Abstract
Background
Population-based studies have revealed 2 to 10% measles vaccine failure rate even after two vaccine doses. While the mechanisms behind this remain unknown, we hypothesized that host genetic factors are likely to be involved.
Methods
We performed a genome-wide association study of measles specific neutralizing antibody and IFNγ ELISPOT response in a combined sample of 2,872 subjects.
Results
We identified two distinct chromosome 1 regions (previously associated with MMR-related febrile seizures), associated with vaccine-induced measles neutralizing antibody titers. The 1q32 region contained 20 significant SNPs in/around the measles virus receptor-encoding CD46 gene, including the intronic rs2724384 (p-value = 2.64x10−09) and rs2724374 (p-value = 3.16x10−09) SNPs. The 1q31.1 region contained nine significant SNPs in/around IFI44L, including the intronic rs1333973 (p-value = 1.41x10−10) and the missense rs273259 (His73Arg, p-value = 2.87x10−10) SNPs. Analysis of differential exon usage with mRNA-Seq data and RT-PCR suggests the involvement of rs2724374 minor G allele in the CD46 STP region exon B skipping, resulting in shorter CD46 isoforms.
Conclusions
Our study reveals common CD46 and IFI44L SNPs associated with measles-specific humoral immunity, and highlights the importance of alternative splicing/virus cellular receptor isoform usage as a mechanism explaining inter-individual variation in immune response after live measles vaccine.
Keywords: Genome-Wide Association Study, Measles, Measles Vaccine, Measles-Mumps-Rubella Vaccine, Immunity, Humoral, Immunity, Cellular, Polymorphism, Single Nucleotide, Alternative Splicing, Genetic Variation, CD46 protein, Human, Adult
Measles still remains a disease of public health concern in the developing world and well-developed countries with multiple outbreaks even among populations with high vaccine coverage. From 2010 to date, the European region registered 135,600 measles cases, and the US experienced 1,381 measles cases in 27 states. (Haralambieva et al. 2015; Haralambieva et al. 2013; Poland and Jacobson 2012; Prevention 2015; Whitaker and Poland 2014) Several population-based studies have estimated that 2 to 10% of vaccine recipients do not develop or sustain measles-specific protective immunity after two doses of MMR vaccine. (Bednarczyk et al. 2016; Haralambieva et al. 2011b; Haralambieva et al. 2013; Poland and Jacobson 2012; Whitaker and Poland 2014) The mechanisms behind vaccine failure are unknown. This knowledge gap is an impediment to controlling future outbreaks or designing improved vaccine candidates.
Measles vaccine-induced humoral immunity is reported to have an extremely high heritability of 88.5%. (Tan et al. 2001) We have performed a series of candidate genetic association studies delineating the effect of HLA alleles and single nucleotide polymorphisms on measles humoral and cellular immune responses, but thus far only approximately 30% of the inter-individual variation in immune response to this vaccine can be explained.(Dhiman et al. 2007; Haralambieva et al. 2015; Haralambieva et al. 2011a; Haralambieva et al. 2013; Haralambieva et al. 2011c; Kennedy et al. 2012a; Ovsyannikova et al. 2011a; Ovsyannikova et al. 2011b; Ovsyannikova et al. 2012)
We report the first GWAS study (on a sample of 2,872 subjects) of measles vaccine-induced humoral and cellular immune response outcomes in children and younger adults, that identify significant SNP associations (in the CD46 and IFI44L genes) underlying the observed inter-individual variability in neutralizing antibody titers after vaccination.
Methods
Additional methods details are available in the Supplementary data.
Study Subjects
We analyzed a large sample of 3,191 healthy children, older adolescents, and healthy adults (age 11 to 40 years) consisting of three independent cohorts: a Rochester cohort (n=1,062); a San Diego cohort (n=1,071); and a US cohort (n=1,058). The demographic and clinical characteristics of these cohorts have been previously published (Haralambieva et al. 2011a; Haralambieva et al. 2011c; Kennedy et al. 2012a; Kennedy et al. 2012b; Kennedy et al. 2012c; Lambert et al. 2015; Ovsyannikova et al. 2011a; Ovsyannikova et al. 2011b; Ovsyannikova et al. 2012).
The Institutional Review Boards of the Mayo Clinic (Rochester, MN) and the NHRC (San Diego, CA) approved the study, and written informed consent was obtained from each subject, i.e., from age-appropriate participants and the parents of all children who participated in the study.
Genotyping and Immune Outcomes
The genome-wide SNP typing was performed using the Infinium Omni 1M-Quad SNP array (Illumina; San Diego, CA) for the Rochester cohort, Illumina Human Omni2.5–8 BeadChip array for the US cohort, and Illumina Infinium HumanHap550v3_A or HumanHap650Yv3 BeadChip arrays for the San Diego cohort. Measles-specific neutralizing antibody and cytokine responses were quantified using a fluorescence-based plaque reduction microneutralization assay (PRMN) and ELISPOT/ELISA assays, as previously described (Haralambieva et al. 2011b). Our humoral immune response phenotype, the 50% end-point titer (Neutralizing Doze, ND50), was calculated using Karber’s formula and transformed into mIU/mL (using the 3rd WHO international measles antibody standard), as described previously (Haralambieva et al. 2011b). The variability of the PRMN assay, calculated as a coefficient of variation (CV) based on the log-transformed ND50 values of the third WHO standard, was 5.7%, as previously published (Haralambieva et al. 2011b).
Next Generation Sequencing (mRNA-Seq) and RT-PCR
Libraries were generated from total RNA (extracted from PBMCs of 30 subjects) using Illumina’s mRNA TruSeq (v1) kit and sequenced (paired end sequencing) on an Illumina HiSeq 2000 (Illumina; San Diego, CA) with Illumina’s TruSeq Cluster kit (v3-cBot-HS) and 51 Cycle Illumina TruSeq SBS Sequencing Kit (v3), as previously published (Haralambieva et al. 2016). One-Step RT-PCR system with Platinum Taq® DNA polymerase (Invitrogen, Carlsbad, CA) and primers allowing CD46 isoform/isoforms discrimination were used, as previously described (Wang et al. 2000).
Statistical methods
GWAS Analysis
To achieve greatest power to detect SNPs associated with measles-specific immune response phenotypes, we pooled our data across genotyping platforms and the three cohorts. To perform the pooled analyses, we first accounted for the effects of potentially confounding factors that vary across ancestry/platform/cohort strata. After thoroughly evaluating the quality of the genotype data, we used the genetic data to define major ancestry groups. We then estimated eigenvectors within each ancestry group to account for the effects of population stratification within ancestry groups. Because the largest ancestry groups were Caucasian and African-American, we restricted our pooled analysis to these groups. After accounting for population stratification, we evaluated the covariates that were available within each of the ancestry-platform-cohort strata to determine if the covariates were associated with the phenotype, in order to regress out the effects of potential confounding factors. This produced residuals (adjusted traits) that were then used for the GWAS analyses. The immune response trait measles-specific IFNγ ELISPOT, as well as measles-specific secreted cytokines, were transformed by normal quantiles of the difference of the mean stimulated and mean unstimulated values. The immune response trait neutralizing antibody titer was transformed as the natural log of the PRMN mIU/mL value (for more details, refer to Supplemental Methods).
