Abstract

This paper introduces a novel bacterial identification assay based on thermal wave analysis through surface-imprinted polymers (SIPs). Aluminum chips are coated with SIPs, serving as synthetic cell receptors that have been combined previously with the heat-transfer method (HTM) for the selective detection of bacteria. In this work, the concept of bacterial identification is extended toward the detection of nine different bacterial species. In addition, a novel sensing approach, thermal wave transport analysis (TWTA), is introduced, which analyzes the propagation of a thermal wave through a functional interface. The results presented here demonstrate that bacterial rebinding to the SIP layer resulted in a measurable phase shift in the propagated wave, which is most pronounced at a frequency of 0.03 Hz. In this way, the sensor is able to selectively distinguish between the different bacterial species used in this study. Furthermore, a dose–response curve was constructed to determine a limit of detection of 1 × 104 CFU mL–1, indicating that TWTA is advantageous over HTM in terms of sensitivity and response time. Additionally, the limit of selectivity of the sensor was tested in a mixed bacterial solution, containing the target species in the presence of a 99-fold excess of competitor species. Finally, a first application for the sensor in terms of infection diagnosis is presented, revealing that the platform is able to detect bacteria in clinically relevant concentrations as low as 3 × 104 CFU mL–1 in spiked urine samples.
Keywords: surface-imprinted polymers, thermal wave transport analysis (TWTA), bacterial identification, cross-selectivity matrix, mixed bacterial solution
Accurate, sensitive, and fast bacterial identification is of major importance due to the risk these microorganisms impose on human health, causing conditions ranging from common illnesses to mortal infections and even pandemics. Portable bacterial assays that are able to detect bacteria in a fast and sensitive manner could be used for a wide range of applications, including the prevention of hospital-acquired infections (HAIs) or the detection of pathogens in food and drinking water. According to the annual report of the Centers for Disease Control and Prevention (CDC) 727,000 people acquired a HAI in 2011 in the United States alone, of which 75,000 patients died during hospitalization.1 The annual costs associated with the five major HAIs amounts were $9.8 billion in 2013.2 In addition, diseases caused by waterborne pathogens cause up to 1.8 million of human deaths annually on a global scale according to the World Health Organization.3 Bacterial assays that can be used on-site for routine screening of, for example, health care surfaces or environmental samples could lead to a faster bacterial identification and detection, which enables the installation of appropriate countermeasures.
Conventional bacterial identification assays are usually based on laboratory techniques and involve fluorescent labeling and microscopic analysis, making them typically slow and limiting their sensitivity and selectivity.4 Molecular techniques based on genetic screening, polymerase chain reaction (PCR), and real-time PCR have been developed more recently and are able to identify bacteria in a fast and sensitive manner. However, their applicability in point-of-care bacterial detection is limited as they require sample preparation and expensive readout devices and need to be used in a laboratory environment by trained professionals.5−7
Over the years, various biosensor platforms for bacterial identification have been developed that offer a fast and low-cost alternative to these traditional assays. The key element for selective detection in these sensors is the biological recognition layer, which in the case of bacterial detection typically consists of enzymes8 or monoclonal antibodies.9 Rebinding of bacteria to the receptor layer usually results in a physical or chemical change at the sensor surface that is translated into a measurable signal by electrochemical,8−11 optical,12−14 or microgravimetrical transducers.15−17 Although biological recognition elements are very selective, their chemical and physical stabilities are poor. Additionally, they have to be isolated from natural sources and purified, making their synthesis typically labor intensive and expensive. Finally, they are not compatible with most transducer surfaces, which necessitates the need for carefully optimized linker surfaces.18,19
Synthetic cell receptors such as surface-imprinted polymers (SIPs) can overcome many of the problems associated with natural receptors while displaying similar degrees of sensitivity and selectivity.20−22 In recent years, SIP-based biomimetic sensor platforms for microbial identification have been developed on the basis of microgravimetric,23−25 optical,26−29 and electrochemical detection principles.30−32
In 2012, a novel readout technology was developed that has shown to be very versatile in terms of biosensing.33,34 This technique, the so-called heat-transfer method (HTM), is based on analyzing thermal transport through functionalized solid–liquid interfaces and has been combined with SIPs for the detection of macrophages, cancer cells, and bacteria.35−39 HTM offers several benefits in comparison to “classical” biosensor readout principles: it is a low-cost platform that requires little instrumentation, which, in turn, allows for miniaturization. In addition, it can be used “on the bench”, and data processing and analysis are relatively straightforward. As thermal currents are analyzed, HTM can also be combined with insulating materials and analyte liquids. Finally, most biosensor readout technologies require some form of temperature control, whereas HTM is in itself essentially a thermal control unit that simultaneously serves as the readout platform.
The major drawbacks associated with HTM are the inherent noise on the thermal resistance signal and the need for comparing thermal resistance values at equilibrium, which limit the sensitivity and the response time of the technique, respectively. Therefore, a similar readout method that analyzes the propagation of a thermal wave through a functionalized interface rather than its thermal resistance was combined with molecularly imprinted polymers (MIPs) for the detection of dopamine in banana juice.40 This technique, thermal wave transport analysis (TWTA), showed to be advantageous over HTM in terms of response time and sensitivity. In this work, TWTA is used to analyze bacterial rebinding to SIP-coated aluminum chips. Polyurethane layers were imprinted with nine different bacterial targets, and dose–response curves were obtained for each SIP, clearly indicating that bacteria can be detected both qualitatively and quantitatively in concentrations as low as 1 × 104 colony-forming units (CFU) mL–1. In addition, cross-selectivity for all species was assessed and detection of Staphylococcus aureus in the presence of a 100-fold excess of eight competitor bacteria was established. In a final set of experiments, a first application in terms of infectious disease diagnosis is presented. To assess the potential of the platform for urinary tract infection (UTI), urine samples were spiked with increasing concentrations of Escherichia coli and analyzed using HTM. These measurements indicate that the sensor was able to detect bacteria in urine in physiologically relevant concentrations as low as 3 × 104 CFU mL–1.41,42
Experimental Methods
Bacterial Culturing and Sample Preparation
The following bacteria were obtained from DSM-Z (Germany) and in a certified biosafety level II laboratory: Escherichia coli (ATCC 8739), Staphylococcus aureus (ATCC 6538), Klebsiella pneumonia (ATCC 4352), Pseudomonas aeruginosa (ATCC 9027), Enterococcus cecorum (ATCC 43198), Staphylococcus epidermidis (ATCC 14990), Acinetobacter baumannii (ATCC 19606), Clostridium difficile (ATCC 9689), and Escherichia coli K12.
