Abstract
Background
The ubiquitin protein E3A ligase gene (UBE3A) gene is imprinted with maternal-specific expression in neurons and biallelically expressed in all other cell types. Both loss-of-function and gain-of-function mutations affecting the dosage of UBE3A are associated with several neurodevelopmental syndromes and psychological conditions, suggesting that UBE3A is dosage-sensitive in the brain. The observation that loss of imprinting increases the dosage of UBE3A in brain further suggests that inactivation of the paternal UBE3A allele evolved as a dosage-regulating mechanism. To test this hypothesis, we examined UBE3A transcript and protein levels among cells, tissues, and species with different imprinting states of UBE3A.
Results
Overall, we found no correlation between the imprinting status and dosage of UBE3A. Importantly, we found that maternal Ube3a protein levels increase in step with decreasing paternal Ube3a protein levels during neurogenesis in mouse, fully compensating for loss of expression of the paternal Ube3a allele in neurons.
Conclusions
Based on our findings, we propose that imprinting of UBE3A does not function to reduce the dosage of UBE3A in neurons but rather to regulate some other, as yet unknown, aspect of gene expression or protein function.
Electronic supplementary material
The online version of this article (doi:10.1186/s13072-017-0134-4) contains supplementary material, which is available to authorized users.
Keywords: Ube3a, Genomic imprinting, Dosage compensation, Angelman syndrome, Ube3a antisense, Evolution
Background
Genomic imprinting is a rare epigenetic phenomenon that leads to the differential expression of paternally and maternally derived alleles of a gene in a parent-of-origin dependent manner [50, 54]. It has been documented only in therian mammals and flowering plants and only at a few loci in mammals, of which fewer than half are imprinted in both mouse and human [1, 4, 8, 39, 42, 50, 52]. Several theories have been proposed to explain the evolution and function of genomic imprinting (e.g., dosage regulation, complementation, parental conflict/kinship, host defense, maternal time-bomb, and developmental plasticity models), but there is currently no unifying theory that can explain the conservation of genomic imprinting across taxa [2, 17–19, 35, 50, 51, 58, 59]. Thus, apart from downregulating or silencing the expression of one allele, the functional significance of imprinting is largely unknown. Nevertheless, imprinting is believed to be an important regulatory mechanism, inasmuch as almost all recognized imprinting abnormalities are associated with pathological states [8, 49].
The ubiquitin protein E3A ligase gene (UBE3A) is located at the distal end of a cluster of imprinted genes on human chromosome 15q11-q13 and a homologous region on mouse chromosome 7C. In neurons of the central nervous system (CNS), UBE3A is imprinted with maternal-allelic expression, whereas in all other cell types, it is expressed from both alleles [11, 43, 62]. Imprinting of UBE3A is regulated by expression of the paternally expressed UBE3A antisense transcript (UBE3A-AS), which comprises the 3′ end of a long polycistronic transcription unit (PTU) containing multiple clusters of C/D box small nucleolar RNAs (snoRNA) and the SNRPN-SNURF bicistronic transcript [7, 29, 30, 44]. In both mouse and human, expression of Ube3a-AS/UBE3A-AS in cis is both necessary and sufficient to silence expression of the paternal Ube3a/UBE3A allele [ [31], [33] ], which, at least in mouse, appears to occur by inhibiting transcriptional elongation, giving rise to a paternally expressed, 5′-truncated transcript of unknown function [33, 38].
The UBE3A gene is highly conserved among vertebrate and invertebrate species [6, 9, 21, 22, 61]; however, imprinting of UBE3A is believed to have evolved in the common ancestor of eutherian mammals after divergence from the metatherian (marsupials) lineage, as studies to date show that UBE3A is biallelically expressed in tammar wallaby, platypus, chicken, and fruit-fly brain, and that there is no orthologous imprinted region detectable in marsupials or monotreme (prototherian) mammals [9, 21, 41], suggesting that imprinting of UBE3A in neurons is somehow advantageous to biallelic expression in eutherian species. The snoRNAs located in 15q11-q13, which serve as the precursors of Ube3a-AS/UBE3A-AS, are also eutherian-specific and appear to have rapidly evolved in a lineage-specific manner [64]; however, the relationship, if any, between these transcripts or the evolution of this region and imprinting of Ube3a/UBE3A is currently unknown.
Mutations or epimutations affecting the expression or function of UBE3A are associated with several neurodevelopmental disorders and psychological conditions. Loss-of-function or loss-of-expression of the maternally inherited UBE3A allele causes Angelman syndrome (AS), which presents with intellectual disability, ataxia, epilepsy, sleep disorders, and an atypical ‘happy’ disposition [26, 32, 60]. Conversely, maternally derived duplications of 15q11-q13 cause dup15q syndrome—a neurodevelopmental disorder distinctly different from AS—involving intellectual disability, ataxia, speech impairment, epilepsy, and autism spectrum disorder (ASD) [16, 20, 36, 40, 47]. Although dup15q syndrome is a contiguous gene disorder, overexpression of UBE3A is believed to be the principal pathological mechanism underlying the syndrome, as UBE3A is the only maternally expressed gene located within the duplication and as the severity of the condition correlates with the number of copies and expression levels of UBE3A [47]. There are also reports of UBE3A gain-of-function mutations (e.g., biochemical and genetic) in individuals with ASD and other psychological conditions [37, 63]. Paternally inherited deletions of 15q11-q13, namely those involving the C/D box snoRNA SNORD116, result in Prader–Willi syndrome (PWS), which is characterized by dysregulated hunger and satiety patterns, abnormal thermoregulation, sleep disorders, and behavioral issues [45].
The notion that imprinting of UBE3A evolved as a dosage-regulating mechanism stems from the role of UBE3A in AS and dup15q syndromes and from observations showing that loss of Ube3a-AS reactivates paternal Ube3a expression, leading to an increase in Ube3a protein levels in the brain [5, 7, 13, 34]. In the present study, we compared UBE3A expression levels (RNA transcript and protein) between cells, tissues, and species with different imprinting states of UBE3A. We also examined parental Ube3a protein levels during the acquisition of the imprint in neurons. Overall, our findings show that the dosage of Ube3a/UBE3A is relatively constant regardless of its imprinting status, suggesting that imprinting does not function to regulate the dosage of Ube3a/UBE3A in neurons.
Results
Ube3a/UBE3A is highly expressed from the maternal allele in the CNS
To determine whether imprinting of Ube3a in neurons of the mouse CNS reduces the dosage of Ube3a relative to other tissues where Ube3a is biallelically expressed (non-CNS), we compared the steady-state levels of Ube3a RNA (hereafter referred to as transcript) and Ube3a protein among tissues in adult wild-type mice. To estimate the relative expression levels of the maternal Ube3a allele, we also examined mice with a paternally inherited mutation in the Ube3a gene [23]. For both Ube3a m+/p+ and Ube3a m+/p− mice, Ube3a transcript levels were significantly higher in CNS (cortex and hippocampus) than in non-CNS (heart, kidney, liver, and lung) tissues [Ube3a m+/p+: F(1, 15.8) = 55.6, p < 0.0001; Ube3a m+/p−: F(1, 14) = 338.6, p < 0.0001 (Fig. 1a)]. Likewise, Ube3a protein levels in both Ube3a m+/p+ and Ube3a m+/p− mice were significantly higher in CNS (cerebellum, cortex, hippocampus) than in non-CNS (heart, liver, and lung) tissues [Ube3a m+/p+: F(1, 19) = 22.8, p < 0.0001; Ube3a m+/p−: F(1, 14) = 118.5, p < 0.001 (Fig. 1b)]. Neither Ube3a RNA or Ube3a protein levels were significantly different in the CNS of Ube3a m+/p+ and Ube3a m+/p− mice (RNA: t = 0.9, p = 0.8; protein: CNS, t = 2.1, p = 0.2), indicating that the relatively high level of Ube3a expression in mouse CNS is primarily attributable to expression of the maternal allele.
Fig. 1.