Analysis of mRNA-Seq Data for Differential Exon Usage
We tested for evidence of differential exon usage in the CD46 and IFI44L genes using the method of Anders et al., implemented in the DEXSeq package (version 1.16.10) in the R programing language (version 3.2.3).(Anders et al. 2012; Team 2009)
Molecular Modeling
In order to evaluate the effect of differential splicing on the dynamics of the CD46 molecular structure, we generated homology models (Roy et al. 2010) of the extracellular domains (SCR1-4 and STP domains) for each isoform and analyzed each using Anisotropic Network Models (ANMs) (Atilgan et al. 2001; Chennubhotla and Bahar 2007; Yang et al. 2009) using multiple templates (see Supplemental Data for details). Interactions between CD46 and MV-H were modeled using the crystal structure PDB 3INB (Santiago et al. 2010) as a template wherein dimeric MV-H is bound symmetrically by the SCR1 and SCR2 domains of two CD46 molecules (see Supplemental Data for details).
Results
Genome-wide analysis results with humoral immunity
Genetic regions associated with variations in measles-specific antibody response after vaccination
The demographic and immune characteristics of our study sample (n=2,872) are summarized in Table 1. We identified two independent gene regions on chromosome 1 associated with antibody response following measles vaccination (Fig. 1, Supplemental Fig. 1). As depicted on the locus zoom plot (Fig. 1A), the right region on chromosome 1 contained multiple SNPs (n=20) in/around the MV receptor-encoding CD46 gene and region (1q32, bp 207917499-208025926, NCBI Build 37/hg19). Analyzing associations between antibody response and SNPs in the two other MV receptors on chromosome 1, SLAM (SLAMF1) and nectin-4 (NECTIN4/PVRL4), did not result in significant findings. The left region (Fig. 1C) on chromosome 1 (1p31.1, bp 79082772-79110518) contained nine significant SNPs in/around the interferon-induced, protein 44-like gene IFI44L.
Table 1.
Demographic and immune characterization of the study population
| African Americane (n=317) | Caucasiane (n=2555) | Total (n=2872) | P- value | |
|---|---|---|---|---|
| Neutralizing Ab titera | <0.001f | |||
| N missing (failed assay QC) | 5 | 49 | 54 | |
| Mean (SDc) | 2065 (2821) | 1271 (1652) | 1359 (1835) | |
| Median (IQRd) | 1180 (550, 2632) | 803 (383, 1607) | 845 (394, 1683) | |
|
| ||||
| IFNγ ELISPOT responseb | 0.427f | |||
| N missing (failed assay QC) | 26 | 228 | 254 | |
| Mean (SDc) | 24.4 (26) | 25.2 (32) | 25.1 (31.4) | |
| Median (IQRd) | 15 (7, 34.5) | 15 (6, 32.5) | 15 (6, 32.7) | |
|
| ||||
| Gender/sex | 0.082g | |||
| Female | 73 (23%) | 707 (27.7%) | 780 (27.2%) | |
| Male | 244 (77%) | 1848 (72.3%) | 2092 (72.8%) | |
|
| ||||
| Race (self-declared) | NAh | |||
| White | 0 (0%) | 2206 (86.3%) | 2206 (76.8%) | |
| Black or African American | 302 (95.3%) | 38 (1.49%) | 340 (11.8%) | |
| American Indian/Alaska Native | 0 (0%) | 29 (1.14%) | 29 (1.01%) | |
| Asian/Hawaiian/Pacific Islander | 1 (0.315%) | 19 (0.744%) | 20 (0.696%) | |
| Multiple | 9 (2.84%) | 100 (3.91%) | 109 (3.8%) | |
| Other | 4 (1.26%) | 119 (4.66%) | 123 (4.28%) | |
| Unknown | 1 (0.315%) | 44 (1.72%) | 45 (1.57%) | |
|
| ||||
| Ethnicity (self-declared) | <0.001g | |||
| Hispanic/Latino | 13 (4.1%) | 387 (15.1%) | 400 (13.9%) | |
| Not Hispanic/Latino | 285 (89.9%) | 2137 (83.6%) | 2422 (84.3%) | |
| Unknown | 19 (5.99%) | 31 (1.21%) | 50 (1.74%) | |
|
| ||||
| Age at enrollment (years) | <0.001f | |||
| Number missing | 49 | 196 | 245 | |
| Mean (SDc) | 24.5 (5.97) | 21.1 (6.06) | 21.4 (6.14) | |
| Median (IQRd) | 24 (21, 28) | 22 (16, 25) | 22 (16, 25) | |
|
| ||||
| Age at last vaccination (years) | <0.001f | |||
| Number missing | 107 | 577 | 684 | |
| Mean (SDc) | 19.8 (7.97) | 16.1 (8.38) | 16.4 (8.41) | |
| Median (IQRd) | 20 (18, 24) | 18 (10, 22) | 18 (11, 23) | |
|
| ||||
| Time from last vaccination to enrollment (years) | 0.031f | |||
| Number missing | 107 | 577 | 684 | |
| Mean (SDc) | 3.7 (3.85) | 4.0 (3.43) | 4.0 (3.47) | |
| Median (IQRd) | 2.6 (0.04, 5.5) | 3.5 (0.03, 6.4) | 3.4 (0.03, 6.4) | |
|
| ||||
| Cohort | <0.001g | |||
| Rochester | 40 (12.6%) | 942 (36.9%) | 982 (34.2%) | |
| San Diego | 164 (51.7%) | 718 (28.1%) | 882 (30.7%) | |
| US | 113 (35.6%) | 895 (35%) | 1008 (35.1%) | |
Neutralizing antibody titer in mIU/mL, measured by the plaque reduction microneutralization assay (PRMN)
IFNγ-positive spot-forming units (SFUs) per 2 x 105 cells (mean of measles virus-specific stimulated response, measured in triplicate, minus the mean unstimulated response, also measured in triplicate).
Standard Deviation.
IQR, inter-quartile range with 25% and 75% quartiles
Genetically classified into African American or Caucasian ancestry group based on STRUCTURE (see Methods)
Kruskal-Wallis Rank Test
Fisher’s Exact Test
Not applicable
Fig. 1. Locus zoom plots of the chromosome 1 regions and effects of SNPs associated with measles-specific neutralizing antibody titers in the combined cohorta.
A and C. Locus zoom plots of the 1q32(CD46, A) and 1q31.1 (IFI44L, C) regions associated with neutralizing antibody titer after measles vaccination (combined cohorta). On the x-axis SNPs are plotted by chromosomal location. The left y-axis reflects the association (−log10P-value) with vaccine-induced measles-specific antibody titer, while the right y-axis reflects recombination rates and LD (r2 color) of each plotted SNP with the most significant SNP (designated by a black diamond).