The bacteria were cultured using growth conditions according to ATCC. All broth and agars were made following the directions stated on the product. The microbes were plated onto an agar plate and were grown overnight at 37 °C. Nonfastidious bacteria were plated onto tryptone soy broth (TSB) agar, whereas fastidious bacteria were plated onto a brain–heart infusions agar. C. difficile was grown in 8 mL of 0.1% l-cysteine-reduced blood brain infusion broth. Afterward, the respective broths were inoculated with a single colony and allowed to grow overnight at 37 °C while shaking. Prior to imprinting, 5 mL of the overnight culture was diluted in 20 mL of the respective broth and allowed to grow at 37 °C until an OD600 of 1. One milliliter of the broth was harvested for cells by centrifugation at 6000 rpm for 1 min. The pellet was washed with sterilized PBS, and resuspended in 1 mL of PBS, resulting in an approximate cell concentration of (5–9) × 108 cells mL–1. This solution was further diluted to achieve desired concentrations for imprinting and measuring.
Preparation of Bacteria-Imprinted Polyurethane Layers
Polyurethane layers were synthesized by dissolving 122 mg of 4,4′-diisocyanatodiphenylmethane, 222 mg of bisphenol A, and 25 mg of phloroglucinol in 500 μL of anhydrous tetrahydrofuran (THF). All reagents had a purity of at least 99.9% and were used as received (Sigma-Aldrich N.V., Diegem, Belgium). The mixture was gently stirred and allowed to polymerize at 65 °C until the gelling point was reached, after which the solution was diluted with anhydrous THF in a 1:5 ratio. Aluminum chips with a mirror finish were coated with polyurethane by spin coating at 2000 rpm for 60 s.
In parallel, PDMS stamps (Sylgard 184 silicone elastomer kit, Mavom N.V., Schelle, Belgium) were covered with a solution of bacteria in phosphate-buffered saline (PBS, concentration 1 × 107 CFU mL–1). After the bacteria had sedimented toward the surface of the stamp for 3 min, the excess buffer was removed by spin coating at 3000 rpm for 60 s, leaving behind a firm monolayer of bacteria on the surface of the stamp. The stamp was gently pressed onto the polyurethane-covered chip, and the polymer was cured at 65 °C overnight under inert atmosphere. The stamp was removed, and template bacteria were washed off using ethanol and PBS, leaving behind microcavities on the surface of the stamp.
Sensor Setup and Measuring Methodology
The sensor setup is shown in Figure 1, and its use in biosensing assays has been described earlier.33,40 SIP-covered aluminum chips were pressed onto a copper block, serving as a heat provider. The temperature of the copper, T1, was monitored by a thermocouple and actively steered by a proportional–integral–derivative controller (P = 8, I = 1, D = 0). The SIP layer was exposed to buffer and cell solutions by means of a poly(methyl methacrylate) (PMMA) flow cell with an inner volume of 110 μL and sealed by an O-ring, defining a contact area of 28 mm2 between the chip and the liquid. The temperature inside the flow cell was measured by a second thermocouple.
Figure 1.
Schematic representation of the TWTA setup. The temperature control unit consists of a thermocouple, a PID controller, and a heating element allowing full control over the temperature of the heat provider, T1. The thermal wave (phase φ) is sent through the functionalized chip, and the output wave and corresponding phase shift φ′ are recorded by a second thermocouple.
At the beginning of each experiment, the system is allowed to stabilize in PBS (pH 7.4) for 60 min. Rebinding is studied by injecting 3 mL of bacterial suspension in PBS into the cell at a controlled flow rate (2.5 mL min–1). After each injection, the signal is allowed to stabilize for 0.5 h. At the end of each plateau a thermal wave is produced with an amplitude of 0.10 °C at an increasing frequency (from 0.01 to 0.05 Hz). The resulting thermal wave is transported through the SIP layer, and the output wave is recorded by a second thermocouple, monitoring the temperature inside the flow cell, T2, in time. The corresponding delay in the phase φ′ and the decrease in amplitude α′ of the wave were analyzed as a function of bacterial rebinding to the SIP layer.
