Abstract
Genetic alterations in the fibroblast growth factor receptor (FGFR) pathway are promising therapeutic targets in many cancers, including intrahepatic cholangiocarcinoma (ICC). The FGFR inhibitor BGJ398 displayed encouraging efficacy in patients with FGFR2 fusion-positive ICC in a phase II trial, but the durability of response was limited in some patients. Here, we report the molecular basis for acquired resistance to BGJ398 in three patients via integrative genomic characterization of cell-free circulating tumor DNA (cfDNA), primary tumors, and metastases. Serial analysis of cfDNA demonstrated multiple recurrent point mutations in the FGFR2 kinase domain at progression. Accordingly, biopsy of post-progression lesions and rapid autopsy revealed marked inter- and intra-lesional heterogeneity, with different FGFR2 mutations in individual resistant clones. Molecular modeling and in vitro studies indicated that each mutation lead to BGJ398 resistance and was surmountable by structurally distinct FGFR inhibitors. Thus, polyclonal secondary FGFR2 mutations represent an important clinical resistance mechanism that may guide development of future therapeutic strategies.
Keywords: BGJ398, Fibroblast Growth Factor Receptor, Intrahepatic Cholangiocarcinoma, Resistance Mutations, cell free DNA
Introduction
The FGFR pathway includes a family of 22 polypeptide ligands (FGFs) and 4 receptor tyrosine kinases (FGFRs) which regulate diverse physiologic processes [reviewed in (1)]. FGFR signaling is activated through recurrent gain-of-function mutations, chromosomal translocations, and amplifications in a variety of cancers including squamous non-small cell lung cancers, head and neck squamous cell carcinomas, breast cancers, urothelial carcinomas, and intrahepatic cholangiocarcinomas (ICCs) [reviewed in (2)]. Thus, the recent clinical development of potent and selective FGFR kinase inhibitors has opened a promising therapeutic avenue in many previously intractable malignancies.
Genomic alterations that activate FGFR2 are particularly common in ICC, a malignancy of the intrahepatic bile ducts that has been rising in incidence both in the United States and globally for decades (3, 4). As an aggressive tumor originating in the liver, ICC often remains asymptomatic until reaching an advanced stage, precluding curative therapy in a majority of cases. Palliative chemotherapy with gemcitabine and a platinum agent offers patients with unresectable or metastatic disease a median survival of less than one year (5). While no targeted agents are currently approved in ICC, the recent discovery of FGFR2 fusions in an estimated 10-20% of patients stands to change the therapeutic paradigm in this disease. Multiple fusion partners have been identified, including BICC1, AHCYL1, TACC3, MGEA5, and PPHLN1 (6-12). In most cases, the 5′ exons 1-17 of FGFR2, containing the intact kinase domain, are fused in-frame to a 3′ partner. The chimeric proteins are believed to undergo ligand-independent receptor dimerization leading to engagement of multiple downstream oncogenic pathways including MAPK and PI3K/AKT signaling (13).
The oral, selective, and ATP-competitive pan-FGFR inhibitor BGJ398 has demonstrated anti-tumor activity in preclinical models and a phase I study of patients with tumors harboring FGFR genetic alterations (14-16). This agent is currently being tested in a phase II multicenter trial in patients with advanced cholangiocarcinoma with FGFR aberrations who have progressed on first line chemotherapy (NCT02150967). The reported preliminary data from this trial highlight an impressive objective response rate of 22% and a median time on treatment of 188 days (17). This efficacy of targeted therapy beyond the first line in ICC is unprecedented. However, as seen with other targeted therapies and kinase inhibitors in particular (18, 19), acquired resistance inevitably develops.
Here, we present the integrative molecular analysis of cell-free circulating tumor DNA (cfDNA), primary tumors, and metastases to define the acquired resistance mechanisms to BGJ398 in three patients with advanced FGFR2-fusion+ ICC. These combined analyses revealed the emergence of secondary kinase mutations that confer BGJ398 resistance in each patient at the time of progression. A striking degree of inter-lesional heterogeneity was observed, with distinct FGFR2 point mutations identified in different metastases from the same patient. Overall, these data suggest that secondary FGFR2 kinase domain mutations are an important mechanism of clinical acquired resistance to FGFR inhibitors, and that next-generation inhibitors capable of overcoming these resistance mutations may be important future clinical strategies for these cancers.
Results
Clinical acquired resistance to BGJ398
Among 32 patients with ICC screened with our internal MGH Solid Fusion Assay (SFA) as part of routine clinical care, nine (28%) tested positive for an FGFR2 fusion and four of them enrolled in the phase II trial of BGJ398. Three of these four patients experienced significant tumor regression between -28 and -50% followed by short interval disease progression. Although this represents a few of the total number of patients enrolled in the trial and thus may not accurately reflect the true efficacy of BGJ398 in this population, we highlight these three patients to report early genetic evidence of acquired resistance to FGFR inhibition with BGJ398.
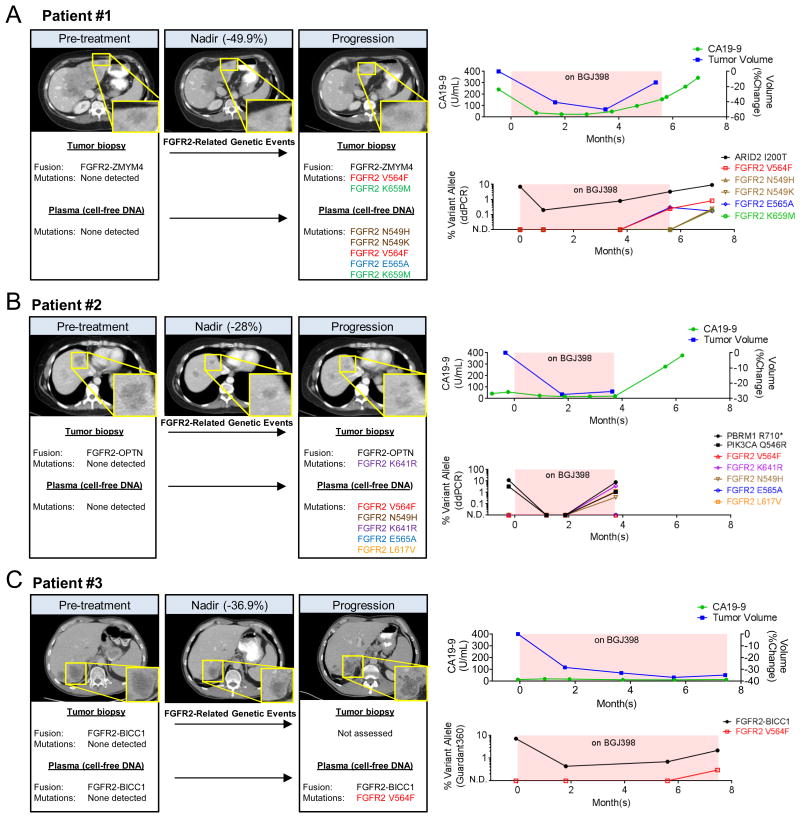
Patient #1 was a 59 year old female with an unresectable ICC, and she was initially treated with gemcitabine and cisplatin for 10 months. Molecular testing of the tumor tissue with the SFA , a clinical test capable of detecting fusion events in over 50 cancer-related genes (20), revealed a novel in-frame fusion between FGFR2 exon 17 and exon 10 of the Zinc Finger MYM-Type protein 4 (ZMYM4) gene. Following progression on chemotherapy, she underwent a repeat liver biopsy (pre-treatment) and then initiated treatment on BGJ398. The patient achieved a -40.6% response by RECISTv1.1 criteria at 2 months and a -49.9% response at four months (Figure 1A, left panel and right-upper panel). However, the 6 month CT scan showed a mixed response with progression of 2 satellite left lobe lesions. The patient's CA 19-9 decreased from 242 to 48, and then rose to 156 U/mL during this time (Figure 1A, right-upper panel).