B and D. Effect of top two CD46 SNPs (B) and IFI44L SNPs (D) on measles-specific antibody response (effect in the combined cohort is presented as Turkey box-and-whisker plots). On the x-axis 0 designates subjects with homozygous major allele genotype, 1 designates heterozygous subjects and 2 designates subjects with homozygous minor allele genotype. On the y-axis neutralizing antibody titer is presented as the natural log of the PRMN mIU/mL value. The top (bottom) of the box indicates the 75th (25th) percentiles, respectively, while the bold line within the box indicates the median, and the whiskers indicate 1.5 times the IQR
an=2872, reduced to 2818 after excluding subjects with immune outcome data that failed QC
CD46 region SNPs associated with variations in measles-specific antibody response after vaccination
The genetic association signal from the 1q32 region was linked to two blocks (97 kb and 8 kb) of 20 genetic variants in and around the CD46 gene (seven intronic CD46 SNPs, four SNPs in the uncharacterized LOC101929385 [currently Gene ID 100128537, C1orf132 chromosome 1 open reading frame 132], and nine intergenic SNPs, including the previously reported rs1318653 (Feenstra et al. 2014), located between CD46 and CD34), which were in high linkage disequilibrium (LD) (Supplemental Fig. 2). The most significant CD46 SNPs, rs2724384 and rs2724374 (in high LD, r2=97), lie in intron 1, and near the boundary of intron 8 (of the reference sequence ENST00000358170, RefSeq NM_002389), respectively. The minor alleles (G) of rs2724384 and rs2724374 were significantly associated (Table 2) with an allele dose-related decrease in measles-specific neutralizing antibody titers after vaccination (46 – 47% decrease in antibody titer in homozygous minor allele genotype subjects compared to homozygous major allele genotype subjects, Table 2, Fig 1B). Due to the high LD (Supplemental Fig. 2), all significant CD46 SNPs displayed similar effects. Most of the1q32 associations remained genome-wide significant (p < 5.0x10−08) in the subjects of Caucasian ancestry, where the most significant SNP was rs2724374 (p-value = 4.88x10−09, Table 3).
Table 2.
Genome-wide significant associations of SNPs with measles-specific neutralizing antibody titers after MMR vaccination (combined analysis, n=2872a)
| SNP IDb | Gene/Locationc | Major allele | Minor allele | MAFd | P-valuee | Obs.0f | Obs.1f | Obs.2f | Median (IQR) 0g | Median (IQR) 1g | Median (IQR) 2g |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1333973 | IFI44L, intron | T | A | 0.32 | 1.41E-10 | 1289 | 1230 | 299 | 1010 (440, 1919) | 772 (363, 1491) | 625 (314, 1304) |
| rs273259 | IFI44L, missense | A | G | 0.33 | 2.87E-10 | 1253 | 1250 | 315 | 1003 (438, 1879) | 777 (364, 1520) | 631 (326, 1304) |
| rs2724384 | CD46, intron | A | G | 0.23 | 2.64E-09 | 1680 | 999 | 139 | 978 (429, 1882) | 710 (363, 1468) | 516 (272, 884) |
| rs2761437 | . | G | A | 0.23 | 3.14E-09 | 1671 | 1003 | 144 | 981 (431, 1881) | 710 (359, 1475) | 517 (283, 884) |
| rs2724374 | CD46, intron | T | G | 0.23 | 3.16E-09 | 1666 | 1006 | 146 | 978 (430, 1881) | 717 (361, 1470) | 526 (277, 884) |
| rs2796265 | . | T | C | 0.23 | 3.70E-09 | 1671 | 1002 | 145 | 981 (431, 1881) | 710 (358, 1470) | 518 (286, 884) |
| rs4650590 | IFI44L, 3′UTR | A | G | 0.33 | 3.72E-09 | 1266 | 1226 | 326 | 1022 (450, 1932) | 755 (361, 1470) | 650 (329, 1327) |
| rs11118612 | LOC101929385 | T | A | 0.23 | 4.06E-09 | 1685 | 991 | 142 | 978 (432, 1882) | 703 (347, 1435) | 545 (300, 972) |
| rs2761434 | . | G | A | 0.23 | 4.97E-09 | 1671 | 1002 | 145 | 981 (431, 1881) | 710 (358, 1463) | 518 (286, 884) |
| rs4844392 | LOC101929385 | C | G | 0.23 | 5.26E-09 | 1688 | 987 | 143 | 978 (432, 1882) | 707 (345, 1443) | 545 (301, 952) |
| rs4844619 | CD46, intron | C | T | 0.23 | 5.40E-09 | 1685 | 993 | 140 | 973 (430, 1879) | 712 (354, 1462) | 539 (291, 891) |
| rs2466572 | CD46, intron | C | A | 0.23 | 6.33E-09 | 1668 | 1005 | 145 | 978 (430, 1880) | 715 (361, 1473) | 533 (286, 884) |
| rs2724360 | CD46, intron | T | C | 0.23 | 6.51E-09 | 1664 | 1009 | 145 | 980 (430, 1880) | 715 (360, 1473) | 533 (286, 884) |
| rs6657476 | CD46, intron | G | T | 0.23 | 6.51E-09 | 1687 | 991 | 140 | 972 (430, 1878) | 710 (354, 1462) | 539 (291, 891) |
| rs56075814 | . | T | C | 0.23 | 9.15E-09 | 1682 | 993 | 143 | 976 (431, 1879) | 708 (347, 1450) | 545 (301, 952) |
| rs6669384 | . | T | C | 0.22 | 1.06E-08 | 1711 | 980 | 127 | 970 (431, 1884) | 709 (349, 1420) | 545 (296, 992) |
| rs55935450 | . | T | A | 0.22 | 1.21E-08 | 1702 | 981 | 135 | 978 (430, 1885) | 703 (348, 1417) | 552 (301, 1013) |
| rs6693207 | IFI44, downstream | G | A | 0.33 | 1.23E-08 | 1273 | 1223 | 322 | 1023 (455, 1932) | 753 (354, 1479) | 650 (340, 1316) |
| rs273255 | IFI44, intron | A | T | 0.32 | 1.24E-08 | 1311 | 1222 | 285 | 1005 (439, 1873) | 771 (362, 1504) | 637 (330, 1299) |
| rs11118668 | . | C | T | 0.22 | 1.30E-08 | 1702 | 981 | 135 | 978 (430, 1885) | 703 (348, 1417) | 552 (301, 1013) |
| rs66532523 | LOC101929385 | A | C | 0.23 | 1.45E-08 | 1683 | 992 | 143 | 974 (432, 1875) | 709 (347, 1455) | 545 (301, 952) |
| rs273261 | IFI44L, intron | G | A | 0.34 | 2.03E-08 | 1223 | 1251 | 344 | 997 (438, 1857) | 774 (363, 1522) | 662 (335, 1366) |
| rs4844620 | LOC101929385 | G | A | 0.21 | 2.18E-08 | 1762 | 937 | 119 | 971 (430, 1876) | 701 (347, 1417) | 495 (280, 884) |
| rs1318653 | . | T | C | 0.22 | 2.94E-08 | 1708 | 976 | 134 | 972 (430, 1882) | 705 (347, 1417) | 565 (303, 1023) |
| rs273256 | IFI44, intron | A | C | 0.36 | 3.15E-08 | 1166 | 1277 | 375 | 1002 (445, 1898) | 774 (362, 1520) | 691 (349, 1398) |
| rs273244 | IFI44L, intron | A | T | 0.36 | 3.30E-08 | 1162 | 1280 | 376 | 1004 (445, 1898) | 772 (362, 1520) | 694 (349, 1417) |
| rs61821293 | . | T | G | 0.22 | 4.14E-08 | 1697 | 984 | 137 | 970 (430, 1879) | 710 (347, 1426) | 561 (302, 992) |
| rs273238 | . | G | A | 0.35 | 4.37E-08 | 1220 | 1249 | 349 | 996 (438, 1845) | 779 (363, 1528) | 658 (338, 1359) |
| rs4844390 | CD46, intron | A | G | 0.21 | 4.63E-08 | 1753 | 940 | 125 | 974 (427, 1883) | 702 (357, 1419) | 495 (274, 880) |
Reduced to 2818 after excluding subjects with immune outcome data that failed QC
SNP identification number
Gene/genetic region and SNP location relative to the gene
Minor allele frequency
P-values, as described in the Online Methods, Statistical Methods
Number of subjects with homozygous major allele genotype (0), heterozygous (1) and homozygous minor allele genotype (2)
Median neutralizing antibody titer (in miU/mL, with 25% and 75% inter-quartile range/IQR) for subjects with homozygous major allele genotype (0), heterozygous (1) and homozygous minor allele genotype (2)
Table 3.
