Abstract
The 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) cycle fixes CO2 in extremely thermoacidophilic archaea and holds promise for metabolic engineering because of its thermostability and potentially rapid pathway kinetics. A reaction kinetics model was developed to examine the biological and biotechnological attributes of the 3HP/4HB cycle as it operates in Metallosphaera sedula, based on previous information as well as on kinetic parameters determined here for recombinant versions of five of the cycle enzymes (malonyl-CoA/succinyl-CoA reductase, 3-hydroxypropionyl-CoA synthetase, 3-hydroxypropionyl-CoA dehydratase, acryloyl-CoA reductase, and succinic semialdehyde reductase). The model correctly predicted previously observed features of the cycle: the 35%–65% split of carbon flux through the acetyl-CoA and succinate branches, the high abundance and relative ratio of acetyl-CoA/propionyl-CoA carboxylase (ACC) and MCR, and the significance of ACC and hydroxybutyryl-CoA synthetase (HBCS) as regulated control points for the cycle. The model was then used to assess metabolic engineering strategies for incorporating CO2 into chemical intermediates and products of biotechnological importance: acetyl-CoA, succinate, and 3-hydroxyproprionate.
Keywords: CO2 fixation, 3-hydroxypropionate, 4-hydroxybutyrate, Metallosphaera sedula
1. Introduction
Concerns over sustainability and global climate change have generated interest in developing biological systems for industrial production of fuels and chemicals, with particular interest in using inorganic carbon feed stocks, such as CO2 (Conrado et al., 2013). To do so a CO2 fixation pathway is needed, six of which are currently known: the 3-hydroxypropionate (3HP) bicycle, the dicarboxylate/4-hydroxybutyrate (DC/4HB) cycle, the reductive citric acid cycle, the reductive acetyl-CoA (Wood-Ljungdahl) pathway, the Calvin-Benson-Bassham cycle, and the 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) cycle (Berg, 2011; Berg et al., 2010; Herter et al., 2002; Huber et al., 2008). There have been efforts to produce chemicals and fuels based on several of these pathways, including the 3HP bicycle, Calvin-Benson-Bassham cycle, reductive acetyl-CoA pathway, and, of interest here, the 3HP/4HB cycle (Keller et al., 2013; Li et al., 2012; Mattozzi et al., 2013; Muller et al., 2013; Ueki et al., 2014; Yuzawa et al., 2012) (see Table 1).
Table 1.
Enzymes of the 3-hydroxypropionate/4-hydroxybutyrate cycle in M. sedula
| Enzyme name | Label | ORF(s) | Reaction(s) Catalyzed | a Reference |
|---|---|---|---|---|
| Acetyl-CoA/Propionyl-CoA carboxylase | ACC | Msed_0147 Msed_0148 Msed_1375 |
a) Acetyl-CoA + HCO3− + ATP ↔ Malonyl-CoA + ADP + Pi b) Propionyl-CoA + HCO3− + ATP ↔ (S)-Methylmalonyl-CoA + ADP + Pi |
NP (Hugler et al., 2003) |
| Malonyl-CoA/Succinyl-CoA reductase (NADPH) | MCR | Msed_0709 | a) Malonyl-CoA + NADPH ↔ Malonic semialdehyde + NADP + CoA b) Succinyl-CoA + NADPH ↔ Succinic semialdehyde + NADP + CoA |
NCE (Kockelkorn and Fuchs, 2009) R (this work) R (S. tokodaii) (Alber et al., 2006) |
| Malonic semialdehyde reductase (NADPH) | MSR | Msed_1993 | Malonic semialdehyde + NADPH ↔ 3-Hydroxypropionate + NADP | R (Kockelkorn and Fuchs, 2009) |
| 3-Hydroxypropionyl-CoA synthetase (AMP-forming) | HPCS | Msed_1456 | 3-Hydroxypropionate + ATP + CoA ↔ 3-Hydroxypropionyl-CoA + AMP + PPi | NP, R (S. tokodaii) (Alber et al., 2008) R (this work) |
| 3-Hydroxypropionyl-CoA dehydratase | HPCD | Msed_2001 | a) 3-Hydroxypropionyl-CoA ↔ Acryloyl-CoA + H2O b) Crotonyl-CoA + H2O ↔ (S)-3-hydroxybutyryl-CoA |
NP (Teufel et al., 2009), R (this work) |
| Acryloyl-CoA reductase (NADPH) | ACR | Msed_1426 | Acryloyl-CoA + NADPH ↔ Propionyl-CoA + NADP | R (S. tokodaii) (Teufel et al., 2009) R (this work) |
| Methylmalonyl-CoA epimerase | MCE | Msed_0639 | (S)-Methylmalonyl-CoA ↔ (R)-Methylmalonyl-CoA | R (Marcheschi et al., 2012) |
| Methylmalonyl-CoA mutase | MCM | Msed_0638 | (R)-Methylmalonyl-CoA ↔ Succinyl-CoA | R (Marcheschi et al., 2012) |
| Msed_2055 | ||||
| Succinic semialdehyde reductase (NADPH) | SSR | Msed_1424 | Succinic semialdehyde + NADPH ↔ 4-Hydroxybutyrate + NADP | R (Kockelkorn and Fuchs, 2009, this work) |
| 4-Hydroxybutyryl-CoA synthetase (AMP-forming) | HBCS | Msed_0406 | a) 4-Hydroxybutyrate + ATP + CoA ↔4-Hydroxybutyryl-CoA + AMP + PPi b) 3-Hydroxypropionate + ATP + CoA ↔ 3-Hydroxypropionyl-CoA + AMP +PPi |
R (Hawkins et al., 2013) |
| 4-Hydroxybutyryl-CoA dehydratase | HBCD | Msed_1321 | 4-Hydroxybutyryl-CoA ↔ Crotonyl-CoA + H2O | R (Hawkins et al., 2014) |
| Crotonyl-CoA hydratase/(S)-3-Hydroxybutyryl-CoA dehydrogenase (NADH) | CCH/HBCD | Msed_0399 | a) Crotonyl-CoA + H2O ↔ (S)-3-Hydroxybutyryl-CoA b) (S)-3-Hydroxybutyryl-CoA + NAD ↔ Acetoacetyl-CoA + NADH |
NP (Ramos-Vera et al., 2011), R (Hawkins et al., 2014) |
| Acetoacetyl-CoA β-ketothiolase | AACT | Msed_0656 | Acetoacetyl-CoA + CoA ↔ 2 Acetyl-CoA | NP (Ramos-Vera et al., 2011), R (Hawkins et al., 2014) |
NCE = native cell extract; NP = native purified; R = recombinant purified
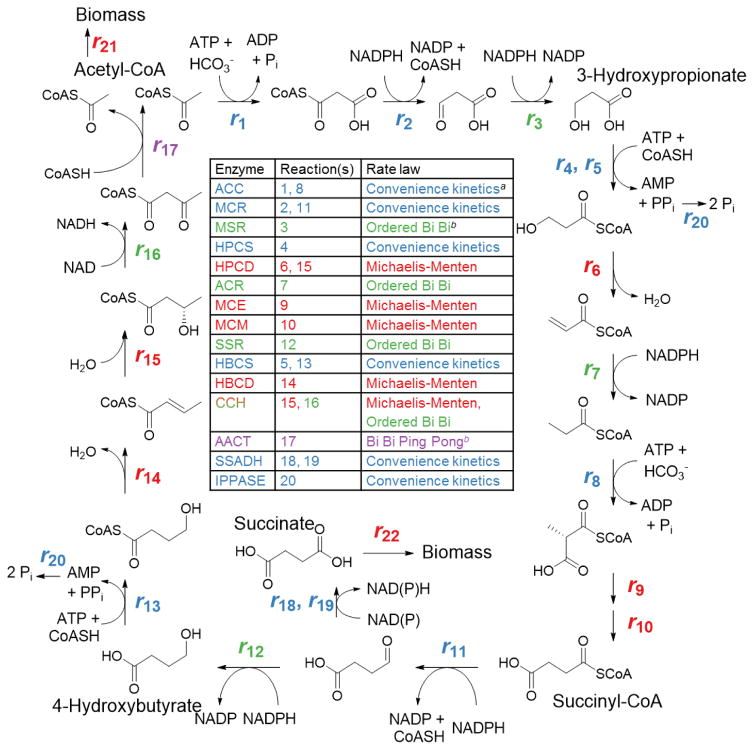
The 3HP/4HB cycle is a promising candidate for microbial production of chemicals from CO2 for several reasons. First, it functions at high temperatures, allowing it to be used in an extremely thermophilic host with concomitant minimal risk of contamination and reduced cooling costs (Keller et al., 2015; Zeldes et al., 2015). Second, the 3HP/4HB cycle can function in either an aerobic or anaerobic host, unlike the DC/4HB and reductive acetyl-CoA pathways, which are found exclusively in anaerobic organisms (Fast and Papoutsakis, 2012). Third, it was shown that the 3HP/4HB cycle can drive rapid autotrophic growth with a doubling time of less than five hours (Hawkins et al., 2013), suggesting the potential for fast pathway kinetics. Components of the 3HP/4HB cycle can be identified in genomes within the crenarchaeal order Sulfolobales (Kockelkorn and Fuchs, 2009), and has been studied most intensively in the extremely thermoacidophilic archaeon Metallosphaera sedula (Topt = 73°C; pHopt = 2.0) (Auernik and Kelly, 2010; Berg et al., 2010; Hawkins et al., 2014). The cycle can be divided into three sub-pathways to track the reduction of CO2 into acetyl-CoA. In the first sub-pathway, acetyl-CoA is carboxylated by acetyl-CoA/propionyl-CoA carboxylase (ACC) and subsequently reduced to the stable intermediate 3HP (Figure 1, reactions 1–3). In the second sub-pathway, 3HP is ligated to coenzyme A (CoA), reduced to propionyl-CoA, carboxylated by ACC, converted to succinyl-CoA, which is further reduced to the second stable intermediate 4HB (Figure 1, reactions 4–12). In the third sub-pathway, 4HB is ligated to CoA and cleaved to regenerate the starting substrate and produce an additional molecule of acetyl-CoA (Figure 1, reactions 13–17). Cellular intermediates used for biomass generation are drawn from the cycle through the intermediate acetyl-CoA, as well as through succinic semialdehyde via oxidation to succinate by succinic semialdehyde dehydrogenase (Figure 1, reactions 18–22) (Estelmann et al., 2011). Isotopic labeling studies in M. sedula have shown that 65% of cellular intermediates are formed via succinate, while the remaining 35% of carbon enters cellular metabolism via acetyl-CoA (Estelmann et al., 2011). Putative cycle enzymes have been previously characterized to various extents, although some remain to be verified and characterized in purified form (Alber et al., 2006; Alber et al., 2008; Han et al., 2012; Hawkins et al., 2014; Hawkins et al., 2013; Hugler et al., 2003; Kockelkorn and Fuchs, 2009; Ramos-Vera et al., 2011; Teufel et al., 2009).
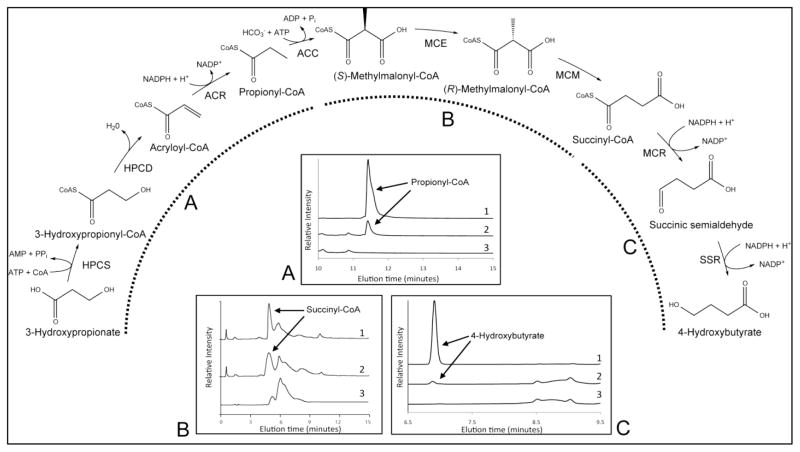
Figure 1. Reactions of the 3-hydroxypropionate/4-hydroxybutyrate carbon fixation cycle.
Reactions are color-coded by rate law. Enzyme abbreviations: ACC, acetyl-CoA/propionyl-CoA carboxylase; MCR, malonyl-CoA/succinyl-CoA reductase; MSR, malonic semialdehyde reductase; HPCS, 3-hydroxypropionyl-CoA synthetase; HBCS, 4-hydroxybutyryl-CoA synthetase; HPCD, 3-hydroxypropionyl-CoA dehydratase; ACR, acryloyl-CoA reductase; MCE, methylmalonyl-CoA epimerase; MCM, methylmalonyl-CoA mutase; SSR, succinic semialdehyde reductase; HBCD, 4-hydroxybutyryl-CoA dehydratase; CCH/HBCD, bifunctional crotonoyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase; AACT, acetoacetyl-CoA β-ketothiolase; SSADH, succinic semialdehyde dehydrogenase; IPPASE, inorganic pyrophosphatase; BM1, biomass production reaction #1; BM2, biomass production reaction #2. For a full list of rate law equations, see Supplementary Table S2.
Outside of its natural context, there are many opportunities to use 3HP/4HB cycle as a route to renewable production of chemicals. The complete cycle for metabolic engineering could enable the production of chemicals directly from CO2, while an alternative is to use portions of the complete cycle for production of chemicals from sugars via the intermediate acetyl-CoA. Prior to introducing all or parts of the 3HP/4HB cycle into a metabolically engineered host organism, it is useful to identify and address potential bottlenecks. To this end, genome-scale flux balance modeling has been used extensively to assist metabolic engineering of model microorganisms, such as Escherichia coli and Saccharomyces cerevisiae (Kerkhoven et al., 2014; McCloskey et al., 2013). However, enzyme kinetics-based models can account for features such that the interplay of biochemical reaction pathways can be considered, making them valuable for testing metabolic engineering strategies (Kerkhoven et al., 2014; Loder et al., 2015). Here, we describe the development of such a model to explore aspects of the 3H/4HB cycle first as it operates in its native form in the model extreme thermoacidophile Metallosphaera sedula and then as a basis for production of bio-based fuels and chemicals, focusing on the cycle intermediates acetyl-CoA, 3HP, and succinate.
