Abstract
Background & objectives:
Severe acute respiratory infection (SARI) is one of the leading causes of death among children worldwide. As different respiratory viruses exhibit similar symptoms, simultaneous detection of these viruses in a single reaction mixture can save time and cost. The present study was done in a tertiary care children's hospital for rapid identification of viruses causing SARI among children less than or equal to five years of age using multiplex real-time reverse transcription polymerase chain reaction (RT-PCR) kit.
Methods:
A total of 155 throat swabs were collected from equal number of children suspected to have SARI and processed for extraction of nucleic acids using automated extraction system. Multiplex real-time RT-PCR was done to identify the viruses in the samples.
Results:
The overall positivity for viruses in the study was found to be 72.9 per cent with a co-infection rate of 19.5 per cent. Human metapneumovirus (HMPV) was the predominant virus detected in 25.7 per cent children followed by influenza A (H1N1)pdm09, human rhinovirus (HRV) and human adenovirus (HAdV) in 19.9, 11.0 and 8.8 per cent children, respectively. The HMPV was at its peak in February 2013, HAdV showed two peaks in March-April, 2012 and November 2012-March 2013 while HRV was detected throughout the year.
Interpretation & conclusions:
Multiplex real-time PCR helped in rapid identification of viruses. Seventeen viruses were detected in SARI cases with overall positivity of 72.9 per cent. HMPV was the most predominant virus. However, for better clinico-virological correlation, studies are required with complete work up of all the aetiological agents, clinical profile of patients and treatment outcome.
Keywords: Human adenovirus, human metapneumovirus, human rhinovirus, influenza A (H1N1)pdm09, multiplex reverse transcription-polymerase chain reaction, respiratory syncytial virus, severe acute respiratory infection
Severe acute respiratory infections (SARIs) are defined as an acute respiratory illness of recent onset (within seven days) manifested by fever (≥38°C), cough and shortness of breath or difficulty in breathing requiring hospitalization1. The ARIs are the leading cause of deaths among children across the globe2 and result in an estimated 1.9 million deaths per year3, of which 70 per cent occur in developing countries4 accounting for 30 per cent of total childhood deaths5. The potential viral pathogens of ARIs include seasonal A and B influenza viruses, the new influenza A (H1N1)pdm09 virus strain, human metapneumovirus (HMPV), human rhinovirus (HRV), human adenovirus (HAdV), human parainfluenza viruses (HPIV), respiratory syncytial virus (RSV), human bocavirus (HBoV), human coronaviruses (HCoVs) and enterovirus (EV)4,6,7,8.
SARIs are usually diagnosed clinically and treated by antibiotics as per bacterial culture and susceptibility tests/empirically and supportive care in the absence of facilities for diagnosing viruses. Diagnosis of specific respiratory viral infection can help in proper management of patients by initiation of antiviral drug oseltamivir for influenza, discontinuation of unnecessary antibiotics, enhance supportive therapy, reduce the cost related to unnecessary investigations and reduce the hospital stay9,10. SARI, particularly in children being a serious condition demands rapid identification of causative agent.
Similar clinical signs and symptoms are caused by various respiratory viral pathogens; hence, rapid simultaneous identification of different viruses is of major epidemiological and clinical interest4. Many methods are available for diagnosis of respiratory viruses such as uniplex polymerase chain reaction (PCR), multiplex PCR, immunofluorescence assay, serology, etc. Immunofluorescence detects viral antigen rapidly, but lacks sensitivity and may require confirmation by viral culture11. Serology is also not useful clinically as antibodies appear after one week and require testing of paired serum samples for confirmation of infection. Multiplex real-time PCR assays allow simultaneous amplification of several viruses in a single mixture as compared to monospecific uniplex PCR assay in which separate amplification of each target has to be done which turns out to be more expensive, resource intensive and time consuming12. The present study was undertaken to rapidly identify the viruses causing SARI in children admitted to a hospital using the multiplex real-time reverse transcription-PCR (RT-PCR) method.
Material & Methods
The present study was conducted in the J. K. Lone Hospital, a paediatric hospital attached to SMS Medical College, Jaipur, India, over a period of 13 months (March, 2012 & March, 2013). Consecutive children aged less than or equal to five years, presenting with fever, cough, shortness of breath, sore throat and nasal catarrh to the hospital were included in the study. Those (n=61) with chronic respiratory ailments, non-consenting caregivers, with history of hospitalization in preceding 14 days, on antibiotics, having positive bacterial culture report or not admitted in hospital, and children aged greater than five years were excluded.