Figure 1. Clinical acquired resistance to BGJ398.
A-C. Left panels. Axial contrast enhanced CT images in the portal venous phase demonstrating initial response lesions that subsequently progressed despite BGJ398 therapy. Inset shows higher magnification of responsive/resistant lesion. FGFR2-related genetic events detected by Solid Fusion Assay (SFA), RNA sequencing, whole-exome sequencing (WES), or the Guardant360 assay are documented in the text below CT images. Right upper panels, Graphs illustrating serial CA 19-9 serum levels and tumor volume measurements by RECIST v1.1 criteria for Patients 1-3, respectively. The pink box indicates time while the patient was receiving BGJ398. Right-lower panels, Percent allele burden in cfDNA of the indicated mutations from Patient #1 (A), Patient #2 (B), and Patient #3 (C) were monitored over time. The pink box indicates time while the patient was receiving BGJ398. N.D. = not detected.
Patient #2 was a 47 year old female who underwent a left hepatectomy for a 10.9 cm, T2N0 ICC, and received adjuvant chemotherapy. Five months later, she developed a solitary biopsy-proven liver metastasis (pre-treatment) and multiple subcentimeter bilateral pulmonary metastases. SFA testing on the liver metastasis revealed a different novel fusion, this time between FGFR2 exon 17 and the Optineurin (OPTN) exon 5. The patient was then enrolled in the BGJ398 trial, achieving a maximum response of -28% at 2 months (Figure 1B, left panel and right-upper panel). The 4 month CT scan revealed a mixed response with a new 1.3 cm lesion appearing in the dome of the liver. The CA 19-9 also initially decreased from 57 to 17, but then increased marginally to 21 U/mL during this period (Figure 1B, right-upper panel). The patient passed away 3 months later.
Patient #3 was a 43 year old man who presented with a 7 cm ICC involving the right hepatic lobe with metastases to the lymph nodes and lungs, and his tumor progressed through initial palliative chemotherapy. SFA testing revealed a fusion between FGFR2 exon 17 and BICC1 exon 3. The patient enrolled in the BGJ398 trial and achieved a response of -28.4% at 2 months and a maximum response of -36.9% at 6 months. The 8 month scan showed a mixed response with growth of multiple liver lesions and lymph nodes (Figure 1C, left panel and right-upper panel). CA19-9 levels remained within normal limits throughout therapy and therefore did not serve as a reliable surrogate for tumor burden in this patient.
Detection of secondary FGFR2 resistance mutations in cfDNA
To identify potential mechanisms of acquired resistance, we analyzed pre-treatment and post-progression cfDNA from all three patients for mutations in 70 cancer-related genes and selected fusions, insertions/deletions and copy number amplifications using the digital sequencing-based Guardant360 assay (21). Remarkably, analysis of the post-progression cfDNA from Patient #1 showed five independent point mutations in FGFR2 (p.N549H, p.N549K, p.V564F, p.E565A and p.K659M), none of which were detected in the baseline plasma (Figure 1A, left panel). Similarly, analysis of cfDNA from Patient #2 also revealed five FGFR2 point mutations (p.N549H, p.V564F, p.E565A, p.L617V, and p.K641R) exclusively in post-progression specimens (Figure 1B, left panel). Finally, cfDNA from Patient #3, who continued on therapy for the longest period of time, contained a single FGFR2 point mutation (p.V564F) at the time of progression (Figure 1C, left panel). Interestingly, the p. V564F point mutation was identified in all three cases, further highlighting its relevance.
We monitored the specific mutant allele frequencies in cfDNA in a set of serially collected plasma samples from patients #1 and #2 (N=5 and N=4 samples, respectively) using droplet digital PCR (ddPCR). Since detection of gene fusions across all cfDNA specimens is technically challenging, we examined ‘truncal’ mutations identified by whole-exome sequencing of pre-treatment tumor biopsy as a surrogate for overall tumor burden in cfDNA (ARID2 p.I200T in Patient #1 and PBRM1 p.R710* in Patient #2). Both alterations decreased initially with treatment but rose with disease progression (Figure 1A and B, lower-right panel), mirroring radiological assessments. By contrast, FGFR2 point mutations were not detected at baseline, but each arose at 2 to 6 months after response, coincident with clinical progression. In patient #2, the FGFR2 p.V564F and p.E565A mutations were detected by Guardant360 at 6 months, but were not yet detected at 4 months by ddPCR, when they were likely present at lower allele frequencies below the assay's limit of detection. In Patient #3, NGS analysis of cfDNA using the Guardant360 assay identified this specific FGFR2-BICC1 fusion in each of three serial samples collected and revealed a decrease upon initiation of BGJ398 and an increase at the time of radiological progression. Concomitantly with progression, an increase of the FGFR2 p.V564F mutant levels was detected (Figure 1C, lower-right panel) with cfDNA NGS. The development of secondary FGFR2 kinase domain mutations in all three patients suggests that such mutations are an important mechanism of acquired resistance to FGFR inhibition.
Whole exome and RNA sequencing of serial tumor biopsies
Paired pre-treatment and post-progression biopsy samples available from Patients #1 and #2 were analyzed by whole-exome sequencing (WES) and RNA sequencing to determine intratumoral genomic changes. Importantly, RNA sequencing confirmed the presence of the fusion alleles in the post-progression biopsies in both cases (Patient #1, FGFR2-ZMYM4; Patient #2, FGFR2-OPTN) (Figures 1A and B, right panels), suggesting that clinical progression resulted from the acquisition of resistance by the FGFR2-fusion+ tumor cells rather than the outgrowth of fusion-negative subclones. A subset of the FGFR2 kinase domain mutations observed in the cfDNA were detected by WES of the post-progression biopsies, whereas none were found in the pre-treatment samples. These secondary mutations included FGFR2 p.V564F and p.K659M (Patient #1) and FGFR2 p.K641R (Patient #2) (Figures 1A and B, left panels). To determine whether these mutations occurred in the full-length FGFR2 allele or the FGFR2 fusion allele, we reverse transcribed either full-length FGFR2 or FGFR2-ZMYM4 mRNA from our post-progression biopsy sample for Patient #1 and subjected both products to Sanger sequencing. Indeed, the p.K569M mutation could only be detected on the FGFR2-ZMYM4 allele, while the full-length FGFR2 had no evidence of mutations (Supplementary Figure S1A-B). Overall, these data are consistent with convergent evolution of multiple individual FGFR2 resistance mutations arising in distinct metastatic lesions or in different tumor cells within the same metastatic lesion. These findings also reinforce the inadequacy of single-lesion tumor biopsy to capture the full heterogeneity of resistance mechanisms present in a single patient and illustrate the potential of cfDNA analysis toward this purpose.