2. Materials and Methods
2.1. Materials
Growth conditions for M. sedula (DSM 5348) and genomic DNA purification were conducted, as reported previously (Auernik and Kelly, 2010). Strains and vectors used for cloning included the pET-46b EK/LIC cloning Kit, pRSF-2 Ek/LIC Vector Kit, pCDF-2 Ek/LIC Vector Kit, Novablue GigaSinglesTM E. coli competent cells (Novagen, San Diego, CA), and RosettaTM (DE3) E. coli competent cells (Stratagene, La Jolla, CA). The reagents and devices used include: n-propionyl-Coenzyme A lithium salt, succinyl-CoA sodium salt, succinic semialdehyde solution, NADPH, ATP, and sodium acrylate (Sigma Chemical Co., St. Louis, MO); GelCode Blue Stain Reagent (Thermo Fisher Scientific Inc., Rockford, IL); Bio-Rad Protein Assay Dye Reagent (Hercules, CA), 3-hydroxypropionic acid (TCI America, http://www.tciamerica.net/); Quickload DNA Ladder (100 bp) (New England Biolabs, Ipswich, MA); QlAquick gel extraction kit and QIAprep Spin Miniprep Kit (QIAGEN, Inc., Valencia, CA); Isopropyl β-D-thiogalactopyranoside (IPTG), Amicon Ultra 10k, 30k, 50k centrifugal filter units (Millipore, Billerica, MA); antibiotics, agar, agarose, sodium chloride, tryptone, sodium acetate, acetic acid, methanol, and K2HPO4, KH2PO4 (Fisher Scientific, Pittsburgh, PA); imidazole (ACROS Organics, Geel, Belgium); HisTrap™ Column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ USA); BenchMark protein ladder (10 to 220 kDa) (Invitrogen); Superdex 75 10/300 GL size exclusion column (GE Healthcare).
2.2. Cloning, expression, and purification of HPCS, HPCD, ACR, MCR, and SSR
The genes hpcs (Msed_1456), hpcd (Msed_2001), acr (Msed_1426), mcr (Msed_0709), and ssr (Msed_1424) were amplified from genomic DNA using the primers listed in Supplementary Table S1. The genes hpcs, acr and ssr were ligated into pET46 Ek/LIC, pRSF-2 Ek/LIC, and pET46 Ek/LIC, respectively. The gene hpcd was ligated with pET46 Ek/LIC and pCDF-2 Ek/LIC. Novablue GigaSinglesTM E. coli cells were transformed with the ligations of pET46-HPCS, pET46-HPCD, pCDF-HPCD, pRSF-ACR, and pET46-SSR, and selected on Luria-Bertani (LB) agar medium with appropriate antibiotics (ampicillin, kanamycin or streptomycin). The colonies were cultured in LB liquid medium with the same antibiotics, and the plasmids were extracted and sequenced by Eton Bioscience, Inc. (Durham, NC).
For protein expression, E. coli Rosetta 2 (DE3) was independently transformed with the plasmids pET46-HPCD and pET46-SSR by heat shock and selected on LB plates supplemented with antibiotics (ampicillin: 100 μg/ml, chloramphenicol: 50 μg/ml). Single colonies were picked and cultured in liquid LB medium at 37°C with the same antibiotics until OD600 = 0.3–0.4. The expression of HPCD was induced with 0.1 mM IPTG, temperature was reduced to 16°C, and cells were cultured for another 15 h at the same conditions. SSR was induced at 37°C with 0.1 mM IPTG and cultured for another 4 h at 37°C.
Co-expression was also conducted for two pairs of enzymes: HPCS and HPCD, ACR and SSR. E. coli Rosetta 2 (DE3) was co-transformed with the plasmids pET46-HPCS and pCDF-HPCD by heat shock and selected by growing on LB medium with antibiotics (ampicillin: 100 μg/ml, streptomycin: 50 μg/ml, chloramphenicol: 50 μg/ml). E. coli Rosetta 2 (DE3) was co-transformed with the plasmids pRSF-ACR and pET46-SSR and selected on LB solid medium with antibiotics (ampicillin: 100 μg/ml, kanamycin: 50 μg/ml, chloramphenicol: 50 μg/ml). Single colonies were selected and cultured in LB liquid medium at 37°C with the same antibiotics. Expression was induced at OD600 = 0.3–0.4 with 0.1 mM IPTG and cultured for another 4 h.
The E. coli cultures expressing the genes of interest were collected by centrifuging at 6,000 × g for 10 min at 4°C, re-suspended in binding buffer (50 mM K2HPO4-KH2PO4, 300 mM NaCl, pH 7.0), and lysed by sonication (S-4000, Misonix Ultrasonic Liquid Processors, Farmingdale, NY) for 10 min with 10 sec off/on pulses. The cell extract was then centrifuged at 10,000 × g for 30 min at 4°C, after which the supernatant and insoluble pellet were separated and analyzed by SDS-PAGE to check for soluble expression.
2.3. Protein purification protocol for HPCS, HPCD, ACR, SSR, and MCR
Before chromatographic purification, E. coli proteins in the supernatant were removed by incubating for 10 min at 65°C and centrifuging at 10,000 × g for 30 min at 4°C. Proteins were further purified using metal affinity chromatography. The heat-treated soluble extract was applied to a HisTrap™ column and eluted (50 mM K2HPO4-KH2PO4, 300 mM NaCl, 300 mM Imidazole, pH 7.5) by monitoring A280 using FPLC (Bio-Rad, BioLogic DuoFlow system, Hercules, CA).
The proteins purified from heat treatment and IMAC were concentrated and exchanged into phosphate buffer (50 mM K2HPO4-KH2PO4, 150 mM NaCl, pH 7.0) by Amicon centrifugal filter. HPCD and SSR were further purified by size exclusion chromatography (Superdex 75 10/300 GL, GE Healthcare). The column was pre-equilibrated with buffer (50 mM K2HPO4-KH2PO4, 150 mM NaCl, pH 7.0) and the sample was added and eluted at a flow rate of 0.5 ml/min while monitoring A280. Extracts containing the co-expressed HPCS/HPCD and ACR/SSR were also loaded onto the Superdex 75 10/300 GL column and eluted in same buffer. Fractions were collected (2 ml/tube) for SDS-PAGE analysis.
The following protein standards were used to make the calibration curve for the Superdex 75: blue dextran (2000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), BSA (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The standard curve was generated by plotting the retention volume versus the logarithm of molecular mass of the protein standards. The apparent molecular weight of each protein was measured by using the calibration curves and elution volume.
The cloning, expression, and purification of MCR was conducted, as described elsewhere, using vector pET21(b) with no His-tag (Alber et al., 2006).
2.4. In vitro measurement of HPCS, HPCD, ACR, ACC, MCR and SSR activities
The activity of HPCS was assayed using the method reported elsewhere (Alber et al., 2008). The reaction mixture included 3-hydroxypropionate (2 mM), tert-butyl-3-hydroxypropionate (2 mM), or acrylate (2 mM) as substrate, along with ATP (3 mM), MgCl2 (2 mM), CoA (0.15 mM), buffer (KHPO4-K2HPO4, 100 mM, pH 7.0), and was started by adding HPCS at 65°C. The reaction was terminated by the addition of HEPES (pH 7.2) – 5,5-dithiobis (2-nitrobenzoic acid) (DTNB, 0.5 mM), and free CoASH was measured at 412 nm. The KM and Vmax values were determined by varying substrate concentration (3-hydroxypropionate: 0.98–62.5 μM, tert-butyl-3-hydroxypropionate: 7.5–250 μM, acrylate: 0.98–62.5 μM), with the concentration of all other substrates kept constant at the same reaction conditions described above.
The activity of HPCD was assayed in a coupled reaction with HPCS and ACR (Teufel et al., 2009). The reaction mixture included 3HP (10 mM), ATP (3 mM), MgCl2 (20 mM), CoA (0.1 mM), buffer (KHPO4-K2HPO4, 100 mM, pH 6.5), and NADPH (0.5 mM). The mixture was first incubated with HPCS and ACR, following which recombinant HPCD was added to start the reaction that was monitored spectrophotometrically at 365 nm for NADPH at 65°C. For kinetic parameter estimation, 3-hydroxypropionate (7.8–62.5 μM) was added separately; the concentrations of the other substrates and reaction conditions were kept constant.
ACR activity was assayed in a coupled reaction with HPCS (Teufel et al., 2009). The reaction mixture included acrylate (10 mM), ATP (3 mM), MgCl2 (10 mM), Coenzyme A (0.1 mM), buffer (KHPO4-K2HPO4, 100 mM, pH 6.5), NADPH (0.5 mM). The mixture was first incubated at 65°C with HPCS, and then the reaction was started by adding ACR and followed spectrophotometrically at 365 nm for NADPH oxidation. To determine kinetic parameters, the concentration of acrylate was varied from 0.98 to 62.5 μM, with the reaction conditions and concentration of other substrates held constant.
MCR activity was measured by incubating the enzyme at 65°C with succinyl-CoA (1 mM), MgCl2 (5 mM), DTT (4 mM), NADPH (0.5 mM), buffer (KHPO4-K2HPO4, 100 mM, pH 6.5); NADPH consumption was followed spectrophotometrically at 365 nm. The KM and Vmax values were determined by varying succinyl-CoA concentration (25–300 μM), with the concentration of all other substrates and reaction conditions kept constant.
SSR activity was measured by incubating the enzyme at 65°C with succinic semialdehyde (1 mM), MgCl2 (5 mM), DTT (5 mM), NADPH (0.5 mM), buffer (KHPO4-K2HPO4, 100 mM, pH 6.5); NADPH consumption was followed spectrophotometrically at 365 nm (Kockelkorn and Fuchs, 2009). The KM and Vmax values of SSR were determined with succinic semialdehyde concentrations between 10–120 μM, with the concentration of all other substrates and reaction conditions kept constant.
2.5. Conversion of 3HP to 4HB in vitro
The sub-pathway for 3HP to 4HB conversion was divided into three parts: conversion of 3HP to propionyl-CoA, conversion of (S)-methylmalonyl-CoA to succinyl-CoA, and conversion of succinyl-CoA to 4HB. For conversion of 3HP to propionyl-CoA, the reaction mixture contained buffer (KHPO4-K2HPO4, 100 mM, pH 6.5), 3HP (10 mM), ATP (3 mM), MgCl2 (20 mM), CoA (0.1 mM), and NADPH (0.5 mM). The reaction mixture was incubated at 65°C for 5 min, ACR was added and incubated for another 5 min, then co-expressed HPCS and HPCD were added and incubated for an additional 5 min followed by HPLC analysis. Conversion of (S)-methylmalonyl-CoA to succinyl-CoA was performed as described previously (Han et al., 2012). Both productions of propionyl-CoA and succinyl-CoA were detected with a Waters HPLC (2487 dual absorbance detector and a 717 plus autosampler) using a 3.9×150 mm NovaPak C18 column equipped with a C18 Sentry guard column (Waters, Milford, MA). The elution profile was monitored at 260 nm and done as follows: 0–12 min: 0–60% buffer B (10% Na-acetate: 100 mM, pH 4.6; 90% methanol); 13–16 min: 60% B; 17–20 min: 100% buffer A (90% Na-acetate: 100 mM, pH 4.6; 10% methanol). Commercial propionyl-CoA and (S)-methylmalonyl-CoA (Sigma Chemical Co., St. Louis, MO) were applied as a standard in HPLC.
For conversion of succinyl-CoA to 4HB, the reaction mixture contained buffer (MOPS-KOH, 100 mM, pH 7.5), succinyl-CoA (2 mM), MgCl2 (2 mM), NADPH (4 mM), and purified recombinant MCR and SSR. The reaction mixture was incubated at 70°C for 20 min and then derivatized with 2,4-dibromoacetophenone (DBAP) to form the phenacyl ester and assayed using reversed-phase HPLC. For each sample, 500 μl of sample was acidified with 50% H2SO4 to pH 2, and extracted with 750 μl diethyl ether to remove salts before the derivatization. The ether fraction was neutralized with 50 μl of 20 mM bicarbonate and the ether was removed by evaporation. The remaining aqueous portion was mixed with 50 μl of acetonitrile and a small amount of pH indicator (0.5 μl of 0.5% phenolphthalein). The solution was then alkalized with 0.1 or 1 M KOH until the sample turned pink (pH ~9–10), after which the following was added: 100 μl of acetonitrile, 50 μl of 1 μM 15-crown-5-ether, and 200 μl of 20 mM 2,4-dibromoacetophenone. The solution was heated to 80°C for 30 min, cooled back to room temperature, and transferred into vials for injection (10 μl) onto the HPLC column. The samples were run on an Atlantis dC18 column (Waters, 3 μm, 4.6 × 150 mm) at 30°C with a flow rate of 1.5 ml/min using a gradient elution profile. The initial mobile phase composition was 65% Buffer A (0.1% formic acid) and 35% Buffer B (acetonitrile). Samples were eluted with a fifteen minute gradient to a final composition of 30% Buffer A and 70% Buffer B. Products were detected by following the absorption spectrum at 254 nm using a single channel of the Waters 2998 photodiode array detector.
2.6. Yeast two-hybrid assay for protein interaction analysis of HPCS-HPCD, and ACR-SSR
Yeast two-hybrid assay was done according to the protocol of Matchmaker GAL4 Two-Hybrid System (Clontech, Mountain View, CA, USA), with two vectors pGBKT7 and pGADT7. HPCS and ACR were ligated into vector pGADT7, and HPCD and SSR were ligated into vector pGBKT7. The genes were cloned from genomic DNA of M. sedula using the primers listed in the Supplementary Table S1. An NdeI restriction site was introduced into the 5’ terminal end of pGADT7-M1456-F, pGBKT7-M2001-F, and pGADT7-M1426-F. A XhoI restriction site was introduced into the 5’ terminal end of pGADT7-M1456-R and pGADT7-M1426-R. An EcoRI restriction site was introduced into the 5’ terminal end of pGBKT7-M2001-R and pGBKT7-M1424-F. Lastly, a SalI restriction site was introduced into the 5’ terminal end of pGBKT7-M1424-R. The cloned genes were digested with the corresponding enzymes and ligated into the aforementioned vectors, digested with the same enzymes. The ligations were transformed into E. coli Novablue competent cells and selected on LB medium with ampicillin. The colonies were picked and cultured in LB liquid medium; the extracted plasmids were sequenced by Eton Biosciences. The sequence confirmed plasmids were transformed into Yeast AH109, using the protocol in reported in Matchmaker Gold Yeast Two-Hybrid System User Manual. Different combinations were applied: pGADT7 and pGBKT7; pGADT7 and pGBKT7-M2001-F; pGADT7-M1456-R and pGBKT7; pGADT7-M1456-R and pGBKT7-M2001-F; pGADT7 and pGBKT7-M1424-R; pGADT7-M1426-R and pGBKT7; pGADT7-M1426-R and pGBKT7-M1424-R. The transformations were viewed on plates containing SC-medium, lacking leucine and tryptophan, and incubated at 30°C for 72 h. Colonies were picked and cultured in the same liquid medium at 30°C for 16 h. The cultures were inoculated onto plates of SC-medium lacking leucine, tryptophan and histidine-HCl, and that contained X-GAL (20 μg/ml). The inoculated plates were incubated at 30°C for 1 week, along with a positive control.