Considering the prevalence of ARIs in under five children in Rajasthan to be 7 per cent as reported by National Family Health Survey-3 (NFHS-3)13, the sample size was calculated as 104 using the formula n=4 pq/l2 [where n=total number of samples; 4 is the factor to achieve the confidence level of 95%; p=known prevalence; q=100-p and l=allowable permissible (absolute) error, set at 5%].
Sample collection and transportation: A total of 155 throat swab samples were collected from equal number of consecutive patients with SARI using a sterile nylon flocked swabs and placed in viral transport medium, labelled and transported on ice at the earliest to the laboratory. Informed consent was obtained from the parents/guardians of the patients. The study protocol was approved by the institutional ethics committee.
Nucleic acid extraction: Viral nucleic acid was extracted from 200 μl sample using NucliSENS EasyMAG automated nucleic acid extractor (Biomeuriex, France) as per manufacturer's instructions. At the final step, the nucleic acid was eluted in a volume of 55 μl and processed immediately for multiplex PCR. Internal control was added to each sample during nucleic acid extraction to validate the process of extraction and PCR technique used in the study.
Multiplex real-time reverse transcription polymerase chain reaction (RT-PCR): Multiplex RT-PCR assays were performed on ABI7500 Fast (Life Technologies, USA) using 1 μl AgPath-IDTM One-Step RT-PCR kit (Ambion, USA), 12.5 μl assay buffer and 1.5 μl primer probe mix (5 sets) of FTD respiratory pathogens 21 kit (Fast-track Diagnosis, Luxembourg) for the detection of (18 viruses, two subtypes RSV A & B, HMPV A & B and Mycoplasma pneumoniae) influenza A, influenza A (H1N1)pdm09, influenza B, HCoVs NL63 (HCoV-NL63), 229E (HCoV 229E), OC43 (HCoV-OC43) and HKU1 (HCoV HKU1), human parainfluenza viruses 1, 2, 3, and 4 (HPIV- 1, 2, 3 and 4), (HMPV A/B), HRV, RSV A/B, HAdV, EV, human parechovirus (HPeV), and HBoV. The multiplex real-time RT-PCR thermal profile was as follows: 50°C for 15 min, 95°C for 10 min, 40 cycles of 95°C for 8 sec, 60°C for 34 sec. The time taken for extraction, amplification and detection was three hours.
Statistical analysis: To determine the significance of the data, Chi-square test was employed.
Results
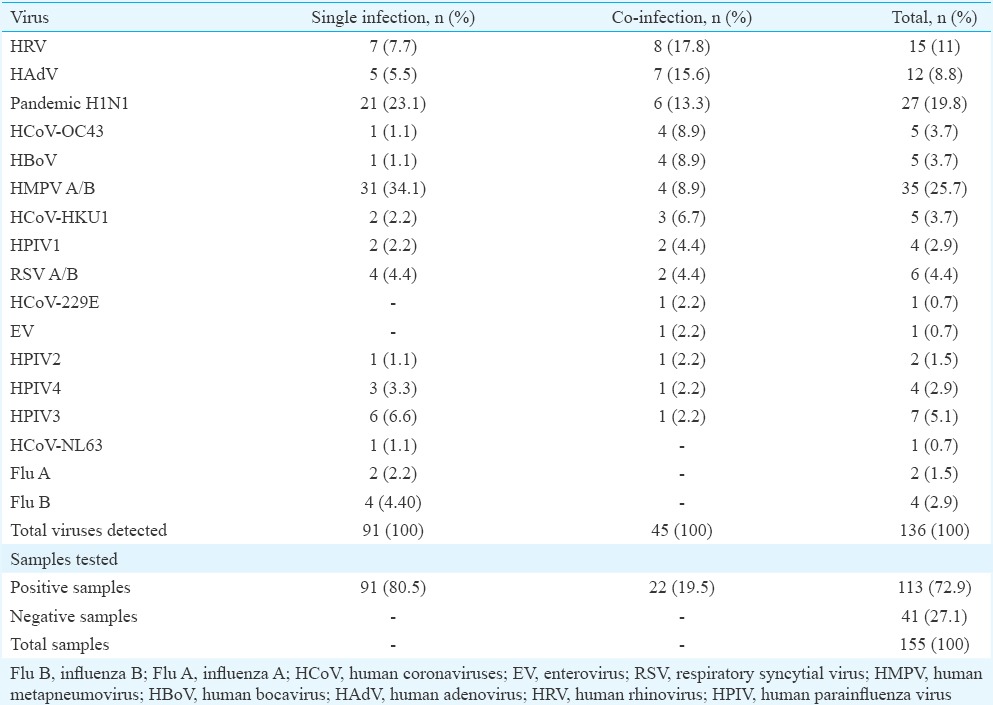
Details of samples collected as per age, months and sex are given in Table I; highest number of samples was collected between September and March (Table I). Of the 155 children, 113 (72.9%) were positive for at least one of the respiratory viruses, among whom co-infections were detected in 22 (19.5%) children. Single viral infections were observed in 91 (80.5%) children (Table II). The major viruses detected in the study (including single and co-infection) were HMPV A/B (25.7%), followed by influenza A (H1N1)pdm09 (19.8%), HRV (11%), HAdV (8.8%), HPIV3 (5.1%), RSVA/B (4.4%), HCoV-HKU1 (3.7%), HCoV-OC43 (3.7%), influenza B (2.9%), HPIV1 (2.9%), HPIV4 (2.9%), HBoV (3.7%), HPIV2 (1.5%), influenza A (1.5%), EV (0.7%), HCoV-229E (0.7%) and HCoV-NL63 (0.7%) (Table II). HMPV was found to be predominant in single infections whereas HRV was found to be predominant in co-infections. No human parechovirus (HPeV) was identified in the study. Four samples (2.6%) were found to be positive for bacterial agent Mycoplasma pneumoniae.