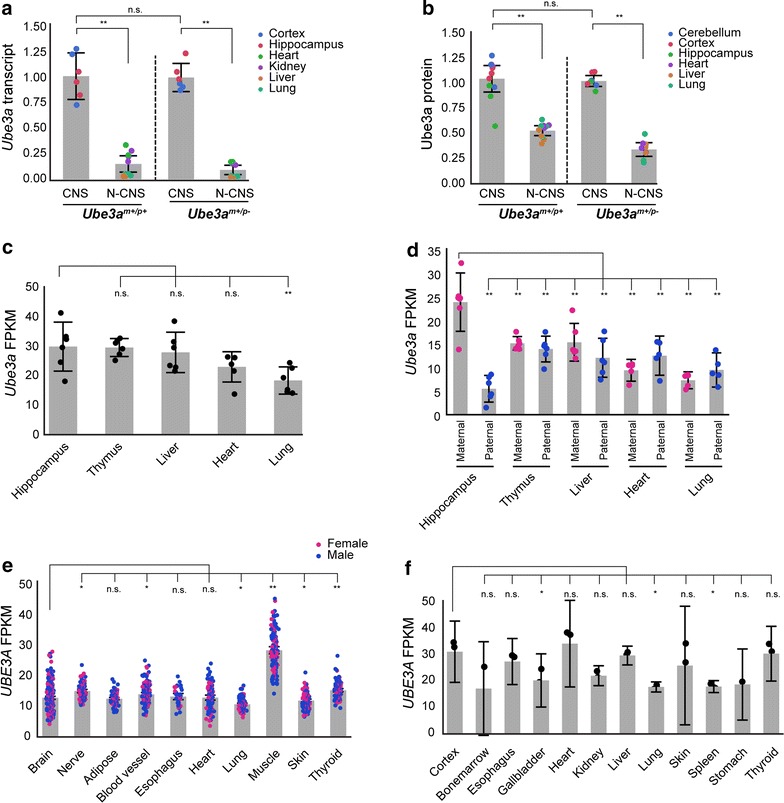
Ube3a/UBE3A is highly expressed in mouse and human CNS tissues. a Ube3a transcript levels in adult Ube3a m+/p+ (n = 4) and Ube3a m+/p− (n = 3) CNS (cortex, hippocampus) and non-CNS (N-CNS; heart, kidney, liver, and lung) tissues. Levels are shown as the ratio of expression in tissues relative to heart. b Ube3a protein levels in adult Ube3a m+/p+ (n = 4) and Ube3a m+/p− (n = 3) CNS (cerebellum, cortex, hippocampus) and non-CNS (heart, liver, and lung). Levels are shown as the ratio of expression in tissues relative to heart. c Normalized FPKM values of total Ube3a transcripts and d maternal and paternal Ube3a transcripts in B6D2F1 mouse hippocampus, thymus, liver, lung, and heart (n = 6). e, f Normalized FPKM values of UBE3A transcripts in adult human tissues (GTEx: n ≥ 30; Fagerberg et al.: n = 3). Abbreviations n.s., not significant; *p < 0.05; **p < 0.001. Individual data points provided with mean (gray bar chart) and 95% confidence intervals
To confirm that the maternal Ube3a allele is highly expressed in the CNS, we used RNA-seq data to compare Ube3a transcript levels expressed from each parental allele within and among CNS (hippocampus) and non-CNS (heart, liver, lung, and thymus) tissues in adult mice [25]. Total Ube3a transcript levels (i.e., those expressed by both maternal and paternal alleles) were significantly different among the tissues [F(4, 41.8) = 8.3, p < 0.0001], with slightly higher, albeit not significant, Ube3a transcript levels in hippocampus compared to those in liver (t = 0.8, p = 0.9) and thymus (t = 0.1, p = 0.9) and significantly higher transcript levels than those in heart (t = 2.9, p = 0.02) and lung [t = 4.7, p < 0.0001 (Fig. 1c)]. Importantly, we found that maternal Ube3a transcript levels in hippocampus were significantly higher than those expressed from either allele in all the other tissues, whereas paternal Ube3a transcript levels in hippocampus were lower than those expressed from either allele in all the other tissues, with significantly lower levels relative to hippocampus, thymus, liver, and heart (paternal allele) (Fig. 1d and Additional file 1), confirming that the relatively high levels of Ube3a expression in the CNS arise from the maternal allele.
To compare UBE3A expression levels between CNS and non-CNS tissues in human, we analyzed UBE3A transcript levels in 10 tissues (n ≥ 30) using RNA-seq data generated by the GTEx consortium [10] and in 12 tissues (n = 3) using RNA-seq data generated by Fagerberg et al. [12]. In the GTEx data set, UBE3A transcript levels were significantly different among the tissues [F(9, 1129.8) = 262.5, p < 0.0001], with no significant effect of sex [F(1, 186.6) = 0.2, p = 0.6] or significant interaction between tissue and sex [F(9, 1129.8] = 0.8, p = 0.6). Relative to brain, UBE3A transcript levels were similar to those in heart, adipose tissue, and esophagus, significantly higher than those in lung and skin, but significantly lower than those in blood vessel, muscle, thyroid, and nerve (tibial) (Fig. 1e and Additional file 2). In the Fagerberg data set, UBE3A transcript levels were also significantly different among the tissues [F(11, 14) = 7.5, p < 0.0004]. Relative to cortex, UBE3A transcript levels were similar to those in bone marrow, esophagus, heart, kidney, liver, skin, and thyroid, and significantly higher than those in gallbladder, lung, spleen, and stomach (Fig. 1f and Additional file 3).
Taken together, these findings show that in both mouse and human cells, expression levels of Ube3a/UBE3A in CNS are generally equal to or higher than those in non-CNS tissues, despite the different imprinting states.
Maternal Ube3a compensates for loss of paternal Ube3a expression during neurogenesis
Our findings that the maternal Ube3a allele is highly expressed in brain prompted us to examine the expression of each parental Ube3a allele during the acquisition of the imprint in neurons. Using the Ube3a YFP reporter mouse model [11], we compared paternal Ube3aYFP (Ube3a m+/pYFP) and maternal Ube3aYFP (Ube3a mYFP/p+) protein levels in neural stem/progenitor cells (NSC) and in NSC-derived neurons over the course of 16 days of differentiation in vitro. In NSC cultures, maternal and paternal Ube3aYFP protein levels were approximately equal [maternal/paternal ratio = 48.3:51.7; F(1, 2) = 0.8, p = 0.5 (Fig. 2a, b)], which was not affected by the parent-of-origin of the Ube3aYFP reporter allele [F(1, 2) = 1.31, p = 0.4] or number of passages in culture (data not shown). In NSC-derived neurons, paternal and maternal Ube3aYFP protein levels were approximately equal at 1 day post-differentiation (DPD; t = 0.9, p = 0.4); however, at 4, 8, and 16 DPD, maternal Ube3aYFP protein levels were significantly higher than paternal Ube3aYFP protein levels (4 DPD: t = 2.9, p = 0.004; 8 DPD: t = 5.7, p < 0.0001; and 16 DPD: t = 11.8, p < 0.0001), with the protein levels produced by each parental allele diverging at a similar rate (slope: maternal = 0.76, paternal = −0.56; Fig. 2c, d). As a result, total Ube3aYFP protein levels in neurons remained relatively constant during the 16 days of differentiation [F(3, 3) = 1.3, p = 0.3 (Fig. 2e)], demonstrating that the maternal Ube3a allele compensates for loss of expression of the paternal Ube3a allele during the acquisition of the imprint in neurons. In contrast, paternal and maternal Ube3aYFP protein levels were approximately equal in NSC-derived astrocytes at 16 DPD [t = 0.3, p = 0.8; maternal/paternal ratio = 45.4:54.6 (Fig. 2f)]. Taken together, these findings indicate that imprinting of Ube3a is initiated upon neuronal differentiation and that the maternal Ube3a allele fully compensates for loss of expression of the paternal allele in neurons.
Fig. 2.