2.7. Reaction kinetics model of the 3HP/4HB cycle
To model the 3HP/4HB cycle, a system of differential mass balance equations was constructed based on known stoichiometry, rate law expressions, and in vitro enzyme parameters for each of the reactions in Figure 1. To provide a sink for acetyl-CoA and succinate produced by the cycle, two reactions were included to simulate the conversion of acetyl-CoA and succinate into biomass. Some parameters are not known for the M. sedula enzymes, particularly KM values for cofactors or reaction products. These unknown parameters were estimated using available literature on the same enzyme from a different organism, or a reasonable value based on literature reports was assumed. For enzymes with known mechanisms, rate law expressions were adapted from Cook and Cleland (Cook and Cleland, 2007). For enzymes with unknown mechanisms or unknown action of inhibitors, convenience kinetics rate laws were used (Liebermeister and Klipp, 2006). All of the reactions are treated as reversible, with the exception of the two biomass-generating reactions. Equilibrium constants were estimated using eQuilibrator (Flamholz et al., 2012) at pH 5.4 and ionic strength of 0.1 M, corresponding to approximate intracellular conditions in M. sedula (Peeples and Kelly, 1995). Reaction Gibbs free energies were calculated according to the equation ΔG′ = −RT ln(Q/Keq), where Q is the reaction quotient and Keq is the equilibrium constant; the temperature used was 25°C because the entropy contribution to most reactions is not known and, therefore, it is not possible to estimate ΔG at alternate temperatures. Cofactor concentrations were held constant at values measured in exponentially-growing E. coli (Bennett et al., 2009) as follows: ATP, 9.6 mM; ADP, 560 μM; AMP, 280 μM; Pi, 1.1 mM; NADPH, 120 μM; NADP+, 2.1 μM; NADH, 83 μM; NAD+, 2.6 mM; CoA, 1.4 mM. Bicarbonate concentration was held constant at 3 mM, corresponding to the equilibrium concentration of bicarbonate in water at 70°C and a CO2 partial pressure of 0.2 bar (Carroll et al., 1991). A complete list of reactions, rate law expressions, and parameter values is found in Supplementary Tables S2 and S3.
The model was constructed and implemented using the SimBiology package in Matlab (version R2014a, Mathworks, Inc.). To determine optimum enzyme ratios, a constrained nonlinear optimization problem was set up to maximize the total biomass production rate after cycle intermediates reached steady-state (r21 + r22). The total mass of all cycle enzymes (excluding BM1 and BM2 which are placeholders for unspecified enzymes) was constrained to a constant value to simulate a constant cellular carbon and energy investment into producing the cycle enzymes. To prevent biologically unrealistic concentrations of cycle intermediates, the steady-state concentrations of these compounds were constrained to less than 10 mM. To avoid local maxima, the optimization problem was run multiple times using quasi-random initial enzyme concentrations determined by Latin hypercube sampling. An average of enzyme concentrations resulting in the top tenth percentile of biomass production rate was used for the final optimized solution.
Enzyme flux control coefficients, cofactor elasticities, and parameter response coefficients were determined for the biomass production rates. The flux control coefficient for enzyme A on reaction B is defined as:
where EA is the enzyme concentration and JB is the reaction flux (Fell, 1997). Cofactor elasticities and parameter response coefficients have analogous definitions with enzyme concentration replaced by cofactor concentration or parameter value, respectively. To evaluate these coefficients, small perturbations to the enzyme concentration, cofactor concentration, or parameter value were made and the model was run to determine the change in biomass production rate.
2.8. Model analysis of metabolic engineering opportunities
To model autotrophic acetyl-CoA production, all cycle enzymes were included except for SSADH and BM2, to ensure biomass produced via acetyl-CoA was the only sink for carbon. To model autotrophic succinate production, all enzymes were included except for BM1, to ensure biomass produced via succinate was the only sink for carbon. In these two cases, the biomass-producing reactions serve as placeholders for additional pathways that convert acetyl-CoA or succinate into products. To model heterotrophic production of succinate, only the enzymes required to convert acetyl-CoA to biomass via succinate were included (ACC, MCR, MSR, HPCS, HPCD, ACR, MCE, MCM, SSADH, and BM2). To model heterotrophic production of 3HP, only the enzymes required to convert acetyl-CoA to 3HP were included (ACC, MCR, and MSR). For heterotrophic production of 3HP or succinate, the concentration of acetyl-CoA was set to 610 μM (Bennett et al., 2009).
3. Results and Discussion
3.1. Characterization of recombinant cycle enzymes involved in 3HP to 4HB conversion
While enzymes of the 3HP/4HB cycle in M. sedula had been characterized to some extent prior to this work, detailed kinetics information was not available in many cases for the purified proteins. This was especially the case for the segment of the cycle converting 3HP to 4HB. To this end, the gene identities of HPCS, HPCD, ACR, MCR, and SSR were confirmed via recombinant expression of active enzymes in E. coli (characterization of MCE and MCM was reported previously (Han et al., 2012)). Kinetic parameters for these enzymes were determined for cycle-relevant reactions, including for M. sedula ACR and for MCR acting on succinyl-CoA.
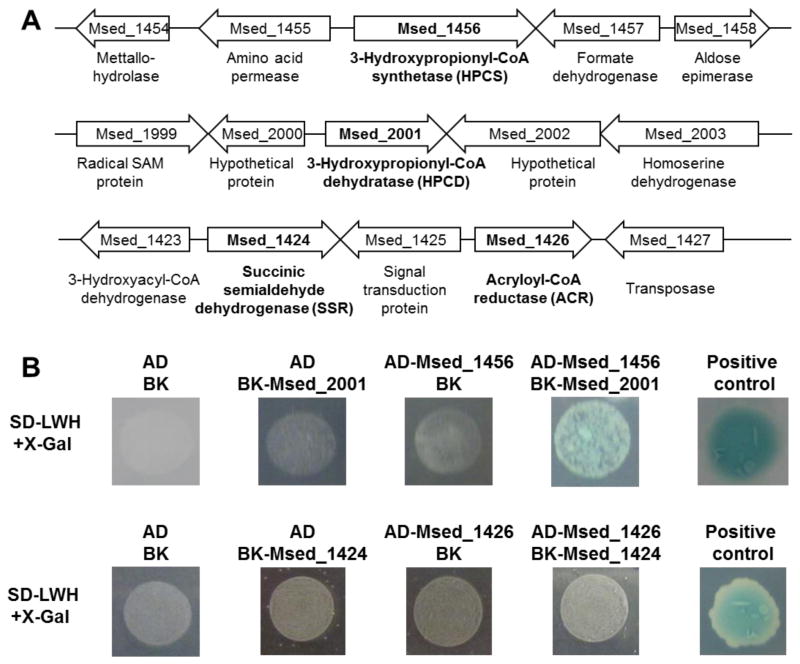
In some cases, there were difficulties in expressing active, recombinant 3HP/4HB cycle enzymes in E. coli, suggesting protein-protein interactions could be important. For example, in order to obtain soluble recombinant HPCS in E. coli, co-expression with HPCD was required (see below). A similar situation was observed for ACR, which required co-expression with SSR. Yeast two-hybrid analysis was used to further investigate the potential for protein-protein interactions between HPCS/HPCD and ACR/SSR. Interaction between HPCS and HPCD or ACR and SSR is expected to result in blue colonies for yeast strains AD-Msed_1456/BK-Msed_2001 or AD-Msed_1426/BK-Msed_1424 grown in the presence of X-Gal, while any strains containing empty AD or BK vectors are expected to be white (Figure 2). The results demonstrate that HPCS and HPCD interact in vivo in yeast, although there is no evidence of interaction between ACR and SSR (Figure 2). Many metabolic pathways involve enzyme complexes held together by non-covalent interactions, viz. metabolons, and can be stable, weakly-interacting, or transient (Zhang, 2011). Metabolons are known to occur in a variety of core metabolic pathways, including the Calvin cycle (Zhang, 2011). Thus, a weak association between HPCS and HPCD is plausible and may be representative of a larger metabolon involving some or all of the 3HP/4HB cycle enzymes.
Figure 2. Protein interaction assay for HPCS and HPCD, ACR and SSR by Yeast Two Hybrid.
(A) Genomic context of HPCS (Msed_1456) and SSR (Msed_2001); ACR (Msed_1426) and SSR (Msed_1424). (B) Yeast two hybrid analyses of HPCS and HPCD, ACR and SSR. For HPCS and HPCD assay, Vector AD and BK, AD and BK-Msed_2001, AD-Msed_1456 and BK were applied as negative control, a commercial positive control was included. For ACR and SSR, Vector AD and BK, AD and BK-Msed_1424, AD-Msed_1426 and BK were applied as negative control.
3.1.1. 3-hydroxypropionyl-CoA dehydratase (HPCD)
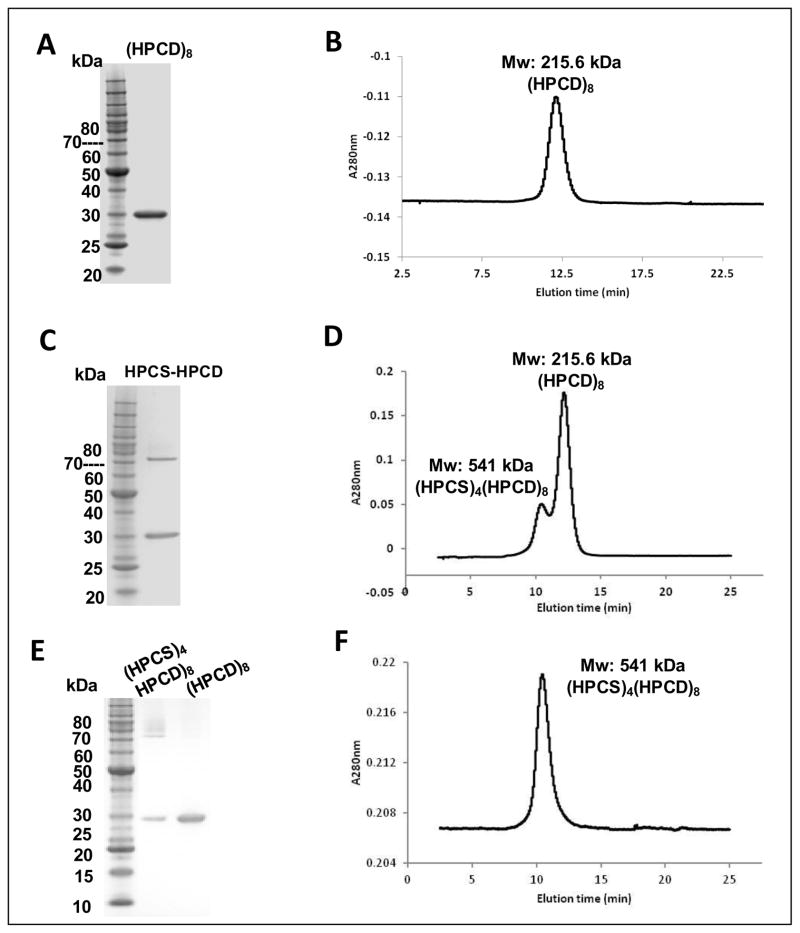
Recombinant HPCD formed a homo-octamer, with an estimated molecular mass of 220 kDa; SDS-PAGE analysis showed that the monomer molecular mass is 28 kDa (Figure 3A, B). The kinetic parameters for HPCD were determined by combining the enzyme with HPCS and ACR (see below). The KM and Vmax of HPCD were 25.1 μM and 272 μmol min−1 mg−1, respectively (Table 2). The Vmax of HPCD is higher than had been reported previously, possibly because of differences in assay pH (Teufel et al., 2009).
Figure 3. Purification, and quaternary structure analysis of HPCS and HPCD.
(A) HPCD was expressed in E. coli, purified by IMAC, and separated by size exclusion chromatography (Superdex 75). (B) Molecular assembly of HPCD as homooctomer. (C) HPCS and HPCD co-expressed in E. coli, purified by IMAC, and viewed on SDS-PAGE. (D) HPCS and HPCD separated by size exclusion chromatography (Superdex 75). (E) SDS-PAGE of two peaks eluted from size exclusion chromatography. (F) (HPCS)4(HPCD)8 determined by size exclusion chromatography.
Table 2.
