Abstract
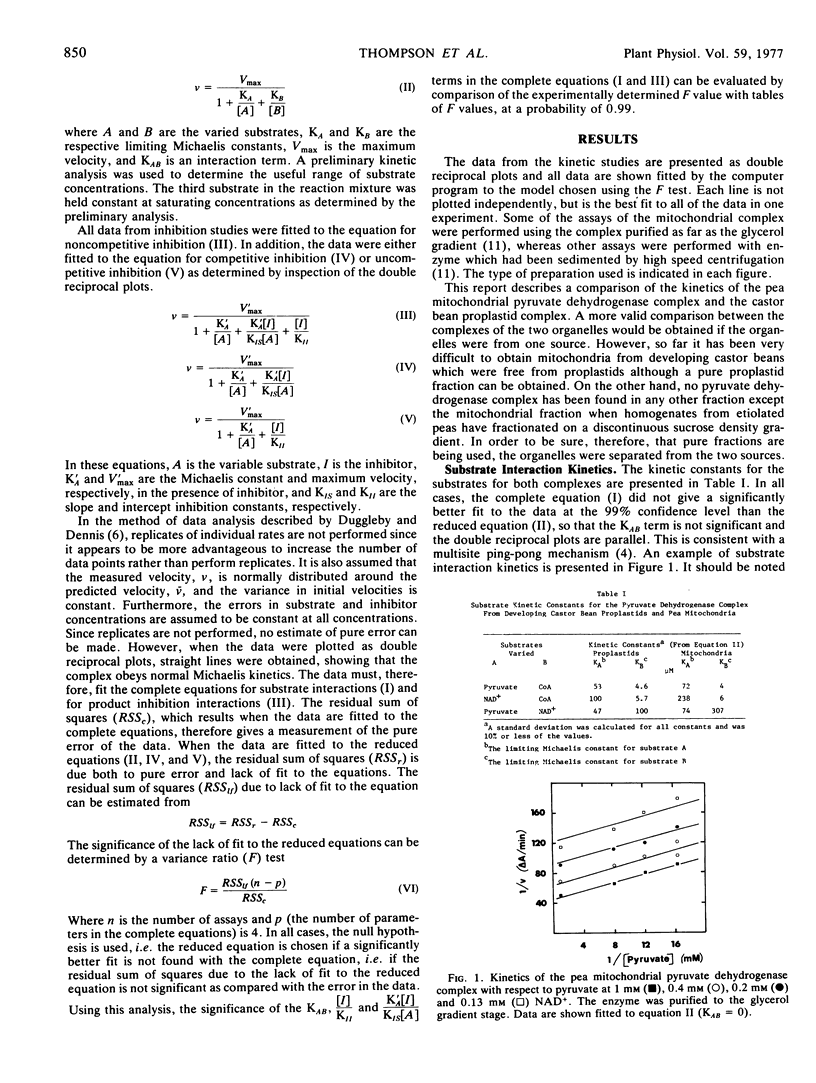
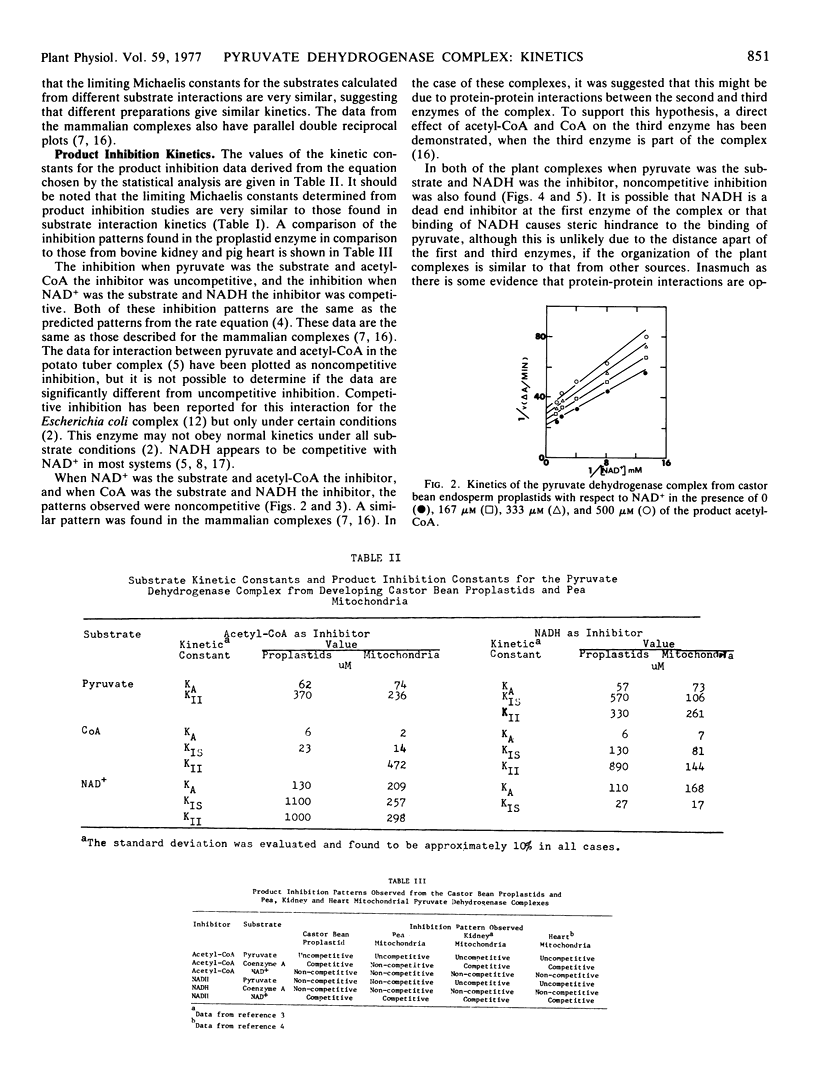
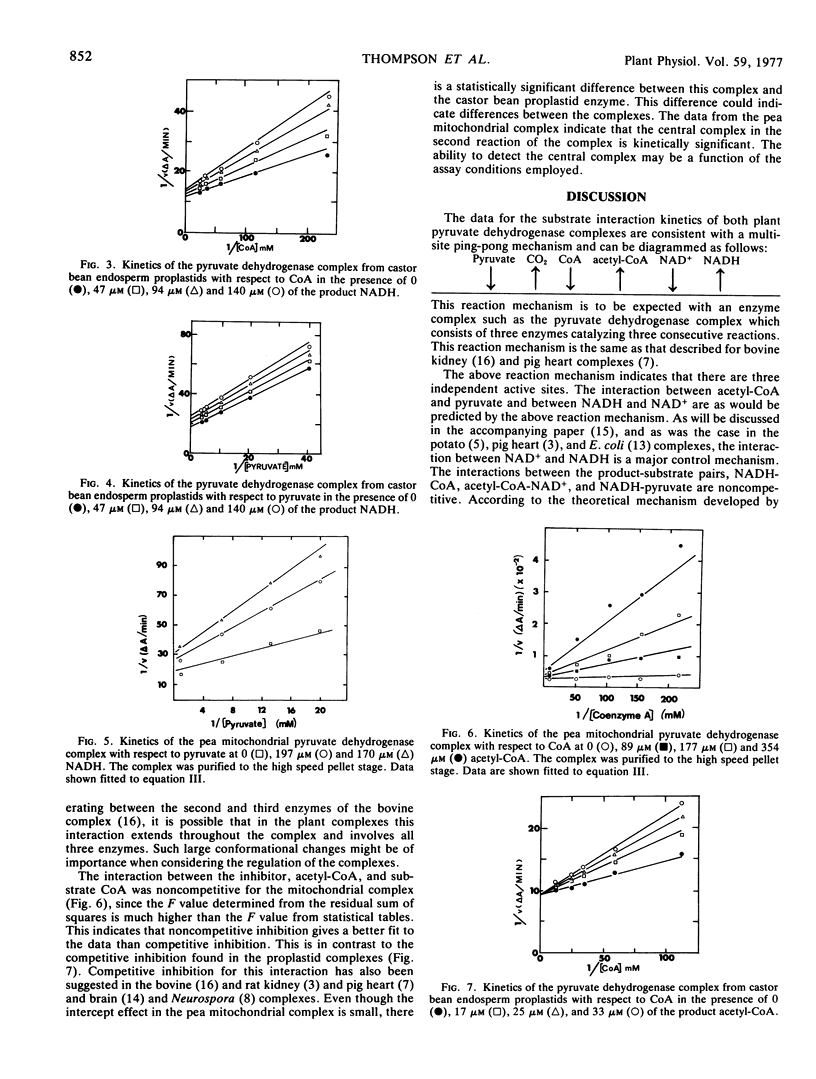
A steady-state kinetic analysis has been performed on the pyruvate dehydrogenase complex from pea (Pisum sativum L.) mitochondria and castor bean (Ricinus communis L.) proplastids. Substrate interaction kinetics for all substrates gave parallel lines consistent with a multisite ping-pong mechanism. Product inhibition studies showed uncompetitive inhibition between acetyl-CoA and pyruvate and competitive inhibition between NADH and NAD+, both of which are also consistent with this mechanism. In the mitochondrial complex, acetyl-CoA showed noncompetitive inhibition versus CoA which suggests that the intermediate complex is kinetically important in the lipoamide transacetylase component of this complex. In contrast, the proplastid complex showed competitive inhibition in this interaction. NADH is a noncompetitive inhibitor versus CoA in both complexes indicating that these complexes, like the mammalian complex, may have protein-protein interactions between the second and third enzymes of the complex. Since NADH also shows noncompetitive inhibition versus pyruvate, this interaction may extend to all components of the complex. Acetyl-CoA shows noncompetitive inhibition versus NAD+ which may also be a result of interaction between the second and third enzymes of the complex. The limiting Michaelis constants for substrates and the inhibitor constants for both complexes were determined.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson D. E., Roach P. J., Schwedes J. S. Metabolite concentrations and concentration ratios in metabolic regulation. Adv Enzyme Regul. 1975;13:393–411. doi: 10.1016/0065-2571(75)90027-8. [DOI] [PubMed] [Google Scholar]