Table I.
Number of samples tested in each age group month-wise
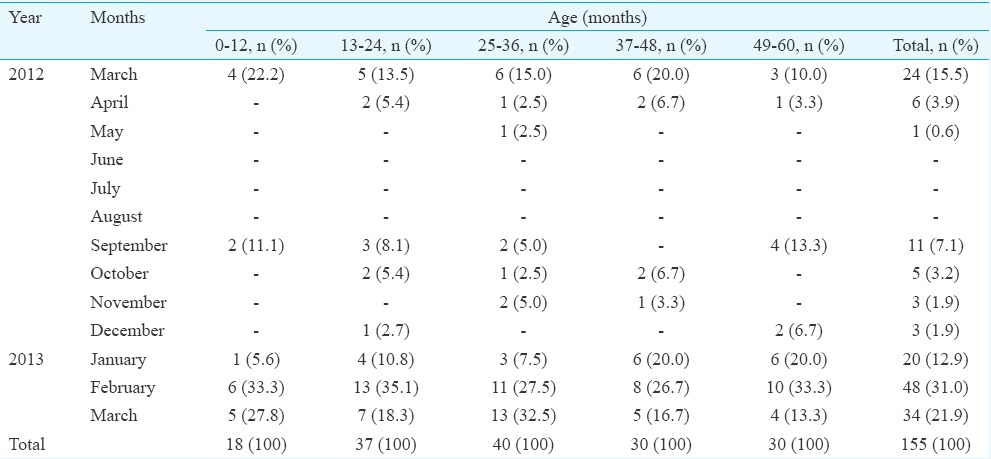
Table II.
Details of single and co-infections of respiratory viruses in children aged less than or equal to five years
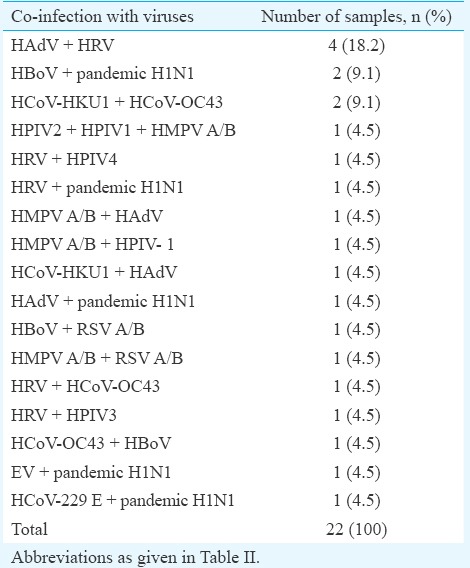
Severe acute respiratory infection (SARI) co-infections: Among the 22 children with co-infections, no co-infections with more than three viruses were observed (Table III). A total of six (27.3%) cases infected with coronaviruses were detected along with other respiratory viruses. Four (18.2%) children were infected with HRV and HAdV. Of the 27 children with influenza A (H1N1)pdm09 infection, co-infections were observed in six samples, with HBoV as the predominant virus.
Table III.