Molecular properties of selected enzymes for 3HP to 4HB conversion
| Enzyme | Substrate | Vmax (μmol min−1 mg−1) | Km (μM) | kcat/Km (105 M−1 s−1) | Mra (kDa) | Molecular assembly | Ref. |
|---|---|---|---|---|---|---|---|
| MCR | Succinyl-CoA | 40b | 150 | 1.75 | 39.3 | NRd | (Kockelkorn and Fuchs, 2009) |
| 15.9 | 191 | 0.545 | (MCR)3 | This work | |||
| NADPH | 13.1 | 79.2 | 1.09 | ||||
| HPCS | 3-Hydroxypropionate | 18 | 180 | 1.24 | 74.4 | (HPCS)4 | (Alber et al., 2008) |
| HPCD | 3-Hydroxypropionyl-CoA | 272 | 25.1 | 51.2 | 28.3 | (HPCD)8 | This work |
| 151 | 60 | 11.9 | (HPCD)1 | (Teufel et al., 2009) | |||
| ACR | Acryloyl-CoA | 7.6 | 9.1 | 5.01 | 36.0 | (ACR)1 | This work |
| 2.9c | NRd | NRd | NR | (Teufel et al., 2009) | |||
| 18.7b | 10 | 11.2 | (ACR)1 | (Teufel et al., 2009) | |||
| SSR | Succinic semialdehyde | 683 | 27 | 160 | 37.9 | (SSR)5 | This work |
| 700 | 70 | 63.2 | (SSR)2 | (Kockelkorn and Fuchs, 2009) |
Predicted from sequence
Recombinant S. tokodaii, expressed in E. coli and purified
Native M. sedula, partially purified
NR, not reported
3.1.2. 3-hydroxypropionyl-CoA synthetase (HPCS)
In previous reports, native HPCS was purified from M. sedula cell extract and compared to HPCS from Sulfolobus tokodaii produced recombinantly in E. coli (Alber et al., 2008). The reported KM of M. sedula HPCS for 3HP was 180 μM with a Vmax of 18 μmol min−1 mg−1, respectively (Table 2). Here, several different approaches were tried to produce soluble recombinant HPCS from M. sedula. Msed_1456 (HPCS) was cloned and ligated into different vectors (pET46, pET21, and pET42) and expressed in E. coli, but in each case the resulting protein was insoluble. Since the reactions catalyzed by HPCS and HPCD are sequential in the 3HP/4HB cycle, it is possible that the two enzymes interact, and this association could impact soluble expression in a heterologous host. As such, the genes encoding HPCS and HPCD were ligated into pET46 and pCDF, respectively, and the resulting constructs were then co-transformed into Rosetta DE3. Only when both enzymes were co-expressed in E. coli could HPCS be obtained in soluble, active form (Figure 3C). The purified protein complex containing HPCS and HPCD was further analyzed by gel filtration chromatography, and two peaks were observed (Figure 3D). When fractions from the two peaks were analyzed by SDS-PAGE, one peak consisting solely of HPCD was observed, while the other peak contained a mixture of both HPCS and HPCD, suggesting a quaternary structure of (HPCS)4(HPCD)8 (Figure 3E and 2F). Several attempts were made to obtain HPCS from the HPCS/HPCD complex using hydrophilic interaction and ion exchange chromatography, but neither column could effectively separate the two enzymes. Note that recombinant HPCS from S. tokodaii was homodimeric, according to gel filtration analysis (Alber et al., 2008).
3.1.3. Succinic semialdehyde reductase (SSR)
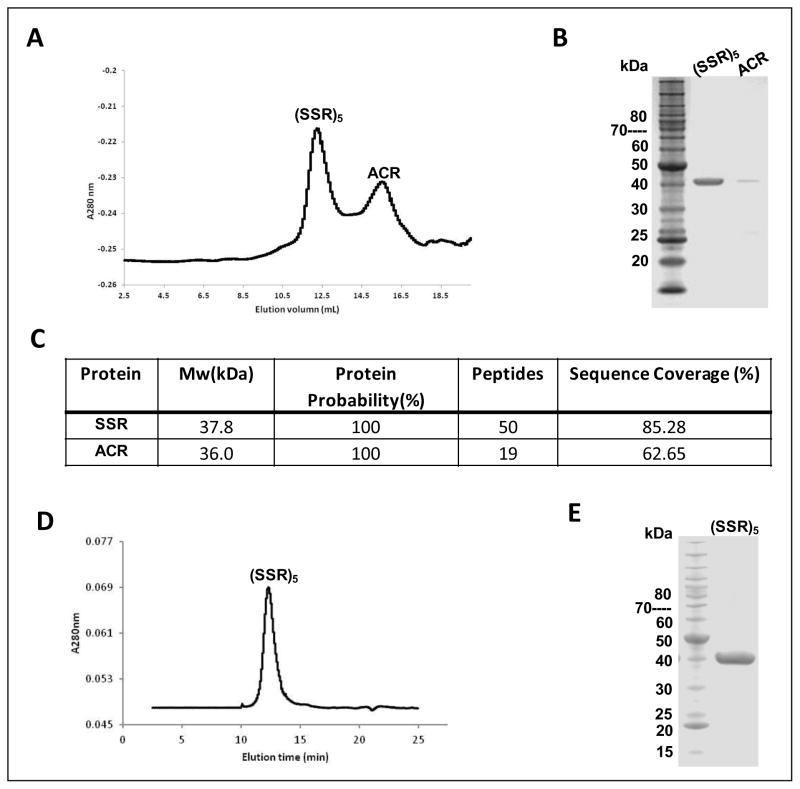
SSR catalyzes the conversion of succinic semialdehyde to 4-hydroxybutyrate. The molecular mass of the SSR monomer is 37.9 kDa, and the enzyme assembles into a homopentamer with Mr of ~200 kDa, based on gel filtration analysis (see Figure 4D). Using succinic semialdehyde as substrate, the KM and Vmax for SSR were 27 μM and 682 μmol min−1 mg−1, respectively, comparable to what was previously reported (Table 2).
Figure 4. Purification and quaternary structure analysis of ACR and SSR.
(A) ACR and SSR were co-expressed in E. coli and recombinant enzymes were purified by IMAC. Quaternary structure was assayed by Superdex 75. (B) The two eluted peaks from (A) were collected and analyzed by SDS-PAGE. (C) NanoLC-MS/MS of the two peaks collected in (A). (D) SSR was expressed in E. coli and recombinant SSR purified by IMAC was analyzed by Superdex 75. (F) The collected peak of recombinant SSR in Superdex 75 was analyzed by SDS-PAGE.
3.1.4. Acryloyl-CoA reductase (ACR)
ACR could not be expressed in soluble, active form in E. coli, even though several vectors and different expression conditions were tried. In the M. sedula genome, the genes for ACR and SSR (Msed_1426 and Msed_1424) are clustered together, separated by a single gene (Msed_1425). In an attempt to obtain soluble, active enzymes, Msed_1426 and Msed_1424 were ligated into pET46 and pRSF, respectively, and co-expressed in E. coli. The molecular masses of ACR and SSR are similar (37.8 and 36.0 kDa, respectively), so that only a single band was observed by SDS-PAGE analysis (see Figure 4A, 4B). To determine the relative amounts of ACR and SSR in the co-expressed samples, the soluble fraction was first purified by metal affinity chromatography, and then the eluted fraction was applied to a gel filtration column. Two peaks were observed (Figure 4A), corresponding to masses of approximately 200 kDa and 40 kDa. To identify these two peaks, the purified co-expressed fraction after metal affinity chromatography was analyzed by NanoLC-MS/MS (Figure 4C) and the results confirmed that both enzymes were present. The two elution peaks from the gel filtration (Figure 4A) were tested for activity and the results showed that peak 1 and peak 2 corresponded to SSR and ACR, respectively. ACR was, therefore, obtained from peak 2 and used for biochemical analysis. For ACR, the Vmax for acryloyl-CoA was 7.6 μmol min−1 mg−1, compared to the previously reported values of 3.0 μmol min−1 mg−1 for the native M. sedula ACR and 18.7 μmol min−1 mg−1 for the version from S. tokodaii (Teufel et al., 2009). Note that although the KM for the recombinant versions of ACR from M. sedula and S. tokodaii were comparable, this is the first report of a KM for the M. sedula enzyme (Table 2).
3.1.5. Malonyl-CoA/succinyl-CoA reductase (MCR)\
MCR was previously partially purified from M. sedula extracts and compared with recombinant MCR from S. tokodaii produced in E. coli (Alber et al., 2006; Kockelkorn and Fuchs, 2009). Here, recombinant M. sedula MCR was obtained by expression of Msed_0745 in E. coli, and the enzyme was purified with metal affinity, ion exchange, and gel filtration chromatography. MCR is a promiscuous enzyme responsible for two reactions in the 3HP/4HB cycle: the reduction of malonyl-CoA and succinyl-CoA to malonic semialdehyde and succinic semialdehyde, respectively, using NADPH as the electron donor (Kockelkorn and Fuchs, 2009). The activity of MCR on succinyl-CoA was verified by following NADPH consumption at 340 nm. The KM and Vmax of MCR with succinyl-CoA were 191 μM and 15.9 μmol min−1 mg−1, respectively. The KM and Vmax of MCR with NADPH were 79.2 μM and 13.1 μmol min−1 mg−1, respectively (Table 2). The catalytic parameters of M. sedula MCR are consistent with previously reported values for recombinant S. tokodaii MCR (Kockelkorn and Fuchs, 2009).
3.1.6. Confirmation of 3HP to 4HB conversion in vitro by recombinant cycle enzymes
Given the availability of the enzymes discussed above, experimental confirmation of their role in the 3HP/4H cycle was addressed. Figure 5 shows that recombinant versions of HPCS, HPCD, ACC (Lian et al., 2016), MCE, MCM, MCR and SSR do indeed catalyze the formation of intermediates the second sub-pathway of the cycle, involved in the conversion of 3HP to 4HB. This information, in conjunction with previous studies (Estelmann et al., 2011; Hawkins et al., 2014; Hawkins et al., 2015; Hawkins et al., 2013; Keller et al., 2013; Lian et al., 2016), forms the basis for the development of a mathematical model describing the 3HP/4HB cycle.
Figure 5. Conversion of 3-hydroxypropionate to 4-hydroxybutyrate by enzymes in the CO2 fixation cycle.
Stepwise confirmation of sub-cycle operation using HPLC: (A) Formation of propionyl-CoA from 3HP by HPCS, HDCD, and ACR – 1: Propionyl-CoA standard, 2: Reaction mixture, 3: Reaction mixture control (no enzymes); (B) Formation of succinyl-CoA from (S)-methylmalonyl-CoA by MCE and MCM – 1: Reaction mixture with both MCE and MCM, 2: Reaction mixture with MCM only, 3: Reaction mixture control (no enzymes); (C) Formation of 4-hydroxybutyrate from succinyl-CoA by MCR and SSR – 1: 4HB standard, 2: Reaction mixture, 3: reaction mixture control (no enzymes).
3.2. Development of an enzyme reaction kinetics model for the 3HP/4HB cycle
A reaction kinetics model of the 3HP/4HB carbon fixation cycle was developed to first understand how the native cycle operates in vivo in extreme thermoacidophiles, such as Metallosphaera sedula. Such a model could then be used to examine ways in which all or part of the cycle could be employed for chemical production via metabolic engineering. The mathematical model represented all 16 enzymatic steps of the cycle, as it is currently understood (Estelmann et al., 2011; Hawkins et al., 2014; Hawkins et al., 2013; Leyn et al., 2015). In addition, side reactions were included for the conversion of succinic semialdehyde to succinate and the conversion of acetyl-CoA or succinate to biomass. Finally, the hydrolysis of pyrophosphate was included, which improves the thermodynamic driving forces for synthesis of 3-hydroxypropionyl-CoA and 4-hydroxybutyryl-CoA (reactions 4, 5, and 13 in Figure 1), despite the relatively low equilibrium constants for these reactions. Reaction rates were modeled using realistic enzyme rate expressions, populated with in vitro kinetic parameters from the literature or determined as part of this study.
3.3. Kinetic modeling and optimization of 3HP/4HB cycle function
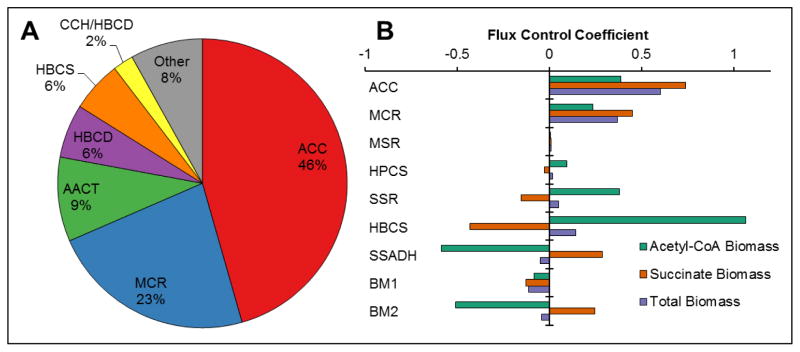
Initially, enzyme concentrations in the model were set to correspond to previously measured activities in M. sedula cell extracts (Berg et al., 2007; Estelmann et al., 2011). However, use of these enzyme concentrations led to the prediction of extremely low specific carbon fixation rates by the model in comparison to a previous study (Estelmann et al., 2011); additionally, comparison of activities in cell extracts with specific activities of purified enzymes implied that the cycle enzymes account for >100% of total cell protein, a physical impossibility. As such, the relative amounts of cycle enzymes in the model were optimized mathematically to give the maximum specific biomass production rate after the concentrations of cycle intermediates reached steady-state. For this optimization, the total enzyme mass was held constant to simulate a fixed carbon and energy investment into the cycle enzymes. The results of the optimization suggest that maximum biomass production is achieved when ACC and MCR make up 46% and 23% of the total enzyme mass of the cycle, respectively, with the remaining enzymes making up significantly smaller portions (Figure 6A). Using these optimized ratios, the model predicts a specific carbon fixation rate of 159 nmol C/min/mg enzyme. The model also predicts that, of the biomass produced, 38.4 ± 2.3% is made via the acetyl-CoA route, with the remaining 61.6 ± 0.6% made via the succinate route. This is in excellent agreement with measurements done by carbon labeling experiments in M. sedula which showed that 65% of carbon tracked the succinate route, with the remaining 35% tracking the acetyl-CoA route (Estelmann et al., 2011). It should be noted that this 38/62% split is predicted a priori by the model, and is not the result of any of the model constraints or the stoichiometry of cycle reactions. This agreement between modeling and experimental results supports the validity of the model. Assays of ACC and MCR activity in M. sedula cell extracts, in comparison with purified enzyme activities, suggest that these two enzymes make up ~2.0% and ~1.0% of total protein, respectively (Alber et al., 2006; Hugler et al., 2003). Thus, the MCR/ACC ratio of ~2 is consistent with model predictions, providing further validation of the model.
Figure 6. Optimized amounts of 3HP/4HB cycle enzymes and flux control coefficients.
(A) Relative enzyme amounts, on a mass basis, for maximum biomass production as predicted by the model for the reactions summarized in Figure 1. (B) Flux control coefficients of each enzyme for biomass production from acetyl-CoA (r21), from succinate (r22), or total (r21 + r22). Only enzymes with flux control coefficients >0.01 are shown.
The specific carbon fixation rate predicted by the model can be used to estimate a growth rate for the organism by assuming a cellular elemental composition of CH2O0.5N0.25 and that cell dry mass is 50% protein (Blanch and Clark, 1997). Using optimized enzyme ratios, the model predicts a maximum growth rate of 0.12 h−1, corresponding with a minimum doubling time of 5.7 h. Observed autotrophic doubling times for M. sedula range from 15 h under microaerophilic conditions to 5 h under hydrogen-, oxygen-, and carbon dioxide-rich conditions (Estelmann et al., 2011; Hawkins et al., 2013). Thus, the model predicts these growth rates could be achieved if a significant fraction of cell protein was made up of cycle enzymes.
