Abstract
Background
The species-rich genus Impatiens is mainly distributed throughout much of tropical Africa, India, southwest Asia, southern China and Japan. There are more than 270 species recorded in China, most of which are restricted to the southwest. An unknown species of Impatiens was collected from Yunnan, southwest China.
Results
Impatiens pandurata Y. H. Tan & S. X. Yu, a new species of Balsaminaceae from Jinping County and Malipo County, Yunnan, China is similar to I. apalophylla and I. clavigera in having racemose inflorescences, 4 lateral sepals, hammer-shaped capsules and ellipsoid seeds, but differs in having leaves with oblanceolate blades aggregated at the top of the stem, 3–5-flowered racemes, a yellow lower sepal without reddish patches, yellowish flowers, and a dorsal petal with stalks at the base. Molecular phylogenetic analyses of sequences from both nuclear ribosomal and plastid genes confirm that this new species is distinct from morphologically similar species previously recorded.
Conclusion
With the support of careful morphological studies and phylogenetic analysis, I. pandurata is a species new to science.
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-015-0108-4) contains supplementary material, which is available to authorized users.
Keywords: Balsaminaceae, Impatiens, Morphology, New species, Phylogeny
Background
The genus Impatiens L. (Balsaminaceae), containing over 1000 species (Grey-Wilson 1980; Fischer 2004; Yu et al. 2015), is mainly distributed throughout much of tropical Africa, India, South-west Asia, southern China and Japan, with only a few species spreading into the north temperate zone of Europe, Russia and China as well as North America (Grey-Wilson 1980). Impatiens species occur in diverse habitats, from sea level to 4000 m in elevation, in forest understories, roadside ditches, valleys, abandoned fields, along streams and in seepage, usually in mesic or wet conditions, although some species can tolerate drier habitats (Yu et al. 2015). Because of its species diversity, the genus has been regarded as ‘the dicot counterpart of the orchid’ (Yuan et al. 2004).
Five diversity hotspots for Impatiens have been recognized, i.e. tropical Africa, Madagascar, southern India and Sri Lanka, the eastern Himalayas, and southeast Asia (Song et al. 2003; Yuan et al. 2004). Impatiens is notoriously difficult to classify morphologically (Hooker 1908; Grey-Wilson 1980) and the semi-succulent stems, fleshy leaves, and extremely fragile flowers make it challenging to prepare well-dried herbarium specimens. The publication of new species each year shows that the genus has been under-collected and under-studied (e.g. Narayanan et al. 2013; Utami 2013; Kuang et al. 2014; Luo et al. 2014).
There are more than 270 species of Impatiens recorded in China (Yu 2012; Chen 2001; Chen et al. 2007), most of them restricted to the southwest. During recent field expeditions in Yunnan, the authors collected several specimens with distinctive morphological characteristics. After careful consultation of the literature and specimens, we concluded that these specimens are morphologically distinct from any described species. After additional molecular phylogenetic analysis, we are confident that this species is new to science.
Methods
Morphology
Characteristics of the leaves, inflorescence and flowers were described and measured on both dried herbarium specimens (from HITBC and PE) and fresh specimens in the field.
Molecular methods
DNA sequences of 151 species of Impatiens were used and three species, Hydrocera triflora (L.) Wight & Arn. (Balsaminaceae), and Marcgravia umbellata L. and Norantea guianensis Aubl. (Marcgraviaceae), were included as outgroups, based on the results of Yuan et al. (2004), Janssens et al. (2006) and Yu et al. (2015). All sequences were downloaded from GenBank, except those of the new species, I. pandurata, which were newly generated for this study (Genbank accession numbers XXXX, XXXX for ITS, XXXX, XXXX for atpB-rbcL and XXXX, XXXX for trnL-F). Vouchers and GenBank accession numbers are listed in Additional file 1: Table S1.
Three molecular markers were used: ITS, atpB-rbcL and trnL-F. Total genomic DNA was extracted from silica gel-dried leaves using a modified CTAB protocol from Doyle and Doyle (1987). Primers and PCR protocols for ITS, atpB-rbcL and trnL-F are derived from White et al. (1990), Janssens et al. (2006) and Taberlet et al. (1991), respectively. PCR products were purified using a GFX™PCR DNA and Gel Band Purification Kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Sequencing reactions were carried out using an ABI Prism Bigdye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Products were analyzed on an ABI3730xl automated DNA sequencer. Sequences were aligned using the default parameters in Clustal X v.1.83 (Thompson et al. 1997) and further adjusted manually in BioEdit v.7.0 (Hall 1999). Four difficult-to-align regions in trnL-F (encompassing 73 sites) and one difficult-to-align region in atpB-rbcL (encompassing 42 sites) were excluded from the analyses.