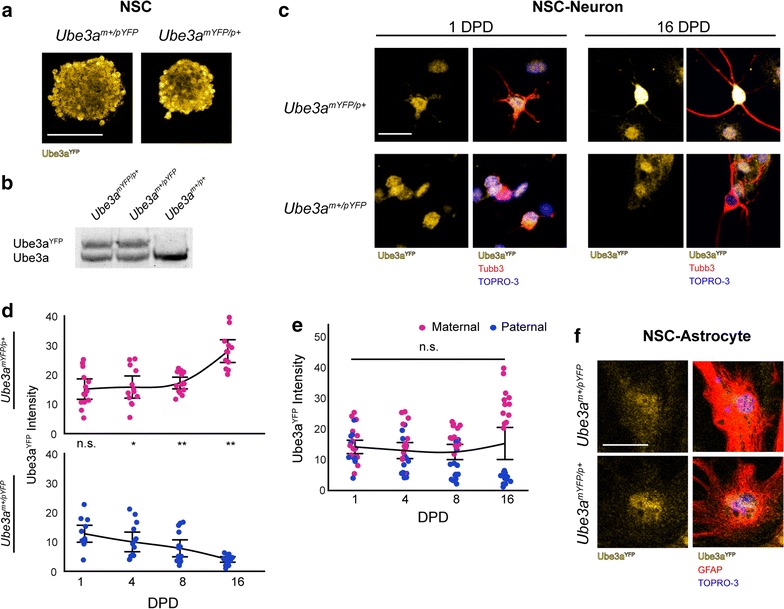
Maternal Ube3a compensates for loss of paternal Ube3a expression during neurogenesis. a Immunofluorescence images of primary neurospheres derived from the hippocampal formation of prenatal Ube3a m+/pYFP and Ube3a mYFP/p+ mice. Scale bar = 100 µm. b Western blot analysis of Ube3a and Ube3aYFP protein in NSC derived from Ube3a m+/pYFP, Ube3a mYFP/p+ and wild-type mice. c Immunofluorescence images of Ube3a m+/pYFP and Ube3a mYFP/p+ NSC-derived neurons at 1 and 16 days post-differentiation (DPD; scale bar = 25 µm). d Ube3aYFP intensity values of paternal Ube3aYFP and maternal Ube3aYFP protein levels in primary neurons at 1 DPD (Ube3a m+/pYFP: n = 13; Ube3a mYFP/p+ n = 14), 4 DPD (Ube3a m+/pYFP: n = 14; Ube3a mYFP/p+: n = 13), 8 DPD (Ube3a m+/pYFP: n = 15; Ube3a mYFP/p+: n = 15), and 16 DPD (Ube3a m+/pYFP: n = 14; Ube3a mYFP/p+: n = 13). Ratios normalized relative to total Ube3aYFP protein levels at 1 DPD. e Total Ube3aYFP protein levels in neurons during neuronal development. f Normalized paternal Ube3aYFP and maternal Ube3aYFP protein levels in Ube3a m+/pYFP and Ube3a mYFP/p+ NSC-derived astrocytes at 16 DPD (Ube3a m+/pYFP: n = 15; Ube3a mYFP/p+: n = 14). Abbreviations TOPRO-3, nuclear stain; n.s., not significant; *p < 0.05; **p < 0.001. Individual data points provided with 95% confidence intervals
Ube3a/UBE3A protein levels in the mouse and opossum brain do not correlate with their imprinting status
The UBE3A gene is highly conserved among vertebrates, but its specific location within the AS-PWS imprinted region in eutherian (placental) mammals suggests that imprinting of UBE3A evolved subsequent to divergence of the eutherian and metatherian (marsupial) lineages. Expression data from unspecified tissues of tammar wallaby (Macropus eugenii) and platypus (Ornithorhynchus anatinus) brain show that UBE3A is not imprinted in these species, consistent with this evolutionary scenario [41]. If genomic imprinting of UBE3A arose in the common ancestor of modern eutherian mammals as an epigenetic mechanism to reduce the dosage of UBE3A in neurons, we should expect levels of UBE3A in the CNS of eutherian mammals to be reduced relative to that in other (non-CNS) cell types, but have no expectation of a similar pattern of relative reduction in CNS cells of non-eutherian mammals. To test this hypothesis, we contrasted Ube3a/UBE3A protein levels in non-CNS tissues and cortex, within and between mouse and the gray, short-tailed opossum (Monodelphis domestica), a metatherian mammal lacking an orthologous AS-PWS region. We first confirmed the non-imprinted imprinting status of UBE3A in opossum cortex using RNA-seq. We next used western blot analysis of UBE3A protein levels to obtain measures of relative expression among opossum tissues and normalized, absolute expression between mouse and opossum cortex.
Analysis of RNA-seq data derived from opossum cortex (n = 4) revealed 4 informative SNVs in UBE3A that were equally represented from each parental allele [SNV-1: X 2 = 0.2, p = 0.7; SNV-2: X 2 = 0.02, p = 0.9; SNV-3: X 2 = 0.9, p = 0.7; SNV-4: X 2 = 2.5, p = 0.1 (Fig. 3a and Additional file 4)]. Combined, the maternal-to-paternal allelic ratio of UBE3A transcripts was 49:51 (95% CI maternal: 46–51, paternal: 51–54), confirming that UBE3A is biallelically expressed in opossum brain. Alignment of the mouse, human, and opossum Ube3a/UBE3A amino acid sequences revealed a high percent identity (93–95% identical) among the three species, indicating that opossum UBE3A is sufficiently similar to the eutherian protein to enable meaningful comparisons by western blot (Additional file 5). Consistent with our findings in mice, UBE3A protein levels in the opossum CNS (cortex and hippocampus) were significantly higher than those in non-CNS tissues [heart and lung; F(1, 11) = 38.6, p < 0.001 (Fig. 3b)], suggesting that an increased level of expression in cortex may be a conserved characteristic of all therian mammals, independent of imprinting. Importantly, direct comparisons between mouse and opossum showed significantly higher levels of Ube3a protein in the mouse cortex (n = 4) than in the opossum cortex [t = 2.6, p < 0.05 (Fig. 3c)]. Thus, despite imprinted expression of Ube3a in the mouse CNS, Ube3a protein levels in cortex are substantially higher than those produced by biallelic expression in the opossum cortex.
Fig. 3.
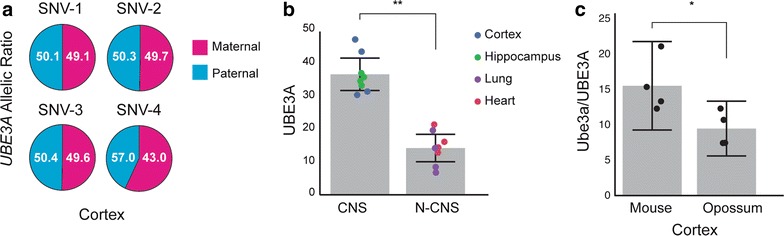
Ube3a protein levels are higher mouse and opossum brain despite different imprinting states of Ube3a/UBE3A. a UBE3A allelic ratio expressed in adult opossum cortex (n = 4) estimated from four single-nucleotide variants (SNV). b Normalized UBE3A protein levels in adult opossum CNS (cortex and hippocampus) and non-CNS (N-CNS; heart and lung) tissues (n = 4). c Normalized UBE3A/Ube3a protein levels in adult mouse (n = 4) and opossum (n = 4) cortex. Abbreviations n.s., not significant; *p < 0.05; **p < 0.001. Individual data points provided with mean (gray bar chart) and 95% confidence intervals
Imprinting of Ube3a during neurogenesis is developmentally regulated
Given our findings that expression of the paternal Ube3a allele gradually decreases during neuronal development in vitro, we decided to examine the developmental timing of the acquisition of the imprint in vivo by examining parental Ube3aYFP protein expression in two neurogenic regions of the adult mouse brain: (1) the subgranular zone of the dentate gyrus (SGZ) and (2) the subventricular zone of the lateral ventricles (SVZ) and rostral migratory stream (RMS). In the SGZ, neural stem cells differentiate into mature granular neurons that integrate into the dentate gyrus through migration to the granular cell layer (GCL), whereas in the SVZ, neural stem cells differentiate while migrating through the RMS to the olfactory bulb, where they differentiate into mature olfactory neurons [28].
In the SVZ of adult mice, paternal Ube3aYFP protein was detected in NSC/progenitor cells located along the lateral ventricles and expressing the polysialylated-neural cell adhesion molecule (PSA-NCAM) and nestin (NES) (Additional File 6). In the RMS, paternal Ube3aYFP protein was detected in immature neurons expressing doublecortin (DCX); however, in the olfactory bulb, paternal Ube3aYFP protein was barely detectable in mature neurons expressing the RNA-binding protein, Fox-1 homolog 3 [RBFOX3 (Fig. 4a)]. In the dentate gyrus, paternal Ube3aYFP protein was only detected in the SGZ, where it was present in radial glia (i.e., putative neural stem cells) expressing glial fibrillary acidic protein (GFAP), progenitor cells expressing PSA-NCAM, and immature neurons expressing DCX but not in mature neurons expressing RBFOX3 in the GCL (Fig. 4b, c). In contrast, maternal Ube3aYFP protein was detected in each neurogenic cell type and in mature neurons throughout the CNS (Additional file 6). Taken together, these observations indicate that expression of the paternal Ube3a allele is silenced at a specific stage of neurogenesis, preceding the developmental maturation of neurons.
Fig. 4.