3.4. Impact of enzyme concentrations, cofactor levels, and parameters on model results
Given the realistic predictions of cycle performance by the model, the impact of relative enzyme ratios on the predicted biomass production rate, and flux control coefficients (FCCs) for biomass production for each enzyme were determined. A key concept in metabolic control analysis theory, FCCs quantify the effect of an enzyme’s concentration on a reaction flux and indicate the importance of the enzyme to the pathway. An FCC near zero indicates that changing the enzyme concentration would not change the reaction flux; an FCC equal to one represents a classical rate-limiting enzyme, where a 1% increase in enzyme concentration is expected to result in a 1% increase in reaction flux (Fell, 1997).
For the 3HP/4HB cycle, nine enzymes are predicted to have FCCs greater than 0.01 (Figure 6B). ACC and MCR both have large positive FCCs for biomass produced from both acetyl-CoA and succinate, consistent with these enzymes making up a large proportion of the total. This indicates that absolute increases in either the amount or kcat of ACC and MCR would increase the total biomass production rate. SSR and HBCS, which come just after the branch point for succinate-derived biomass, have large positive FCCs for acetyl-CoA-derived biomass but negative FCCs for succinate-derived biomass. SSADH, representing the other side of the succinate branch point, has the opposite effect as SSR and HBCS. This suggests the potential of SSR, HBCS, and SSADH as key control points in driving the proportion of carbon that is directed to biomass through succinate versus the proportion directed through the final part of the cycle to form acetyl-CoA. Finally, the placeholder enzymes BM1 and BM2 have moderately large FCCs, highlighting the impact of the rate at which acetyl-CoA and succinate are drawn away from the cycle on the biomass production.
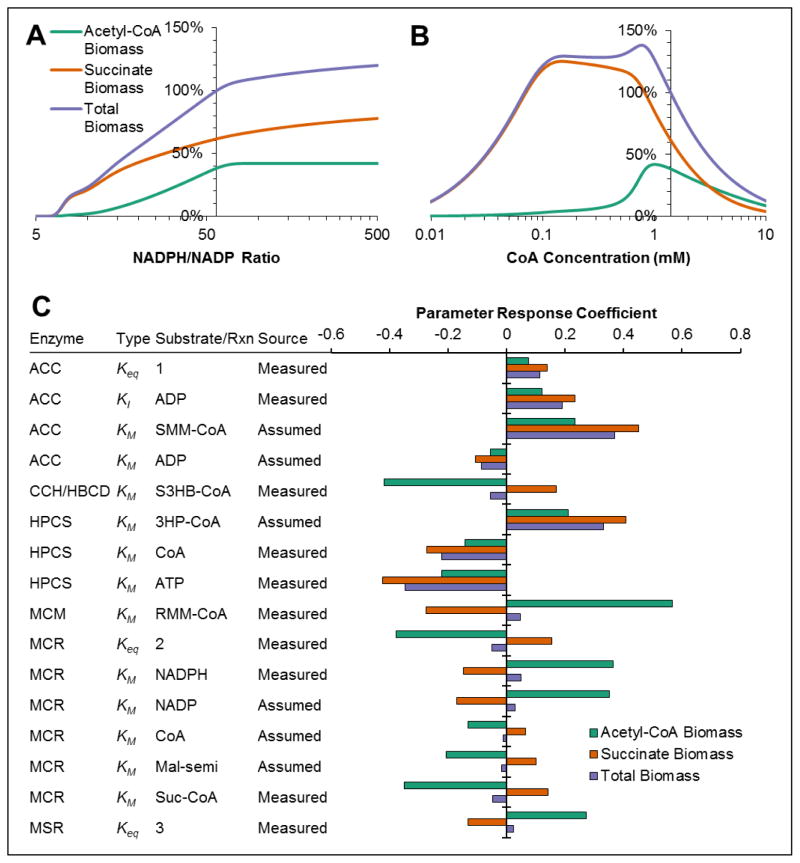
The kinetic model of the 3HP/4HB cycle used cofactor concentrations as measured in glucose-fed, exponentially-growing E. coli (Bennett et al., 2009). But, cofactor concentrations in chemolithoautotrophically-growing M. sedula may be significantly different. To evaluate the effect of cofactor concentrations on biomass production rates, cofactor elasticities were determined for ATP, NADPH, NADH, and coenzyme A. Elasticities are analogous to flux control coefficients, and quantify the impact of a species concentration on a reaction flux. ATP and NADH concentrations had low elasticity for total biomass production (<0.05), indicating that increased or decreased concentrations of these cofactors have little effect on biomass production. Because the ATP concentration used in the model is much greater than enzyme KM values for ATP, ATP does not begin to have large effects on biomass production until its concentration decreases by 10-fold, at which point the elasticity value is 0.44. However, NADPH and CoA elasticity coefficients were significant, with values of 0.38 for NADPH and −0.80 for CoA, indicating that increased NADPH concentration or decreased CoA concentration would increase the total biomass production rate. To further examine the effects of NADPH and CoA, biomass production rates using optimized enzyme ratios were modeled for a wide range of NADPH and CoA concentrations (Figure 7A, 7B). Results indicate that a 20% increase in biomass production rate could be achieved by increasing NADPH concentration, but this would require an NADPH/NADP ratio of nearly 500, well out of the range of typical NADPH/NADP ratios (Bennett et al., 2009). There appears to be an optimum CoA concentration at 0.77 mM that results in an increase in biomass production by 38%, driven by an increase in biomass production from acetyl-CoA. This is most likely driven by the inhibition of ACC and/or AACT by CoA, both of which have KI values below the CoA concentration used.
Figure 7. Effect of cofactor concentrations and parameters on 3HP/4HB cycle.
(A) Effect of NADPH concentration on biomass production rates, relative to the biomass production rate predicted after optimization. The y-intercept represents the concentration used for optimization. (B) Effect of CoA concentration on biomass production rates. (C) Parameter response coefficients for biomass production rates. Only parameter response coefficients >0.01 are shown.
Although many kinetic parameters in the model were obtained from literature reports on the M. sedula enzymes, it was still necessary to make assumptions or estimates from limited data for a number of parameters, mostly for those describing the reverse reaction (see Supplementary Table S3). To evaluate the potential impact of these assumptions, parameter response coefficients for biomass production were determined for all parameters in the model (except for kcat values, which have the same effect as increasing or decreasing enzyme concentration and are, therefore, better-represented as flux control coefficients). Several of these assumed parameters did indeed have large impacts on biomass production (Figure 7C); note that all of these parameters were KM values for reaction products. However, several parameters with measured values also had a large impact on biomass production. These large parameter response coefficients highlight the importance for high-quality measurement of kinetic parameters in interpreting model results, but suggest that errors in parameter estimation do not cause any more uncertainty in the results than do inaccuracies in measured parameters.
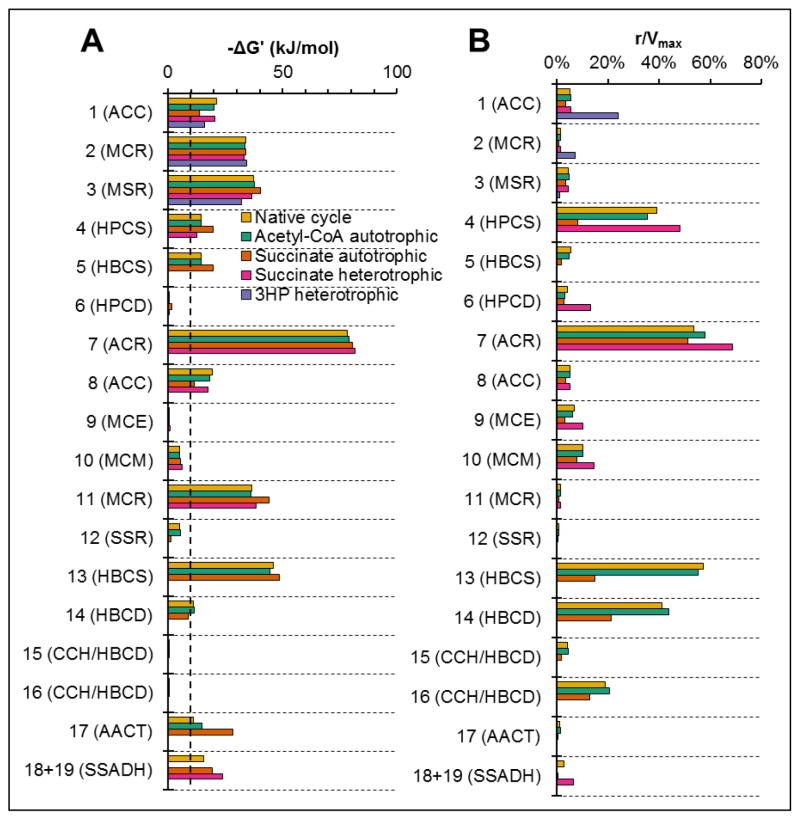
Another perspective from which to view the model is to look for thermodynamic and kinetic limitations on the carbon fixation rate. The thermodynamic driving force and kinetic utilization of each reaction enzyme were determined to evaluate this (Figure 8). The thermodynamic driving force, represented by the Gibbs free energy of reaction at pH 5.4 and model-predicted metabolite concentrations, has little effect on the reaction rate above 10 kJ/mol, at which point the reverse reaction rate is less than 1% of the total reaction rate (Noor et al., 2014). There are few reactions with a driving force of <10 kJ/mol; for these reactions, the small actual reaction rate compared to the maximum velocity of the enzyme (r/Vmax) is a result of an equilibrium limitation for that reaction. More interesting are reactions with large driving forces but small r/Vmax, implying that these reactions are limited by enzyme kinetics. Many of the reactions in the pathway appear to be kinetically-limited, including the reactions ACC and MCR, which make up most of the total enzyme mass (Figure 8). For ACC, the kinetic limitation is primarily a result of inhibition by CoA and succinyl-CoA, for which the KI and concentration have similar values (Supplementary Tables S3 and S4). For MCR, the kinetic limitation is caused by a combination of substrate limitation (for malonyl-CoA/succinyl-CoA and NADPH) and product inhibition (for CoA). These results support the cofactor elasticity and parameter response coefficient results (Figure 7) in suggesting that increased NADPH, decreased CoA, and decreased KM values for MCR would allow increased carbon fixation rates.
Figure 8. Thermodynamic landscape and kinetic utilization of 3HP/4HB cycle enzymes.
(A) −ΔG’, the Gibbs energy of reaction at steady-state metabolite concentrations and pH 5.4, represents the thermodynamic driving force of each reaction. The dotted line represents a driving force of 10 kJ/mol, above which the forward reaction accounts for >99% of the total enzyme reaction rate. (B) The ratio of actual reaction rate to maximum reaction rate for the enzyme (r/Vmax), representing the kinetic utilization of each enzyme.
Because most of the cycle reactions are predicted to be kinetically-limited, changes in the values of ΔG′° are not expected to have large impacts on the model predictions. Values of ΔG′° at 25°C were used because the entropy contribution to ΔG′° is unknown for the cycle reactions, making it impossible to calculate ΔG′° at 65°C. Amend and Shock caution against using values of ΔG′° at 25°C when analyzing reactions for thermophiles (Amend and Shock, 2001). However, a change in the value of ΔG′° as the temperature increases from 25°C to 65°C would impact the reaction rate only for reactions that are thermodynamically-limited. The model predicts that most of the reactions are far from equilibrium and are instead limited by kinetic factors (Figure 8). In particular, reactions catalyzed by enzymes with the largest FCCs, such as ACC, MCR, and HBCS, are predicted to have thermodynamic driving forces of >20 kJ/mol. Therefore, a change in ΔG′° of >10 kJ/mol would be required before thermodynamic effects on these rate-controlling reactions become non-negligible. For the reduction of CO2 to succinic acid (4 CO2 + 7 H2 ↔ Succinic acid + 4 H2O), the primary overall reaction for carbon fixation carried out by M. sedula, ΔG′° changes from −273 kJ/mol at 70°C to −253 kJ/mol at 25°C, a change of 20 kJ/mol (Amend and Shock, 2001). By comparison, a change in ΔG′° of >10 kJ/mol for any of the individual reactions in the cycle seems unlikely. Therefore, changes in values of ΔG′° are not expected to have a large impact on the model results.
Enzymes with large flux control coefficients are likely candidates for regulation. Two of the enzymes predicted to have large flux control coefficients are ACC and HBCS. ACC is strongly up-regulated at the transcriptional level under autotrophic compared to heterotrophic conditions, and recent comparative genomics work indicates that ACC is part of a regulon that includes 13 of the 3HP/4HB cycle genes and is controlled by the transcription factor HhcR (Hawkins et al., 2014; Leyn et al., 2015). In addition, ACC is known to be post-translationally regulated through inhibition by malonyl-CoA, succinyl-CoA, ADP, and CoA (Hugler et al., 2003). The FCCs for HBCS are interesting in that the FCC is positive for the acetyl-CoA branch but negative for the succinate branch, suggesting it could serve to regulate the relative flux between these two branches of the cycle. This provides further evidence for the role of HBCS as a major control point for the distribution of carbon to the acetyl-CoA and succinate branches (Hawkins et al., 2014). Both ACC and HBCS as control points make sense from an energetic standpoint, as they require an ATP investment and are the first steps toward the succinate and acetyl-CoA branches for biomass production, respectively.
4. Metabolic engineering analysis of the 3HP/4HB cycle
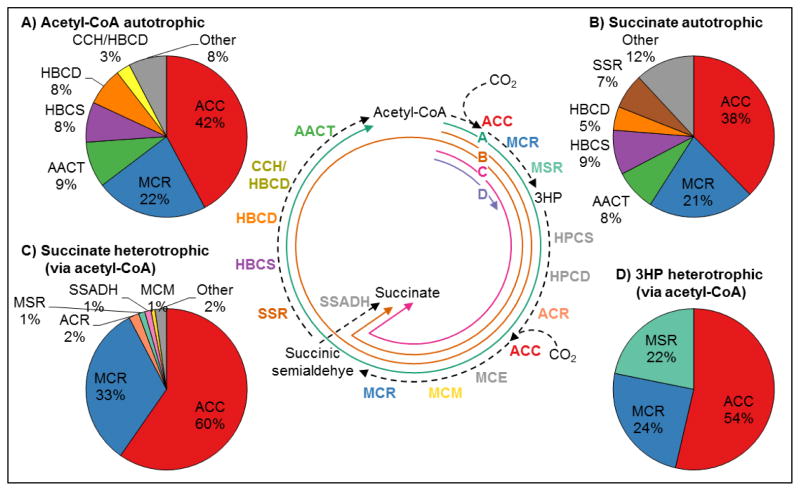
To examine ways in which the 3HP/4HB cycle could be used for metabolic engineering, different pathways to three products were analyzed using the reaction kinetics model. These pathways, composed of subsets of the cycle enzymes, were for the production of acetyl-CoA from CO2 (autotrophic growth), succinate from CO2 (autotrophic growth), 3HP from acetyl-CoA (heterotrophic growth), and succinate from acetyl-CoA (heterotrophic growth). The first two routes require all of the carbon to be derived from CO2, while the second two routes assume two carbons come from sugar catabolism, with the remaining one (for 3HP) or two carbons (for succinate) derived from CO2. The portion of the 3HP/4HB cycle that makes up each pathway is shown in Figure 9.