Top significant associations of SNPs with measles-specific neutralizing antibody titers after MMR vaccination (Caucasians, n=2555a)
| SNP IDb | Gene/Locationc | Major allele | Minor allele | MAFd | P-valuee | Obs.0f | Obs.1f | Obs.2f | Antibody titer Median (IQR) 0g | Antibody titer Median (IQR) 1g | Antibody titer Median (IQR) 2g |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2724374 | CD46, intron | T | G | 0.23 | 4.88E-09 | 1462 | 915 | 129 | 924 (414, 1799) | 703 (352, 1418) | 516 (274, 843) |
| rs2761437 | G | A | 0.23 | 6.98E-09 | 1467 | 912 | 127 | 924 (414, 1797) | 702 (349, 1422) | 515 (280, 825) | |
| rs2796265 | T | C | 0.23 | 8.25E-09 | 1467 | 911 | 128 | 924 (414, 1797) | 702 (349, 1420) | 515 (283, 847) | |
| rs2466572 | CD46, intron | C | A | 0.23 | 9.49E-09 | 1464 | 914 | 128 | 924 (414, 1798) | 703 (350, 1418) | 517 (283, 847) |
| rs4844619 | CD46, intron | C | T | 0.23 | 1.05E-08 | 1468 | 914 | 124 | 916 (414, 1794) | 705 (348, 1420) | 526 (283, 864) |
| rs6657476 | CD46, intron | G | T | 0.23 | 1.06E-08 | 1468 | 914 | 124 | 916 (414, 1794) | 705 (348, 1420) | 526 (283, 864) |
| rs2724360 | CD46, intron | T | C | 0.23 | 1.18E-08 | 1461 | 917 | 128 | 924 (414, 1798) | 703 (349, 1418) | 517 (283, 847) |
| rs2761434 | G | A | 0.23 | 1.19E-08 | 1467 | 910 | 129 | 924 (414, 1797) | 702 (349, 1418) | 516 (286, 858) | |
| rs2724384 | CD46, intron | A | G | 0.23 | 1.27E-08 | 1466 | 914 | 126 | 924 (414, 1798) | 702 (354, 1418) | 506 (277, 854) |
| rs4844620 | LOC101929385 | G | A | 0.23 | 1.64E-08 | 1490 | 899 | 117 | 913 (416, 1772) | 701 (345, 1405) | 495 (286, 884) |
| rs4844392 | LOC101929385 | C | G | 0.23 | 1.69E-08 | 1461 | 917 | 128 | 919 (416, 1793) | 702 (344, 1418) | 539 (291, 883) |
| rs11118612 | LOC101929385 | T | A | 0.23 | 1.98E-08 | 1461 | 918 | 127 | 918 (416, 1793) | 702 (346, 1417) | 533 (290, 884) |
| rs1333973 | IFI44L, intron | T | A | 0.33 | 2.10E-08 | 1122 | 1106 | 278 | 942 (422, 1768) | 759 (361, 1448) | 616 (306, 1283) |
| rs66532523 | LOC101929385 | A | C | 0.23 | 2.27E-08 | 1461 | 917 | 128 | 918 (416, 1793) | 702 (345, 1417) | 539 (291, 883) |
| rs56075814 | T | C | 0.23 | 2.34E-08 | 1460 | 918 | 128 | 917 (416, 1794) | 703 (346, 1417) | 539 (291, 883) | |
| rs273259 | IFI44L, missense | A | G | 0.33 | 2.62E-08 | 1118 | 1104 | 284 | 942 (424, 1765) | 757 (356, 1455) | 628 (319, 1284) |
| rs273261 | IFI44L, intron | G | A | 0.34 | 3.41E-08 | 1110 | 1106 | 290 | 950 (428, 1768) | 754 (355, 1441) | 631 (323, 1287) |
| rs4844390 | CD46, intron | A | G | 0.23 | 3.48E-08 | 1478 | 907 | 121 | 919 (414, 1795) | 702 (352, 1417) | 495 (274, 858) |
| rs6669384 | T | C | 0.23 | 3.75E-08 | 1484 | 909 | 113 | 916 (414, 1797) | 703 (349, 1410) | 545 (293, 884) | |
| rs273256 | IFI44L, intron | A | C | 0.35 | 5.10E-08 | 1052 | 1134 | 320 | 958 (435, 1806) | 743 (347, 1448) | 646 (329, 1301) |
| rs273244 | IFI44L, intron | A | T | 0.35 | 6.35E-08 | 1050 | 1136 | 320 | 964 (434, 1807) | 740 (348, 1438) | 646(329, 1311) |
| rs4650590 | IFI44L, 3′UTR | A | G | 0.35 | 6.58E-08 | 1065 | 1131 | 310 | 945 (432, 1808) | 739 (352, 1440) | 646 (329, 1316) |
| rs273238 | G | A | 0.34 | 6.91E-08 | 1108 | 1105 | 293 | 950 (427, 1766) | 758 (355, 1436) | 630 (327, 1299) | |
| rs55935450 | T | A | 0.23 | 6.93E-08 | 1477 | 910 | 119 | 916 (414, 1793) | 700 (347, 1407) | 545 (296, 898) | |
| rs11118668 | C | T | 0.23 | 7.38E-08 | 1477 | 910 | 119 | 916 (414, 1793) | 700 (347, 1407) | 545 (296, 898) | |
| rs61821293 | T | G | 0.23 | 8.51E-08 | 1475 | 910 | 121 | 911 (414, 1790) | 702 (345, 1415) | 561 (299, 884) | |
| rs6693207 | IFI44L, downstream | G | A | 0.35 | 9.28E-08 | 1069 | 1131 | 306 | 956 (434, 1807) | 740 (346, 1447) | 643 (337, 1306) |
| rs273255 | IFI44L, intron | A | T | 0.32 | 1.01E-07 | 1149 | 1095 | 262 | 945 (426, 1769) | 735 (348, 1417) | 634 (328, 1287) |
| rs1318653 | T | C | 0.23 | 1.04E-07 | 1480 | 907 | 119 | 909 (414, 1776) | 702 (345, 1414) | 561 (301, 898) |
Reduced to 2506 after excluding subjects with immune outcome data that failed QC
SNP identification number
Gene/genetic region and SNP location relative to the gene
Minor allele frequency
P-values, as described in the Online Methods, Statistical Methods
Number of subjects with homozygous major allele genotype (0), heterozygous (1) and homozygous minor allele genotype (2)
Median neutralizing antibody titer (in miU/mL, with 25% and 75% inter-quartile range/IQR) for subjects with homozygous major allele genotype (0), heterozygous (1) and homozygous minor allele genotype (2)
IFI44L region SNPs associated with variations in measles-specific antibody response after vaccination
The 1p31.1 region signal was linked to a 21-kb block of 8 IFI44L SNPs and one intergenic SNP, in high LD (Supplemental Fig. 2). The two most significant IFI44L SNPs, rs1333973 and rs273259 (Table 2) were located in IFI44L intron 2 (boundary) and IFI44L exon 2, respectively. The missense SNP rs273259 (His73Arg, Ensembl transcript ENST00000370751), as well as the other significant SNPs, demonstrated an allele dose-related decrease in measles-specific neutralizing antibody titers (Table 2, Fig 1D). Three of the nine SNPs remained genome-wide significant (p < 5.0x10−08) in the subjects of Caucasian ancestry (Table 3).