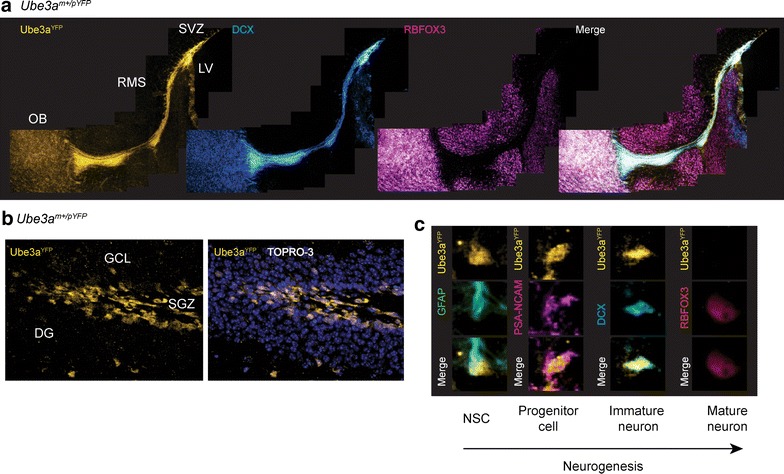
Imprinting of Ube3a is developmentally regulated. a Immunofluorescence images of paternal Ube3aYFP protein expression in the subventricular zone, rostral migratory stream, and olfactory bulb of Ube3a m+/pYFP mice. Immature and mature neurons were identified by DCX and RBFOX3 staining, respectively (×10 magnification; scale bar = 100 µm). b Immunofluorescence images of paternal Ube3aYFP protein expression in the dentate gyrus of Ube3a m+/pYFP mice (×10 magnification; scale bar = 20 µm). c Immunofluorescence images of paternal Ube3aYFP in putative neural stem cells (GFAP labeled radial glia), neural progenitor cells (PSA-NCAM), immature neurons (DCX), and mature neurons (RBFOX3) of the subgranular zone (×43 magnification; scale bar = 20 µm). Abbreviations OB, olfactory bulb; RMS, rostral migratory stream; SVZ, subventricular zone; LV, lateral ventricle; DG, dentate gyrus; GCL, granular cell layer; SGZ, subgranular zone; TOPRO-3, nuclear stain; NSC, neural stem cell
Ube3a is biallelically expressed in myenteric neurons of the PNS
Lastly, we asked whether Ube3a is also imprinted in neurons of the peripheral nervous system (PNS). To test this hypothesis, we examined parental Ube3aYFP protein levels in myenteric neurons in Auerbach’s ganglia in the colon. In adult Ube3a mYFP/p+ and Ube3a m+/pYFP mice, maternal Ube3aYFP and paternal Ube3aYFP protein were detected in mature myenteric neurons expressing RBFOX3 (Fig. 5a), in contrast to hippocampal granular neurons in which paternal Ube3aYFP protein was almost undetectable (Fig. 5b). The levels of Ube3aYFP protein expressed from each allele in myenteric neurons, however, was skewed, with higher levels of maternal Ube3aYFP protein albeit not significantly different (maternal/paternal ratio = 60:40; F(1, 4.2) = 6.3, p = 0.06) (Fig. 5b). Although studies of additional PNS neurons are needed, these findings suggest that imprinting of Ube3a is likely specific to neurons of the CNS.
Fig. 5.
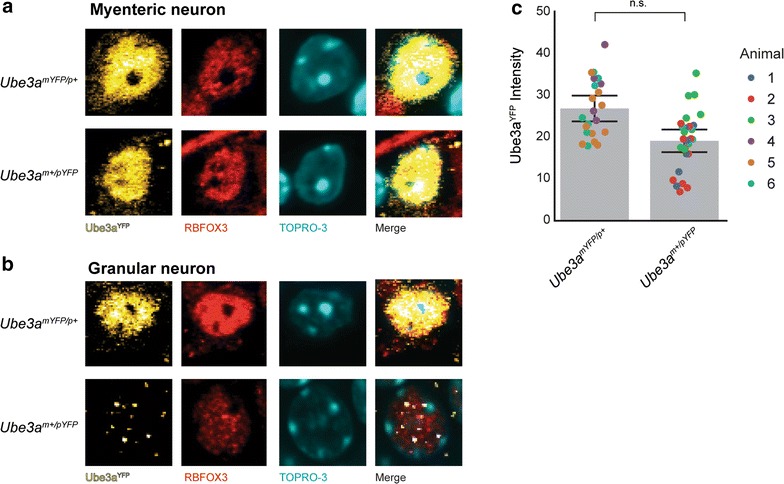
Ube3a is biallelically expressed in myenteric neurons of the PNS. a Immunofluorescence images of neurons in the myenteric ganglia of the peripheral nervous system of Ube3a m+/pYFP and Ube3a mYFP/p+ mice. b Immunofluorescence images of granular neurons in the dentate gyrus of Ube3a m+/pYFP and Ube3a mYFP/p+ mice. c Normalized intensity values of Ube3aYFP protein levels in myenteric neurons of Ube3a m+/pYFP and Ube3a mYFP/p+ mice (n = 3 per genotype; Ube3a m+/pYFP: n = 22 neurons; Ube3a mYFP/p+: n = 28). Abbreviations TOPRO-3, nuclear stain; n.s., not significant. Individual data points provided with mean (gray bar chart) and 95% confidence intervals
Discussion
In this study, we examined whether imprinting functions to reduce the dosage of Ube3a/UBE3A (i.e., levels of transcripts and/or protein) in neurons. Our results show that imprinting of the mouse Ube3a gene in neurons is achieved through increasing expression of the maternal Ube3a allele in step with decreasing expression of the paternal Ube3a allele that occurs prior to a specific developmental time point during neurogenesis. These compensating allelic expression states maintain a relatively constant level of Ube3a protein in neurons and relatively high levels of Ube3a RNA and protein in the CNS. Overall, we found that the expression levels of Ube3a/UBE3A are relatively constant among tissues and between eutherian and metatherian mammals, despite different imprinting states. Taken together, these findings suggest that the acquisition of UBE3A imprinting in eutherian mammals coincided with increased expression levels, supporting the conclusion that imprinting of UBE3A did not evolve as a dosage-regulating mechanism in neurons.
The dosage model of genomic imprinting suggests that imprinting evolved at some loci to reduce the expression levels of the gene by half [50]. For example, the paternal allele of the Murr1 gene is imprinted in neurons during brain development by the paternally expressed U2af1-rs1 antisense transcript, leading to a substantial reduction in Murr1 RNA levels in the postnatal brain [57]. Additionally, differential imprinting (i.e., monoallelic and biallelic) of the Igf2 gene in neurogenic niches of the adult mouse brain has been shown to regulate autocrine and paracrine roles of Igf2 in a dose-dependent manner [14]. In contrast to these two genes, we show that the levels of Ube3a/UBE3A RNA and protein are remarkably stable among tissues and species with different imprinting states, which stems from increased expression of the maternal Ube3a allele during the acquisition of the imprint. These findings are consistent with previous studies showing relatively constant levels of Ube3a/UBE3A transcripts during mouse brain development and among human tissues [15, 27] and the notion that dosage compensation may in fact be a common feature of imprinted genes, as suggested previously [3].
Given the importance of UBE3A in the brain and the conservation of the imprint in mouse and human neurons, and perhaps all placental mammals, it is likely that imprinting of UBE3A is a functionally relevant process in neurons that is somehow linked to the neuron-specific expression of the PWS polycistronic transcription unit (PWS-PTU). For example, imprinting of UBE3A might have evolved as a means to permit simultaneous expression of both UBE3A and the PWS-PTU in neurons, similar to that proposed in the complementation model of genomic imprinting [24]. Alternatively, transcription of UBE3A-AS across UBE3A or, perhaps, UBE3A-AS itself might expand the functional repertoire of UBE3A isoforms (coding and non-coding) expressed in neurons. Unlike most imprinted genes regulated by an antisense transcript, studies in mouse show that Ube3a-AS inhibits transcriptional elongation of the paternal Ube3a allele, resulting in a paternally expressed, 5′-truncated transcript expressed exclusively in neurons [33, 38]. Given recent findings that mouse Ube3a expresses a non-coding isoform that is important for synapse development [55], it is tempting to speculate that the 5′-truncated Ube3a transcript is in fact a functional transcript rather than just a by-product of the imprinting mechanism.