Details of co-infections with respiratory viruses in children aged less than or equal to five years
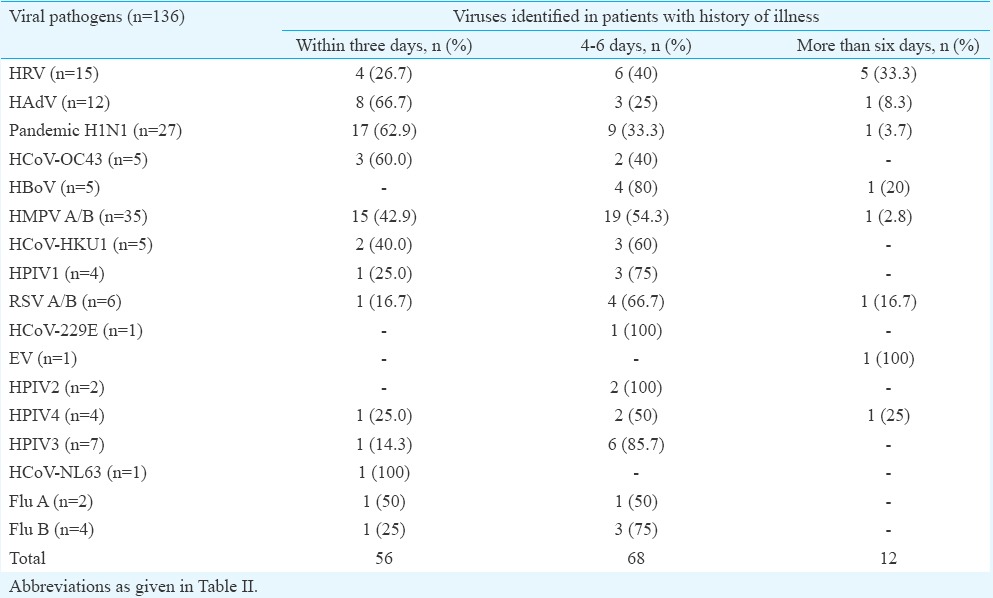
Clinical features among different viruses: Most of the patients presented with fever and cough followed by nasal catarrh, sore throat and shortness of breath (Table IV). The HMPV was predominant in cases presenting with fever and cough followed by influenza A (H1N1)pdm09 as compared to other viruses in the study. A total of 56 (41.2%) viruses were found in the samples collected within three days of illness, 68 (50%) in three to six days of illness and 12 (8.8%) in samples collected after more than six days of illness (Table V).
Table IV.
Clinical features in association with different respiratory viruses
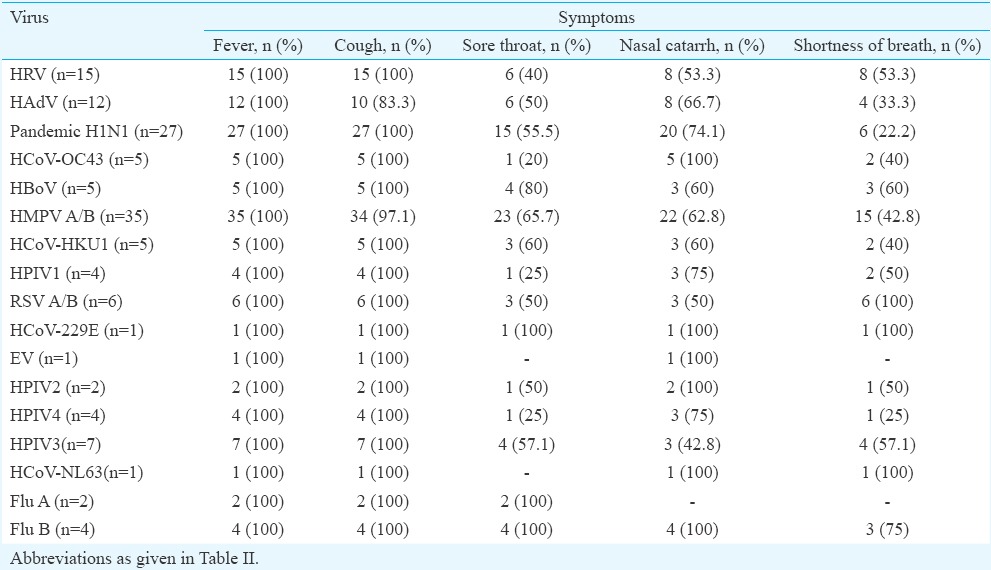
Table V.
