 3 mg/kg of body weight. Macaque sera were harvested before injection (0 h) and after injection (1, 2, 4, 6, 8, 12, 18, 24, 36, 48, 60, and 72 h). The anti-HIV activity of all the sera was determined with the HIV-1NL4-3 isolate in a single-cycle infection assay. The 50% effective concentration was defined as the fold serum dilution that inhibited 50% of virus infection.
Therapeutic efficacy of LP-19 in SHIV-infected rhesus monkeys.
Twenty adult Chinese rhesus macaques were screened and found to be negative for SIV, herpesvirus B, and simian T-lymphotropic virus. SHIVs (SF162P3) were expanded on macaque peripheral blood mononuclear cells (PBMCs), and the TCID50 was quantitated. Macaques were intravenously inoculated with 1,000 TCID50 of viruses and randomly assigned to 5 treatment groups (4 monkeys/group). For treatment of acute infection (groups A, B, and E), macaques were subcutaneously injected with 1 ml of a 0.9% saline solution, LP-19 (3 mg/kg), or T-20 (3 mg/kg) once daily for 4 weeks from day 11 after virus inoculation. For treatment of chronic infection, the same doses of LP-19 or controls were subcutaneously given once daily for 4 weeks from day 118 after virus inoculation.
Measurement of plasma viral loads by qRT-PCR.
Plasma viral loads were determined by a quantitative real-time reverse transcription-PCR (qRT-PCR) assay for SIV gag477 with the upstream primer GCAGAGGAGGAAATTACCCAGTAC, the downstream primer CAATTTTACCCAGGCATTTAATGTT and the probe FAM (6-carboxyfluorescein)-ACCTGCCATTAAGCCCGA-MGB on a Perkin-Elmer ABI 7500 instrument. Viral RNA was extracted and purified from cell-free plasma by using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). RNA was eluted in 20 μl of nuclease-free water and frozen immediately at −80°C until analysis. The SIV gag477 cDNA sequence was used as a positive control for PCR. The limits of detection were 100 copy equivalents of RNA per ml of plasma. Triplicate test reactions were performed for each sample.
Ethics statement.
Protocols for the ex vivo studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the Beijing Institute of Microbiology and Epidemiology (approval no. 2016-15). Protocols for the in vivo studies were approved by the IACUC (51) at the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences (approval no. ILAS-VL-2015-004). All monkeys were housed and fed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility. The study of animals was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Science to ensure personnel safety and animal welfare. All animals were anesthetized with ketamine hydrochloride (10 mg/kg) prior to the procedures. The experiments were performed in a biosafety level 3 laboratory.
ACKNOWLEDGMENTS
We thank Zene Matsuda at the Institute of Medical Science, University of Tokyo, for providing the DSP1–7 and DSP8–11 plasmids for the cell fusion assay; Nuno Taveira at the Faculty of Pharmacy of the University of Lisbon for providing a molecular clone encoding the HIV-2 genome; and Jianqing Xu at the Shanghai Public Health Clinical Center and Institutes of Biomedical Sciences of Fudan University for providing the plasmids encoding SIV Env.
This work was supported by grants from the National Natural Science Foundation of China (81630061, 81473255, 8167348, and 81271830) and the National Science and Technology Major Project of China (2014ZX10001001-001).