Functional modeling of secondary FGFR2 resistance mutations
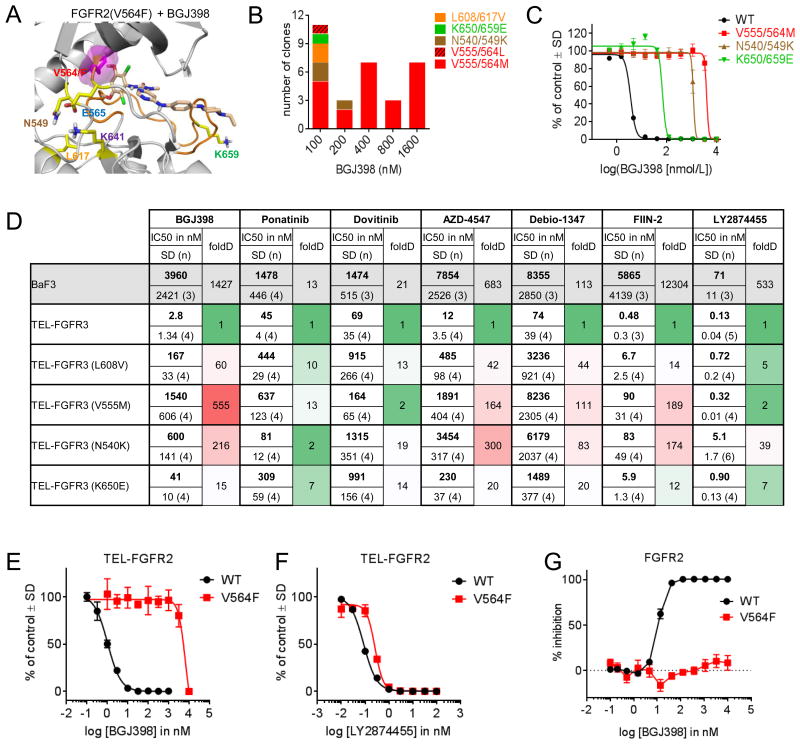
All together, six amino acids were affected across the three patients. Notably, structural modeling predicts that each mutation compromises inhibition by BGJ398. The p.V564F gatekeeper mutation, which was common to all three patients, confers resistance by inducing a steric clash with BGJ398 in its FGFR2 binding pocket (Figure 2A). V564 is in direct contact with the di-chloro, di-methoxy phenyl ring of BGJ398. A residue with a bulkier side chain clashes with this part of the inhibitor.
Figure 2. FGFR point mutations confer resistance to BGJ398 and other FGFR inhibitors.
A. In silico model of BGJ398 in binding pocket of FGFR2, demonstrating steric clash in the context of a V564F mutation. B and C. Mutagenesis screen in BaF3 cells which were engineered to express a TEL-FGFR3 fusion protein and subjected to increasing doses of BGJ398. The bar graph in B indicates the number of BGJ398-resistant clones isolated with indicated FGFR3 point mutations, with higher doses of BGJ398 resulting exclusively in colonies harboring the V555M gatekeeper mutation. Proliferation was quantified in C after 3 days with Alamar blue. Corresponding FGFR2 amino acids are indicated after the ‘/’. D. IC50 values for BaF3 cells expressing the indicated constructs and treated with a variety of inhibitors. E and F. Proliferation assays with BaF3 cells expressing the FGFR2 V564F constructs and treated with increasing doses of either BGJ398 (E) or LY287445 (F). G. Phospho-FGFR2 ELISA demonstrating that FGFR2 V564F is resistant to inhibition by BGJ398 compared to wild-type.
Interestingly, although none of the other five residues are in direct contact with BGJ398, their mutation destabilizes the inactive conformation of the kinase to which BGJ398 binds. The p.N549, p.E565, and p.K641 triad define a molecular brake that maintains the kinase in an inactive conformation. Thus, mutation of these residues leads to activation of the kinase (22). Mutation of the p.K659 residue also leads to FGFR2 kinase activation by stabilizing the active conformation of its activation loop (23). Similarly, the effect of the p.L617 mutation is to weaken a stabilizing interaction of this residue with the phenylalanine residue of the FGFR2 Asp-Phe-Gly (DFG) motif, which normally adopts a special conformation favoring the binding of inhibitors in the BGJ398 class. Thus, acquired resistance to BGJ398 is associated with the emergence of recurrent and functionally relevant FGFR2 kinase domain mutations in cfDNA.
To model the development of resistance mutations within the FGFR2 kinase domain following exposure to BGJ398, we performed a mutagenesis screen using BaF3 cells engineered to express a TEL-FGFR2 fusion protein. Under growth factor restricted conditions, these cells were dependent on FGFR2 signaling and were thus sensitive to BGJ398 inhibition. Nevertheless, exposure to lethal doses resulted in the rapid development of resistant cells. Sanger sequencing of pooled resistant clones identified FGFR2 p.N549D and p.V564I mutations (data not shown), both corresponding to amino acid residues observed in the setting of clinical resistance. Since FGFR2 and FGFR3 share 89% amino acid homology in their kinase domains (Supplementary Figure S2A-C), all six corresponding residues in FGFR3 would be expected to have the same role in either inducing steric clash with BGJ398 or stabilizing the active kinase conformation. Accordingly, exposure of TEL-FGFR3-expressing BaF3 cells to lethal doses of BGJ398 resulted in the rapid formation of resistant colonies harboring five distinct FGFR3 mutations, all of which resulted in amino acids changes that corresponded to those seen in FGFR2 in Patients #1-3. At the lowest dose (100 nM), 10 individual clones were isolated, harboring FGFR3 p.N540K, p.V555L, p.V555M, p.L608V, and p.K650E mutations, corresponding to FGFR2 p.N549, p.V564, p.L617, and p.K659, respectively. By contrast, higher doses of BGJ398 resulted exclusively in colonies harboring the p.V555M gatekeeper mutation (Figure 2B). These mutations decreased the sensitivity of BaF3 cells by 10-1000 fold, with p.V555M conferring the greatest degree of resistance, consistent with the appearance of the corresponding FGFR2 mutation in all three patients and the predominance of this resistant clone at higher doses of BGJ398 (Figure 2C).