To determine if there were multiple SNPs in each of the CD46 and IFI44L regions associated with the neutralizing antibody, with the effects of the SNPs adjusted for each other, we used elastic net to select SNPs. An advantage of elastic net is that it can select highly correlated SNPs, although it cannot give p- values for selected SNPs. Hence, after selection, we evaluated the selected SNPs in linear regression models. This resulted in the selection of two SNPs in the CD46 region and three SNPs in the IFI44L region, based on the Caucasian subjects. For the CD46 region, we found rs2724374 to be the most significant and the primary driving SNP (p=4.3x10−8), with rs11806810 having much less association (p=0.02) when the effects of the SNPs were adjusted for each other. For the IFI44L region, the SNPs rs12026737, rs1333973, and rs273259 were selected. However, rs273259 and rs1333973 were highly correlated (r2 = .99), making it difficult to disentangle their effects and fit both in a regression model. When choosing rs1333973 (top significant SNP) over rs273259, we found rs1333973 (p=2x10−8) and rs12026737 (p=6.0x10−4) to have strong statistical associations, with effects adjusted for each other. Furthermore, the CD46 and IFI44L regions have associations that are independent of each other (since when the aforementioned SNPs were modeled together, the results were not significantly different from what was obtained when the regions were modeled separately).
Genome-wide analysis results with measles vaccine cellular immunity
The GWAS analyses did not reveal significant SNP associations with cellular immunity after vaccination, as measured by MV-specific IFNγ ELISPOT (Supplemental Table 1, Supplemental Fig. 3). Analyses of all chromosome 1 SNPs with MV-specific secreted cytokines in 625 Caucasian subjects (for whom we had available cytokine data) demonstrate suggestive associations between CD46 SNPs (including rs2724374 and rs2724384) and the secretion of IFNα; however, an allele-dose-dependency of IFNα secretion was not noted. (Supplemental Table 2).
Analysis of differential usage of exons using mRNA-Seq data
In the CD46 analyses, we observed a highly significant per-exon estimate (q=2.96E−07) with lower exon B (in the CD46 STP region) expression (exon skipping) in subjects with rs2724374 minor G allele (Fig. 2A). These results were further confirmed by RT-PCR analysis of common CD46 isoforms in PBMCs. In this analysis, the predominant “lower band” CD46 isoform phenotype (C2, associated with exon B skipping) was clearly more pronounced in the homozygous minor G allele genotype subjects (Fig. 2B). We also observed differential exon usage based on IFI44L rs1333973/rs273259 genotypes (Fig. 2C)
Fig. 2. Differential exon/isoform CD46 and IFI44L usage.
A. Estimated exon usage in the CD46 gene comparing individuals with at least one minor allele (G for rs2724374, i.e., all the heterozygous subjects plus the one homozygous minor allele subject combined, total n=9) (blue) to individuals that are homozygous major (T for rs2724374) allele, n=19 (red). Even with a relatively small number of subjects possessing the minor allele genotype, differential exon usage with a highly significant p-value was observed for the STP exon B (genomic ID 207941124-207941168) (p=2.96E−07).
B. RT-PCR analysis of common CD46 isoforms was performed in PBMCs of rs2724374 homozygous major allele genotype individuals (n=10) compared to homozygous minor allele genotype individuals (n=10) and heterozygous individuals (n=10). The presented figure is representative of the patterns observed in all 30 subjects (10 subjects per genotype group) with the experiment replicated twice.
C. Estimated exon usage in IFI44L rs1333973/rs273259 homozygous minor allele genotype subjects (A for rs1333973, n=5 [blue]) vs. homozygous major allele subjects (T for rs1333973, n=11 [red]). The results demonstrate significant per-exon estimates for several IFI44L exons, the most significant being exon 2 (genomic ID 79093591-79094078) (q=2.37E−150).