REFERENCES
- 1.Este JA, Telenti A. 2007. HIV entry inhibitors. Lancet 370:81–88. doi: 10.1016/S0140-6736(07)61052-6. [DOI] [PubMed] [Google Scholar]
- 2.Fatkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD, Rockstroh JK, Dezube BJ, Jenkins TM, Medhurst C, Sullivan JF, Ridgway C, Abel S, James IT, Youle M, van der Ryst E. 2005. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med 11:1170–1172. doi: 10.1038/nm1319. [DOI] [PubMed] [Google Scholar]
- 3.Lalezari JP, Henry K, O'Hearn M, Montaner JS, Piliero PJ, Trottier B, Walmsley S, Cohen C, Kuritzkes DR, Eron JJ Jr, Chung J, DeMasi R, Donatacci L, Drobnes C, Delehanty J, Salgo M, TORO 1 Study Group . 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N Engl J Med 348:2175–2185. doi: 10.1056/NEJMoa035026. [DOI] [PubMed] [Google Scholar]
- 4.Chan DC, Kim PS. 1998. HIV entry and its inhibition. Cell 93:681–684. doi: 10.1016/S0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 5.Chan DC, Chutkowski CT, Kim PS. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci U S A 95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 7.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Chong H, Yao X, Qiu Z, Sun J, Zhang M, Waltersperger S, Wang M, Liu SL, Cui S, He Y. 2013. Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1. FASEB J 27:1203–1213. doi: 10.1096/fj.12-222547. [DOI] [PubMed] [Google Scholar]
- 9.Chong H, Yao X, Sun J, Qiu Z, Zhang M, Waltersperger S, Wang M, Cui S, He Y. 2012. The M-T hook structure is critical for design of HIV-1 fusion inhibitors. J Biol Chem 287:34558–34568. doi: 10.1074/jbc.M112.390393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong H, Yao X, Qiu Z, Qin B, Han R, Waltersperger S, Wang M, Cui S, He Y. 2012. Discovery of critical residues for viral entry and inhibition through structural insight of HIV-1 fusion inhibitor CP621-652. J Biol Chem 287:20281–20289. doi: 10.1074/jbc.M112.354126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong H, Qiu Z, Su Y, He Y. 2015. The N-terminal T-T motif of a third-generation HIV-1 fusion inhibitor is not required for binding affinity and antiviral activity. J Med Chem 58:6378–6388. doi: 10.1021/acs.jmedchem.5b00109. [DOI] [PubMed] [Google Scholar]
- 12.Chong H, Qiu Z, Su Y, Yang L, He Y. 2015. Design of a highly potent HIV-1 fusion inhibitor targeting the gp41 pocket. AIDS 29:13–21. doi: 10.1097/QAD.0000000000000498. [DOI] [PubMed] [Google Scholar]
- 13.Chong H, Yao X, Qiu Z, Sun J, Qiao Y, Zhang M, Wang M, Cui S, He Y. 2014. The M-T hook structure increases the potency of HIV-1 fusion inhibitor sifuvirtide and overcomes drug resistance. J Antimicrob Chemother 69:2759–2769. doi: 10.1093/jac/dku183. [DOI] [PubMed] [Google Scholar]
- 14.Chong H, Qiu Z, Sun J, Qiao Y, Li X, He Y. 2014. Two M-T hook residues greatly improve the antiviral activity and resistance profile of the HIV-1 fusion inhibitor SC29EK. Retrovirology 11:40. doi: 10.1186/1742-4690-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong S, Borrego P, Ding X, Zhu Y, Martins A, Chong H, Taveira N, He Y. 2017. A helical short-peptide fusion inhibitor with highly potent activity against human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus. J Virol 91:e01839-16. doi: 10.1128/JVI.01839-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong H, Wu X, Su Y, He Y. 2016. Development of potent and long-acting HIV-1 fusion inhibitors. AIDS 30:1187–1196. doi: 10.1097/QAD.0000000000001073. [DOI] [PubMed] [Google Scholar]