Importantly, the potential for a convergent resistance mechanism involving the FGFR2 kinase domain may have major implications for the selection of subsequent treatment strategies, as irreversible or structurally distinct FGFR inhibitors have been designed to overcome specific FGFR resistance mutations in vitro (24) and are already in clinical trials. Accordingly, we compared the sensitivity of individual resistant clones to a panel of five FGFR inhibitors that are currently in clinical trials, as well as the irreversible inhibitor FIIN-2. Each drug displayed a different profile, with LY2874455 showing the smallest reduction in activity against the different resistance mutations overall, and with ponatinib and dovitinib showing the smallest change against the N540/549K mutation in particular (Figure 2D). Interestingly, LY2874455, rather than FIIN-2, was by far the most potent compound tested against all of the different resistance mutations.
To validate the effects of the key gatekeeper mutations in FGFR2, we introduced p.V564F mutation into a retroviral vector bearing TEL-FGFR2 by site-directed mutagenesis and infected BaF3 cells, rendering them IL-3 independent. Consistent with our observations in the FGFR3 system, this mutation conferred resistance to BGJ398 but retained sensitivity to LY2874455 (Figure 2E and 2F). Similarly, BGJ398 was unable to inhibit phosphorylation of FGFR2 p.V564F when overexpressed in 293T cells (Figure 2G). Thus, different FGFR inhibitors may have unique abilities to overcome specific secondary mutations arising in cancers with FGFR2 or FGFR3 alterations.
Rapid autopsy reveals inter-lesional and intra-lesional heterogeneity of resistance
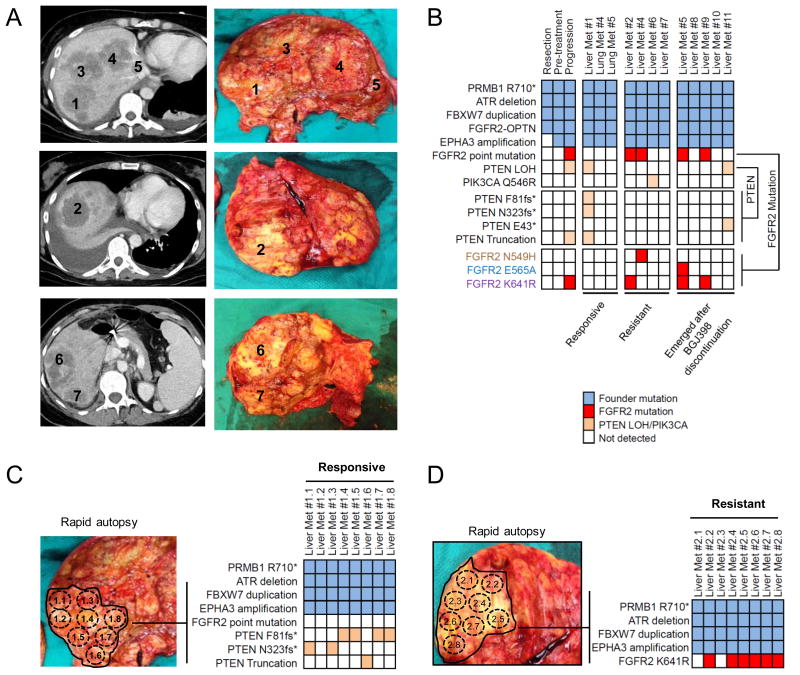
The identification of five concurrent FGFR2 resistance mutations in the cfDNA of Patients #1 and #2 suggests marked inter- and/or intra-lesional heterogeneity associated with acquired resistance. Since single-lesion tumor biopsies performed at disease progression exhibited only a subset of these mutations, we performed a rapid autopsy on Patient #2 to obtain a more complete assessment of the genetic alterations harbored by resistant tumors. This patient passed away three months after discontinuation of BGJ398 treatment, without any intervening anti-cancer therapies and exhibited a substantial tumor burden with >25 discrete metastatic tumors. We collected 10 liver metastases and two lung metastases that were of adequate tumor cellularity for molecular analysis. Quantitative RT-PCR (qRT-PCR) detected the FGFR2-OPTN fusion in all samples (Supplementary Figure S3A-B), reinforcing the conclusion that resistant tumor cells retain the FGFR2 fusion and that progression is not likely to be driven by the emergence of fusion negative clones.
Autopsy specimens along with the original resection specimen and the pre-treatment and post-progression biopsies were analyzed by targeted deep-sequencing using the FoundationOne assay, which assesses the mutational status of 315 cancer-related genes with >500 fold coverage (Figure 3A and 3B). The autopsy specimens exhibited three of the FGFR2 kinase domain mutations that were identified in this patient's post-progression cfDNA (p.N549H, p.E565A, and p.K641R), whereas only p.K641R was seen in the post-progression tumor biopsy. These mutations were found in four of the 12 distinct metastases analyzed, with one metastasis harboring two concurrent mutations (Figure 3B). Despite this extensive analysis of multiple tumor lesions, two FGFR2 mutations identified in cfDNA were still not detected. This was likely due to the fact that this patient's extensive tumor burden distributed throughout the liver did not allow for a comprehensive sampling of all metastatic sites. We next sought to relate the responsiveness of a given lesion to the presence of specific genetic alterations. Spatial correlation of autopsy specimens with the patient's most recent CT scan identified three lesions that were clearly shrinking at the time of progression (‘responsive’) and four lesions that were either growing or appeared as new lesions while the patient was on BGJ398 (‘resistant’). Another five lesions appeared following discontinuation of BGJ398 and therefore could not be correlated with clinical response. The characterization of these tumors at the time of autopsy as either clearly ‘resistant’ or ‘responsive’ was limited by the fact that the autopsy occurred three months after the discontinuation of BGJ398, which may have allowed resistant tumor cells to seed previously responsive tumors or sensitive tumor cells to divide in the absence of selective pressure. Nevertheless, we identified FGFR2 mutations in two of the four ‘resistant’ tumors (p.N549H and p.K641R) and in two of the five lesions that emerged after discontinuation of BGJ398 (p.K641R alone or concurrently with p.E565A), but in none of the “responsive” lesions. Truncal mutations in PRBM1, ATR, and FBXW7 were observed in all specimens, confirming a common clonal origin. A number of additional mutations were also detected that were specific to different tumors, although their clinical significance remains unclear (Supplementary Figure S4).
Figure 3. Rapid autopsy reveals inter-lesional and intra-lesional heterogeneity of resistance.
A. Axial contrast-enhanced CT images and autopsy photographs of seven liver metastases from Patient #'2. B. Heatmap illustrating mutations detected in the indicated autopsy lesions. Three distinct FGFR2 point mutations and four distinct PTEN mutations were identified. C and D. Corresponding images of Liver Met #1 (C) and Liver Met #2 (D) taken from Patient #2's autopsy including heatmaps indicating mutations identified in eight spatially distinct pieces isolated from Responsive Liver Met#1 and eight spatially distinct pieces isolated from Resistant Liver Met #2.