Conclusions
Although future studies are needed to resolve the functional significance of imprinting of UBE3A in neurons, the findings presented here provide evidence of an evolutionarily constrained, developmentally regulated process that maintains the dosage of UBE3A despite its monoallelic expression in neurons. Understanding the function of imprinting of UBE3A is directly relevant to understanding the function of UBE3A in the brain, and understanding the imprinted regulation of UBE3A is critically important for approaches aimed at reactivating expression of the paternal UBE3A allele as a therapy for individuals with Angelman syndrome.
Methods
Analysis of Ube3a/UBE3A expression levels in tissues
Animals
C57BL/6J mice were obtained from The Jackson Laboratories (http://www.jax.org). The Ube3a m−/p+ mouse model was obtained from the laboratory of Dr. Arthur Beaudet (Baylor College of Medicine). Ube3a m−/p+ and animals were generated by crossing wild-type C57BL/6J males with Ube3a m+/p− females, and Ube3a m+/p− animals were generated by crossing Ube3a m+/p− males with wild-type C57BL/6J females. Animals were maintained by the Texas A&M University Comparative Medicine Program. All procedures involving animals were approved by the Texas A&M Institutional Animal Care and Use Committee.
Quantitative RT-PCR
Total RNA was extracted from tissue samples using the PureLink RNA Mini Kit (Life Technologies, Carlsbad, CA). First-strand cDNA synthesis was performed using: (1) the Superscript III First-Strand Synthesis kit and oligo-dT primers (messenger RNA [mRNA]; Life Technologies) and (2) the High Capacity RNA to cDNA kit and random hexamer primers (total RNA [toRNA]; Life Technologies). Real-time PCR was performed using the TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays per manufacturer’s protocol (Life Technologies). Beta-2 microglobulin (TaqMan Assay #Mm00437762_m1) was used as an internal control. TaqMan Assay #Mm00839910_m1 was used to assess Ube3a toRNA and mRNA levels. The primer and probe set targets an amplicon of 121 base pairs that spans exons 6 and 7 of Ube3a isoforms 1 and 3 and exons 8 and 9 of Ube3a isoform 2. The reactions were performed using an ABI 7900HT real-time PCR machine.
Statistical analysis
Measurements for inferential statistics were taken using normalized ΔCt values , as outlined previously [46]. A Shapiro–Wilk goodness-of-fit test was used to test normality of sample distribution. A mixed-effect model was used to examine the effect of tissue on Ube3a RNA transcript levels, with sample ID as a random effect to account for repeated measurements of individual. A Tukey HSD multiple comparison test was used for pair-wise comparisons of tissues. The tissues were then grouped as CNS or non-CNS, and a mixed-effect model was used to examine the effect of tissue type on Ube3a RNA levels, with sample ID as a random effect. Descriptive statistics consist of ΔΔCt values ; target = Ube3a, internal control = beta-2 microglobulin).
Western blot analysis
Total protein was isolated by homogenizing tissue samples with a 1% Nonidet P40/0.01% SDS lysis buffer and protease inhibitors (Roche, Indianapolis, IN). The resulting lysates were mixed 1:1 with Laemmli loading buffer (Bio-Rad, Hercules, CA) and heated to 95 °C for 5 min. The samples were then resolved on a SDS-PAGE gel (7.5%) at 25 V for approximately 12 h and then transferred to nitrocellulose membranes at 100 V for 2 h. To normalize samples, the membranes were treated with Ponceau stain (Sigma-Aldrich) and digitally photographed. The membranes were then blocked in 5% milk in Tris-buffered saline plus Tween 20 (T-TBS) for 1 h at room temperature. Ube3a primary antibody (Additional file 7) was diluted in 2.5% milk/T-TBS and incubated on the membrane for 1 h at room temperature. After three 15 min washes in T-TBS, the secondary antibody (Additional file 7) was diluted 1:2000 in 2.5% milk/T-TBS and incubated on the membrane for 1 h at room temperature. Three 15 min washes in T-TBS were performed before developing with Clarity Western ECL Substrate (Bio-Rad), according to the manufacturer’s protocol. Membranes were imaged using the FluorChem system.
Statistical analysis
Digital images of western blot membranes (16bit.tif) were imported into ImageJ, and Ube3a protein levels were quantified using the Gel Analysis feature. Protein levels were quantified (as percentage) and then normalized by the amount of total protein per sample using a Ponceau stain. A Shapiro–Wilk goodness-of-fit test was used to test normality of sample distribution. A linear mixed-effects model was used to examine the association of tissue on Ube3a protein levels, with tissue modeled as a fixed, categorical effect and sample ID modeled as a random effect to account for repeated measurements of individual. A Tukey HSD multiple comparison test was used for pair-wise comparisons of tissues. The tissues were then grouped as CNS or non-CNS, and a linear mixed-effects model was used to examine the effect of tissue type on Ube3a protein levels, with sample ID included as a random effect. Descriptive statistics consist of Ube3a protein levels relative to heart.
RNA-sequencing analysis in mouse tissues
Data from Keane et al. [25]: Samples consisted of 76 bp paired-end (PE) libraries generated from total RNA isolated from heart, liver, lung, thymus, and hippocampus of 8-week-old B6D2F1 hybrid mice (C57BL/6 male X DBA female; n = 6). FASTQ files were downloaded from the NCBI GEO SRA (accession: ERP000591) and aligned to the mouse reference genome (mm9) using the default settings in TopHat, with the following option: -b2 sensitive.
Normalized RNA steady-state levels of the RefSeq gene annotation were estimated for each sample using Cuffnorm with the following option: -u. The FPKM value of Ube3a transcripts for each sample was extracted from the output file and used for descriptive and inferential statistics.
Statistical analysis
The parental allelic ratio of Ube3a for each tissue was determined using single-nucleotide polymorphisms (SNPs) located in Ube3a exon 5 (SNP-1 = chr7:66,527,581) and 8 (SNP-2 = chr7:66,541,539). Raw counts of the 2 SNPs were determined using the CLC Genomics Workbench quality-based variant detection module with the following settings: neighborhood radius = 5; maximum gap and mismatch count = 2, minimum neighborhood quality = 15, minimum central quality = 20, ignore non-specific matches, minimum coverage = 20, minimum variant % = 1. The raw counts of each SNP for each sample and tissue were then used to estimate the fragments per thousand per million (FPKM) values of Ube3a transcripts of the parental alleles using the following equations:
A mixed-effect model was used to examine the effect of tissue and allele on Ube3a transcript levels (full factorial), with sample ID as a random effect to account for repeated measurements of individual. A Dunnett’s–Hsu multiple comparison test was used to compare total Ube3a transcript levels between hippocampus and each tissue, and a Dunnett’s–Hsu multiple comparison test was used to compare maternal and paternal Ube3a transcript levels between hippocampus and each allele of each tissue.
RNA-sequencing analysis in human tissues
Data from Fagerberg et al. [12]: Samples consisted of 100 bp PE sequencing libraries generated from mRNA isolated from 12 human tissues. Three samples per tissue were used for the analysis. FASTQ files were downloaded (http://www.ebi.ac.uk/arrayexpress/; accession: E-MTAB-1733) and aligned to the human reference genome (hg19) using the default settings in TopHat. Normalized FPKM values of the RefSeq gene annotation were estimated using Cuffnorm using the default settings and the following option: -u. The FPKM value of UBE3A for each sample was extracted from the output file and used for descriptive and inferential statistics.
Data from GTEx: The FPKM values and accompanying information were downloaded for UBE3A from the GTEx Portal (http://www.gtexportal.org/home/GTEx_Analysis_V4_RNA-seq_RNA-SeQCv1.1.8_gene_rpkm.gct) [10, 53]. The initial data set consisted of 25 different histologic samples derived from 175 individuals (1518 tissue samples). Only tissues for which at least 30 samples were available were included in the inferential analyses (1227 tissue samples from 10 tissues): adipose tissue (n = 111), brain (n = 296), blood vessels (n = 138), esophagus (n = 34), heart (n = 103), lung (n = 113), muscle (n = 132), nerve (n = 86), skin (n = 114), and thyroid (n = 100). A total of 291 tissue samples from 14 tissues were excluded. Blood samples were also excluded from inferential analysis.
Statistical analysis
A mixed-effect model was used to examine the effect of tissue on UBE3A transcript levels, with sample ID as a random effect to account for repeated measurements of individual. For the GTEx data set, tissue and sex were modeled as fixed effects (full factorial), with sample ID as a random effect. A Dunnett’s–Hsu multiple comparison test was used to compare UBE3A transcript levels among tissues relative to the brain. Goodness-of-fit of models was assessed by AIC and BIC values and visual inspection of diagnostic residual plots.