Maximum parsimony (MP) and Bayesian inference (BI) were used to analyze the ITS and plastid data sets. The MP analyses were carried out in PAUP* v.4.0b10 (Swofford 2003). Heuristic searches were conducted with 1000 replicates of random addition, one tree held at each step during stepwise addition, tree-bisection-reconnection (TBR) branch swapping, MulTrees in effect, and steepest descent off. Bootstrapping was conducted with 1000 replicates with 10 random taxon additions and heuristic research options. The BI analyses were carried out in MrBayes v.3.0b4 (Ronquist and Huelsenbeck 2003). Each of the three regions (ITS, atpB-rbcL, and trnL-F) was assigned its own model of nucleotide substitution, as determined by the Akaike information criterion (AIC) in Modeltest v.3.06 (Posada and Crandall 1998).
Results and discussion
Impatiens pandurata Y. H. Tan & S. X. Yu, sp. nov.
This species is similar to I. apalophylla and I. clavigera in having racemose inflorescences, 4 lateral sepals, hammer-shaped capsules and ellipsoid seeds, but differs in having leaves with oblanceolate blades aggregated at the top of the stem, 3–5-flowered racemes, a yellow lower sepal without reddish patches, yellowish flowers, and a dorsal petal with stalks at the base.
Type: CHINA. Yunnan, Malipo County, Tianbao, Bajiaoping, limestone forests, elev. 1250 m, 22°57′44″ N, 104°51′45″ E, 30 Oct 2012, Yun-Hong Tan 5728 (holotype: HITBC; isotype: PE). Paratype: China. Yunnan: Malipo County, Tianbao Town, elev. 1200 m, 23° 01′02.47″ N, 104°49′34.19″ E, 20 Nov 2014, Xin–Xin Zhu 0001 (CSH)
Fig. 1.
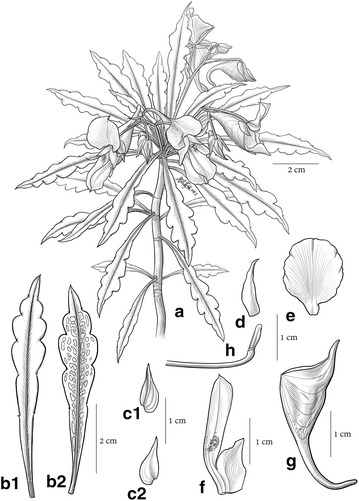
Impatiens pandurata Y. H. Tan & S. X. Yu. a Habit; b, b 1, b 2 Leaf, adaxial surface and abaxial surface; c, c 1, c 2 Outer lateral sepal, abaxial surface and adaxial surface; d Inner lateral sepal; e dorsal petal; f lateral united petal; g lower sepal, lateral view; h capsule, immature. All from Tan 5728 (HITBC) and drawn by Yun-Xi Zhu
Fig. 2.

Impatiens pandurata Y. H. Tan & S. X. Yu. a Habitat; b habit; c leaf adaxial surface; d leaf abaxial surface; e flower, lateral view; f flower, different parts in separation, front view. All from Tan 5728 (HITBC)
Herb perennial, 20–30 cm tall, glabrous. Stems fleshy, erect, simple or branched; inferior nodes unapparent. Leaves alternate, aggregated at stem apex, blades oblanceolate to linear oblanceolate, 5–7 cm long, 1–1.5 cm wide, apex acuminate, base cuneate, deep green above, pale green beneath, sometime with grey patches, margin deeply crenate, with spinose teeth. Veins unapparent. Petioles 0.8–1.2 cm. Racemes solitary in the upper axils, 5–7 cm long, 2–3 (−5)-flowered. Pedicels thin, 15–20 mm long. Bracts ovate to lanceolate, 7–9 mm long, acute. Flowers yellowish or cream. Lateral sepals 4, the outer 2 large, ovate to lanceolate, inaequilateral, 2–3 veined, yellowish-green, base rounded, apex acuminate to caudate, 7.8–9.1 × 3.3–3.7 mm; the inner 2 small, 10.7–11.4 × 1.2–1.6 mm, inaequilateral, apex acuminate. Lower sepal 2.5–3.0 cm long excluding spur, saccate, spur 5–5.6 mm. Dorsal petal 12.5–13.5 mm long, 11.6–12.3 mm wide, orbicular, apex rounded, base broadly cuneate and abruptly constricted into a stalk, midrib obvious, with a slight dorsal crest. Lateral united petals 2.1–2.4 cm long, the lower lobes 11.5–12.5 mm long, 5–5.5 mm wide, oblong, the upper ones 21–24 mm long, 4.5–5.5 wide, elliptic, apex emarginate, middle of inner margin without appendage. Stamens 5, filaments linear, 2–3 mm long, anthers obtuse. Ovary clavate, superior part inflated. Capsule hammer-shaped, seed ellipsoid.