Beyond FGFR2 point mutations, several alterations in the PTEN/PI3K pathway were detected, including a PTEN truncation in the post-progression biopsy from Patient #2, resulting from an apparent fusion between PTEN and the COL17A1 locus (Supplementary Figure S5A). WES analysis revealed single copy loss across the entire long (‘q’) arm of chromosome 10 including the PTEN gene in both pre-treatment and post-progression biopsies from this patient (Supplementary Figure S5B). Thus, the PTEN truncation could represent loss of heterozygosity (LOH) in a subset of tumor cells. Similarly, PTEN nonsense or frameshift mutations were also observed in one responsive lesion and one lesion that emerged after discontinuation of treatment. A single resistant lesion harbored a PIK3CA p.Q546R mutation, however its significance is uncertain since this mutation was detected in the patient's plasma prior to BGJ398 treatment and decreased in prevalence during therapy (Figure 1B, left-lower panel). Thus, it is possible that alteration of the PTEN/PI3K pathway may represent an FGFR2-independent mechanism of resistance to FGFR2 inhibition, although further study is needed.
Finally, we sought to assess the degree of intra-lesional genomic heterogeneity by conducting multi-region sequencing of two autopsy specimens from Patient #2. One ‘responsive’ (Liver Met #1) and one ‘resistant’ (Liver Met #2) lesion were each transected into eight individual pieces (Figure 3C and D). The responsive Liver Met #1 harbored three distinct PTEN loss-of-function mutations, each found in separate adjacent pieces, suggesting a potential selective advantage of PTEN inactivation in this particular lesion. In the resistant Liver Met #2, the FGFR2 p.K641R mutation was detected in six of eight tumor pieces, and no other point mutation was observed (Figure 3B). By contrast, multi-region sequencing of the original resection specimen revealed no detectable FGFR2 point mutations or PTEN alterations (Supplementary Figure S6). Taken together, our findings suggest that recurrent secondary FGFR2 kinase domain mutations and molecular heterogeneity confer clinical acquired resistance to FGFR inhibitors in FGFR2-fusion+ ICC, and that therapeutic strategies capable of overcoming these mechanisms of resistance may be critical components of future treatments for these cancers.
Discussion
To our knowledge, this study is the first to define mechanisms underlying clinical acquired resistance to FGFR inhibitor therapy. The observation that all three FGFR2-fusion+ ICC patients developed secondary FGFR2 kinase domain mutations, with multiple mutations emerging in two patients, suggests that these alterations are major drivers of clinical resistance to FGFR inhibition. The p.V564 gatekeeper mutation, which was common to all three patients, and the additional six FGFR2 point mutations identified were predicted to confer resistance by previous in vitro studies and structural modeling (22-26). These findings may have more general significance since equivalent FGFR2 point mutations exist de novo in many cancers (26). Moreover, FGFR3 fusions are present in several malignancies, including glioblastoma multiforme, urothelial carcinoma, and lung adenocarcinoma (27), and our in vitro data suggest that mutations affecting corresponding amino acids in FGFR3 may cause resistance to FGFR inhibitors in this context. Importantly, the discoveries reported here have immediate clinical implications since covalent FGFR inhibitors designed to overcome such mutations have recently been developed, including TAS-120, an irreversible FGFR inhibitor that has already entered phase I testing (NCT02052778) (28).
The rapid acquisition of polyclonal resistance can undoubtedly create challenges for the establishment of more effective therapeutic strategies. Polyclonal resistance to targeted therapy within a single patient has been described in multiple solid tumors including ALK positive lung cancer, BRAF mutant melanoma, KRAS wildtype colon cancer, and cKIT mutant GIST (29-34). Encouragingly, however, prior studies have illustrated that convergent evolution of resistance through multiple secondary kinase mutations in ALK fusion-positive non-small cell lung cancers can be overcome clinically by structurally optimized inhibitors (35), suggesting a similar strategy may be effective in tumors harboring FGFR aberrations. Notable in this regard, we found that a collection of structurally and functionally distinct clinical and preclinical FGFR inhibitors displayed varying abilities to overcome specific resistance mutations. LY2874455, for instance, appeared to be the compound least affected by the panel of resistance mutations, although it still showed reduced activity in the p. N540/p.549K mutated model (Figure 2F). Thus, our study highlights that future development of FGFR inhibitors should focus on agents capable of surmounting multiple common secondary resistance mutations.
It is also possible that FGFR2-independent mechanisms may contribute to acquired resistance and further limit the efficacy of next-generation FGFR inhibitors. In our study, patient #2 had a PTEN loss of function mutation in the biopsy tissue of a lesion that was clearly progressing on BGJ398 and had additional PI3K/PTEN pathway mutations in three of twelve metastases analyzed at rapid autopsy (Figure 3A). It is notable that three distinct inactivating PTEN mutations were detected in the residual tissue from a single “responsive” tumor, suggesting there was a significant selective advantage granted by PTEN LOH in this lesion. Interestingly, knockdown of PTEN was recently found to confer resistance to loss of FGFR2 signaling in FGFR2-amplified cell lines (36). The PI3KCA p.Q546R mutation, however, was detectable in patient #2's cfDNA prior to initiation of therapy (Figure 1B, left-lower panel), and levels of this mutation tracked with those of a truncal PBRM1 nonsense mutation, implying that this mutation did not emerge during therapy. Thus, future studies will be needed to determine whether activation of the PI3K/PTEN pathway serves as an FGFR-independent resistance mechanism in FGFR-driven cancers.
Our findings also highlight the potential advantages of cfDNA analysis in the monitoring and clinical management of patients undergoing FGFR inhibitor therapy. While cfDNA analysis detected multiple distinct resistant clones emerging concurrently in different tumors within the same patient, a single post-progression biopsy or even extensive tumor sampling at rapid autopsy was unable to identify the full spectrum of resistance mutations detected in the plasma of these patients. cfDNA, which may reflect DNA shed from multiple metastatic sites, can thus offer a non-invasive and efficient approach to capturing intra- and inter-lesional heterogeneity in the setting of resistance (37). Furthermore, since structurally distinct FGFR inhibitors display varying abilities to overcome different secondary FGFR2 resistance mutations, real-time detection and monitoring of clonal evolution of individual mutations may be a valuable tool to guide the selection of the appropriate inhibitors for patient management. Limitations of ctDNA analysis include lack of spatial specificity for anatomically critical and clinically relevant lesions, inadequate shedding of ctDNA by certain tumors, and the lack of prospective validation for clinical practice for a majority of tumors. Additionally, current commercially available ctDNA assays assess a less comprehensive panel of genes compared to tissue-based panels, and they have limited sensitivity for detecting fusion events as evidenced by the fact the FGFR2 fusion events were only detected in one out of three of our patients by ctDNA analysis. Thus, a combined approach integrating cfDNA analysis with direct tumor sampling may represent an important approach to guide clinical decision-making.