Analysis of allelic Ube3aYFP in neural stem cells and differentiated neurons
Animals
The Ube3a YFP mouse model was obtained from the laboratory of Dr. Arthur Beaudet (Baylor College of Medicine). Ube3a mYFP/p+ animals were generated by crossing wild-type C57BL/6J males with Ube3a m+/pYFP females, and Ube3a m+/pYFP animals were generated by crossing Ube3a m+/pYFP males with wild-type C57BL/6J females. Wild-type mice consisted of siblings lacking the Ube3a YFP allele. PCR to determine the genotypes of mice (i.e., Ube3a m+/p+ or Ube3a YFP) were performed using methods described previously [11].
Neural stem cell cultures
Neural stem cell cultures were established from Ube3a m+/p+, Ube3a mYFP/p+, and Ube3a m+/pYFP mice using methods described previously [48]. Briefly, the hippocampal formation (HF) was removed from fetuses at embryonic day 17.5 (E17.5), digested using a 10× Trypsin–EDTA solution, triturated into a single cell suspension, and then seeded in neural stem cell (NSC) medium, consisting of DMEM-F12 (Invitrogen, Carlsbad, CA), B-27 supplement (Invitrogen), progesterone, putrescine (Sigma-Aldrich), epidermal growth factor (Sigma-Aldrich), glucose, penicillin/streptomycin (Invitrogen), insulin–transferrin–sodium selenite (Sigma-Aldrich), HEPES, and heparin. The NSC cultures were maintained in humidified incubators at 37 °C and 5% CO2. Every 3–4 days, the neurospheres were dissociated with TrypLE (Invitrogen) for 20 min and then resuspended in NSC media.
Western blot analysis
Western blot analysis of Ube3a and Ube3aYFP protein levels in NSC cultures was performed using methods described above.
Statistical analysis
A mixed-effect model was used to examine the effect of allele (i.e., Ube3a or Ube3a YFP) and parent-of-origin (i.e., maternal or paternal) on Ube3a and Ube3aYFP protein levels (full factorial), with sample ID as a random effect to account for repeated measurements.
Neural stem cell differentiation
To differentiate NSC cultures, neurospheres were first dissociated using TrypLE (Invitrogen) following the manufacturer’s protocol. The dissociated cells were then resuspended in neuronal growth media (Neurobasal-A [Invitrogen], B-27 supplement [Invitrogen], and Glutamax [Invitrogen]) and plated on glass coverslips coated with poly-ornithine [Sigma-Aldrich] and laminin [Invitrogen] at a density of 380,000 cells per well in a 12-well, cell-culture plate; the plates were maintained in humidified incubators at 37 °C and 5% CO2.
Immunofluorescence imaging of Ube3aYFP
Immunofluorescence imaging was used to quantify Ube3aYFP protein levels in differentiated neurons as previously described [11]. Briefly, at 1, 4, 8, and 16 days post-differentiation (DPD; day of differentiation = 0 DPD), differentiated cells were washed twice with 1× PBS, fixed in 4% paraformaldehyde in PBS for 15 min, and then washed three times in 1× PBS. The cells were blocked in 0.3% Triton-X100 in PBS (T-PBS) plus 5% goat or donkey serum for 1–2 h at room temperature with gentle agitation. Cells were incubated with the primary antibodies (Additional file 7) for 24 h at 4 °C with gentle agitation. Cells were washed 3 times in 0.1% Tween 20 1× PBS for 15 min each and then incubated with fluorescently labeled secondary antibodies (Additional file 7) for 24 h at 4 °C in the dark. Cells were then washed 4 times in 0.1% Tween 20 1× PBS for 15 min each. Nuclei were labeled using TOPRO-3(Invitrogen) at a dilution of 1:1000 in the third wash. Coverslips were mounted on glass slides with Vectashield (Vector Laboratories, Burlingame, CA) mounting reagent. Confocal images were obtained using a LSM 510 META NLO multiphoton microscope (Zeiss, Oberkochen, Germany). Images of the individual neurons were taken at 43× magnification. Images were imported into ImageJ and converted to the RGB color format. The Plot Profile feature was then used to determine the median gray value of Ube3aYFP in individual neurons. Neurons and astrocytes were identified by immunoreactivity with the Tubb3 and GFAP antibodies, respectively.
Statistical analysis
To examine the effect of day on Ube3aYFP protein levels, a linear regression model was used with normalized intensity values of Ube3aYFP as the dependent variable and days post-differentiation (DPD) and allele (i.e., maternal and paternal) as fixed effects. To compare maternal and paternal Ube3aYFP protein levels at each time point, a least square means contrast linear regression model was used with normalized gray values of Ube3aYFP protein levels as the dependent variable and DPD (dummy variable) and allele as fixed effects.
Analysis of opossum Ube3a/UBE3A expression
Animals
For the analysis of UBE3A protein levels, opossum (Monodelphis domestica) tissues (cortex, hippocampus, heart, and lung) samples were obtained from a colony at the Comparative Medicine Program facilities at Texas A&M University. All animals were derived from an outbred stock (LL1) that was founded using wild animals captured in Eastern Brazil. All procedures involving animals were approved by the Texas A&M Institutional Animal Care and Use Committee. For the allelic expression analysis of UBE3A, opossum fetal brain (cortex) samples were obtained at 13 days post-copulation (d.p.c.) from F1 animals derived from reciprocal crosses of two semi-inbred stocks (LL1 and LL2) [56].
RNA-seq analysis
The brain tissues were homogenized in TRI Reagent (Invitrogen) and total RNA was extracted using BCP (1-bromo-2 chloropropane), precipitated with isopropanol, and resuspended in RNase-free water. Potential DNA contamination was removed by both DNase I treatment and DNA removal columns in Qiagen RNeasy Plus Mini kit (Qiagen, CA). RNA-seq libraries were made using the Illumina TruSeq RNA Sample Prep Kit (Illumina Inc., CA), and sequenced on an Illumina HiSeq 2000 instrument (Illumina Inc., CA). Details on RNA-seq data analysis could be found in Wang et al. [56].
Statistical analysis
A Pearson’s two-sided Chi-square test of the allelic counts of each SNV was used to determine the whether the allelic ratios of UBE3A were equal (Ho: maternal/paternal = 50:50; Ha: maternal/paternal = 50:50).
Western blot analysis
Total protein was isolated from adult (21-week-old) opossum tissues (n = 4) and adult (8-week-old) mouse cortex (n = 4) as described above. For the analysis of UBE3A protein levels among opossum tissues, a single western blot was performed for each animal for all tissues. UBE3A protein levels were estimated and normalized as described above. For the comparison of Ube3a/UBE3A protein levels between opossum and mouse cortex, samples were run on a single western blot, and Ube3a/UBE3A protein levels were normalized and compared as described above.
Statistical analysis
To compare UBE3A protein levels among opossum tissues, a mixed-effect model was used with normalized UBE3A protein levels as the dependent variable, tissue as a fixed effect, and sample ID as a random effect to account for repeated measurements of individual. To compare Ube3a/UBE3A protein levels between mouse and opossum cortex, a two-tailed Welsh t test was performed.
Analysis of Ube3aYFP in neurogenic niches of the adult mouse brain
Immunofluorescence imaging of Ube3aYFP
Ube3a m+/pYFP and Ube3a mYFP/p+ adult mice (6-week-old) were anesthetized with 0.5–1.0 mL of 20 mg/mL Avertin (Sigma-Aldrich, St. Louis, MO) via intraperitoneal injection. Mice were perfused with ice-cold phosphate-buffered saline (PBS) and 4% paraformaldehyde. Dissected brains were post-fixed in 4% paraformaldehyde solution for approximately 12 h and then cryoprotected in 30% sucrose solution. Fifty µm sections (sagital and coronal) were cut on a cryostat and stored in PBS. For immunostaining, sections were blocked in 0.3% Triton-X100 in PBS (T-PBS) plus 5% serum (goat or donkey) for 1–2 h at room temperature in a humidified chamber with gentle agitation. Primary antibodies (Additional file 7) were incubated with sections for 48 h at 4 °C with gentle agitation. Sections were washed 3 times in 0.1% Tween 20 1× PBS for 15 min each and then incubated with fluorescently labeled secondary antibodies (Additional file 7) for 24 h at 4 °C in the dark with gentle agitation. Sections were washed 4 times in 0.1% Tween 20 plus 1× PBS for 15 min each. Nuclei were labeled by adding TOPRO-3 at 1:1000 dilution in the third wash. Sections were mounted on glass slides with Vectashield (Vector Laboratories, Burlingame, CA) mounting reagent. Confocal images were obtained using a LSM 510 META NLO multiphoton microscope (Zeiss, Oberkochen, Germany). Images were taken using 10× and 43× (oil) objectives. Rostral migratory stream images were taken at 10× magnification, and then images were then stitched together. For resolution of individual cell images in vivo Z-stack images of 8–11 slices were used.