- Biswanger H., Henning U. Regulatory properties of the pyruvate-dehydrogenase complex from Escherichia coli. Positive and negative cooperativity. Eur J Biochem. 1971 Dec;24(2):376–384. doi: 10.1111/j.1432-1033.1971.tb19696.x. [DOI] [PubMed] [Google Scholar]
- Bremer J. Pyruvate dehydrogenase, substrate specificity and product inhibition. Eur J Biochem. 1969 Apr;8(4):535–540. doi: 10.1111/j.1432-1033.1969.tb00559.x. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. Derivation of rate equations for multisite ping-pong mechanisms with ping-pong reactions at one or more sites. J Biol Chem. 1973 Dec 25;248(24):8353–8355. [PubMed] [Google Scholar]
- Crompton M., Laties G. G. The regulatory function of potato pyruvate dehydrogenase. Arch Biochem Biophys. 1971 Mar;143(1):143–150. doi: 10.1016/0003-9861(71)90194-9. [DOI] [PubMed] [Google Scholar]
- Duggleby R. G., Dennis D. T. Nicotinamide adenine dinucleotide-specific glyceraldehyde 3-phosphate dehydrogenase from Pisum sativum. Assay and steady state kinetics. J Biol Chem. 1974 Jan 10;249(1):167–174. [PubMed] [Google Scholar]
- Hamada M., Koike K., Nakaula Y., Hiraoka T., Koike M. A kinetic study of the alpha-keto acid dehydrogenase complexes from pig heart mitochondria. J Biochem. 1975 May;77(5):1047–1056. doi: 10.1093/oxfordjournals.jbchem.a130805. [DOI] [PubMed] [Google Scholar]
- Harding R. W., Caroline D. F., Wagner R. P. The pyruvate dehydrogenase complex from the mitochondrial fraction of Neurospora crassa. Arch Biochem Biophys. 1970 Jun;138(2):653–661. doi: 10.1016/0003-9861(70)90393-0. [DOI] [PubMed] [Google Scholar]
- Reid E. E., Lyttle C. R., Canvin D. T., Dennis D. T. Pyruvate dehydrogenase complex activity in proplastids and mitochondria of developing castor bean endosperm. Biochem Biophys Res Commun. 1975 Jan 6;62(1):42–47. doi: 10.1016/s0006-291x(75)80402-5. [DOI] [PubMed] [Google Scholar]