Phenology Flowering and fruiting from September to December.
Ecology This new species grows under evergreen broad leaf forest; 1000–1200 m.
Distribution Impatiens pandurata is known from Jinping County and Malipo County, Yunnan, China.
Etymology The specific epithet ‘pandurata’ refers to the leaf shape of the new species.
Discussion
The phylogenetic topologies obtained with ITS and atpB-rbcL + trnL-F are congruent with those of previous studies (Yu et al. 2015). Both ITS and atpB-rbcL + trnL-F indicate that I. pandurata is a distinct member of the basal clade, subgenus Clavicarpa Yu et al. (2015) (Figs. 3, 4, Additional file 2: Figure S1, Additional file 3: Figure S2). The morphological characters, including perennial herb, racemose inflorescence, 4 lateral united petals, 4-carpellate ovary and one ovule per carpel, also support membership of I. pandurata in this subgenus. Although the ITS data shows that I. pandurata belongs to the basal clade, the relationships among the species in this clade are unclear. In the atpB-rbcL + trnL-F tree, I. pandurata and other species form a large polytomy, so the relationships among subgenus Clavicarpa are also unresolved. However, both nuclear ribosomal and plastid genes agree with the morphological evidence that I. pandurata is a new and distinct species.
Fig. 3.
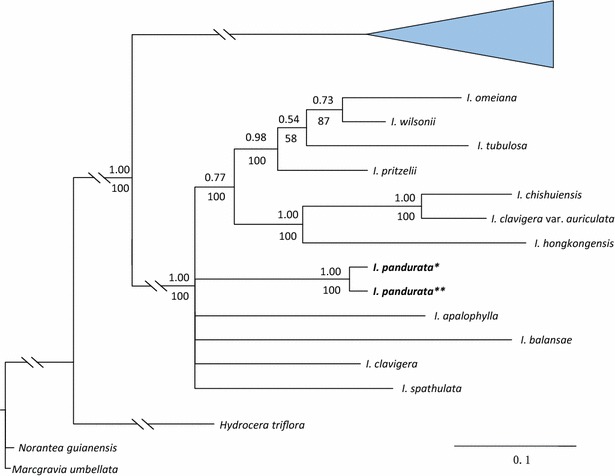
Partial Bayesian consensus phylogram based on the branch lengths of the ITS data. Numbers above and below branches are Bayesian posterior probabilities (>0.5) and bootstrap percentages (>50 %), respectively. “−” indicates nodes not supported
Fig. 4.
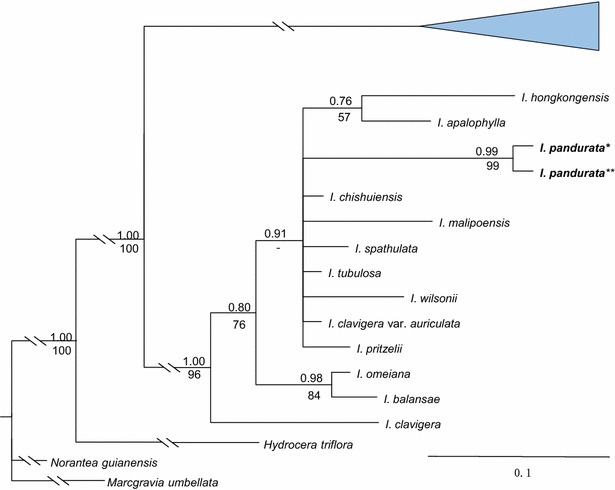
Partial Bayesian consensus phylogram based on the branch lengths of the cpDNA data (atpB-rbcL + trn L-F). Numbers above and below branches are Bayesian posterior probabilities (>0.5) and bootstrap percentages (>50 %), respectively. “−” indicates nodes not supported
The diagnostic morphological characters that distinguish I. pandurata from its allies are the oblanceolate leaves aggregated on the stem apex, with white macula beneath, the narrow lanceolate bracteole, and the subrotund dorsal petal with apparent stalk. Four species with similar morphological characters, I. pandurata, I. apalophylla, I. clavigera, and I. spathulata, are compared with each other, on the basis of their reproductive and vegetative characters in Table 1.