Simulation of the structural differences between CD46 isoforms (with or without the STP exon B)
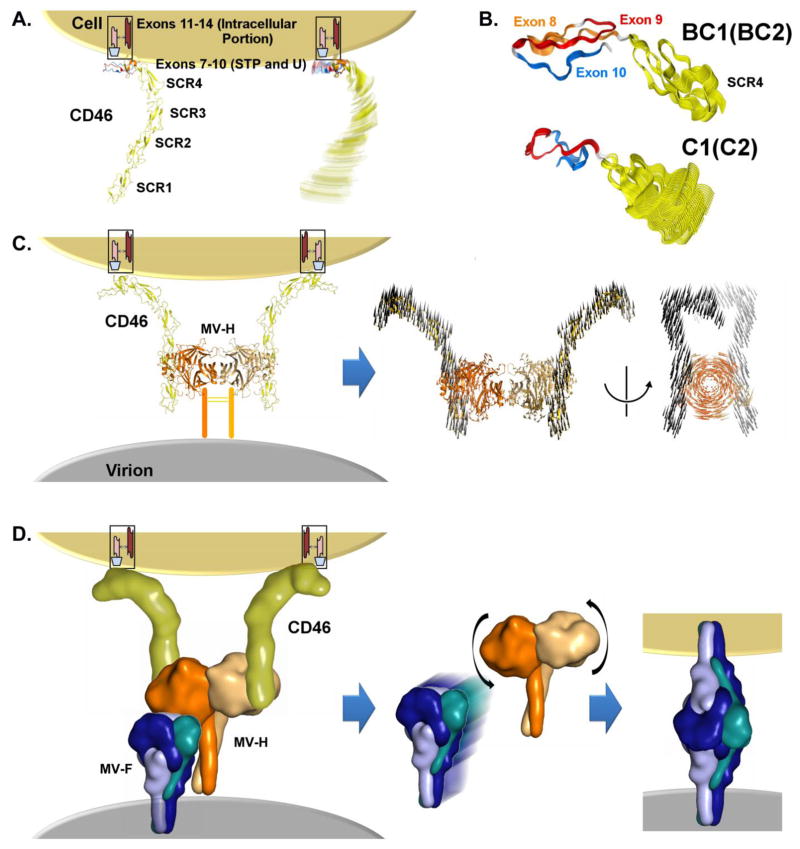
We have generated structural models of the predominant CD46 isoforms (extracellular portion): isoforms with the STP exons B and C (i.e., BC1 and BC2) and those skipping exon B (i.e., C1 and C2) (see Fig. 3, Fig. 4, Supplemental Fig. 4). The longest isoform, ABC1, generates a high-quality homology model (Supplemental Fig. 4). Interestingly, the protein sequences encoded by exons A (exon 7) and B (exon 8) have their N- and C-terminal ends close to each other. The amino acids encoded by exon A (exon 7) make a short beta strand within the STP domain. Deletion of these residues by exon skipping removes this strand from the beta-sheet, but the architecture of the domain is not significantly altered. Exon B (8) encodes amino acids making up two additional strands. In the isoforms C1 and C2, only two strands encoded by exons 9 and 10 remain. Computing the dynamics of each isoform’s atomic model, smaller STP domain isoforms exhibited greater flexibility and less collective motion (i.e., in the C1/C2 isoforms when compared to BC1/BC2 of the common isoforms). Monitoring the domain-domain angles about the SCR4-STP hinge as each structure is deformed about its low-frequency normal modes, the shorter isoform/isoforms exhibit a greater range of flexibility (Supplemental Fig. 4). As illustrated in Fig. 4, based on data from our molecular models, we propose that the differential isoform flexibility may influence the rate of formation of the CD46-MV-hemagglutinin (H) encounter complex, and/or influence the propensity for MV-H to undergo the conformational change necessary to trigger MV fusion protein.
Fig. 3. Structure of CD46.
The extracellular portion of CD46 consists of four N-glycosylated conserved short consensus repeats SCR1-4 (SCR1 and SCR2 containing binding sites for MV); a STP region that is O-glycosylated (encoded by exons 7, 8 and 9, designated as A, B and C); and a region of unknown function (U), followed by a hydrophobic transmembrane segment (H), basic amino acid anchor (A), and a cytoplasmic tail (CYT1 of 16 amino acids if exon 13 is present, or CYT2 of 23 amino acids if exon 13 is alternatively spliced).). Of the 14 known CD46 isoforms resulting from alternative splicing, four are commonly found in most human tissues and are designated based on the present STP exon/exons and the cytoplasmic tail: BC1 and BC2 (with B and C exons/domains in the STP and with either CYT1 or CYT2), and C1 and C2 (with C exon/domain in the STP and with either CYT1 or CYT2). The effect of CD46 rs2724374 on CD46 isoform prevalence (exon B presence or skipping), interaction between CD46 and MV, and immune response following measles vaccination is also summarized.
Fig. 4. Flexibility of CD46 isoforms and impact on MV-CD46 interactions and MV fusion.
A. CD46 is a cell surface receptor affected by isoform differences. Due in part to its elongated structure, the extracellular domains will naturally exhibit flexibility within the physiologic environment (represented as rotational blurring of the reference isoform ABC1/ABC2). SCR domain residues are colored in yellow. The modeled segment of CD46 includes the extracellular domains and is shown in cartoon representation.
B. Molecular modeling of the extracellular portion of BC1 (BC2) and C1 (C2) isoforms (the four most common isoforms) through SCR4, followed by generation of mechanics-based models (ANM; see Methods), indicates a difference in the intrinsic flexibility between the isoforms (see Supplemental Fig. 4 for details). The amino acids encoded by exons 8–10 are indicated by different colors. We show representative motion from our ANM models for the BC1 and C1 isoforms by showing multiple conformational states superimposed.
C. CD46 on the cell surface interacts with MV-H on the virion surface. Two CD46 molecules interacting with a MV-H dimer are depicted, as described previously. (Santiago et al. 2010) Using ANM, we quantified the intrinsic flexibility of the MV-H dimer. The dominant motion is an anti-correlated twisting or ratcheting of the monomers with respect to one another. Projection the effect of this motion onto the bound CD46 molecules is represented by black arrows on each residue. As the C-terminus of CD46 is anchored in the cell membrane, activation of this motion would require flexibility of the CD46 molecule - flexibility that may differ by isoform (panel B). Presented is a second view of the motion rotated 90°, omitting the cartoon representation for clarity – only the motion-vectors are shown to emphasize the twisting of the MV-H dimer and its effect on CD46.
D. Proposed molecular mechanism: MV-H and the fusion protein MV-F are normally associated with one another on the virus surface. Proteins are represented by smoothed molecular surfaces; see Supplemental Methods. The encounter complex between CD46 and MV-H leads to MV-F disassociation. The disassociation is influenced by the MV-H conformational change and motion in the context of the CD46-H dimer complex.(Navaratnarajah et al. 2011) The free MV-F undergoes a substantial conformational change, the molecular details of which are not fully resolved, leading to bridging between the virus and target cell. We believe that the degree of flexibility exhibited by CD46 may influence 1) the ease with which the complex may undergo conformational changes (motion in the context of MV-H-CD46 complex) leading to MV-F triggering and fusion, and 2) the rate of encounter complex formation with MV-H.
Discussion
This work is the first GWAS to reveal significant associations of two distinct chromosome 1 regions (CD46 and IFI44L) with variations in neutralizing antibody response after measles vaccination.