Figure 9. Optimized enzyme amounts for metabolic engineering applications of 3HP/4HB cycle enzymes.
Relative amounts predicted by model, on a mass basis, shown for each pathway derived from the 3HP/4HB cycle. The pathways are (A) acetyl-CoA autotrophic, (B) succinate autotrophic, (C) succinate heterotrophic (via acetyl-CoA), and (D) 3HP heterotrophic (via acetyl-CoA). Enzyme abbreviations: Acetyl-CoA/propionyl-CoA carboxylase (ACC), Malonyl-CoA/succinyl-CoA reductase (MCR), Malonic semialdehyde reductase (MSR), 3-Hydroxypropionyl-CoA synthetase (HPCS), 4-Hydroxybutyryl-CoA synthetase (HBCS), 3-Hydroxypropionyl-CoA dehydratase (HPCD), Acryloyl-CoA reductase (ACR), Methylmalonyl-CoA epimerase (MCE), Methylmalonyl-CoA mutase (MCM), Succinic semialdehyde reductase (SSR), 4-Hydroxybutyryl-CoA dehydratase (HBCD), Bifunctional crotonoyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase (CCH/HBCD), Acetoacetyl-CoA β-ketothiolase (AACT), Succinic semialdehyde dehydrogenase (SSADH).
4.1. Optimum enzyme ratios
The model was used to predict enzyme ratios for each of the pathways that maximize their specific productivity. Enzyme ratios for the two pathways for autotrophic production of acetyl-CoA or succinate look similar to that of the native cycle, with the majority of enzyme mass being made up of ACC and MCR in a ~2:1 ACC:MCR ratio, and most of the rest being made up of enzymes required for the conversion of 4HB to acetyl-CoA (HBCS, HBCD, and AACT) (Figure 9A, 9B). For maximum production of succinate, SSR makes up 7% of the enzyme mass, in contrast to acetyl-CoA production where it makes up less than 2% (Figure 9B). SSR is present in greater quantities in the autotrophic succinate production pathway because it competes with SSADH for succinic semialdehyde in the acetyl-CoA production pathway.
To produce succinate from organic carbon sources such as sugar, nine of the cycle enzymes are required, plus the reaction “BM2” which serves as a sink for succinate to represent export from the cell or conversion to a different final product. Optimum enzyme ratios indicate that ACC and MCR make up >90% of the total enzyme mass. This reflects the need for ACC and MCR to each catalyze two reactions in the succinate pathway, while other enzymes present in significant amounts in the autotrophic pathways are not required for succinate production from sugar.
The pathway to produce 3HP from sugar catabolism, via acetyl-CoA as a precursor metabolite, is much simpler than any of the other pathways as it only requires three enzymes. ACC and MCR make up >75% of the protein mass, again in a ~2:1 ratio, with MSR making up the remaining amount.
4.2. Impact of parameter uncertainty on pathway flux predictions
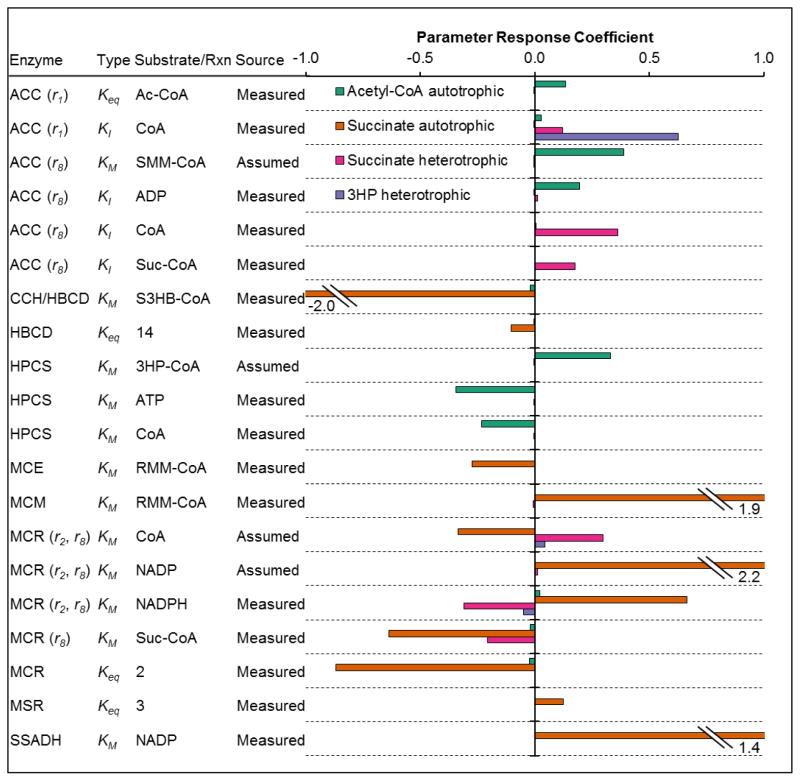
For the pathways considered, several parameter values were assumed, primarily KM values for reaction products or cofactors. To evaluate the impact of uncertainty in measured and assumed parameters on the predicted pathway fluxes, parameter response coefficients were calculated for all parameters in the model. The majority of parameters (81%) had response coefficients with magnitude less than 0.1 for all of the pathways, indicating that errors in these parameters have little impact on the model predictions. The remaining 19% of parameters, with larger expected effects on pathway flux, are shown in Figure 10.
Figure 10. Parameter response coefficients for metabolic engineering applications of 3HP/4HB cycle at optimum enzyme ratios predicted by model.
Only parameters with response coefficients >0.1 are shown.
For all of the pathways, except for autotrophic succinate production, errors in certain parameter estimates are expected to have moderate, but not severe, impacts on pathway flux predictions. The autotrophic acetyl-CoA pathway has three parameters with magnitudes of ~0.3, indicating that if, for example, the KM of HPCS for 3HP-CoA increased by 30%, the pathway flux would be expected to increase by ~10% (to a linear approximation). Thus, errors in parameter estimates or assumptions are not expected to have a major impact on the flux or optimum enzyme ratios predicted for the acetyl-CoA pathway. A similar story holds for the heterotrophic succinate pathway. For the heterotrophic 3HP pathway, only one parameter, the KI of ACC for CoA, had a large impact, with a value of 0.63 (Figure 10), suggesting that if this parameter increased by 30%, a 21% increase in pathway flux would be expected. The autotrophic succinate pathway, however, is very sensitive to changes in multiple parameters, with four parameters having response coefficient magnitudes greater than one. This indicates that errors of 30% in these parameter estimates would have up to a 66% impact on succinate production rate. Therefore, there is less confidence in the model predictions for this pathway than for the other pathways.
4.3. Impact of enzyme and cofactor concentrations on pathway flux
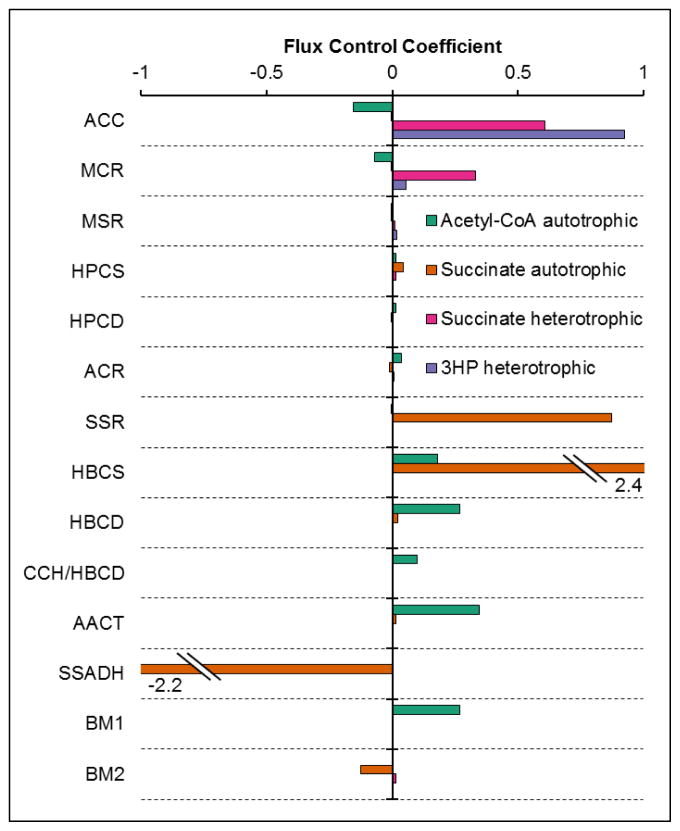
To evaluate the impact of changes in enzyme and cofactor concentrations on the flux of each pathway, flux control coefficients (FCCs) and elasticities for each cofactor were calculated by the model. Flux control coefficients for acetyl-CoA production from CO2 indicate that is the most stable pathway with respect to changes in enzyme concentrations (Figure 11). The flux is controlled primarily by the concentrations of enzymes in the last part of the cycle (HBCS, HBCD, and AACT), similar to the control of acetyl-CoA production from biomass for the native cycle. In contrast, succinate production from CO2 seems to be the least stable of the pathways with respect to enzyme production, with large FCCs for SSR, HBCS, and SSADH.
Figure 11. Flux control coefficients for metabolic engineering applications of 3HP/4HB cycle at optimum enzyme ratios predicted by model.
Only enzymes with FCCs >0.01 are shown.
FCCs for succinate production from sugar indicate that succinate production rate is controlled by both ACC and MCR, consistent with these enzymes making up >90% of the total protein for this pathway. For 3HP production, the flux is controlled almost exclusively by ACC.
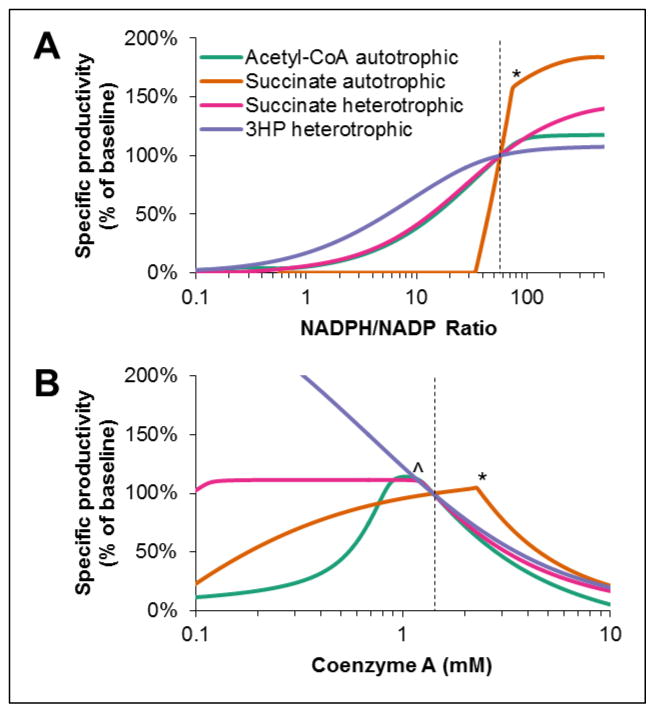
Similar to the native pathway, pathway fluxes were insensitive to a broad range of NADH or ATP, but CoA and NADPH had larger effects. The predicted pathway fluxes over a range of NADPH and CoA concentrations are shown in Figure 12. In general, increasing NADPH concentration and decreasing CoA concentration allows for higher pathway flux. The autotrophic succinate pathway appears to have a discontinuity in CoA elasticity around 2 mM and a discontinuity in NADPH elasticity around an NADPH/NADP ratio of 75. These sharp changes in response to NADPH and CoA are a result of the buildup of pathway intermediates above 2 mM CoA and NADPH/NADP of 75 (data not shown), and are further indicative of the instability of the autotrophic succinate pathway. The heterotrophic succinate pathway also has a discontinuity in CoA elasticity as CoA decreases below 1.1 mM, and also indicates a buildup of pathway intermediates as ACC inhibition by CoA is alleviated. Re-optimizing the enzyme ratios for lower concentrations of CoA would likely result in a curve for the succinate pathway that resembles that of the 3HP pathway (Figure 12B).
Figure 12. Effect of (A) NADPH and (B) coenzyme A concentrations on specific carbon fixation rate for metabolic engineering applications of 3HP/4HB cycle at optimum enzyme ratios predicted by model.
Dotted lines indicate the baseline NADPH and CoA concentrations at which enzymes ratios were optimized. *Sharp corners in curves for autotrophic succinate pathway represent points above which buildup of pathway intermediates prevents reaching steady-state. ^Sharp corner in heterotrophic succinate pathway represents the point below which buildup of pathway intermediates prevents reaching steady-state.
Predicted specific productivities for each pathway, with the native cycle productivity for comparison, are shown in Table 3. The productivity can be evaluated on the basis of moles of product, moles of carbon, or moles of CO2 fixed. The productivity on a carbon basis is much higher for succinate and 3HP produced from sugar than for any of the autotrophic pathways, because two of the carbons in the heterotrophic pathways are derived from the acetyl-CoA supplied to the pathway. The CO2 fixation rate is the best value for comparison between all the pathways because it is independent of whether carbon is supplied to the pathway via CO2, acetyl-CoA, or both. Carbon fixation rates correlate with how many “turns” of the cycle are required for each product. Succinate produced from CO2 has the lowest carbon fixation rate at one and one half turns of the cycle for each succinate produced, followed by acetyl-CoA from CO2 which requires one turn, then succinate from sugar which requires one half turn, and finally 3HP from sugar which requires less than half a turn and has the highest carbon fixation rate.
Table 3.