Results
Quantitative Detection of Bacteria
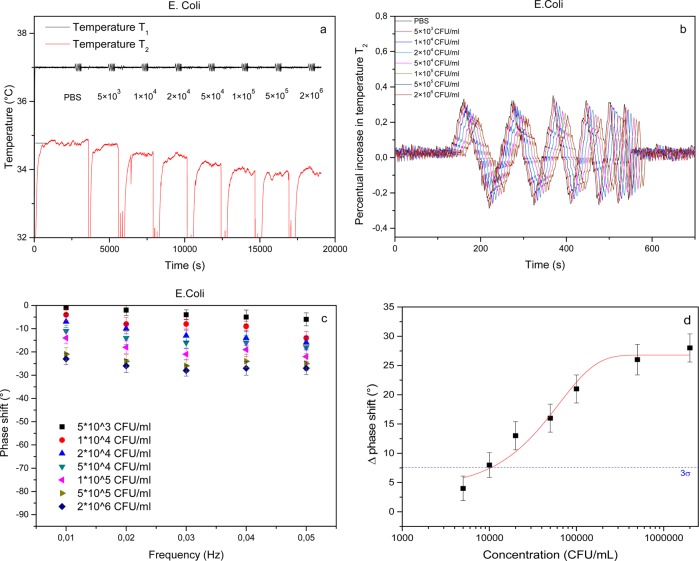
To test if it was possible to detect bacteria in a quantifiable manner using TWTA, SIPs were imprinted with E. coli cells and exposed to an increasing amount of target bacteria in PBS. On the basis of results obtained with a similar platform, a dilution series of bacteria was made with a concentration range that corresponds to the linear response regime of the sensor.39 Therefore, a stock solution with a bacterial concentration of 1 × 107 CFU mL–1 was diluted 5, 20, 100, 200, 500, 1000, and 2000 times. The SIP layer was exposed to an increasing concentration of bacteria, after which the signal was allowed to stabilize for 30 min. At the end of each stabilization plateau, a thermal wave was transmitted through the sample, and the phase and amplitude of the transmitted wave were analyzed. Between each addition step, the flow cell was consecutively flushed with ethanol and PBS to remove any bound bacteria from the SIPs and thereby regenerate the sensor surface as demonstrated in previous work.39 The results of this experiment are summarized in Figure 2.
Figure 2.
TWTA of an E. coli-SIP in response to an increasing concentration of target bacteria. The time-dependent temperature profile indicates that the temperature inside the flow cell decreases as a function of E. coli concentration (a). Analysis of the transmitted wave at all frequencies reveals a phase shift that becomes more pronounced at increasing E. coli concentrations (b). The bode plot reveals that the resolution is optimal at 0.03 Hz (c). A dose–response curve was constructed by plotting the absolute value of the phase shift as a function of the concentration on a logarithmic scale. The data were fitted with a sigmoidal fit (R2 = 0.98). The blue dashed line indicates the 3σ value that determines the limit of detection (d).
The time-dependent temperature data (Figure 2a) indicate that T2 decreases as a function of the concentration of bacteria present in the flow cell. These findings are in agreement with previous experiments and indicate that the thermal resistance of the solid–liquid interface increases due to bacteria binding to the SIP.39 Zooming in on the transmitted thermal wave (Figure 2b), it can be seen that the phase shift increases as a function of concentration at frequencies 0.01–0.04 Hz. The data reveal that no phase shift is observed at 0.05 Hz due to distortion of the temperature signal. The phase shift is calculated for each concentration and frequency and summarized in a “temperature bode-plot” (Figure 2c). From these bode-plots it can be observed that the measurement resolution appears to be optimal at 0.03 Hz with a clear phase shift observed at low concentrations that becomes more pronounced with increasing concentration of target bacteria. These results confirm the results obtained in previous research, using TWTA for dopamine detection in buffer solution and banana juice.40 The phase shift increases with increasing concentrations until it finally saturates at concentrations >5 × 105 CFU mL–1. On the basis of these results, 0.03 Hz was chosen as measuring frequency from here on. To determine the limit of detection of the methodology, these data were used to construct a dose–response curve (Figure 2d). To this extent, the absolute value of the phase shift was plotted as a function of the concentration (logarithmic) scale. The data were fitted using a standard sigmoidal dose–response function (R2 = 0.98), and the limit of detection was defined as 3 times the maximal standard error on the data (blue dashed line) at 1.01 × 104 CFU mL–1.
Similar analyses were performed for SIPs imprinted with A. baumannii, E. coli K12, K. pneumoniae, S. epidermidis, S. aureus, C. difficile, E. cecorum, and P. aeruginosa. The results of these experiments are summarized in Table 1 (detailed information can be found in the Supporting Information, section S1) and reveal a similar pattern for all bacterial species: a concentration-dependent phase shift that can be observed in the transmitted wave and saturates at higher concentrations and a limit of detection between 1 × 105 and 2 × 105 CFU mL–1.
Table 1. Concentration-Dependent TWTA Response for SIPs Imprinted with Eight Different Bacterial Speciesa.
| phase shift (deg) |
||||||||
|---|---|---|---|---|---|---|---|---|
| target | 5 × 103 CFU mL–1 | 1 × 104 CFU mL–1 | 2 × 104 CFU mL–1 | 5 × 104 CFU mL–1 | 1 × 105 CFU mL–1 | 5 × 105 CFU mL–1 | 2 × 106 CFU mL–1 | LoD (CFU mL–1) |
| A. baumannii | 2 ± 2.1 | 7 ± 2.1 | 11 ± 2.4 | 12 ± 2.3 | 18 ± 2.4 | 20 ± 2.7 | 21 ± 2.4 | 1.9 × 104 |
| E. coli K12 | 4 ± 2.1 | 7 ± 2.6 | 11 ± 2.4 | 15 ± 2.6 | 20 ± 2.4 | 25 ± 2.6 | 27 ± 2.4 | 1.4 × 104 |
| K. pneumoniae | 3 ± 2.1 | 7 ± 2.5 | 12 ± 2.5 | 13 ± 2.4 | 19 ± 2.5 | 21 ± 2.4 | 22 ± 2.4 | 1.5 × 104 |
| S. epidermidis | 4 ± 2.1 | 7 ± 2.6 | 12 ± 2.4 | 15 ± 2.4 | 21 ± 2.4 | 26 ± 2.6 | 27 ± 2.4 | 1.5 × 104 |
| S. aureus | 3 ± 2.1 | 6 ± 2.5 | 11 ± 2.5 | 15 ± 2.4 | 18 ± 2.4 | 21 ± 2.3 | 22 ± 2.4 | 1.7 × 104 |
| C. difficile | 4 ± 2.5 | 8 ± 2.5 | 13 ± 2.2 | 14 ± 2.4 | 20 ± 2.2 | 22 ± 2.8 | 23 ± 2.6 | 1.5 × 104 |
| E. cecorum | 3 ± 1.7 | 7 ± 1.7 | 12 ± 2.0 | 15 ± 2.1 | 20 ± 2.2 | 25 ± 2.5 | 27 ± 2.2 | 1.3 × 104 |
| P. aeruginosa | 4 ± 1.7 | 9 ± 1.7 | 13 ± 2.0 | 16 ± 1.9 | 21 ± 2.2 | 26 ± 2.5 | 27 ± 2.2 | 1.4 × 104 |
The absolute value of the phase shift is shown for each bacterium and each concentration. A clear correlation between phase shift and concentration can be established up to a concentration of 5 × 105 CFU mL–1, after which the signal saturates.