The important implications of our findings are supported by the genome-wide identification of genetic variants in the same two loci (in an independent state-of-the-art GWAS study) as risk variants for the development of adverse events/febrile seizures following MMR vaccination, but not for MMR-unrelated febrile seizures (suggesting the influence of these loci on measles vaccine virus entry/propagation). (Feenstra et al. 2014)
Most of the significant SNP-association signals with measles-specific antibody response were from a region in and around the CD46 gene on 1q32. The encoded glycoprotein CD46 (structure is summarized in Fig. 3) is ubiquitously expressed and serves as a regulator of complement activation (protecting the cells from complement and antibody-mediated lysis). CD46 also serves as a cellular receptor for measles virus attenuated (vaccine) strains, as well as for other pathogens (group B and D adenoviruses, human herpesvirus 6, bovine viral diarrhea virus, pathogenic Neisseria and Streptococcus pyogenes). (Cattaneo 2004) Four CD46 isoforms, resulting from alternative splicing, are commonly found in most human tissues and are designated based on the present STP exon/exons and the cytoplasmic tail: BC1 and BC2 (with B and C exons/domains in the STP and with either CYT1 or CYT2), and C1 and C2 (with C exon/domain in the STP and with either CYT1 or CYT2). (Liszewski et al. 1994; Post et al. 1991; Russell et al. 1992)(Fig. 3)
Our significant GWAS findings in the CD46 region consisted only of non-coding SNPs in high LD; the most significant were rs2724384 in CD46 intron 1, and rs2724374 in CD46 intron 8. The previously observed (Feenstra et al. 2014) genetic association of intergenic rs1318653 with MMR-related febrile seizures is in the same region and was also significant in our genome-wide association study with measles-specific neutralizing antibody titers (p-value = 2.94x10−08, Table 2), although this association exhibited a slightly weaker signal in the subset analysis of subjects of Caucasian ancestry (p-value = 1.04x10−07, Table 3). Feenstra et al. also reported the major CD46 rs2724384 allele A as a risk allele for MMR-related febrile seizures (Feenstra et al. 2014), which relates to higher measles vaccine-induced antibody titers in our GWAS and in three other candidate gene studies. (Clifford et al. 2012; Dhiman et al. 2007; Haralambieva et al. 2015; Ovsyannikova et al. 2011a) Furthermore, CD46 rs2724384 has been associated with overall CD46 gene expression and isoform abundance in lymphoblastoid cell lines (Lappalainen et al. 2013), and with variations in measles-specific IL-6, TNFα and IFNα secretion after in vitro viral stimulation of human PBMCs.(Ovsyannikova et al. 2011a) In this study, both of our top CD46 GWAS hits (rs2724384 and rs2724374) exhibited suggestive associations with IFNα (see Supplemental Table 2).
The second CD46 genetic variant with plausible functional consequences is rs2724374 (located in intron 8, near the intron-exon boundary with exon 8/STP B), which is the most significant SNP in our analysis of subjects of Caucasian ancestry and is identified by elastic net modeling as being predictive of neutralizing antibody response. CD46 rs2724374 has been reported as a genetic variant, which is highly correlated with the splicing/skipping of exon B (i.e., a sQTL/splicing quantitative trait locus) in a study assessing the splicing patterns of 250 exons and their associations with DNA polymorphisms.(Hull et al. 2007) Similarly, a study assessing sQTLs from RNA-Seq data reported CD46 rs2724374 as one of the most significant sQTLs (p=3.55x10−11) associated with alternative splicing (i.e., the skipping of CD46 exon B) (Zhao et al. 2013), a finding validated by our differential exon usage analysis of mRNA-Seq data and RT-PCR analysis of CD46 isoforms in PBMCs. Thus, the minor allele G of CD46 rs2724374 is significantly associated (in a dose-response dependent manner) with lower measles-specific neutralizing antibody titer after vaccination (approximately 45% reduction in antibody titer), and with the skipping of exon B to preferentially yielding CD46 isoforms with a shorter (and less O-glycosylated) STP region (i.e., preferentially yielding C1/C2 vs. BC1/BC2 isoforms). Although some tissues preferentially express specific CD46 isoforms (sperm, brain, salivary gland, kidney, placenta), the abundance ratio of CD46 isoforms in most human tissues (e.g., PBMCs) is considered to be constant for each individual. (Liszewski et al. 1994; Post et al. 1991; Russell et al. 1992) These CD46 genetic and phenotypic inter-individual variations and their implications for host-pathogen interactions and vaccine/pathogen-induced immunity still remain unknown.
Since innate and adaptive immune responses rely on recognition, virus entry and propagation into susceptible cells, it is tempting to speculate about the mechanisms by which CD46 isoform usage can affect MV binding/fusion and propagation at the site of injection and the associated lymphoid tissue during measles vaccination and immune response priming. It is generally accepted that all CD46 isoforms can serve as MV receptors (primarily for attenuated MV strains) and confer susceptibility to infection (Manchester et al. 1994); however, several studies report differences in MV binding and fusion in CD46 BC1/BC2 vs. C1/C2 isoforms, where longer BC isoforms support superior virus binding and shorter C isoforms support superior fusion competence. (Buchholz et al. 1996a; Buchholz et al. 1996b; Iwata et al. 1994) It is likely that the complex dynamics of interactions between MV H dimers/tetramers, F trimers and cross-linked CD46 molecules at the virus-cell surface interface (Navaratnarajah et al. 2011; Persson et al. 2010) are dependent in part on the varying flexibility conferred by different CD46 isoforms. Our molecular modeling (Fig. 4 and Supplemental Fig. 4) provides evidence for differences in the flexibilities of common CD46 isoforms, including the STP exon B (BC1/BC2 exhibiting decreased flexibility) and those excluding exon B (C1/C2 exhibiting increased flexibility). We hypothesize that the increased flexibility for the C1/C2 isoforms relative to BC1/BC2 isoforms allows increased motion between the components of the CD46-H dimer complex (needed for F triggering, as proposed in the model by Navaratnarajah et al., (Navaratnarajah et al. 2011; Persson et al. 2010)) and increased fusogenicity. It is also possible that the increased flexibility of the C1/C2 isoforms (relative to BC1/BC2) may increase the encounter rate between CD46 and H protein. Thus, our proposed mechanistic hypothesis (informed by molecular modeling) is in concert with--and adds to--the prior literature, and suggests potential molecular mechanisms linking the top GWAS CD46 hits to differences in splicing/CD46 isoform flexibility with potential effects on MV response.
The CYT1 and CYT2-containing CD46 isoforms (generated by alternative splicing of exon 13) are reported to differentially co-stimulate T helper 1 effector cells, affect T helper 1 switching to IL-10 producing T regulatory cells, and modulate cellular immunity, inflammation, B cell responses and signal transduction. (Fuchs et al. 2009; Marie et al. 2002; Wang et al. 2000) Although the differential usage of CYT1 vs. CYT2-containing isoforms cannot be inferred from our data, the involvement of such mechanisms in regulating vaccine-induced immunity cannot be excluded. Interestingly, we did not observe significant associations between polymorphisms in the other known MV receptor genes (SLAM and nectin-4) and vaccine-induced humoral immunity. This finding suggests the preferential usage of CD46 (over other receptors) by MV vaccine strains and/or insufficient expression of SLAM and Nectin-4 receptors at the site of injection and immune response priming during vaccination.