Table 1.
Comparison among Impatiens pandurata, I. apalophylla, I. clavigera and I. spathulata
| Characters | I. pandurata | I. apalophylla | I. clavigera | I. spathulata |
|---|---|---|---|---|
| Plant | Glabrous | Glabrous | Glabrous | Puberulous |
| Shape of leaf | Oblanceolate | Ovate to oblanceolate | Obovate to oblanceolate | Obovate |
| Size of leaf | 5–7 cm × 1–1.5 mm | 10–22 cm × 4–8 cm | 5–15 cm × 3–5 cm | 6–11 cm × 2.5–3.5 cm |
| Inflorescence | 3–5-flowered | 4–10-flowered | 5–9-flowered | 2–4-flowered |
| Floral color | Yellow | Yellow with reddish patches | Yellow | Pink |
| Outer lateral sepals | Oblique, ovate-orbiculate, aequilateral, apex acuminate | Oblique ovate, apex acuminate | Oblique ovate, apex acuminate | Oblique ovate to ovate, apex acuminate |
| Inner lateral sepals | Linear | Lanceolate | Linear-lanceolate | Linear-lanceolate |
| Lower sepal | Absent patch | With redish patches | Absent patch | Absent patch |
| Spur | Long, 1–1.5 cm | Long, 1.5–2 cm | Short, 5–6 mm | Long, 1.5–1.8 cm |
| Lower lobe of united petals | 8–12 mm long, elliptic | 1–1.5 cm long, oblong | 1–1.2 cm long, oblong | 1–1.2 cm long, oblong |
| Upper lobe of united petal | 2–2.5 cm long, oblong, yellow, with slightly reddish patches | 2.5–2.7 cm long, oblong, yellow, with apparent reddish patches | 2.5–2.6 cm long, oblong, yellowish-green, absent patch | 2.3–2.5 cm long, oblong, pink, absent patch |
| Dorsal petal | Suborbicular | Elliptic | Obovate | Elliptic to ovate |
Conclusion
With the support of morphological studies and molecular phylogenetic analysis, I. pandurata is a species new to science. Detailed descriptions, line drawings, color plates, phylogenetic analysis and comparisons with phenetically similar species are provided to aid in identification.
Additional files
Additional file 1: Table S1. Species, GenBank accession numbers and vouchers for the sequences used in this study.
Additional file 2: Figure S1. Bayesian consensus phylogram based on the branch length of the ITS data. Numbers above and below branches are Bayesian posterior probabilities (> 0.5) and bootstrap percentages (> 50%), respectively. “-” indicates nodes not supported.
Additional file 3: Figure S2. Bayesian consensus phylogram based on the branch length of the cpDNA data (atpB-rbcL + trnL-F). Numbers above and below branches are Bayesian posterior probabilities (> 0.5) and bootstrap percentages (> 50%), respectively. “-” indicates nodes not supported.
Authors’ contributions
YHT, XXZ and WZ collected the new species. Morphological studies were carried out by YHT, HJ and SXY. Molecular studies were carried out by YNL. YXZ prepared the line drawing. YHT, SXY and YNL prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to thank the keepers of the herbaria of Paris (P) and Royal Botanic Garden, Kew (K) for loan of type specimens, Mr Yun-Xi Zhu for making the drawing, and Professor Richard T Corlett for polishing the language. We are also grateful to Dr. Wei Wang and Xiao-Guo Xiang for checking the manuscript. This work was supported by National Natural Science Foundation of China (31170177), National Natural Science Foundation of China (Grant no. 31110103911), Main Direction Program of Knowledge Innovation of Chinese Academy of Sciences (Grant no. KSCX2-EW-Z-1) and the special program for public health of traditional Chinese medicine named “Investigation and monitoring in the raw material resources of Chinese medicine needed in the National basic medicine” (Fiscal agency [2011] No. 76) and Special program for Chinese pharmaceutical industry named “Protection and utilization of regional representative Chinese medicine resources”(201207002).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40529-015-0108-4) contains supplementary material, which is available to authorized users.
Contributor Information
Yun-Hong Tan, Email: tyh@xtbg.org.cn.