The duration of illness on sampling day and viruses detected
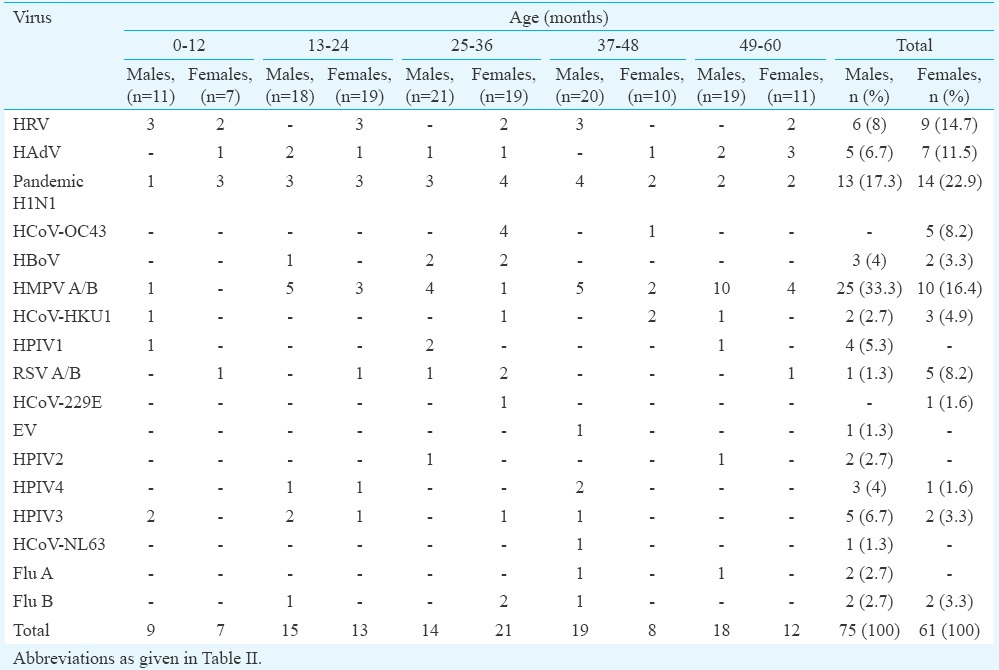
Infections among different age groups: Positivity of samples for respiratory viruses was found to be more in the age group 49-60 months in 27 (23.9%) children followed by 25-36 months in 26 (23%), 37-48 months in 23 (20.4%), 13-24 months in 23 (20.4%) and 14 (12.4%) children in the group 0-12 months. Of the viruses observed in various age groups, rhinovirus was the most predominant virus in the age group 0-12 months (5/16, 31.3%), whereas in the age groups 13-24, 37-48 and 49-60 months, HMPV was the most predominant virus with 8/28 (28.6%), 7/27 (23.3%) and 14/30 (46.7%), respectively. In the age group of 25-36 months, the most predominant virus was influenza A (H1N1)pdm09 [7/35 (20%)] (Fig. 1 & Table VI).
Fig. 1.
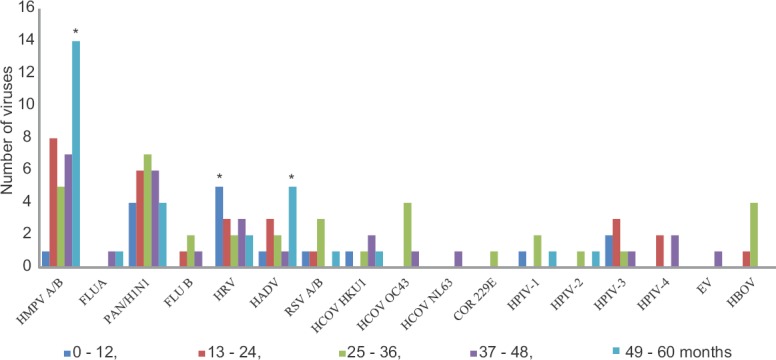
Graph showing the distribution of different respiratory viruses among children of different age groups (in months). *P < 0.05 (χ2 test, comparison among the different age groups). HMPV A/B, human metapneumovirus A/B; FLUA, influenza A; PAN/H1N1, Pandemic H1N1; FLUB, influenza B; HRV, human rhinovirus; HADV, human adenovirus; RSV A/B, respiratory syncytial virus A/B; HCOV HKU1, human coronavirus HKU1; HCOV OC43, human coronavirus OC43; HCOV NL63, human coronavirus NL63; COR 229E, human coronavirus COR 229E; HPIV-1, human parainfluenza virus 1; HPIV-2, human parainfluenza virus 2; HPIV-3, human parainfluenza virus 3; HPIV-4, human parainfluenza virus 4; EV, enterovirus; HBOV, human bocavirus.
Table VI.
Gender-wise break up of viruses detected and the number of children enrolled in each age group
The HMPV (P<0.001) and HAdV were found to be predominantly positive in the age group of 49-60 months (P<0.05) whereas rhinovirus was predominant in the age group 0-12 months (P=0.01) stating that the infections caused were highly significant among the children when age-wise intergroup comparisons were made for each virus (Fig. 1).