Cross-Selectivity Matrix
To address the selectivity of the platform, each of the SIPs was consecutively exposed to eight analogue bacteria and, finally, the target. At each exposure step a bacterial suspension in PBS (pH 7.4, concentration 1 × 107 CFU mL–1) was injected into the flow cell. Upon stabilization of the signal, the setup was flushed with buffer solution to remove any unbound material. The time-dependent temperature profile and TWTA for an E. coli SIP are shown in Figure 3. The addition of analogue cells to the flow cell leads to a decrease in T2, which can be readily reversed by flushing with buffer (Figure 3a), which corresponds to the results that were obtained previously.39 However, upon addition of E. coli K12 cells the signal does not fully return back to baseline and stabilizes at an intermediate value. Addition of target cells further decreases the signal to a minimum, after which the signal stays constant upon flushing with buffer (Figure 3a). These findings are confirmed by TWTA, indicating that the other strains do not cause a phase shift in the transmitted wave compared to the input wave, but exposure to both the analogue E. coli K12 and the target E. coli cells results in a measurable phase shift with a maximum being achieved in the latter case (Figure 3b). A similar experiment on an E. coli K12 SIP confirms these results (Figure 3c,d).
Figure 3.
Cross-selectivity experiment showing the time-dependent temperature data for an E. coli SIP exposed to eight competitor bacteria and the target consecutively (a). The TWTA at 0.03 Hz is also shown (b). Both the temperature profile and TWTA data show some degree of cross-selectivity between the two E. coli strains. The data obtained from a similar experiment with an E. coli K12 SIP are shown in panels c and d.
In addition to these experiments, SIPs were imprinted for each of the bacteria under study and exposed consecutively to target and analogue bacteria. The data, summarized in Supporting Information, S2, indicate that no cross-selectivity is observed in similar experiments using these SIPs. The results of these experiments combined with the findings from Figure 3 are summarized in Table 2.
Table 2. Cross-Selectivity Matrix for the Proposed Platform Exposing SIPs Imprinted with Nine Different Bacteria to Nine Different Target Bacteriaa.
| target |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| SIP | A. b. | K. p. | S. e. | S. a. | C. d. | E. cec. | P. a. | E. c. K12 | E. c. |
| A. baumannii | ● | / | / | / | / | / | / | / | / |
| K. pneumoniae | / | ● | / | / | / | / | / | / | / |
| S. epidermidis | / | / | ● | / | / | / | / | / | / |
| S. aureus | / | / | / | ● | / | / | / | / | / |
| C. difficile | / | / | / | / | ● | / | / | / | / |
| E. cecorum | / | / | / | / | / | ● | / | / | / |
| P. aeruginosa | / | / | / | / | / | / | ● | / | / |
| E. coli K12 | / | / | / | / | / | / | / | ● | ○ |
| E. coli | / | / | / | / | / | / | / | ○ | ● |
The results indicate that each SIP binds its target with only limited or moderate (different E. coli strains) cross-selectivity observed. ● = specific cell binding, ○ = nonspecific cell binding, / = no cell binding.
Sensor Performance in a Complex Matrix: A Model Study
The results described in previous sections indicate that the sensor platform is able to selectively discriminate between various types of bacteria in buffer in a quantitative manner. However, the performance of the setup was assessed only in solutions containing either analogue or competitor bacteria. When aiming at on-site bacterial detection and identification, one can imagine that a real-life sample would contain an excess of competitor molecules and cells in addition to a trace amount of the target. Therefore, a S. aureus SIP was chosen for a progressive enrichment experiment exposing the SIP to a mixture of bacteria containing S. aureus in the presence of an excess of eight nontarget bacteria (1:99 ratio with a total concentration of 1 × 107 CFU mL–1). The SIP was exposed to the mixture five consecutive times; flushing with buffer between each of the exposure events removed any unbound bacteria from the receptor layer.
The time-dependent temperature profile is shown in Figure 4a. To validate TWTA as a measuring technique for SIP-based bacteria detection, HTM was used as a reference technique. To this extent, the thermal resistance (Rth) was calculated from the temperature profile according to the formula
with T1 being the temperature of the copper block underneath the sample (black curve in Figure 4a), T2 the transmitted temperature measured inside the flow cell (red curve in Figure 4a), and P the power over the adjustable heat source. The thermal resistance data are shown as the black curve in Figure 4b with the red line corresponding to a median filter that was applied as a guide to the eye. To visualize the effect of progressive enrichment more clearly, the saturation level of the Rth response was calculated for each exposure cycle (consisting of cell exposure followed by flushing) by dividing the net effect size after flushing by the maximal effect size upon addition of the mixture to the flow cell. These results indicate that the net signal gradually increases with each exposure cycle until the limit of detection is reached after four to five exposure runs. The limit of detection value is indicated as the dashed line in Figure 4b and is defined as 3 times the maximal error on the Rth signal throughout the measurement (the 3σ method). The TWTA data show a similar trend. The net phase shift observed in the transmitted wave after each exposure cycle increases gradually, and after the third exposure cycle, the signal reaches the limit of detection.