- 17.Ashkenazi A, Viard M, Unger L, Blumenthal R, Shai Y. 2012. Sphingopeptides: dihydrosphingosine-based fusion inhibitors against wild-type and enfuvirtide-resistant HIV-1. FASEB J 26:4628–4636. doi: 10.1096/fj.12-215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingallinella P, Bianchi E, Ladwa NA, Wang YJ, Hrin R, Veneziano M, Bonelli F, Ketas TJ, Moore JP, Miller MD, Pessi A. 2009. Addition of a cholesterol group to an HIV-1 peptide fusion inhibitor dramatically increases its antiviral potency. Proc Natl Acad Sci U S A 106:5801–5806. doi: 10.1073/pnas.0901007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pessi A. 2015. Cholesterol-conjugated peptide antivirals: a path to a rapid response to emerging viral diseases. J Pept Sci 21:379–386. doi: 10.1002/psc.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y, Chong H, Xiong S, Qiao Y, Qiu Z, He Y. 2015. Genetic pathway of HIV-1 resistance to novel fusion inhibitors targeting the Gp41 pocket. J Virol 89:12467–12479. doi: 10.1128/JVI.01741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su Y, Chong H, Qiu Z, Xiong S, He Y. 2015. Mechanism of HIV-1 resistance to short-peptide fusion inhibitors targeting the Gp41 pocket. J Virol 89:5801–5811. doi: 10.1128/JVI.00373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, Lu H, Jing W, Jiang S, Zhang L. 2008. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J Biol Chem 283:11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Liu S, Li J, Lu H, Qi Z, Liu Z, Debnath AK, Jiang S. 2008. Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J Virol 82:11129–11139. doi: 10.1128/JVI.01060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Liu S, Jing W, Lu H, Cai D, Chin DJ, Debnath AK, Kirchhoff F, Jiang S. 2007. Conserved residue Lys574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J Biol Chem 282:25631–25639. doi: 10.1074/jbc.M703781200. [DOI] [PubMed] [Google Scholar]
- 26.He Y, Cheng J, Lu H, Li J, Hu J, Qi Z, Liu Z, Jiang S, Dai Q. 2008. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc Natl Acad Sci U S A 105:16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y, Cheng J, Li J, Qi Z, Lu H, Dong M, Jiang S, Dai Q. 2008. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. J Virol 82:6349–6358. doi: 10.1128/JVI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao X, Chong H, Zhang C, Qiu Z, Qin B, Han R, Waltersperger S, Wang M, He Y, Cui S. 2012. Structural basis of potent and broad HIV-1 fusion inhibitor CP32M. J Biol Chem 287:26618–26629. doi: 10.1074/jbc.M112.381079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urbanowicz RA, Lacek K, Lahm A, Bienkowska-Szewczyk K, Ball JK, Nicosia A, Cortese R, Pessi A. 2015. Cholesterol conjugation potentiates the antiviral activity of an HIV immunoadhesin. J Pept Sci 21:743–749. doi: 10.1002/psc.2802. [DOI] [PubMed] [Google Scholar]
- 30.Hollmann A, Matos PM, Augusto MT, Castanho MA, Santos NC. 2013. Conjugation of cholesterol to HIV-1 fusion inhibitor C34 increases peptide-membrane interactions potentiating its action. PLoS One 8:e60302. doi: 10.1371/journal.pone.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wexler-Cohen Y, Ashkenazi A, Viard M, Blumenthal R, Shai Y. 2010. Virus-cell and cell-cell fusion mediated by the HIV-1 envelope glycoprotein is inhibited by short gp41 N-terminal membrane-anchored peptides lacking the critical pocket domain. FASEB J 24:4196–4202. doi: 10.1096/fj.09-151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Bulaj G. 2012. Converting peptides into drug leads by lipidation. Curr Med Chem 19:1602–1618. doi: 10.2174/092986712799945003. [DOI] [PubMed] [Google Scholar]
- 33.Lim SB, Banerjee A, Onyuksel H. 2012. Improvement of drug safety by the use of lipid-based nanocarriers. J Control Release 163:34–45. doi: 10.1016/j.jconrel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Madsen K, Knudsen LB, Agersoe H, Nielsen PF, Thogersen H, Wilken M, Johansen NL. 2007. Structure-activity and protraction relationship of long-acting glucagon-like peptide-1 derivatives: importance of fatty acid length, polarity, and bulkiness. J Med Chem 50:6126–6132. doi: 10.1021/jm070861j. [DOI] [PubMed] [Google Scholar]