Seasonal trends of respiratory viruses: HMPV was the predominant isolate in the study with peak incidence in February 2013. HAdV was found to be circulating between March-April 2012 with a second peak between November 2012 and March 2013. HRV was circulating throughout the year with a peak at the end of winter in March 2013. RSV A/B was detected during winter from September-March and showed a peak in September 2012. The HPIV3 showed a peak in March 2013. Influenza A (H1N1)pdm09 started to appear in September 2012 with the onset of winter and showed a peak during February and March 2013 with the end of winter season (Fig. 2).
Fig. 2.
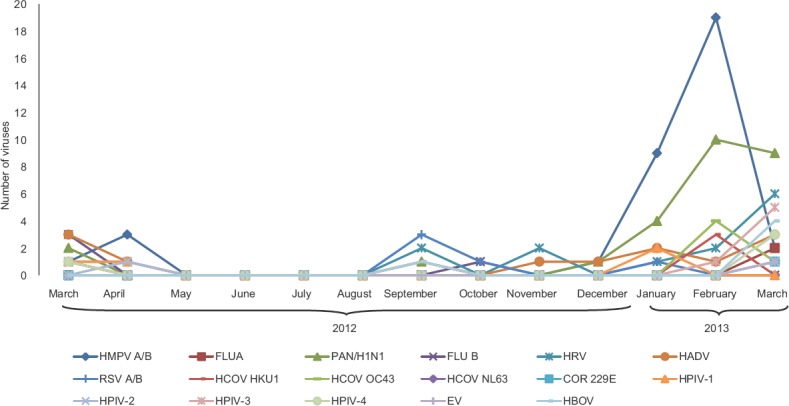
Seasonal distribution of different respiratory viruses among children. Abbreviations as given in Fig.1.
Gender-wise occurrence of respiratory viral infections: The occurrence of respiratory viruses in males was found to be higher (75/136; 55.1%) compared to that in females (61/136; 44.8%). Human metapneumovirus was found to be predominantly occurring in males as compared to females. In contrast, influenza A (H1N1)pdm09, HRV, HAdV, RSV, HCoV-HKU1, HCoV-229E, HCoV-OC43 were found to be more in females than in males (Table VI).
Discussion
The predominant isolate in the present study was found to be HMPV with 25.7 per cent of children showing positivity. Studies by Maggi et al14 from Italy reported that HMPV infections ranged between 7 and 43 per cent over a period of three years, whereas other studies revealed that HMPV infections were secondary to other ARI, and had low positivity of 8.03 per cent infections3 and 9.4 per cent in hospitalised children15. Influenza A (H1N1)pdm09 was the second most predominant virus detected in the present study with a positivity of 19.9 per cent during post-pandemic period. This was high when compared to other studies done in Madagascar (1.70%) and China (2.80%) during pandemic period of 2009-20102,16. However, the results were similar to an Indian study done at Ballabgarh which reported a positivity of 17.3 per cent during the pandemic period17. The increased positivity of influenza A (H1N1)pdm09 during post-pandemic period in the present study may be due to survival of the virus for prolonged period due to climatic conditions.
The HRV and adenovirus were detected in a total of 11.0 and 8.8 per cent patients, respectively. Rhinovirus positivity in the present study varied when compared to earlier study which reported 18.4 per cent in hospitalized patients with varying degree of respiratory illness18, whereas the positivity of the adenovirus was similar with earlier studies which reported between 6.72 and 10.2 per cent2,19 in patients with influenza-like illness (ILI) and lower respiratory infection (LRI). The presence of RSV in the present study was found to be low (4.41%) when compared with other studies reporting 13.1-21.3 per cent2,20 in patients with different degrees of respiratory illness and acute LRI. The reason might be due to low number of children enrolled in the age group of 0-12 months as RSV infection was more common in them. Further, use of throat swab only for collecting samples might have limited the detection of RSV. Addition of nasopharyngeal swab could have increased the yield21.
Coronavirus infections in the present study were found to be 8.8 per cent. Papenburg et al15 in hospitalized patients with acute respiratory tract infections revealed a total of 6 per cent coronaviral infections. The parainfluenza virus infection was seen in 17 (12.5%) children. The results from another study revealed similar pattern with a total of 11.8 per cent children infected with parainfluenza viruses16. However, an Indian study reported no positivity of HPIV1-4 in hospitalized children with ALRI20. A total of 22 (19.5%) co-infections were identified in the present study which was identical with an earlier study20. The frequency of respiratory viral co-infections varied widely in different studies globally (10 - 41%)22,23,24,25.
Fever was the major clinical feature in the present study followed by cough and nasal catarrh as also reported in other studies5,26. However, respiratory infections caused by these viruses usually present clinical features that are nearly indistinguishable5,27.