Specific productivity of pathways constructed from 3HP/4HB cycle
| Pathway | Growth mode | Productivity (nmol product/min/mg) | CO2 fixation rate (nmol C/min/mg) | Compared to native cycle (%) | Potential productivity (nmol product/min/mg) |
|---|---|---|---|---|---|
| Native cycle | - | 159 | 159 | 100 | 181a |
| Acetyl-CoA | autotrophic | 75 | 149 | 94 | 85b |
| Succinate | autotrophic | 23 | 90 | 57 | 26b |
| Succinate | heterotrophic | 107 | 215 | 135 | 61b |
| 3HP | heterotrophic | 421 | 421 | 265 | 140b |
Assuming a 5-h doubling time and 50% cell dry weight as protein
Assuming similar expression levels of pathway enzymes as in M. sedula
Because overall pathway flux is limited by the rates of its constituent enzymes, it is useful to identify factors limiting the enzyme rates. There are two main limitations to the rate at which an enzyme catalyzes its forward reaction: thermodynamic limitations and kinetic limitations. Thermodynamic limitations can be quantified by the thermodynamic driving force (negative Gibbs free energy change) of the reaction. A driving force of zero represents a reaction at equilibrium, at which point half of the enzyme capacity is used for the forward reaction and half for the reverse reaction (Noor et al., 2014). As the driving force increases, an increasing amount of enzyme capacity is used for the forward reaction, and above 10 kJ/mol the vast majority of enzyme capacity (>99%) is used for the forward reaction, representing a negligible effect of thermodynamics on the reaction rate. Kinetic limitations depend on the catalytic properties of the enzyme, and can be affected by concentrations of substrates, products, and inhibitors. To examine the effects of thermodynamic and kinetic limitations on each pathway reaction, the driving force and enzyme utilization factor (r/Vmax) were determined. A few of the reactions are close to equilibrium, and these are catalyzed by HPCD, MCE, MCM, SSR, and CCH/HBCD (Figure 8A). As expected, the enzyme utilization factors for these enzymes are low as a result of the thermodynamic limitation (Figure 8B). However, many reactions have a high driving force but still a low enzyme utilization factor, indicating these reactions are kinetically-limited. In particular, ACC and MCR have utilization factors below 5% for the reactions they catalyze (with the exception of ACC in the 3HP pathway), driven by product inhibition, substrate limitation for MCR, and allosteric inhibition by CoA and succinyl-CoA for ACC (see Supplementary Tables S3 and S4).
4.4. Metabolic engineering scenarios based on the 3HP/4HB cycle
The kinetics and energetics features of the 3HP/4HB cycle as considered for metabolic engineering applications provide a basis for how all or part of it might be implemented in a microbial host. Four such scenarios are considered here with an eye toward the merits of using specific strategies for incorporation of CO2 into bio-based chemicals and fuels.
4.4.1. Autotrophic products from acetyl-CoA
Autotrophic production of acetyl-CoA-derived products has similar reaction kinetics as for the native cycle, implying it could be used effectively for chemical production. Of course, autotrophic chemical production requires that energy and reducing equivalents be obtained via lithotrophy, suggesting that the acetyl-CoA pathway would be best applied in a thermophilic chemolithotroph. Ideally, this pathway would be engineered by modifying the existing 3HP/4HB cycle in an organism such as M. sedula, by knocking-down the expression of SSADH and engineering a pathway for chemical production from acetyl-CoA. While reports of genetic manipulation of M. sedula exist (Maezato et al., 2012), current tools are not sufficient for pathway engineering. An alternative strategy would be to engineer a related organism with available genetic tools, such as Sulfolobus acidocaldarius. This presents certain difficulties, however, because while S. acidocaldarius was initially reported as having the capability for sulfur chemolithoautotrophy, currently used strains with genetic systems are obligate heterotrophs (Wheaton et al., 2015). This suggests that the 3HP/4HB cycle and/or sulfur lithotrophy would need to be repaired prior to using this organism as a host for autotrophic chemical production.
Application of the autotrophic 3HP pathway in a chemolithotrophic host would involve tuning enzyme expression levels such that they approximately match the mass ratios predicted by the model (Figure 6A). In addition, because certain enzymes have a disproportionate impact on pathway flux, it would be desirable to express them in a regulated, or at least inducible, fashion. Dynamic balancing of pathway enzymes in metabolic engineering has been found to be crucial to obtaining high product yields (Zhang et al., 2012). For example, FCCs for the autotrophic acetyl-CoA pathway suggest that because HBCS, HBCD, and ACCT all have a similar impact on pathway flux (Figure 6B), they should be expressed as a single operon driven by a regulated promoter (perhaps regulated in response to acetyl-CoA concentration).
4.4.2. Autotrophic succinate production
Although autotrophic succinate production is attractive from a renewables standpoint, the pathway model suggests that it has a low potential flux (Table 3). Additionally, results for FCCs, parameter response coefficients, and cofactor elasticity for the autotrophic succinate pathway all suggest that this pathway is inherently unstable. This pathway has two enzymes with FCCs of magnitude greater than 2 (HBCS and SSADH; see Figure 6B), unusually large for FCCs (Fell, 1997), indicating that extremely tight control over expression of these enzymes would be required to prevent buildup of pathway intermediates to toxic levels. Similarly, this pathway has four parameter response coefficients with magnitudes greater than one (Figure 7). Moderate changes in NADPH or CoA concentration prevented the autotrophic succinate pathway from reaching steady-state (Figure 12), in contrast to the native cycle, acetyl-CoA pathway, and 3HP pathway, which maintained steady-state solutions over large ranges of NADPH and CoA concentrations (data not shown). This suggests that the presence of a sink for acetyl-CoA, as is the case in the native cycle, provides stability to the overall cycle such that changes in enzyme amounts do not greatly affect succinate production rate. The instability of the autotrophic succinate pathway, combined with its low specific productivity relative to the other pathways (Table 3), suggests that autotrophic production of succinate would be difficult to apply in vivo.
4.4.3. Heterotrophic succinate production
The heterotrophic succinate pathway is predicted to have higher specific productivity than the autotrophic pathways, which is explained by the fact that only half the carbon in succinate is derived from CO2 (Table 3). For successful in vivo application of this pathway, enzymes should be expressed in ratios shown in Figure 9. Also because the pathway flux is controlled almost exclusively by ACC and MCR (Figure 6), it would be ideal to express these two enzymes in an inducible or regulated fashion.
4.4.4. Heterotrophic 3HP production
The heterotrophic 3HP pathway also has a high potential productivity, and requires significantly less ATP per CO2 fixed than any of the other pathways (Table 4). With a low ATP requirement but still high requirement for reducing equivalents (Table 4), 3HP is a promising candidate for production in an anaerobe, particularly if the host can be engineered such that 3HP production is the sole electron sink and is thus coupled with growth. In fact, this 3HP production pathway has already been engineered into the anaerobic fermentative hyperthermophile Pyrococcus furiosus, with engineered strains producing up to 0.3 g/L at productivities of up to 11 mg/L/h, corresponding to a specific productivity of 40 nmol 3HP/min/mg protein (Hawkins et al., 2015; Keller et al., 2013; Thorgersen et al., 2014). The kinetic model of this pathway predicts a specific productivity 2.6-fold higher than that of the native cycle (Table 3). M. sedula has been shown to fix carbon at a rate of up to 181 nmol/min/mg protein (for doubling time of 5 h) (Estelmann et al., 2011; Hawkins et al., 2014). Assuming that the 3HP pathway enzymes could be expressed in P. furiosus at similar levels as the native cycle enzymes in M. sedula, 3HP production rates of up to 480 nmol 3HP/min/mg protein could be achieved (Table 3), a 10-fold improvement over current strains. These increases might be realized by strain engineering to tune enzyme expression levels to be more in line with the optimum ratios predicted by the model (Figure 9D). Additionally, because 3HP production rate is controlled almost exclusively by ACC (Figure 11), this enzyme would ideally be dynamically regulated to balance acetyl-CoA consumption for 3HP with acetyl-CoA requirements of the host organism.
Table 4.
Overall stoichiometry for pathways constructed from 3HP/4HB cycle enzymes
| Pathway | Reaction | ATP/CO2 | NAD(P)H/CO2 |
|---|---|---|---|
| Native cycle | 3.3 CO2 + 8.6 ATP + 7.6 NADPH + 1.65 NAD → 3.3 Biomass (C1) + 8.6 ADP + 7.6 NADP + 1.65 NADH | 2.6 | 1.8 |
| Acetyl-CoA(autotrophic) | 2 CO2 + 6 ATP + 5 NADPH + NAD + CoA → Acetyl-CoA + 6 ADP + 5 NADP + NADH | 3.0 | 2.0 |
| Succinate(autotrophic) | 4 CO2 + 10 ATP + 9 NADPH + 2 NAD → Succinate + 10 ADP + 9 NADP + 2 NADH | 2.5 | 1.8 |
| Succinate(heterotrophic) | Acetyl-CoA + 2 CO2 + 4 ATP + 4 NADPH + NAD → Succinate + 4 ADP + 4 NADP + NADH | 2.0 | 1.5 |
| 3HP(heterotrophic) | Acetyl-CoA + CO2 + ATP + 2 NADPH → 3HP + ADP + 2 NADP + CoA | 1.0 | 2.0 |
4.5. Current status of biological 3HP and succinate production
Both 3HP and succinate have been identified as candidates for bio-based production, and significant effort has been made to produce these chemicals in mesophilic hosts. Succinate production through metabolic engineering has been successful enough for commercialization in some cases (Chung et al., 2015). The highest succinate titer demonstrated to date was achieved by engineering Corynebacterium glutamicum, which produced 146 g/L succinate from glucose via the TCA cycle, with a specific productivity of 87 nmol/min/mg protein (Okino et al., 2008). Given the low potential productivity of the succinate pathway proposed here (61 nmol/min/mg protein; Table 3), combined with the relatively high energy and reducing equivalent requirement (Table 4), it is unlikely that producing succinate from sugar using 3HP/4HB cycle enzymes could be competitive with current biological routes to succinate.
The highest reported 3HP titer to date was achieved by engineering Klebsiella pneumoniae to convert glycerol to 3HP via a two-step pathway, resulting in a specific productivity of 141 nmol 3HP/min/mg protein with a final titer of 48.9 g/L (Huang et al., 2013; Kumar et al., 2013). 3HP production from sugar has also been achieved in E. coli using malonyl-CoA reductase from Chloroflexus aurantiacus (similar to the pathway proposed here), resulting in a titer of 0.2 g/L (Rathnasingh et al., 2012). With a potential specific productivity of 140 nmol/min/mg (Table 3), the kinetics of the 3HP pathway proposed here indicate it could be competitive as an alternate means for biological 3HP production, especially if the pathway enzymes could be overexpressed above the levels at which they exist in M. sedula.
5. Concluding remarks
Assessment of potential strategies for in vivo applications of the 3HP/4HB cycle indicates advantages and disadvantages relative to previously described efforts (Table 5). The autotrophic acetyl-CoA and heterotrophic 3HP pathways have the most promise, the former because it offers the opportunity to produce chemicals directly from CO2, and the latter because of its high flux potential along with the potential for reduced CO2 emissions. Both pathways for succinate production (autotrophic and heterotrophic) would be challenging to implement in vivo with great success. While the heterotrophic succinate pathway has promising kinetics (Table 3), it has a relatively high energy requirement (Table 4) which limits its usefulness as a heterotrophic pathway. Additionally, heterotrophic production of succinate has already had great success in mesophilic hosts, and achieving similar success using the pathway proposed here seems unlikely. Although autotrophic succinate production is attractive from a renewables standpoint, its low potential flux and extreme sensitivity to the quantity of several enzymes indicates it would be nearly impossible to use successfully for metabolic engineering. A final consideration for improving the potential flux of any of these pathways is to find ways to overcome kinetic limitations on many of the enzymes, particularly ACC and MCR. Some of these limitations are driven by product and allosteric inhibition, the effects of which are sensitive to KM/KI values and were assumed for some parameters. Better estimates of pathway productivity could be obtained by first measuring these enzyme parameters, and may alleviate some kinetic limitations. Also, because ACC is significantly limited by allosteric inhibition from CoA, one possibility would be to engineer ACC to reduce the effect of this inhibition, since CoA concentration is tightly regulated in most organisms.
Table 5.
Potential chemical production pathways constructed from 3HP/4HB cycle enzymes
| Product | Carbon from incorporated CO2 (%) | Potential productivity (nmol/min/mg protein) a | Highest reported productivity (nmol/min/mg protein) | Pros | Cons | Enzymes to regulate |
|---|---|---|---|---|---|---|
| “Acetyl-CoA” | 100 | 85 | 133b |
|
|
HBCS/HBCD/AACT (single operon) |
| Succinate | 100 | 26 |
|
|
SSR/HBCS (single operon) SSADH |
|
| Succinate | 50 | 61 | 87c |
|
|
ACC/MCR (single operon) |
| 3HP | 33 | 140 | 141d |
|
|
ACC (rate-controlling enzyme) |
Assuming similar expression levels of pathway enzymes as in M. sedula.
Autotrophic ethanol production by Clostridium ljungdahlii (Nybo et al., 2015)
In Corynebacterium glutamicum (Okino et al., 2008)
In Klebsiella pneumoniae (Huang et al., 2013)
In summary, pathways for constructed from enzymes of the 3HP/4HB carbon fixation cycle have promise for renewable chemical production, and kinetic models suggest concrete strategies for applying these pathways in vivo. Metabolic engineering of extreme thermophiles, driven by tools such as kinetic modeling, and made practicable by genetics tools for hosts such as P. furiosus and S. acidocaldarius (Lipscomb et al., 2011; Wagner et al., 2012), can become an option for bio-based fuels and chemicals.
Supplementary Material
Acknowledgments
This work was supported by grants to RMK and MWWA by the US Department of Energy Research ARPA-E Electrofuels Program (DE-AR0000081), the US National Science Foundation (CBET-1264052, CBET-1264053), and the US Air Force Office of Scientific Research (AFOSR) (FA9550-13-1-0236). ABH acknowledges support from a US Department of Education GAANN Fellowship and AJL acknowledges support from an NIH Biotechnology Traineeship (2T32GM008776).