Yan-Nan Liu, Email: liuyannan@whu.edu.cn.
Hong Jiang, Email: h.tsiang@gmail.com.
Xin-Xin Zhu, Email: zhuzhu8niuniu@126.com.
Wei Zhang, Email: zhangwei@mail.kib.ac.cn.
Sheng-Xiang Yu, Email: yushengxiang@ibcas.ac.cn.
References
- Chen Y-L. Balsaminaceae. In: Chen Y-L, editor. Flora Reipublicae Popularis Sinica. Beijing: Science Press; 2001. pp. 1–243. [Google Scholar]
- Chen Y-L, Akiyama S, Ohba H. Balsaminaceae. In: Wu Z-Y, Raven PH, editors. Flora of China. Beijing, St. Louis: Science Press, Missouri Botanical Garden Press; 2007. pp. 43–113. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Fischer E. Balsaminaceae. In: Kubitzki K, editor. The Families and Genera of Vascular Plants. Berlin: Springer; 2004. pp. 20–25. [Google Scholar]
- Grey-Wilson C. Impatiens of Africa. Rotterdam: Balkema; 1980. pp. 1–57. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hooker JD. Les espèces du genre “Impatiens” dans l’herbier du Museum de Paris. Nov Arch Mus Nat Hist Paris. 1908;10(4):233–272. [Google Scholar]
- Janssens SB, Geuten K, Yuan Y-M, Song Y, Küpfer P, Smets E. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Syst Bot. 2006;31:171–180. doi: 10.1600/036364406775971796. [DOI] [Google Scholar]
- Kuang R-P, Duan L-D, Gu J-Z, Cai X-Z, Cong Y-Y, Liu K-M. Impatiens liboensis sp. nov (Balsaminaceae) from Guizhou, China. Nord J Bot. 2014;32:463–467. doi: 10.1111/j.1756-1051.2013.00194.x. [DOI] [Google Scholar]
- Luo Q, Wang J, Zhao H. Impatiens menghuochengensis Q Luo, a new species of Impatiens (Balsaminaceae) from Sichuan, China. Nord J Bot. 2014;6:839–843. doi: 10.1111/njb.00503. [DOI] [Google Scholar]
- Narayanan MKR, Joseph JP, Kumar NA, Sivadasan M, Alfarhan AH. Impatiens theuerkaufiana (Balsaminaceae), a new scapigerous species from the Western Ghats, India. Phytotaxa. 2013;83:54–60. doi: 10.11646/phytotaxa.83.1.3. [DOI] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Song Y, Yuan Y-M, Küpfer P. Chromosomal evolution in Balsaminaceae, with cytological observations on 45 species from Southeast Asia. Caryologia. 2003;56:463–481. doi: 10.1080/00087114.2003.10589359. [DOI] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), v4.0b10. Sunderland: Sinauer Associates; 2003. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utami N. Impatiens kerinciensis (Balsaminaceae), a new species from Sumatra, Indonesia. Kew Bull. 2013;68:687–688. doi: 10.1007/s12225-013-9468-7. [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a Guide to Methods and Applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Yu S-X. Balsaminaceae of China. Beijing: Peking University Press; 2012. [Google Scholar]
- Yu S-X, Janssens SB, Zhu X-Y, Lidén M, Gao T-G, Wang W. Phylogeny of Impatiens (Balsaminaceae): integrating molecular and morphological evidence into a new classification. Cladistics. 2015 doi: 10.1111/cla.12119. [DOI] [PubMed] [Google Scholar]
- Yuan Y-M, Song Y, Geuten K, Rahelivololona E, Wohlhauser S, Fischer E, Smets E, Küpfer P. Phylogeny and biogeography of Balsaminaceae inferred from ITS sequence data. Taxon. 2004;53:391–403. doi: 10.2307/4135617. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Species, GenBank accession numbers and vouchers for the sequences used in this study.
Additional file 2: Figure S1. Bayesian consensus phylogram based on the branch length of the ITS data. Numbers above and below branches are Bayesian posterior probabilities (> 0.5) and bootstrap percentages (> 50%), respectively. “-” indicates nodes not supported.
Additional file 3: Figure S2. Bayesian consensus phylogram based on the branch length of the cpDNA data (atpB-rbcL + trnL-F). Numbers above and below branches are Bayesian posterior probabilities (> 0.5) and bootstrap percentages (> 50%), respectively. “-” indicates nodes not supported.



 Figs.
Figs.