Analysis of Ube3aYFP in myenteric neurons
Immunofluorescence imaging of Ube3aYFP
Ube3a mYFP/p+ and Ube3a m+/pYFP adult mice (6-week-old; n = 3/genotype) were processed and immunostained as described above. Confocal images of Auerbach’s ganglia (8–10 per animal) were obtained using a 43× (oil) objective. Images were imported into ImageJ and converted to the RGB color format. The Plot Profile feature was then used to determine the median gray value of Ube3aYFP in individual neurons (RBFOX3 positive) as described above.
Statistical analysis
A mixed-effect model was used to examine the effect of allele (maternal and paternal) on Ube3aYFP protein levels, with sample ID as a random effect to account for repeated measurements of neurons within an individual animal.
Statistics
Inferential analyses were performed using JMP Pro® (version 12). Mixed-effect models were used for analyses involving repeated measures of cells/tissues from an individual animal. The degrees of freedom were calculated using the Kenward–Roger correction.
Charts
Charts were generated using JMP Pro® (version 12) and formatted in Adobe Illustrator.
Additional files
Additional file 1: Table S1. Pair-wise comparisons of maternal and paternal Ube3a transcript levels.
Additional file 2: Table S2. Pair-wise comparisons of UBE3A transcript levels.
Additional file 3: Table S3. Pair-wise comparisons of UBE3A transcript levels.
Additional file 4: Table S4. RNA-seq analysis of UBE3A allelic expression in opossum brain.
Additional file 5: Figure S1. Alignment of mouse, opossum, and human Ube3a/UBE3A amino acid sequences. (A) ClustalW alignment of amino acid sequences of human UBE3A isoform 3, mouse Ube3a isoform 2, and opossum UBE3A. (B) Percent identity values of pair-wise comparisons of UBE3A/Ube3a among human, mouse, and opossum. Values represent conservative, semiconservative, non-conserved, and insertion/deletion percent identity.
Additional file 6: Figure S2. (A-C) Immunofluorescence images of paternal Ube3aYFP protein expression in the lateral ventricle of adult Ube3a +/YFP mice. (D) Immunofluorescence image of maternal Ube3aYFP protein expression in the lateral ventricle of adult Ube3a YFP/+ mice. (E) Immunofluorescence images of maternal Ube3aYFP protein expression in the lateral ventricle, subventricular zone, rostral migratory stream, olfactory bulb, and adjacent cortical neurons in Ube3a YFP/+ mice. (F) Immunofluorescence images of maternal Ube3aYFP protein expression in the granular cell layer and subgranular zone of Ube3a YFP/+ mice.
Additional file 7: Table S5. List of antibodies used for western blot and immunofluorescence analyses.
Authors’ contributions
PRH, SGBC and SVD designed the study; PRH, SGBC, KD, and XW performed experiments; PRH, SGBC, XW, RD, KK, NDC and SVD collected and analyzed data; PBS provided technical support and conceptual advice; and PRH, SGBC, PBS, and SVD wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank the Foundation for Angelman Syndrome Therapeutics for funding this project. We also thank Arline Rector for help with the cell-culture studies, Jean Kovar for help with maintaining the Ube3a YFP and Ube3a −/+ mouse colonies, and Rola Barhoumi Mouneimne for help with the confocal imaging and quantification of immunofluorescence images. Confocal microscopy was performed in the Texas A&M University College of Veterinary Medicine & Biomedical Sciences Image Analysis Laboratory, supported by NIH-NCRR (1 S10 RR22532-01).
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
Data used for this study were publicly available at the Gene Expression Omnibus (GEO) under accession ERP000591, ArrayExpress under accession E-MTAB-1733, and the GTEx Portal.
Ethical approval and consent to participate
All procedures involving animals were approved by the Texas A&M Institutional Animal Care and Use Committee. Analyses involving human samples were performed using publicly available data from the Genotype-Tissue Expression Consortium. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 10/13/2015.
Funding
Foundation for Angelman Syndrome Therapeutics.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- UBE3A
ubiquitin protein E3A ligase gene
- CNS
central nervous system
- PTU
polycistronic transcription unit
- UBE3A-AS
UBE3A antisense transcript
- SNRPN-SNURF
SNRPN upstream reading frame—small nuclear ribonucleoprotein polypeptide N
- snoRNA
small nucleolar RNAs
- AS
Angelman syndrome
- Dup15q
duplication 15q syndrome
- PWS
Prader–Willi syndrome
- ASD
autism spectrum disorder
- Ube3aYFP
Ube3a yellow fluorescent protein
- DPD
days post-differentiation
- SVZ
subventricular zone
- RMS
rostral migratory stream
- SGZ
subgranular zone
- PSA-NCAM
polysialylated-neural cell adhesion molecule
- NES
nestin
- DCX
doublecortin
- RBFOX3
NA-binding protein, Fox-1 homolog 3
- GFAP
glial fibrillary acidic protein
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13072-017-0134-4) contains supplementary material, which is available to authorized users.
Paul R. Hillman and Sarah G. B. Christian contributed equally to this work
Contributor Information
Paul R. Hillman, Email: Paul.Hillman@uth.tmc.edu
Sarah G. B. Christian, Email: schristian@cvm.tamu.edu
Ryan Doan, Email: Ryan.Doan@childrens.harvard.edu.
Noah D. Cohen, Email: ncohen@cvm.tamu.edu
Kranti Konganti, Email: konganti@tamu.edu.
Kory Douglas, Email: kodouglas@tamu.edu.
Xu Wang, Email: xw54@cornell.edu.
Paul B. Samollow, Email: PSamollow@cvm.tamu.edu
Scott V. Dindot, Phone: (979) 458-4725, Email: sdindot@cvm.tamu.edu
References
- 1.Ager EI, Pask AJ, Gehring HM, Shaw G, Renfree MB. Evolution of the CDKN1C-KCNQ1 imprinted domain. BMC Evol Biol. 2008;8:163. doi: 10.1186/1471-2148-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babak T, DeVeale B, Tsang EK, Zhou Y, Li X, Smith KS, Kukurba KR, Zhang R, Li JB, van der Kooy D, et al. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat Genet. 2015;47:544–549. doi: 10.1038/ng.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baran Y, Subramaniam M, Biton A, Tukiainen T, Tsang EK, Rivas MA, Pirinen M, Gutierrez-Arcelus M, Smith KS, Kukurba KR, et al. The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 2015;25:927–936. doi: 10.1101/gr.192278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baroux C, Spillane C, Grossniklaus U. Genomic imprinting during seed development. Adv Genet. 2002;46:165–214. doi: 10.1016/s0065-2660(02)46007-5. [DOI] [PubMed] [Google Scholar]
- 5.Bressler J, Tsai TF, Wu MY, Tsai SF, Ramirez MA, Armstrong D, Beaudet AL. The SNRPN promoter is not required for genomic imprinting of the Prader–Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty M, Paul BK, Nayak T, Das A, Jana NR, Bhutani S. The E3 ligase ube3a is required for learning in Drosophila melanogaster. Biochem Biophys Res Commun. 2015;462:71–77. doi: 10.1016/j.bbrc.2015.04.110. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain SJ, Brannan CI. The Prader–Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 8.Charalambous M, Ferron SR, da Rocha ST, Murray AJ, Rowland T, Ito M, Schuster-Gossler K, Hernandez A, Ferguson-Smith AC. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15:209–221. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colosi DC, Martin D, More K, Lalande M. Genomic organization and allelic expression of UBE3A in chicken. Gene. 2006;383:93–98. doi: 10.1016/j.gene.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Consortium GT Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- 12.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics: MCP. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 14.Ferron SR, Radford EJ, Domingo-Muelas A, Kleine I, Ramme A, Gray D, Sandovici I, Constancia M, Ward A, Menheniott TR, et al. Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat Commun. 2015;6:8265. doi: 10.1038/ncomms9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galiveti CR, Raabe CA, Konthur Z, Rozhdestvensky TS. Differential regulation of non-protein coding RNAs from Prader–Willi syndrome locus. Sci Rep. 2014;4:6445. doi: 10.1038/srep06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haig D. Intragenomic conflict and the evolution of eusociality. J Theor Biol. 1992;156:401–403. doi: 10.1016/S0022-5193(05)80683-6. [DOI] [PubMed] [Google Scholar]