Figure 4.
Sensor performance exposing a S. aureus SIP to a mixed cell solution containing a trace amount of target cells in the presence of an excess of nontarget bacterial strains. The time-dependent temperature profile (a) is shown, in addition to an HTM analysis (b) and the phase shift response at 0.03 Hz obtained from a TWTA experiment (c). In all three cases, the signal shows a maximum effect upon exposure of the SIP to the mixture, after which the signal falls back upon flushing. The net effect size will increase with each exposure cycle and eventually reaches the limit of detection (dashed line).
Proof of Application: Detection of Bacteria in Urine
The model study presented in the previous section demonstrates that the sensor would be able to detect bacteria in relatively low concentrations despite the presence of other bacteria in the same sample, illustrating both the selectivity of the SIP and the sensitivity of the TWTA transducing principle. However, the presence of a wider variety of proteins and chemical compounds present in patient samples could severely interfere with the noise on the signal and the rebinding of bacteria to the SIP layer. To further examine this problem and to illustrate a possible application for the sensor in terms of infectious disease diagnosis, clean urine samples of a healthy individual were obtained and spiked with increasing concentrations of E. coli in an analogous manner to the dose–response experiment described earlier. Prior to injecting the spiked urine samples, the signal was left to stabilize for 60 min in clean urine. After each addition, the signal was allowed to stabilize and the temperature profile and phase shifts were recorded. Between each addition the sensor was regenerated by rinsing with 70% ethanol and clean urine. The results of this experiment are shown in Figure 5.
Figure 5.
TWTA of an E. coli-SIP in response to urine samples spiked with an increasing concentration of target bacteria. The time-dependent temperature profile indicates that the temperature inside the flow cell decreases as a function of E. coli concentration (a). Analysis of the transmitted wave at all frequencies reveals a phase shift that becomes more pronounced at increasing E. coli concentrations (b). The bode plot reveals that the resolution is optimal at 0.03 Hz (c). A dose–response curve was constructed by plotting the absolute value of the phase shift as a function of the concentration on a logarithmic scale. The data were fitted with a sigmoidal fit (R2 = 0.98). The blue dashed line indicates the 3σ value that determines the limit of detection (d).
The results in Figure 5 illustrate that the temperature inside the flow cell decreases (Figure 5a) and the phase shift of the transmitted wave (Figure 5b) increases as a function of an increasing concentration of E. coli in a manner similar to the experiment performed in buffer. However, at low concentrations the temperature drop and phase shift are less pronounced, whereas the signals already saturate at concentrations >1 × 105 CFU mL–1. The dose–response curve can also be fitted using a sigmoidal dose–response function, but due to a higher amount of noise on the sample the limit of detection in urine is only 3 × 104 CFU mL–1.
Discussion
The results shown in Figure 2a confirm that binding of bacteria to the SIP layer will lead to an increase in thermal resistance. Bacteria will partially block the heat flow through the SIP layer, which results in a decreased temperature inside the flow cell, which cannot be reversed by flushing with buffer. These findings are in accordance with the previously demonstrated concept of HTM-based bacterial detection. However, the drawback associated with HTM is the high amount of noise on the thermal resistance signal, which limits the sensitivity of the proposed platform.35−39 Therefore, the authors used a novel detection technique based on the propagation of a thermal wave, the so-called thermal wave transport analysis or TWTA principle. The data in Figure 2b illustrate that a thermal wave will experience a delay when propagated through the sample. This observed phase shift will increase as a function of the concentration of bacteria present in the measuring chamber. This might be explained by the fact that the increase in thermal resistance, previously observed by HTM,39 will also result in a delayed propagation of the thermal wave through the sample, which can be registered by TWTA. The results summarized in Figure 2c,d were used to determine the LoD of the methodology at 1.01 × 104 CFU mL–1. These findings imply that TWTA is advantageous over HTM as sensitivity is improved by at least a factor of 2 in comparison to the previously reported LoD for a HTM-based bacterial detection platform.39,43 The data summarized in Table 1 show that the technique can be used to detect at least nine different bacterial strains in a quantitative manner. Furthermore, the absolute concentration-dependent response is similar for all species and strains under study, which could be beneficial for calibrating the sensor in the future.
The experiments summarized in Figure 3 and Table 2 show that the proposed platform is able to discriminate between different bacterial strains in a remarkably selective manner. However, a moderate degree of selectivity is apparent in Figure 3 when trying to discriminate between two different E. coli strains. In this case, the analogue cells will bind to the microcavities on the SIP surface and cannot be fully washed out by rinsing the layer with buffer, as evidenced by the intermediate phase shift observed upon an exposure cycle with the analogue strain. In previous work, it was demonstrated that SIP-based recognition in polyurethane is influenced by the presence or absence of certain sugar moieties on transmembrane proteins.36 This might explain the observed cross-selectivity between both strains as their protein expression patterns will be more similar in comparison to that of different bacterial species. Although it is possible to distinguish between both E. coli strains in this experiment in buffer, the observed cross-selectivity might be troublesome when trying to selectively screen for one specific bacteria strain in a complex matrix or a real-life sample and would require optimization of the synthesis and imprinting protocol. On the other hand, the selectivity limit can also be beneficial for routine screening of environmental samples as it implies that it is not necessary to synthesize a SIP for each individual strain of the same bacteria.
The model study in Figure 4 illustrates the sensor’s performance in a complex matrix. Due to the excess of competitor bacteria in the mixture, only a small amount of target bacteria can bind to the SIP. Therefore, both the response in thermal resistance and phase shift are less pronounced. As the number of exposure events increases, the response gradually increases and eventually reaches the limit of detection level. As the noise on the signal for HTM is significantly higher, the LoD is reached only after four or even five consecutive cycles, whereas a measurable signal that can be regarded as significant is already apparent after two to three cycles when using TWTA as a measurement technique. These findings indicate that due to the low amount of noise on the thermal wave, the development of the TWTA principle can be considered as a valuable advance in thermal bacterial identification.