Overall, these studies indicate that recurrent secondary FGFR2 kinase domain mutations are a common mechanism of clinical resistance to FGFR inhibition. Importantly, our data also suggest that resistant clones harboring such mutations are likely to retain dependence on FGFR2 signaling, implying that structural optimization of next-generation FGFR inhibitors is needed to promote more durable remissions in FGFR2-fusion+ ICC and other cancers harboring alterations in the FGFR signaling pathway.
Methods
Patients
Patients provided written informed consent to treatment on the phase II trial of BGJ398 (NCT02160041) at the Massachusetts General Hospital (MGH) Cancer Center. They received a 100mg capsule and a 25mg capsule of BGJ398 daily for days 1-21 of each 28 day cycle, and all dose reductions and safety assessments were performed per protocol. Computed tomography and/or magnetic resonance imaging scans were performed at baseline and every 8 weeks to assess for tumor response by RECIST version 1.1 criteria. All biopsies, tumor specimens, and peripheral blood draws for plasma isolation were collected and analyzed in accordance with Institutional Review Board-approved protocols, to which patients provided written informed consent, and all studies were conducted in accordance with the Declaration of Helsinki. Rapid autopsy on patient #2 was performed within the first three hours post mortem.
Solid Fusion Assay (SFA)
Our internal tumor profiling assay was performed on RNA extracted from formalin-fixed paraffin-embedded (FFPE) specimens as part of routine clinical care. The SFA is a targeted RNA sequencing method of Anchored Multiplex PCR (AMP) to detect FGFR2 fusions, and the methodology has been previously described (20). Mutational profiling was performed at the Clinical Laboratory Improvement Amendments–certified Translational Research Laboratory at the Massachusetts General Hospital Cancer Center.
Targeted Sequencing of Circulating Cell-free Tumor DNA
Cell-free DNA was extracted from whole blood, and 5ng-30ng of cfDNA was isolated. Sequencing libraries were prepared with custom in-line barcode molecular tagging, and complete sequencing at 15,000× read depth of the critical exons in a targeted panel of 70 genes was performed at a Clinical Laboratory Improvement Act (CLIA)-certified, College of American Pathologists-accredited laboratory (Guardant Health) (38). A summary of all data from these analyses is provided in Supplementary Table 1.
Targeted Sequencing of Tumor Tissue
DNA derived from the primary tumor, serial biopsies of liver metastases, autopsy specimens from liver and lung metastases, and matched normal muscle tissue were analyzed using deep-coverage targeted sequencing of key cancer-associated genes. Tumor samples sent for analysis had ≥ 10% tumor cellularity as determined by review of a hematoxylin and eosin slide by a board certified pathologist (V.D.). Targeted sequencing was performed via the FoundationOne platform, version T5, and the methodology has been previously described (39). A summary of all data from these analyses is provided in Supplementary Table 1.
Quantitative Polymerase Chain Reaction on Tumor Tissue for Detection of the FGFR-OPTN Fusion on Autopsy Samples
Autopsy specimens of Patient #2 were homogenized with a tissue homogenizer and total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer's recommended protocol. cDNA synthesis was performed using the QuantiTect Reverse Transcription Kit (Qiagen) with 1 ug total RNA. All qPCR reactions were performed using the iTaq Universal SYBR Green Supermix (Bio-Rad) and CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The following primers were used to detect the FGFR2-OPTN fusion breakpoint: 5′-TGATGATGAGGGACTGTTGG-3′ and 5′-GCCCAGGACTATGCTTGATT-3′. Relative expression levels of the fusion protein were normalized to expression of RNA18S5: 5′- CGTCTGCCCTATCAACTTTCG-3′ and 5′- TGCCTTCCTTGGATGTGGTAG-3′.
Plasma cfDNA isolation and quantification of genome equivalents
At least 10 mL of whole blood were collected by blood draw using EDTA as anticoagulant. Plasma was separated within 5 hours through 2 different centrifugation steps (the first at room temperature for 10 minutes at 1,600 × g and the second at 3,000 × g for the same time and temperature), obtaining up to 3 mL of plasma. Plasma was stored at -80°C until cfDNA extraction. cfDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (QIAGEN) according to the manufacturer's instructions. 6 μl of cfDNA were used as template for each reaction. All samples were analyzed in triplicate. PCR reactions were performed using 10 μl final volume containing 5 μl GoTaq® qPCR Master Mix, 2× with CXR Reference Dye) (Promega) and LINE-1 [12,5 μmol] forward and reverse primers. DNA at known concentrations was also used to build the standard curve. Primer sequences are available upon request.
Droplet digital PCR
8 to 10 μl of DNA template was added to 10 μl of ddPCR™ Supermix for Probes (Bio-Rad) and 2 μl of the custom primer/probe mixture. This reaction mix was added to a DG8 cartridge together with 60μl of Droplet Generation Oil for Probes (Bio-Rad) and used for droplet generation. Droplets were then transferred to a 96 well plate (Eppendorf) and then thermal cycled with the following conditions: 5 minutes at 95°C, 40 cycles of 94°C for 30s, 55°C for 1 minute followed by 98°C for 10 minutes (Ramp Rate 2°C/sec). Droplets were analyzed with the QX200™ Droplet Reader (Bio-Rad) for fluorescent measurement of FAM and HEX probes. Gating was performed based on positive and negative controls, and mutant populations were identified. The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad) to obtain Fractional Abundance of the mutant DNA alleles in the wild-type/normal background. The quantification of the target molecule was presented as number of total copies (mutant plus WT) per sample in each reaction. Fractional Abundance is calculated as follows: F.A. % = (Nmut/(Nmut+Nwt))*100), where Nmut is number of mutant events and Nwt is number of WT events per reaction. ddPCR analysis of normal control plasma DNA (from cell lines) and no DNA template controls were always included. Probe and primer sequences are available upon request.
In vitro studies
The V564F gatekeeper mutation was introduced in pcDNA3.1 plasmid carrying wild type human FGFR2 cDNA or a pMSCV plasmid carrying TEL-FGFR2 by Site-Directed Mutagenesis (Q5 Site-Directed Mutagenesis Kit – New England Biolabs). PCR primers for mutation introduction and amplification of plasmid (5′-CTATGTCATAtTTGAGTATGCCTCTAAAG-3′ and 5′-AGAGGCCCATCCTGTGTG-3′) were designed using NEBaseChanger program (NEB). Mutant FGFR2 constructs were selected and prepared from positive clones and targeted sequence mutation was confirmed by Sanger sequencing.