- 18.Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc Biol Sci. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haig D. Genomic imprinting and kinship: how good is the evidence? Annu Rev Genet. 2004;38:553–585. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- 20.Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hope KA, LeDoux MS, Reiter LT. The Drosophila melanogaster homolog of UBE3A is not imprinted in neurons. Epigenetics. 2016;11:637–642. doi: 10.1080/15592294.2016.1214783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jana NR. Understanding the pathogenesis of Angelman syndrome through animal models. Neural Plast. 2012;2012:710943. doi: 10.1155/2012/710943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/S0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko-Ishino T. The regulation and biological significance of genomic imprinting in mammals. J Biochem. 2003;133:699–711. doi: 10.1093/jb/mvg090. [DOI] [PubMed] [Google Scholar]
- 25.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 27.Kohama C, Kato H, Numata K, Hirose M, Takemasa T, Ogura A, Kiyosawa H. ES cell differentiation system recapitulates the establishment of imprinted gene expression in a cell-type-specific manner. Hum Mol Genet. 2012;21:1391–1401. doi: 10.1093/hmg/ddr577. [DOI] [PubMed] [Google Scholar]
- 28.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaSalle JM, Reiter LT, Chamberlain SJ. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics. 2015;7:1213–1228. doi: 10.2217/epi.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins-Taylor K, Hsiao JS, Chen PF, Glatt-Deeley H, De Smith AJ, Blakemore AI, Lalande M, Chamberlain SJ. Imprinted expression of UBE3A in non-neuronal cells from a Prader-Willi syndrome patient with an atypical deletion. Hum Mol Genet. 2014;23:2364–73. doi: 10.1093/hmg/ddt628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 33.Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum Mol Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng L, Person RE, Huang W, Zhu PJ, Costa-Mattioli M, Beaudet AL. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013;9:e1004039. doi: 10.1371/journal.pgen.1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet: TIG. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 36.Moreno-De-Luca D, Sanders SJ, Willsey AJ, Mulle JG, Lowe JK, Geschwind DH, State MW, Martin CL, Ledbetter DH. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry. 2013;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, Scherer SW, Stockley T, Vincent JB, Mendoza-Londono R, Stavropoulos DJ. 15q11.2 duplication encompassing only the UBE3A gene is associated with developmental delay and neuropsychiatric phenotypes. Hum Mutat. 2015;36:689–693. doi: 10.1002/humu.22800. [DOI] [PubMed] [Google Scholar]
- 38.Numata K, Kohama C, Abe K, Kiyosawa H. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Res. 2011;39:2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell MJ, Loughran NB, Walsh TA, Donoghue MT, Schmid KJ, Spillane C. A phylogenetic approach to test for evidence of parental conflict or gene duplications associated with protein-encoding imprinted orthologous genes in placental mammals. Mamm Genome. 2010;21:486–498. doi: 10.1007/s00335-010-9283-5. [DOI] [PubMed] [Google Scholar]
- 40.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rapkins RW, Hore T, Smithwick M, Ager E, Pask AJ, Renfree MB, Kohn M, Hameister H, Nicholls RD, Deakin JE, et al. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2006;2:e182. doi: 10.1371/journal.pgen.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renfree MB, Suzuki S, Kaneko-Ishino T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120151. doi: 10.1098/rstb.2012.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 44.Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 45.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 47.Scoles HA, Urraca N, Chadwick SW, Reiter LT, Lasalle JM. Increased copy number for methylated maternal 15q duplications leads to changes in gene and protein expression in human cortical samples. Mol Autism. 2011;2:19. doi: 10.1186/2040-2392-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shetty AK. Progenitor cells from the CA3 region of the embryonic day 19 rat hippocampus generate region-specific neuronal phenotypes in vitro. Hippocampus. 2004;14:595–614. doi: 10.1002/hipo.10206. [DOI] [PubMed] [Google Scholar]
- 49.Soellner L, Begemann M, Mackay DJ, Gronskov K, Tumer Z, Maher ER, Temple IK, Monk D, Riccio A, Linglart A, et al. Recent advances in imprinting disorders. Clin Genet. 2017;91:3–13. doi: 10.1111/cge.12827. [DOI] [PubMed] [Google Scholar]
- 50.Solter D. Differential imprinting and expression of maternal and paternal genomes. Annu Rev Genet. 1988;22:127–146. doi: 10.1146/annurev.ge.22.120188.001015. [DOI] [PubMed] [Google Scholar]
- 51.Spencer HG, Clark AG. Non-conflict theories for the evolution of genomic imprinting. Heredity. 2014;113:112–118. doi: 10.1038/hdy.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stringer JM, Suzuki S, Pask AJ, Shaw G, Renfree MB. GRB10 imprinting is eutherian mammal specific. Mol Biol Evol. 2012;29:3711–3719. doi: 10.1093/molbev/mss173. [DOI] [PubMed] [Google Scholar]
- 53.Sudmant PH, Alexis MS, Burge CB. Meta-analysis of RNA-seq expression data across species, tissues and studies. Genome Biol. 2015;16:287. doi: 10.1186/s13059-015-0853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Surani MA. Genomic imprinting: developmental significance and molecular mechanism. Curr Opin Genet Dev. 1991;1:241–246. doi: 10.1016/S0959-437X(05)80077-2. [DOI] [PubMed] [Google Scholar]
- 55.Valluy J, Bicker S, Aksoy-Aksel A, Lackinger M, Sumer S, Fiore R, Wust T, Seffer D, Metge F, Dieterich C, et al. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci. 2015;18:666–673. doi: 10.1038/nn.3996. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Douglas KC, Vandeberg JL, Clark AG, Samollow PB. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 2014;24:70–83. doi: 10.1101/gr.161919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, Okinami S, Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol Cell Biol. 2004;24:270–9. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 59.Wilkins JF, Ubeda F, Van Cleve J. The evolving landscape of imprinted genes in humans and mice: conflict among alleles, genes, tissues, and kin. BioEssays. 2016;38:482–489. doi: 10.1002/bies.201500198. [DOI] [PubMed] [Google Scholar]
- 60.Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, Laan LA, Magenis RE, Moncla A, Schinzel AA, Summers JA, et al. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet Part A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 61.Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, Fischer JA. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci USA. 2008;105:12399–12404. doi: 10.1073/pnas.0805291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamasaki K. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- 63.Yi JJ, Berrios J, Newbern JM, Snider WD, Philpot BD, Hahn KM, Zylka MJ. An autism-linked mutation disables phosphorylation control of UBE3A. Cell. 2015;162:795–807. doi: 10.1016/j.cell.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang YJ, Yang JH, Shi QS, Zheng LL, Liu J, Zhou H, Zhang H, Qu LH. Rapid birth-and-death evolution of imprinted snoRNAs in the Prader–Willi syndrome locus: implications for neural development in euarchontoglires. PLoS ONE. 2014;9:e100329. doi: 10.1371/journal.pone.0100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Pair-wise comparisons of maternal and paternal Ube3a transcript levels.
Additional file 2: Table S2. Pair-wise comparisons of UBE3A transcript levels.
Additional file 3: Table S3. Pair-wise comparisons of UBE3A transcript levels.
Additional file 4: Table S4. RNA-seq analysis of UBE3A allelic expression in opossum brain.
Additional file 5: Figure S1. Alignment of mouse, opossum, and human Ube3a/UBE3A amino acid sequences. (A) ClustalW alignment of amino acid sequences of human UBE3A isoform 3, mouse Ube3a isoform 2, and opossum UBE3A. (B) Percent identity values of pair-wise comparisons of UBE3A/Ube3a among human, mouse, and opossum. Values represent conservative, semiconservative, non-conserved, and insertion/deletion percent identity.
Additional file 6: Figure S2. (A-C) Immunofluorescence images of paternal Ube3aYFP protein expression in the lateral ventricle of adult Ube3a +/YFP mice. (D) Immunofluorescence image of maternal Ube3aYFP protein expression in the lateral ventricle of adult Ube3a YFP/+ mice. (E) Immunofluorescence images of maternal Ube3aYFP protein expression in the lateral ventricle, subventricular zone, rostral migratory stream, olfactory bulb, and adjacent cortical neurons in Ube3a YFP/+ mice. (F) Immunofluorescence images of maternal Ube3aYFP protein expression in the granular cell layer and subgranular zone of Ube3a YFP/+ mice.
Additional file 7: Table S5. List of antibodies used for western blot and immunofluorescence analyses.