Although the sensor’s performance in a bacterial mixture indicates that the sensor is able to identify bacteria in the presence of an excess of competitor species, natural samples are often more complex than buffer in terms of chemical composition. Bacterial identification in spiked urine samples appears to be more difficult when the results in Figure 5 are analyzed. The sensitivity of the device in urine is about 3 times lower and the signal saturates more rapidly, narrowing the dynamic range of the device. However, these results indicate that the platform can be used for, for example, a fast, on-site diagnosis for urinary tract infection. To date, a definitive diagnosis for UTI usually requires a positive urine culture. As urine is an excellent culture medium for most aerobic microorganisms, ≥105 CFU mL–1 for most infecting bacterial species should be isolated from the urine for most UTI patients.41,42
Conclusion
The sensor platform that was developed within this study was shown to be able to detect up to nine different bacterial strains in buffer with a remarkable degree of selectivity. The experiments performed in this work do not merely describe a follow-up study of a concept that has been developed recently but also introduce a novel sensing principle, the so-called thermal transport analysis method or TWTA. The results obtained in the sensitivity experiment clearly demonstrate that TWTA is advantageous over the previously developed sensing technique in terms of sensitivity and response time. In addition, a first proof of application is provided, illustrating the sensor’s performance in urine samples. Future research opportunities will be focused on further optimizing this novel readout technique by, for example, tuning the parameters controlling the transmitted wave, redesigning the flow cell, and electronic noise reduction to achieve a higher degree of sensitivity and to further increase the response time.
Acknowledgments
This work was financed by the Maastricht Science Programme of Maastricht University, The Edmund Hustinx Foundation, and the C1 project “Smart Cellular Scaffolds” from KU Leuven (C14/15/067). Technical support by I. Muijrers-Chen, W. Stilman, and J. Dassen was greatly appreciated.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.7b00037.
Additional data on bacterial sensing (PDF)
Author Contributions
E.S.R. and K.E. contributed equally to this work. The thermal wave transport device was designed by B.v.G. and T.J.C. TWTA measurements were performed by E.S.R., B.v.G., B.F., and SD. All cell-imprinted polymer layers were synthesized by O.A., J.R., K.N., K.E., and E.S.R., and input on the synthesis and template removal protocol was provided by M.P., H.D., and T.J.C. Data processing and interpretation were done by B.v.G., K.E., E.S.R., and T.J.C. P.C. developed and optimized the readout software and data processing unit. Input on possible medical/biotechnological applications and assistance regarding microbial issues were provided by K.E., E.S.R., and P.W. The manuscript was jointly written by K.E., B.v.G., and E.S.R. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Healthcare-Associated Infections (HAI) Progress Report; Centers for Disease Control and Prevention, 2016.
- Zimlichman E.; Henderson D.; Tamir O.; Franz C.; Song P.; Yamin C. K.; Keohane C.; Denham R.; Bates D. W. (2013) Health Care-Associated Infections: A Meta-analysis of Costs and Financial Impact on the US Health Care System. JAMA Int. Med. 173, 2039–2046. 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- Burden of Disease and Cost-Effectiveness Estimates; World Health Organization, 2014.
- Fournier P.; Drancourt M.; Colson P.; Rolain J.; Scola B. L.; Raoult D. (2013) Modern Clinical Microbiology: New Challenges and Solutions. Nat. Rev. Microbiol. 11, 574–585. 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A.; Law R. J.; Scholz R.; Keeney K. M.; Wlodarska M.; Finlay B. B. (2013) Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham C. A.; Carroll K. C. (2013) Diagnosis of Clostridium difficile Infection: An Ongoing Conundrum for Clinicians and for Clinical Laboratories. Clin. Microbiol. Rev. 26, 604–630. 10.1128/CMR.00016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espy M. J.; Uhl J. R.; Sloan L. M.; Buckwalter S. P.; Jones M. F.; Vetter E. A.; Yao J. D.; Wengenack N. L.; Rosenblatt J. E.; Cockerill F. R. III; Smith T. F. (2006) Real-Time PCR in Clinical Microbiology: Applications for Routine Laboratory Testing. Clin. Microbiol. Rev. 19, 165–256. 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.; Schwartz-Mittelmann A.; Biran D.; Ron E. Z.; Rishpon J. (2003) Combined Phage Typing and Amperometric Detection of Released Enzymatic Activity for the Specific Identification and Quantification of Bacteria. Anal. Chem. 75, 580–585. 10.1021/ac026083e. [DOI] [PubMed] [Google Scholar]