For phospho-FGFR2 studies, HEK293 cells grown in 96-well plates were transiently transfected with pcDNA3.1, pcDNA3.1-FGFR2 WT or pcDNA3.1-FGFR2 V564F using Fugene 6 (Promega #E2691). Cells were treated in triplicates for 40 min with serial dilutions of NVP-BGJ398 or DMSO. The phospho-Tyr content of FGFR2 was measured by capture ELISA assay using the anti-FGFR2 monoclonal antibody (R&D MAB6841) as a capturing antibody and an alkaline phosphatase-coupled anti-phospho-Tyr antibody as secondary antibody (Zymed PY20 #03-7722). CDP-Star® Emerald II (Applied Biosystems #T2216) was used as alkaline phosphatase substrate and luminescence measured using a Tecan reader. Upon subtracting the background value (value obtained from pcDNA3.1-transfected HEK293 cells treated with DMSO) the IC50 was calculated using XLFit software. The IC50 value refers to the compound concentration needed to inhibit 50% of the signal obtained from FGFR2-transfected HEK293 cells treated with DMSO. In this assay, the concentration of NVP-BGJ398 required to reduce by 50% wild type FGFR2 Tyr-phosphorylation was found to be 11.7 ± 2 nM (average of three independent assays). Instead, NVP-BGJ398 was inactive against mutant FGFR2 V564 up to the concentration of 10,000 nM.
For BaF3 cell studies with TEL-FGFR2, Retrovirus was produced using the pMSCV constructs (TEL-FGFR2 wild type and V564F) by transient transfection into EcoPack 2-293 cells (Clontech). Parental BaF3 cells grown in medium containing 10 ng/mL recombinant murine IL-3 were then infected with retrovirus-containing supernatants using a spin infection protocol and selected with 1 μg/mL puromycin and IL-3 withdrawal. Cultures with at least 85% viable cells were used in proliferation assays as described.
For mutagenesis studies, BaF3 TEL-FGFR2 or BaF3 TEL-FGFR3 cells were mutagenized with the alkylating agent N-ethyl-N-nitrosourea, and resistant clones were selected in presence of different concentrations of BGJ398 as previously described (40). In brief, mutagenized cells were distributed into 96-well plates with 100 / 200 / 400 / 800 / 1600 nM BGJ398 (3 plates each), placing 105 cells in 100 μL media into each well. Outgrowing clones were picked and expanded during days 13 to 24 with the original concentration of BGJ398. Mutations in the FGFR2 and FGFR3 kinase domain were determined by PCR and Sanger sequencing. Sequencing traces suggested that pure clones with single point mutations were obtained in all cases. Although it is an N-terminal fusion protein, rather than a C-terminal fusion as seen in ICC patients, the TEL-FGFR fusion construct was used since it is a well-established system for rapidly and accurately evaluating kinase inhibitor sensitivity in Baf3 cells. The precise effect of the multiple distinct fusion partners and C-terminal versus N-terminal fusions on kinase inhibitor sensitivity will require further study.
For proliferation assays with various FGFR inhibitors, BaF3 cells were seeded on 96-well-plates in triplicates at 10000 cells per well and incubated with various concentrations of FGFR inhibitors for 72 hours followed by quantification of viable cells using a resazurin sodium salt dye reduction readout (commercially known as AlamarBlue assay). IC50 values were determined with the XLFit Excel Add-In (ID Business Solutions) using a 4-parameter dose response model. BGJ398 was synthesized at Novartis and other FGFR inhibitors were obtained from commercial sources.
Homology Models of FGFR2 and FGFR3 in complex with BGJ398
The sequences of the human FGFR2 and FGFR3 kinases were obtained from SWISS-PROT (41), entries P21802 and P22607, respectively. The crystallographic structure of the FGFR1 kinase in complex with the inhibitor BGJ398 (PDB code 3TT0 from the Protein Data Bank) was chosen as template. The sequences were aligned using T-Coffe (42). On the basis of the resulting alignment, the 3D structures of the FGFR2 and FGFR3 kinases were modeled using the What If program (43) with the default parameters (PIRPSQ module, BLDPIR command). The figures for the structural models were prepared using PYMOL (Schrodinger, Inc.).
Whole-exome sequencing and RNA sequencing
Whole-exome and RNA sequencing was performed by the Broad Institute sequencing platform. Whole-exome sequencing of matched pre-treatment and post-progression biopsies and normal blood was performed as previously described (44). All BAM files will be deposit in dbGAP prior to publication. Detailed methods for RNA sequencing are as follows. RNA was extracted from patient tumor biopsies using the AllPrep DNA/RNA kit (Qiagen) according to the manufacturer's instructions. All samples were quantified using Nanodrop and quality was evaluated using Agilent's Bioanalyzer 2100.
RNA samples and two positive controls (K-562) were assessed for quality using Agilent's Bioanalyzer 2100. The percentage of fragments with a size greater than 200 nucleotides (DV200) was calculated using the Agilent software. Samples with a DV200 score less than 30% were not included, as the likelihood of success is dramatically reduced with these more fragmented samples (45).
100ng of RNA was used as the input for first strand cDNA synthesis using Superscript III reverse transcriptase (Life Technologies, Cat. #18080044) and Illumina's TruSeq Stranded Total RNA Sample Prep Kit (Illumina, Cat. #RS-122-2201). The fragmentation step prior to cDNA synthesis was omitted in the FFPE RNA samples, only the K-562 positive control samples were fragmented at 94°C for 8 minutes. Synthesis of the second strand of cDNA was followed by indexed adapter ligation. Subsequent PCR amplification enriched for adapted fragments. The amplified libraries were quantified using a Qubit assay (Life Technologies, Cat. #Q3285) and assessed for quality on an Agilent Technologies 2100 Bioanalyzer (DNA 1000 chip).
200ng of each cDNA library, not including controls, were combined into two 4-plex pools. Illumina's Coding Exome Oligos (Illumina, Part #15034575) that target the exome were added, and hybridized on a thermacycler with the following conditions: 95°C for 10 minutes, 18 cycles of 1 minute incubations starting at 94°C, then decreasing 2°C per cycle, then 58°C for 90 minutes. Following hybridization, streptavidin beads were used to capture the probes that were hybridized in the previous step. Two wash steps effectively remove any non-specifically bound products. These same hybridization, capture and wash steps are repeated to assure high specificity.
A second round of amplification enriches the captured libraries. qPCR (Kapa Biosystems, Cat. #KK4600) was performed on the pooled libraries and normalized to 2nM. The normalized, pooled libraries were loaded onto HiSeq2500 for a target of 50 million 2×76bp paired reads per sample. Data have been uploaded to dbGAP, accession number phs000803.v2.p1.
Supplementary Material
Significance.
We report the first genetic mechanisms of clinical acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive ICC. Our findings can inform future strategies for detecting resistance mechanisms and inducing more durable remissions in ICC and in the wide variety of cancers where the FGFR pathway is being explored as a therapeutic target.
Acknowledgments
The authors acknowledge Guardant Health, Richard Lanman, and Rebecca Nagy for assistance with cfDNA analysis with the Guardant360 assay.