- Reid E. E., Thompson P., Lyttle C. R., Dennis D. T. Pyruvate dehydrogenase complex from higher plant mitochondria and proplastids. Plant Physiol. 1977 May;59(5):842–848. doi: 10.1104/pp.59.5.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E. R., Old L. O., Reed L. J. Regulatory properties of pyruvate dehydrogenase from Escherichia coli. Biochem Biophys Res Commun. 1968 May 10;31(3):495–500. doi: 10.1016/0006-291x(68)90504-4. [DOI] [PubMed] [Google Scholar]
- Shen L. C., Atkinson D. E. Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J Biol Chem. 1970 Nov 25;245(22):5974–5978. [PubMed] [Google Scholar]
- Siess E., Wittmann J., Wieland O. Interconversion and kinetic properties of pyruvate dehydrogenase from brain. Hoppe Seylers Z Physiol Chem. 1971 Mar;352(3):447–452. doi: 10.1515/bchm2.1971.352.1.447. [DOI] [PubMed] [Google Scholar]
- Thompson P., Reid E. E., Lyttle C. R., Dennis D. T. Pyruvate dehydrogenase complex from higher plant mitochondria and proplastids: regulation. Plant Physiol. 1977 May;59(5):854–858. doi: 10.1104/pp.59.5.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. S., Burgett M. W., Reed L. J. Alpha-keto acid dehydrogenase complexes. XX. A kinetic study of the pyruvate dehydrogenase complex from bovine kidney. J Biol Chem. 1973 Dec 25;248(24):8348–8352. [PubMed] [Google Scholar]
- Wais U., Gillmann U., Ullrich J. Isolation and characterisation of pyruvate dehydrogenase complex from brewer's yeast. Hoppe Seylers Z Physiol Chem. 1973 Oct-Nov;354(10-11):1378–1388. doi: 10.1515/bchm2.1973.354.2.1378. [DOI] [PubMed] [Google Scholar]
- Wieland O. H., Hartmann U., Siess E. A. Neurospora crassa pyruvate dehydrogenase: interconversion by phosphorylation and dephosphorylation. FEBS Lett. 1972 Nov 1;27(2):240–244. doi: 10.1016/0014-5793(72)80630-6. [DOI] [PubMed] [Google Scholar]