- Boehm D. A.; Gottlieb P. A.; Hua S. Z. (2007) On-Chip Microfluidic Biosensor for Bacterial Detection and Identification. Sens. Actuators, B 126, 508–514. 10.1016/j.snb.2007.03.043. [DOI] [Google Scholar]
- Maalouf R.; Fournier-Wirth C.; Coste J.; Chebib H.; Saikali Y.; Vittori O.; Errachid A.; Cloarec J. P.; Martelet C.; Jaffrezic-Renault N. (2007) Label-Free Detection of Bacteria by Electrochemical Impedance Spectroscopy: Comparison to Surface Plasmon Resonance. Anal. Chem. 79, 4879–4886. 10.1021/ac070085n. [DOI] [PubMed] [Google Scholar]
- de la Rica R.; Baldi A.; Fernandez-Sanchez C.; Matsui H. (2009) Selective Detection of Live Pathogens via Surface-Confined Field Perturbations on Interdigitated Silicon Transducers. Anal. Chem. 81, 3830–3835. 10.1021/ac9001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohk S. H.; Koo O. K.; Sen T.; Yamamoto C. M.; Bhunia A. K. (2010) Antibody-Aptamer Functionalized Fibre-Optic Biosensor for Specific Detection of Listeria monocytogenes from Food. J. Appl. Microbiol. 109, 808–817. 10.1111/j.1365-2672.2010.04709.x. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Knoll W.; Dostalek J. (2012) Bacterial Pathogen Surface Plasmon Resonance Biosensor Advanced By Long Range Surface Plasmons and Magnetic Nanoparticle Assays. Anal. Chem. 84, 8345–8350. 10.1021/ac301904x. [DOI] [PubMed] [Google Scholar]
- Tripathi S. M.; Bock W. J.; Mikulic P.; Chinnappan R.; Ng A.; Tolba M.; Zourob M. (2012) Long Period Grating Based Biosensor for the Detection of Escherichia coli Bacteria. Biosens. Bioelectron. 35, 308–312. 10.1016/j.bios.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Hao R.; Wang D.; Zuo G.; Wei H.; Yang R.; Zhang Z.; Cheng Z.; Guo Y.; Cui Z.; Zhou Y.; Zhang X. (2009) Rapid Detection of Bacillus anthracis Using Monoclonal Antibody Functionalized QCM Sensor. Biosens. Bioelectron. 24, 1330–1335. 10.1016/j.bios.2008.07.071. [DOI] [PubMed] [Google Scholar]
- Jiang X.; Wang R.; Wang Y.; Su X.; Ying Y.; Wang J.; Li Y. (2011) Evaluation of Different Micro/Nanobeads Used as Amplifiers in QCM Immunosensor for More Sensitive Detection of E. coli O157:H7N. Biosens. Bioelectron. 29, 23–28. 10.1016/j.bios.2011.07.059. [DOI] [PubMed] [Google Scholar]
- Salam F.; Uludag Y.; Tothill I. E. (2013) Real-Time and Sensitive Detection of Salmonella Typhimurium Using an Automated Quartz Crystal Microbalance (QCM) Instrument with Nanoparticles Amplification. Talanta 115, 761–767. 10.1016/j.talanta.2013.06.034. [DOI] [PubMed] [Google Scholar]
- Sapsford K. E.; Bradburne C.; Delehanty J. B.; Medintz I. L. (2008) Sensors for Detecting Biological Agents. Mater. Today 11, 38–49. 10.1016/S1369-7021(08)70018-X. [DOI] [Google Scholar]
- Kryscio D. R.; Peppas N. A. (2012) Critical Review and Perspective of Macromolecularly Imprinted Polymers. Acta Biomater. 8, 461–473. 10.1016/j.actbio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Alexander C.; Vulfson E. V. (1997) Spatially Functionalized Polymer Surfaces Produced via Cell-Mediated Lithography. Adv. Mater. 9 (9), 751–755. 10.1002/adma.19970090916. [DOI] [Google Scholar]
- Dickert F. L.; Hayden O.; Halikias K. P. (2001) Synthetic Receptors as Sensor Coating for Molecules and Living Cells. Analyst 126 (6), 766–771. 10.1039/b009893k. [DOI] [PubMed] [Google Scholar]
- Hayden O.; Dickert F. L. (2001) Selective Microorganism Detection with Cell Surface Imprinted Polymers. Adv. Mater. 13, 1480–1483. . [DOI] [Google Scholar]
- Tokonami S.; Nakadoi Y.; Takahashi M.; Ikemizu M.; Kadoma T.; Saimatsu K.; Dung L. Q.; Shiigi H.; Nagaoka T. (2013) Label-Free and Selective Bacteria Detection Using a Film with Transferred Bacterial Configuration. Anal. Chem. 85, 4925–4929. 10.1021/ac3034618. [DOI] [PubMed] [Google Scholar]
- Findeisen A.; Wackerlig J.; Samardzic R.; Pitkänen J.; Anttalainen O.; Dickert F. L.; Lieberzeit P. A. (2012) Artificial Receptor Layers for Detecting Chemical and Biological Agent Mimics. Sens. Actuators, B 170, 196–200. 10.1016/j.snb.2011.08.025. [DOI] [Google Scholar]
- Schirhagl R.; Qian J.; Dickert F. L. (2012) Immunosensing with Artificial Antibodies in Organic Solvents or Complex Matrices. Sens. Actuators, B 173, 585–590. 10.1016/j.snb.2012.07.036. [DOI] [Google Scholar]
- Zhang Z.; Guan Y.; Li M.; Zhao A.; Ren J.; Qu X. (2015) Highly Stable and Reusable Imprinted Artificial Antibody Used for in Situ Detection and Disinfection of Pathogens. Chem. Sci. 6, 2822–2826. 10.1039/C5SC00489F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T.; Starosvetsky J.; Cheruti U.; Armon R. (2010) Whole Cell Imprinting in Sol-Gel Thin Films for Bacterial Recognition in Liquids: Macromolecular Fingerprinting. Int. J. Mol. Sci. 11 (4), 1236–1252. 10.3390/ijms11041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altintas Z.; Gittens M.; Guerreiro A.; Thompson K.; Walker J.; Piletsky S.; Tothill I. E. (2015) Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers. Anal. Chem. 87 (13), 6801–6807. 10.1021/acs.analchem.5b00989. [DOI] [PubMed] [Google Scholar]