Financial Support: L.G. is supported by Massachusetts General Hospital (MGH) Executive Committee on Research Fund for Medical Discovery Award, Cholangiocarcinoma Foundation Research Fellowship, and NIH Loan Repayment Program Grant. S.K.S. is supported by a NCI Mentored Clinical Scientist Research Career Development Award (1K08CA194268-01). D.J. is supported by the HMS Laboratory for Systems Pharmacology Grant (P50GM107618) and the Susan Eid Tumor Heterogeneity Initiative. N.B. is the holder of the Gallagher Chair in Gastrointestinal Cancer Research at MGH. N.B. and A.Z. are supported by the TargetCancer Foundation and by a Translational Research Grant Award from V Foundation for Cancer Research. This work was also supported by grants from the NIH/NCI Gastrointestinal Cancer SPORE P50 CA127003 (to L.G., S.K.S, R.B.C, and N.B.). R.B.C acknowledges support from a Damon Runyon Clinical Investigator Award and NIH/NCI 1K08CA166510 and 1R01CA208437. L.G., N.B., and A.Z. are supported by the Jonathan Kraft Translational Award. A.B. is supported by Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2011, Ministero della Salute and AIRC IG n. 16788.
S.B., B.S., L.B., C.S., P.F., D.G.P., and R.T. are employees and shareholders of Novartis. A.B. is a consultant/advisory board member for Trovagene, Horizon Discovery and Biocartis. R.B.C. has received research funding from AstraZeneca and is a consultant/advisory board member for Genentech, Merrimack Pharmaceuticals, Astex Pharmaceuticals, N-of-one, Taiho Pharmaceuticals, and Avidity Biosciences. D.J. has received research funding from Novartis and is a consultant/advisory board member for Novartis, EMD Serono, Natera and Eisai. A.Z. has received research funding from Novartis and Lilly and is a consultant/advisory board member for Novartis.
Footnotes
Conflicts of Interest: All other authors have no conflicts to disclose.
References
- 1.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 2.Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2014;25:552–63. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. The oncologist. 2016;21:594–9. doi: 10.1634/theoncologist.2015-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Seminars in liver disease. 2004;24:115–25. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 5.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–81. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 6.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–34. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 7.Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Human pathology. 2014;45:1630–8. doi: 10.1016/j.humpath.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. The oncologist. 2014;19:235–42. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia D, Losic B, Moeini A, Cabellos L, Hao K, Revill K, et al. Massive parallel sequencing uncovers actionable FGFR2-PPHLN1 fusion and ARAF mutations in intrahepatic cholangiocarcinoma. Nature communications. 2015;6:6087. doi: 10.1038/ncomms7087. [DOI] [PubMed] [Google Scholar]
- 10.Voss JS, Holtegaard LM, Kerr SE, Fritcher EG, Roberts LR, Gores GJ, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44:1216–22. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer discovery. 2013;3:636–47. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS genetics. 2014;10:e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Ding X, Wang S, Moser CD, Shaleh HM, Mohamed EA, et al. Antitumor effect of FGFR inhibitors on a novel cholangiocarcinoma patient derived xenograft mouse model endogenously expressing an FGFR2-CCDC6 fusion protein. Cancer Letters. 2016;380:163–73. doi: 10.1016/j.canlet.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guagnano V, Furet P, Spanka C, Bordas V, Le Douget M, Stamm C, et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. Journal of medicinal chemistry. 2011;54:7066–83. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 15.Guagnano V, Kauffmann A, Wöhrle S, Stamm C, Ito M, Barys L, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer discovery. 2012;2:1118–33. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 16.Nogova L, Sequist LV, Perez Garcia JM, Andre F, Delord JP, Hidalgo M, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol. 2016:JCO2016672048. doi: 10.1200/JCO.2016.67.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Javle MM, Shroff RT, Zhu AX, Sadeghi S, Choo S, Borad MJ, et al. A phase 2 study of BGJ398 in patients (pts) with advanced or metastatic FGFR-altered cholangiocarcinoma (CCA) who failed or are intolerant to platinum-based chemotherapy. J Clin Oncol; Gastrointestinal Cancers Symposium; San Francisco, CA. 2016. 2015. [Google Scholar]
- 18.Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer discovery. 2012;2:214–26. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 19.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature medicine. 2014;20:1479–84. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 21.Kim ST, Lee WS, Lanman RB, Mortimer S, Zill OA, Kim KM, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget. 2015;6:40360–9. doi: 10.18632/oncotarget.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, et al. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Molecular cell. 2007;27:717–30. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z, Chen H, Blais S, Neubert TA, Li X, Mohammadi M. Structural mimicry of a-loop tyrosine phosphorylation by a pathogenic FGF receptor 3 mutation. Structure. 2013;21:1889–96. doi: 10.1016/j.str.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan L, Wang J, Tanizaki J, Huang Z, Aref AR, Rusan M, et al. Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4869–77. doi: 10.1073/pnas.1403438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byron SA, Chen H, Wortmann A, Loch D, Gartside MG, Dehkhoda F, et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia. 2013;15:975–88. doi: 10.1593/neo.121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patani H, Bunney TD, Thiyagarajan N, Norman RA, Ogg D, Breed J, et al. Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use. Oncotarget. 2016;7:24252–68. doi: 10.18632/oncotarget.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nature communications. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochiiwa H, Fujita H, Itoh K, Sootome H, Hashimoto A, Fujioka Y, et al. Abstract A270: TAS-120, a highly potent and selective irreversible FGFR inhibitor, is effective in tumors harboring various FGFR gene abnormalities. American Association for Cancer Research. 2013;12:A270–A. [Google Scholar]
- 29.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–9. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 30.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liegl B, Kepten I, Le C, Zhu M, Demetri GD, Heinrich MC, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216:64–74. doi: 10.1002/path. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim KH, Huang MJ, Chen LT, Wang TE, Liu CL, Chang CS, et al. Molecular analysis of secondary kinase mutations in imatinib-resistant gastrointestinal stromal tumors. Med Oncol. 2008;25:207–13. doi: 10.1007/s12032-007-9014-2. [DOI] [PubMed] [Google Scholar]
- 35.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6:1118–33. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer discovery. 2016;6:838–51. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer discovery. 2016;6:147–53. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PloS one. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiedt R, Degenkolbe E, Furet P, Appleton BA, Wagner S, Schoepfer J, et al. A drug resistance screen using a selective MET inhibitor reveals a spectrum of mutations that partially overlap with activating mutations found in cancer patients. Cancer research. 2011;71:5255–64. doi: 10.1158/0008-5472.CAN-10-4433. [DOI] [PubMed] [Google Scholar]
- 41.Bairoch A, Boeckmann B. The SWISS-PROT protein sequence data bank: current status. Nucleic acids research. 1994;22:3578–80. [PMC free article] [PubMed] [Google Scholar]
- 42.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 43.Vriend G. WHAT IF: a molecular modeling and drug design program. Journal of molecular graphics. 1990;8:52–6. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 44.Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer discovery. 2015;5:358–67. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evaluating RNA Quality from FFPE Samples. Available from: http://www.illumina.com/documents/products/technotes/technote-truseq-rna-access.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.