There were seasonal variations in the trends of infections of viruses, with two peaks in a year. The present study revealed the circulation of HRV throughout the year which was similar to an earlier study6. However, the seasonal trends of the respiratory viruses seen in the present study varied from other studies28,29. The variations in the seasonal distribution of different respiratory viruses may be attributed to the different geographical locations, viral aetiological agents and the methodologies used in the study.
Most of the respiratory viruses were found in the age group 49-60 months followed by 25-36 months old children. Similar findings have been made previously with predominant infections in the children of age less than five years21. The distribution of respiratory viruses in males was found to be higher as compared to females. Khor et al29 also showed higher proportion of infection in male children over females.
The present study had a few limitations, such as detailed clinico-virological correlation was not done with the severity of disease, duration of hospitalization, use of invasive or non-invasive ventilator, outcome of treatment, mortality, etc. in relation to virus identified. SARI can also be caused by various bacterial agents that are fastidious and not isolated in routine bacterial culture such as Chlamydia pneumoniae, Legionella spp., Bordetella spp. etc.30; moreover, Streptococcus pneumoniae and Haemophilus influenzae may fail to grow on culture very often. Therefore, testing all these bacteria also by PCR may help in increasing yield and rapid diagnosis. Other rare viruses such as influenza C, cytomegalovirus may also be tested in samples negative for the first panel. Though throat swab has been reported to give good yield for all viruses, positivity can be further improved by also taking nasopharyngeal swab, nasal swab, tracheal aspirate, bronchoalveolar lavage21.
The present results provided preliminary understanding of the viral agents associated with SARI. High positivity of 72.9 per cent was found, detecting 17 viruses with HMPV being the predominant virus, followed by HRV and HAdV and overall co-infectivity of 19.5 per cent. There is a need for conducting further studies to detect broader range of viruses and other fastidious organisms, to undertake detailed clinical work up for better clinico-virological correlation, and to carry out additional follow up bacterial culture to understand role of secondary bacterial infection.
Acknowledgment
The authors acknowledge the Indian Council of Medical Research (ICMR), New Delhi, for financial support to the first author (BM) through ICMR Grade-I Virology Laboratory and Senior Research Fellowship to the second author (MAS).
Footnotes
Conflicts of Interest: None.
References
- 1.World Health Organization. Regional Office for Europe. Overview of sentinel systems for hospitalized severe acute respiratory infections (SARI) represented in the weekly EuroFlu surveillance bulletin (as of 10 February 2013) [accessed on May 5, 2013]. Available from: http://www.euro.who.int/__data/assets/pdf_file/0005/186863/Overview-of-SARI-Surveillance-Systemsfinal.pdf .
- 2.Li H, Wei Q, Tan A, Wang L. Epidemiological analysis of respiratory viral etiology for influenza-like illness during 2010 in Zhuhai, China. Virol J. 2013;10:143. doi: 10.1186/1743-422X-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parveen S, Sullender WM, Fowler K, Lefkowitz EJ, Kapoor SK, Broor S. Genetic variability in the G protein gene of group A and B respiratory syncytial viruses from India. J Clin Microbiol. 2006;44:3055–64. doi: 10.1128/JCM.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002;2:25–32. doi: 10.1016/s1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- 5.Bharaj P, Sullender WM, Kabra SK, Mani K, Cherian J, Tyagi V, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89. doi: 10.1186/1743-422X-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujitsuka A, Tsukagoshi H, Arakawa M, Goto-Sugai K, Ryo A, Okayama Y, et al. A molecular epidemiological study of respiratory viruses detected in Japanese children with acute wheezing illness. BMC Infect Dis. 2011;11:168. doi: 10.1186/1471-2334-11-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renois F, Talmud D, Huguenin A, Moutte L, Strady C, Cousson J, et al. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription-PCR DNA microarray systems. J Clin Microbiol. 2010;48:3836–42. doi: 10.1128/JCM.00733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J Virol Methods. 2009;156:111–6. doi: 10.1016/j.jviromet.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xuan C, Yan L, Zegang W. Rapid detection of acute respiratory virus and atypical bacteria infections in children. Jundishapur J Microbiol. 2013;6:e6236. [Google Scholar]
- 10.Loeffelholz M, Chonmaitree T. Advances in diagnosis of respiratory virus infections. Int J Microbiol 2010. 2010 doi: 10.1155/2010/126049. 126049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, Wesselingh S. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol. 2001;39:2779–83. doi: 10.1128/JCM.39.8.2779-2783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Templeton KE, Scheltinga SA, Beersma MFC, Kroes ACM, Claas ECJ. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J Clin Microbiol. 2004;42:1564–9. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Family Health Survey (NFHS-3) September. 2007. [accessed on May 5, 2013]. Available from: http://rchiips.org/nfhs/NFHS-3%20Data/VOL-1/India_volume_I_corrected_17oct08.pdf .