- Altintas Z.; Pocock J.; Thompson K.; Tothill I. E. (2015) Comparative Investigations for Adenovirus Recognition and Quantification: Plastic or Natural Antibodies?. Biosens. Bioelectron. 74, 996–1004. 10.1016/j.bios.2015.07.076. [DOI] [PubMed] [Google Scholar]
- Qi P.; Wan Y.; Zhang D. (2013) Impedimetric Biosensor Based on Cell-Mediated Bioimprinted Films for Bacterial Detection. Biosens. Bioelectron. 39, 282–288. 10.1016/j.bios.2012.07.078. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang Z.; Jain J.; Yi J.; Mueller S.; Sokolov J.; Liu Z.; Levon K.; Rigas B.; Rafailovich M. H. (2010) Potentiometric Sensors based on Surface Molecular Imprinting: Detection of Cancer Biomarkers and Viruses. Sens. Actuators, B 146 (1), 381–387. 10.1016/j.snb.2010.02.032. [DOI] [Google Scholar]
- Cai D.; Ren L.; Zhao H.; Xu C.; Zhang L.; Ying Y.; Wang H.; Lan Y.; Roberts M. F.; Chuang J. H.; Naughton M. J.; Ren Z.; Chiles T. C. (2010) A Molecular-Imprint Nanosensor for Ultrasensitive Detection of Proteins. Nat. Nanotechnol. 5, 597–601. 10.1038/nnano.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grinsven B.; Vanden Bon N.; Strauven H.; Grieten L.; Murib M. S.; Jimenez Monroy K. L.; Janssens S. D.; Haenen K.; Schöning M. J.; Vermeeren V.; Ameloot M.; Michiels L.; Thoelen R.; De Ceuninck W.; Wagner P. (2012) Heat-Transfer Resistance at Solid-Liquid Interfaces: A Tool for the Detection of Single-Nucleotide Polymorphisms in DNA. ACS Nano 6, 2712–2721. 10.1021/nn300147e. [DOI] [PubMed] [Google Scholar]
- van Grinsven B.; Eersels K.; Peeters M.; Losada-Pérez P.; Vandenryt T.; Cleij T. J.; Wagner P. (2014) The Heat-Transfer Method: A Versatile Low-Cost, Label-Free, Fast and User-Friendly Readout Platform for Biosensor Applications. ACS Appl. Mater. Interfaces 6, 13309–13318. 10.1021/am503667s. [DOI] [PubMed] [Google Scholar]
- Eersels K.; van Grinsven B.; Ethirajan A.; Timmermans S.; Jiménez Monroy K. L.; Bogier J. F. J.; Punniyakoti S.; Vandenryt T.; Hendriks J. J. A.; Cleij T. J.; Daemen M. J. A. P.; Somers V.; De Ceuninck W.; Wagner P. (2013) Selective Identification of Macrophages and Cancer Cells Based on Thermal Transport through Surface-Imprinted Polymer Layers. ACS Appl. Mater. Interfaces 5, 7258–7267. 10.1021/am401605d. [DOI] [PubMed] [Google Scholar]
- Bers K.; Eersels K.; van Grinsven B.; Daemen M.; Bogie J. F.; Hendriks J. J.; Bouwmans E. E.; Püttmann C.; Stein C.; Barth S.; Bos G. M.; Germeraad W. T.; De Ceuninck W.; Wagner P. (2014) Heat-Transfer Resistance Measurement Method (HTM)-Based Cell Detection at Trace Levels Using a Progressive Enrichment Approach with Highly Selective Cell-Binding Surface Imprints. Langmuir 30, 3631–3639. 10.1021/la5001232. [DOI] [PubMed] [Google Scholar]
- Eersels K.; van Grinsven B.; Khorshid M.; Somers V.; Püttmann C.; Stein C.; Barth S.; Diliën H.; Bos G. M. J.; Germeraad W. T. V.; Cleij T. J.; Thoelen R.; De Ceuninck W.; Wagner P. (2015) Heat-Transfer-Method-Based Cell Culture Quality Assay through Cell Detection by Surface Imprinted Polymers. Langmuir 31, 2043–2050. 10.1021/la5046173. [DOI] [PubMed] [Google Scholar]
- Eersels K.; van Grinsven B.; Vandenryt T.; Jiménez-Monroy K. L.; Peeters M.; Somers V.; Püttmann C.; Stein C.; Barth S.; Bos G. M. J.; Germeraad W. T. V.; Diliën H.; Cleij T. J.; Thoelen R.; De Ceuninck W.; Wagner P. (2015) Improving the Sensitivity of the Heat-Transfer Method (HTM) for Cancer Cell Detection with Optimized Sensor Chips. Phys. Status Solidi A 212, 1320–1326. 10.1002/pssa.201431709. [DOI] [Google Scholar]
- van Grinsven B.; Eersels K.; Akkermans O.; Ellermann S.; Kordek A.; Peeters M.; Diliën H.; Steen Redeker E.; Wagner P.; Cleij T. (2016) Label-Free Detection of Escherichia coli Based on Thermal Transport Through Surface Imprinted Polymers. ACS Sensors 1, 1140–1147. 10.1021/acssensors.6b00435. [DOI] [Google Scholar]
- Peeters M. M.; van Grinsven B.; Foster C. W.; Cleij T. J.; Banks C. E. (2016) Introducing Thermal Wave Transport Analysis (TWTA): A Thermal Technique for Dopamine Detection by Screen-Printed Electrodes Functionalized with Molecularly Imprinted Polymer (MIP) Particles. Molecules 21, 552. 10.3390/molecules21050552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolle L. E. (2013) Urinary Tract Infection. Crit. Care Clin. 29, 699–715. 10.1016/j.ccc.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Kelley S. O. (2017) What Are Clinically Relevant Levels of Cellular and Biomolecular Analytes?. ACS Sensors 2, 193–197. 10.1021/acssensors.6b00691. [DOI] [PubMed] [Google Scholar]
- Stilman W.; Jooken S.; Wackers G.; Cornelis P.; Khorshid M.; Yongabi D.; Akkermans O.; Dyson S.; van Grinsven B.; Cleij T.; van Ijzendoorn L.; Wagner P.; Eersels K. (2017) Optimization and Characterization of a Flow Cell for Heat-Transfer-Based Biosensing. Phys. Status Solidi A 1600758. 10.1002/pssa.201600758. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.