Abbreviations
Enzyme abbreviations
- 3HP/4HB
3-hydroxypropionate/4-hydroxybutyrate cycle
- ACC
Acetyl-CoA/propionyl-CoA carboxylase
- MCR
Malonyl-CoA/succinyl-CoA reductase
- MSR
Malonic semialdehyde reductase
- HPCS
3-Hydroxypropionyl-CoA synthetase
- HBCS
4-Hydroxybutyryl-CoA synthetase
- HPCD
3-Hydroxypropionyl-CoA dehydratase
- ACR
Acryloyl-CoA reductase
- MCE
Methylmalonyl-CoA epimerase
- MCM
Methylmalonyl-CoA mutase
- SSR
Succinic semialdehyde reductase
- HBCD
4-Hydroxybutyryl-CoA dehydratase
- CCH/HBCD
Bifunctional crotonoyl-CoA hydratase/(S)-3-hydroxybutyryl-CoA dehydrogenase
- AACT
Acetoacetyl-CoA β-ketothiolase
- SSADH
Succinic semialdehyde dehydrogenase
- IPPASE
Inorganic pyrophosphatase
Footnotes
Author contributions
R.M.K. and M.W.W.A. conceived of and managed the research. A.J.L. developed the mathematical model. A.J.L., Y.H., H.L., A.B.H. performed enzyme analysis, G.J.S. and G.L.L. provided advice on biochemical and genetic experiments. A.J.L., Y.H., A.B.H, M.W.W.A., and R.M.K. wrote the manuscript.
References Cited
- Alber B, Olinger M, Rieder A, Kockelkorn D, Jobst B, Hugler M, Fuchs G. Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp. J Bacteriol. 2006;188:8551–9. doi: 10.1128/JB.00987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber BE, Kung JW, Fuchs G. 3-Hydroxypropionyl-coenzyme A synthetase from Metallosphaera sedula, an enzyme involved in autotrophic CO2 fixation. J Bacteriol. 2008;190:1383–9. doi: 10.1128/JB.01593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend JP, Shock EL. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev. 2001;25:175–243. doi: 10.1111/j.1574-6976.2001.tb00576.x. [DOI] [PubMed] [Google Scholar]
- Auernik KS, Kelly RM. Physiological versatility of the extremely thermoacidophilic archaeon Metallosphaera sedula supported by transcriptomic analysis of heterotrophic, autotrophic, and mixotrophic growth. Appl Environ Microbiol. 2010;76:931–5. doi: 10.1128/AEM.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–9. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–6. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- Berg IA, Ramos-Vera WH, Petri A, Huber H, Fuchs G. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology. 2010;156:256–69. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- Blanch HW, Clark DS. Biochemical Engineering. Marcel Dekker, Inc; New York, NY: 1997. [Google Scholar]
- Bobik TA, Rasche ME. Purification and partial characterization of the Pyrococcus horikoshii methylmalonyl-CoA epimerase. Appl Microbiol Biotechnol. 2004;63:682–5. doi: 10.1007/s00253-003-1474-5. [DOI] [PubMed] [Google Scholar]
- Carroll JJ, Slupsky JD, Mather AE. The solubility of carbon dioxide in water at low pressure. J Phys Chem Ref Data. 1991;20:1201. [Google Scholar]
- Chung H, Yang JE, Ha JY, Chae TU, Shin JH, Gustavsson M, Lee SY. Bio-based production of monomers and polymers by metabolically engineered microorganisms. Curr Opin Biotechnol. 2015;36:73–84. doi: 10.1016/j.copbio.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Conrado RJ, Haynes CA, Haendler BE, Toone EJ. Electrofuels: A new paradigm for renewable fuels. In: Lee JW, editor. Advanced Biofuels and Bioproducts. Springer New York; New York, NY: 2013. pp. 1037–1064. [Google Scholar]
- Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. Taylor & Francis; New York, NY: 2007. [Google Scholar]
- Eisenthal R, Danson MJ, Hough DW. Catalytic efficiency and kcat/KM: A useful comparator? Trends Biotechnol. 2007;25:247–9. doi: 10.1016/j.tibtech.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Esser D, Kouril T, Talfournier F, Polkowska J, Schrader T, Bräsen C, Siebers B. Unraveling the function of paralogs of the aldehyde dehydrogenase super family from Sulfolobus solfataricus. Extremophiles. 2013;17:205–216. doi: 10.1007/s00792-012-0507-3. [DOI] [PubMed] [Google Scholar]
- Estelmann S, Hugler M, Eisenreich W, Werner K, Berg IA, Ramos-Vera WH, Say RF, Kockelkorn D, Gad’on N, Fuchs G. Labeling and enzyme studies of the central carbon metabolism in Metallosphaera sedula. J Bacteriol. 2011;193:1191–200. doi: 10.1128/JB.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast AG, Papoutsakis ET. Stoichiometric and energetic analyses of non-photosynthetic CO2-fixation pathways to support synthetic biology strategies for production of fuels and chemicals. Curr Opin Chem Eng. 2012;1:380–395. [Google Scholar]
- Fell D. Understanding the Control of Metabolism. Portland Press; London, UK: 1997. [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, Milo R. eQuilibrator--the biochemical thermodynamics calculator. Nucleic Acids Res. 2012;40:D770–5. doi: 10.1093/nar/gkr874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Hawkins AS, Adams MW, Kelly RM. Epimerase (Msed_0639) and mutase (Msed_0638 and Msed_2055) convert (S)-methylmalonyl-coenzyme A (CoA) to succinyl-CoA in the Metallosphaera sedula 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl Environ Microbiol. 2012;78:6194–202. doi: 10.1128/AEM.01312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T, Urbanke C, Leppanen VM, Goldman A, Brandenburg K, Schafer G. The extreme thermostable pyrophosphatase from Sulfolobus acidocaldarius: Enzymatic and comparative biophysical characterization. Arch Biochem Biophys. 1999;363:135–47. doi: 10.1006/abbi.1998.1072. [DOI] [PubMed] [Google Scholar]
- Hawkins AB, Adams MW, Kelly RM. Conversion of 4-hydroxybutyrate to acetyl-coenzyme A and its anapleurosis in the Metallosphaera sedula 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway. Appl Environ Microbiol. 2014;80:2536–45. doi: 10.1128/AEM.04146-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins AB, Lian H, Zeldes BM, Loder AJ, Lipscomb GL, Schut GJ, Keller MW, Adams MW, Kelly RM. Bioprocessing analysis of Pyrococcus furiosus strains engineered for CO2-based 3-hydroxypropionate production. Biotechnol Bioeng. 2015;112:1533–43. doi: 10.1002/bit.25584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins AS, Han Y, Bennett RK, Adams MW, Kelly RM. Role of 4-hydroxybutyrate-CoA synthetase in the CO2 fixation cycle in thermoacidophilic archaea. J Biol Chem. 2013;288:4012–22. doi: 10.1074/jbc.M112.413195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter S, Fuchs G, Bacher A, Eisenreich W. A bicyclic autotrophic CO2 fixation pathway in Chloroflexus aurantiacus. J Biol Chem. 2002;277:20277–83. doi: 10.1074/jbc.M201030200. [DOI] [PubMed] [Google Scholar]
- Huang Y, Li Z, Shimizu K, Ye Q. Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour Technol. 2013;128:505–12. doi: 10.1016/j.biortech.2012.10.143. [DOI] [PubMed] [Google Scholar]
- Huber H, Gallenberger M, Jahn U, Eylert E, Berg IA, Kockelkorn D, Eisenreich W, Fuchs G. A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc Natl Acad Sci U S A. 2008;105:7851–6. doi: 10.1073/pnas.0801043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugler M, Krieger RS, Jahn M, Fuchs G. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Eur J Biochem. 2003;270:736–44. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- Keller M, Loder A, Basen M, Izquierdo J, Kelly RM, Adams MWW. Production of lignofuels and electrofuels by extremely thermophilic microbes. Biofuels. 2015;5:499–515. [Google Scholar]
- Keller MW, Schut GJ, Lipscomb GL, Menon AL, Iwuchukwu IJ, Leuko TT, Thorgersen MP, Nixon WJ, Hawkins AS, Kelly RM, Adams MWW. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci U S A. 2013;110:5840–5. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoven EJ, Lahtvee P-J, Nielsen J. Applications of computational modeling in metabolic engineering of yeast. FEMS Yeast Res. 2014;15 doi: 10.1111/1567-1364.12199. [DOI] [PubMed] [Google Scholar]
- Kockelkorn D, Fuchs G. Malonic semialdehyde reductase, succinic semialdehyde reductase, and succinyl-coenzyme A reductase from Metallosphaera sedula: Enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in Sulfolobales. J Bacteriol. 2009;191:6352–62. doi: 10.1128/JB.00794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Ashok S, Park S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol Adv. 2013;31:945–61. doi: 10.1016/j.biotechadv.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Leyn SA, Rodionova IA, Li X, Rodionov DA. Novel transcriptional regulons for autotrophic cycle genes in Crenarchaeota. J Bacteriol. 2015;197:2383–91. doi: 10.1128/JB.00249-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC. Integrated electromicrobial conversion of CO2 to higher alcohols. Science. 2012;335:1596. doi: 10.1126/science.1217643. [DOI] [PubMed] [Google Scholar]
- Lian H, Zeldes BM, Lipscomb GL, Hawkins AB, Han Y, Loder AJ, Nishiyima D, Adams MWW, Kelly RM. Ancillary contributions of heterologous biotin protein ligase and carbonic anhydrase for CO2 incorporation into 3-hydroxypropionate by metabolically engineered Pyrococcus furiosus. Biotechnol Bioeng. 2016 doi: 10.1002/bit.26033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermeister W, Klipp E. Bringing metabolic networks to life: Convenience rate law and thermodynamic constraints. Theor Biol Med Model. 2006;3:41. doi: 10.1186/1742-4682-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE, Scott RA, Adams MWW, Westpheling J. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: Construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77:2232–8. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loder AJ, Zeldes BM, Garrison GD, 2nd, Lipscomb GL, Adams MW, Kelly RM. Alcohol selectivity in a synthetic thermophilic n-butanol pathway is driven by biocatalytic and thermostability characteristics of constituent enzymes. Appl Environ Microbiol. 2015;81:7187–200. doi: 10.1128/AEM.02028-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezato Y, Johnson T, McCarthy S, Dana K, Blum P. Metal resistance and lithoautotrophy in the extreme thermoacidophile Metallosphaera sedula. J Bacteriol. 2012;194:6856–63. doi: 10.1128/JB.01413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheschi RJ, Li H, Zhang K, Noey EL, Kim S, Chaubey A, Houk KN, Liao JC. A synthetic recursive “+1” pathway for carbon chain elongation. ACS Chem Biol. 2012;7:689–97. doi: 10.1021/cb200313e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattozzi M, Ziesack M, Voges MJ, Silver PA, Way JC. Expression of the sub-pathways of the Chloroflexus aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: Toward horizontal transfer of autotrophic growth. Metab Eng. 2013;16:130–9. doi: 10.1016/j.ymben.2013.01.005. [DOI] [PubMed] [Google Scholar]
- McCloskey D, Palsson BO, Feist AM. Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol Syst Biol. 2013;9:661. doi: 10.1038/msb.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton B. The kinetic mechanism and properties of the cytoplasmic acetoacetyl-coenzyme A thiolase from rat liver. Biochem J. 1974;139:109–21. doi: 10.1042/bj1390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, MacEachran D, Burd H, Sathitsuksanoh N, Bi C, Yeh YC, Lee TS, Hillson NJ, Chhabra SR, Singer SW, Beller HR. Engineering of Ralstonia eutropha H16 for autotrophic and heterotrophic production of methyl ketones. Appl Environ Microbiol. 2013;79:4433–9. doi: 10.1128/AEM.00973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor E, Bar-Even A, Flamholz A, Reznik E, Liebermeister W, Milo R. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput Biol. 2014;10:e1003483. doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybo SE, Khan NE, Woolston BM, Curtis WR. Metabolic engineering in chemolithoautotrophic hosts for the production of fuels and chemicals. Metab Eng. 2015;30:105–20. doi: 10.1016/j.ymben.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–64. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- Peeples TL, Kelly RM. Bioenergetic response of the extreme thermoacidophile Metallosphaera sedula to thermal and nutritional stresses. Appl Environ Microbiol. 1995;61:2314–21. doi: 10.1128/aem.61.6.2314-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Fuchs G. Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota. J Bacteriol. 2011;193:1201–11. doi: 10.1128/JB.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasingh C, Raj SM, Lee Y, Catherine C, Ashok S, Park S. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J Biotechnol. 2012;157:633–40. doi: 10.1016/j.jbiotec.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Teufel R, Kung JW, Kockelkorn D, Alber BE, Fuchs G. 3-hydroxypropionyl-coenzyme A dehydratase and acryloyl-coenzyme A reductase, enzymes of the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle in the Sulfolobales. J Bacteriol. 2009;191:4572–81. doi: 10.1128/JB.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Evans PR, Leadlay PF. Protection of radical intermediates at the active site of adenosylcobalamin-dependent methylmalonyl-CoA mutase. Biochemistry. 2000;39:9213–9221. doi: 10.1021/bi0004302. [DOI] [PubMed] [Google Scholar]
- Thorgersen MP, Lipscomb GL, Schut GJ, Kelly RM, Adams MWW. Deletion of acetyl-CoA synthetases I and II increases production of 3-hydroxypropionate by the metabolically-engineered hyperthermophile Pyrococcus furiosus. Metab Eng. 2014;22:83–88. doi: 10.1016/j.ymben.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Ueki T, Nevin KP, Woodard TL, Lovley DR. Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii. mBio. 2014;5:e01636–14. doi: 10.1128/mBio.01636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasie M, Chowdhury S, Banerjee R. Importance of the histidine ligand to coenzyme B-12 in the reaction catalyzed by methylmalonyl-CoA mutase. J Biol Chem. 2002;277:18523–18527. doi: 10.1074/jbc.M111809200. [DOI] [PubMed] [Google Scholar]
- Wagner M, van Wolferen M, Wagner A, Lassak K, Meyer BH, Reimann J, Albers SV. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front Microbiol. 2012;3:214. doi: 10.3389/fmicb.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton G, Counts J, Mukherjee A, Kruh J, Kelly R. The confluence of heavy metal biooxidation and heavy metal resistance: Implications for bioleaching by extreme thermoacidophiles. Minerals. 2015;5:397–451. [Google Scholar]
- Yuzawa S, Chiba N, Katz L, Keasling JD. Construction of a part of a 3-hydroxypropionate cycle for heterologous polyketide biosynthesis in Escherichia coli. Biochemistry. 2012;51:9779–81. doi: 10.1021/bi301414q. [DOI] [PubMed] [Google Scholar]
- Zeldes BM, Keller MW, Loder AJ, Straub CT, Adams MW, Kelly RM. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol. 2015;6:1209. doi: 10.3389/fmicb.2015.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- Zhang YH. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol Adv. 2011;29:715–25. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.