- 14.Maggi F, Pifferi M, Vatteroni M, Fornai C, Tempestini E, Anzilotti S, et al. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J Clin Microbiol. 2003;41:2987–91. doi: 10.1128/JCM.41.7.2987-2991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papenburg J, Hamelin MÈ, Ouhoummane N, Carbonneau J, Ouakki M, Raymond F, et al. Comparison of risk factors for human metapneumovirus and respiratory syncytial virus disease severity in young children. J Infect Dis. 2012;206:178–89. doi: 10.1093/infdis/jis333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann J, Rabezanahary H, Randriamarotia M, Ratsimbasoa A, Najjar J, Vernet G, et al. Viral and atypical bacterial etiology of acute respiratory infections in children under 5 years old living in a rural tropical area of Madagascar. PLoS One. 2012;7:e43666. doi: 10.1371/journal.pone.0043666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broor S, Sullender W, Fowler K, Gupta V, Widdowson MA, Krishnan A, et al. Demographic shift of influenza A(H1N1)pdm09 during and after pandemic, rural India. Emerg Infect Dis. 2012;18:1472–5. doi: 10.3201/eid1809.111847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bicer S, Giray T, Çöl D, Erdag GÇ, Vitrinel A, Gürol Y, et al. Virological and clinical characterizations of respiratory infections in hospitalized children. Ital J Pediatr. 2013;39:22. doi: 10.1186/1824-7288-39-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwofie TB, Anane YA, Nkrumah B, Annan A, Nguah SB, Owusu M. Respiratory viruses in children hospitalized for acute lower respiratory tract infection in Ghana. Virol J. 2012;9:78. doi: 10.1186/1743-422X-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh AK, Jain A, Jain B, Singh KP, Dangi T, Mohan M, et al. Viral aetiology of acute lower respiratory tract illness in hospitalised paediatric patients of a tertiary hospital: one year prospective study. Indian J Med Microbiol. 2014;32:13–8. doi: 10.4103/0255-0857.124288. [DOI] [PubMed] [Google Scholar]
- 21.Do AH, van Doorn HR, Nghiem MN, Bryant JE, Hoang TH, Do QH, et al. Viral etiologies of acute respiratory infections among hospitalized Vietnamese children in Ho Chi Minh City, 2004-2008. PLoS One. 2011;6:e18176. doi: 10.1371/journal.pone.0018176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esper FP, Spahlinger T, Zhou L. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J Infect. 2011;63:260–6. doi: 10.1016/j.jinf.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand HK, de Groot R, Galama JM, Brouwer ML, Teuwen K, Hermans PW, et al. Infection with multiple viruses is not associated with increased disease severity in children with bronchiolitis. Pediatr Pulmonol. 2012;47:393–400. doi: 10.1002/ppul.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huijskens EG, Biesmans RC, Buiting AG, Obihara CC, Rossen JW. Diagnostic value of respiratory virus detection in symptomatic children using real-time PCR. Virol J. 2012;9:276. doi: 10.1186/1743-422X-9-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida LM, Suzuki M, Yamamoto T, Nguyen HA, Nguyen CD, Nguyen AT, et al. Viral pathogens associated with acute respiratory infections in central vietnamese children. Pediatr Infect Dis J. 2010;29:75–7. doi: 10.1097/INF.0b013e3181af61e9. [DOI] [PubMed] [Google Scholar]
- 26.Debbia EA, Schito GC, Zoratti A, Gualco L, Tonoli E, Marchese A. Epidemiology of major respiratory pathogens. J Chemother. 2001;13:205–10. doi: 10.1179/joc.2001.13.Supplement-2.205. [DOI] [PubMed] [Google Scholar]
- 27.Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–7. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed JA, Katz MA, Auko E, Njenga MK, Weinberg M, Kapella BK, et al. Epidemiology of respiratory viral infections in two long-term refugee camps in Kenya, 2007-2010. BMC Infect Dis. 2012;12:7. doi: 10.1186/1471-2334-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khor CS, Sam IC, Hooi PS, Quek KF, Chan YF. Epidemiology and seasonality of respiratory viral infections in hospitalized children in Kuala Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatr. 2012;12:32. doi: 10.1186/1471-2431-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization Regional Office for Africa Brazzaville 2015. Protocol for the investigation of acute respiratory illness outbreaks of unknown etiology. [accessed on May 5, 2016]. Available from: http://www.afro.who.int/index.php?option=com_docman&task=doc_download&gid=10087&Itemid=2593 .