- 35.Lalezari JP, Bellos NC, Sathasivam K, Richmond GJ, Cohen CJ, Myers RA Jr, Henry DH, Raskino C, Melby T, Murchison H, Zhang Y, Spence R, Greenberg ML, Demasi RA, Miralles GD, T1249-102 Study Group. 2005. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J Infect Dis 191:1155–1163. doi: 10.1086/427993. [DOI] [PubMed] [Google Scholar]
- 36.Eron JJ, Benoit SL, Jemsek J, MacArthur RD, Santana J, Quinn JB, Kuritzkes DR, Fallon MA, Rubin M. 1995. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N Engl J Med 333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- 37.Adkins JC, Noble S. 1998. Efavirenz. Drugs 56:1055–1064; discussion 1065–1056. doi: 10.2165/00003495-199856060-00014. [DOI] [PubMed] [Google Scholar]
- 38.Arasteh K, Clumeck N, Pozniak A, Lazzarin A, De Meyer S, Muller H, Peeters M, Rinehart A, Lefebvre E, TMC114-C207 Study Team. 2005. TMC114/ritonavir substitution for protease inhibitor(s) in a non-suppressive antiretroviral regimen: a 14-day proof-of-principle trial. AIDS 19:943–947. doi: 10.1097/01.aids.0000171408.38490.01. [DOI] [PubMed] [Google Scholar]
- 39.Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med 4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 40.Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F. 2007. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell 129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 41.Forssmann WG, The YH, Stoll M, Adermann K, Albrecht U, Tillmann HC, Barlos K, Busmann A, Canales-Mayordomo A, Gimenez-Gallego G, Hirsch J, Jimenez-Barbero J, Meyer-Olson D, Munch J, Perez-Castells J, Standker L, Kirchhoff F, Schmidt RE. 2010. Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide. Sci Transl Med 2:63re3. doi: 10.1126/scitranslmed.3001697. [DOI] [PubMed] [Google Scholar]
- 42.Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, Vandamme AM. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc Natl Acad Sci U S A 100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu DJ, Dondero TJ, Rayfield MA, George JR, Schochetman G, Jaffe HW, Luo CC, Kalish ML, Weniger BG, Pau CP, Schable CA, Curran JW. 1996. The emerging genetic diversity of HIV: the importance of global surveillance for diagnostics, research, and prevention. JAMA 275:210–216. doi: 10.1001/jama.1996.03530270050031. [DOI] [PubMed] [Google Scholar]
- 44.Ntemgwa ML, d'Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. 2009. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob Agents Chemother 53:3611–3619. doi: 10.1128/AAC.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. 2004. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther 9:57–65. [PubMed] [Google Scholar]
- 46.Hizi A, Tal R, Shaharabany M, Currens MJ, Boyd MR, Hughes SH, McMahon JB. 1993. Specific inhibition of the reverse transcriptase of human immunodeficiency virus type 1 and the chimeric enzymes of human immunodeficiency virus type 1 and type 2 by nonnucleoside inhibitors. Antimicrob Agents Chemother 37:1037–1042. doi: 10.1128/AAC.37.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borrego P, Calado R, Marcelino JM, Bartolo I, Rocha C, Cavaco-Silva P, Doroana M, Antunes F, Maltez F, Caixas U, Barroso H, Taveira N. 2012. Baseline susceptibility of primary HIV-2 to entry inhibitors. Antivir Ther 17:565–570. doi: 10.3851/IMP1996. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. doi: 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 49.Kondo N, Miyauchi K, Meng F, Iwamoto A, Matsuda Z. 2010. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J Biol Chem 285:14681–14688. doi: 10.1074/jbc.M109.067090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao X, Chong H, Zhang C, Waltersperger S, Wang M, Cui S, He Y. 2012. Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J Biol Chem 287:6788–6796. doi: 10.1074/jbc.M111.317883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Research Council 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]