Even more interesting is the association of the 1p31.1 IFI44L genetic locus with measles-specific antibody titers, since the function of the encoded protein is largely unknown. Among the top significant SNPs in this region (grouped in a 21-kb LD block) are the intronic rs1333973 and the coding missense rs273259 (Table 2 and Table 3); the latter has been reported to be a genetic variant significantly associated with febrile seizures after MMR vaccination. (Feenstra et al. 2014) The rs273259 risk allele (A) for MMR-related febrile seizures was associated with increased measles-specific antibody titer in our study (while the minor allele G was associated with decreased antibody titer). In addition, rs273259 allele A was previously found to correlate with both the reduced expression of IFI44L exon 2 and differences in IFI44L isoform/isoforms abundance (Lappalainen et al. 2013), a finding also confirmed by our mRNA-Seq differential exon usage analysis (Fig. 2C). In their study, Feenstra et al. did not observe direct and/or differential antiviral activity of IFI44L rs273259 allelic variant proteins against MV in human fibroblasts lacking STAT1.(Feenstra et al. 2014) It is likely that IFI44L requires a specific microenvironment (e.g., partners, signaling events, cell/tissue-specific milieu, etc.) to exert its antiviral action or its function is associated with specific isoforms, as suggested by our data and other studies (i.e., SNP rs1333973 has been reported as a significant sQTL for IFI44L (Coulombe-Huntington et al. 2009; Fraser and Xie 2009; Zhao et al. 2013)).
The proteins encoded by IFI44L and its partner IFI44 are both stimulated by interferon type I and thus are likely to be involved in innate immunity. A screen of more than 380 interferon stimulated genes/ISGs for in vitro antiviral activity against different viruses identified IFI44L as an important antiviral effector of type I interferon response against hepatitis C virus. (Schoggins et al. 2011) Importantly, a study assessing susceptibility to viral myocarditis in mice (by Coxsackievirus B3) noted that genetic variants in the H28 (IFI44L) locus are likely involved in infection susceptibility.(Wiltshire et al. 2011) These data from the literature suggest that IFI44L is an important effector of innate antiviral immunity with a plausible role in immune response priming and protection against disease.
The strengths of our study include the use of a relatively large combined sample of three well-characterized cohorts (from different geographic areas) and the detailed and reliable demographic, clinical, immune phenotyping and genomic data to classify study participants into genetically defined ancestry categories. Despite the popularity of the discovery/replication study design, we analyzed available cohorts together (as this is the currently accepted approach (McCarthy et al. 2008; Skol et al. 2006), and taking advantage of the full set of genotyping data), which resulted in greater power and provided more accurate estimates of the magnitude of SNP associations and genomic localization. Although our significant findings (i.e., the involvement of CD46 and IFI44L in the host response to measles vaccine) are supported by a different state-of-the-art GWAS study (Feenstra et al. GWAS study of febrile seizures (Feenstra et al. 2014)), functional mechanistic studies are warranted to elucidate the underlying molecular mechanisms and the link between CD46 and IFI44L genetic variants and humoral immunity to MV.
In conclusion, our study identified common genetic variants associated with inter-individual variations in measles-specific antibody response following MMR vaccination. Ultimately, our study significantly advances the science of measles vaccine immunology/immunogenetics and provides knowledge of the most critical genomic features influencing measles vaccine immune response and their potential molecular mechanisms – a foundational information for the development of better measles vaccine candidates and vaccination approaches. Our study also provides more general insights and a proof of concept for the critical role of genomic variability in virus/pathogen cellular receptors and receptor isoform usage for the development of humoral immunity following vaccination.
Detailed Methods
The methods described herein are similar or identical to those published for our previous studies. [1–12]
Study Subjects
Subjects from previously described cohorts were used for this study [1–5, 9–11, 13]
The study set was a large sample of 3,191 healthy children, older adolescents, and healthy adults (age 11 to 40 years) consisting of three independent cohorts: a Rochester cohort (n=1,062); a San Diego cohort (n=1,071); and a US cohort (n=1,058). The recruitment efforts, demographic and clinical characteristics of these cohorts have been previously published [1–5, 9–11, 13].
The Rochester cohort comprised 1,062 individuals enrolled into three age-stratified cohorts of healthy, school-age children and young adults from all socioeconomic strata in Rochester, MN, recruited between 2001–2009, as described previously.[4, 5, 7, 13, 14] Parental consent was obtained for all participants and each subject had written records of having received two doses of MMR vaccine. Of the 1,062 individuals for this study, 982 (93%) were successfully genotyped and assayed for immune outcomes.
The San Diego cohort consisted of 1,071 healthy older adolescents and healthy adults (age 18 to 40 years) from armed forces personnel in San Diego, CA, enrolled by the US Naval Health Research Center (NHRC) between 2005–2006, as previously published.[6] The US cohort consisted of an additional 1,058 healthy older adolescents and healthy adults recruited from the US armed forces, enrolled between 2010–2011.[8]. Of the 1,071 and 1,058 subjects recruited into this study as the San Diego or US cohort (respectively), 882 (82%) subjects for the San Diego cohort and 1,008 (95%) subjects for the US cohort had provided consent for use of their samples/data in other studies, and were successfully genotyped and assayed for measles-specific immune outcomes for this study. The Institutional Review Boards of the Mayo Clinic (Rochester, MN) and the NHRC (San Diego, CA) approved the study, and written informed consent was obtained from each subject, from the parents of all children who participated in the study, as well as written assent from age-appropriate participants.
Genome-wide SNP typing and QC
The methodology used for this study is identical to that used in our previously published papers. [1–5, 9–11, 13]
Briefly, DNA was extracted from each subject’s blood specimen using the Gentra Puregene Blood kit (Gentra Systems Inc.; Minneapolis, MN) and quantified by Picogreen (Molecular Probes; Carlsbad, CA). The genome-wide SNP typing was performed using the Infinium Omni 1M-Quad SNP array (Illumina; San Diego, CA) for the Rochester cohort, Illumina Human Omni2.5–8 BeadChip array for the US cohort, and Illumina Infinium HumanHap550v3_A or HumanHap650Yv3 BeadChip arrays for the San Diego cohort. DNA samples underwent amplification, fragmentation, and hybridization onto each BeadChip, which were imaged on an Illumina BeadArray reader. Genotype calls based on clustering of the raw intensity data were made using BeadStudio 2 software.
For the 758 subjects in the San Diego cohort with the Illumina 550 array genotyping data, 54 subjects were removed due to high SNP missing rates (more than 5% of their SNPs missing). There were 561,303 unique measured SNPs, of which 508,199 SNPs passed QC. For the 313 subjects in the San Diego cohort with the Illumina 650 array genotyping data, 13 subjects were removed due to high SNP missing rates. There were 660,755 unique measured SNPs, of which 630,240 passed QC. In addition, subjects were further excluded from the San Diego cohort if not of Caucasian or African-American ancestry (determined by STRUCTURE), or if they were missing values for covariates used in the analysis. This resulted in a total of 882 subjects from this cohort used in the study (718 Caucasians and 164 African-Americans, see Table 1). For the 1,058 subjects in the US cohort with the Illumina Omni 2.5 array genotyping data, there were 2,376,105 measured SNPs, of which 2,116,447 passed QC. Subjects were excluded from the US cohort if not of Caucasian or African-American ancestry as determined by STRUCTURE, or had high SNP missing rates, or if values for covariates were missing; this resulted in a total of 1,008 subjects (895 Caucasians and 113 African Americans, Table 1). For the 1,062 subjects in the Rochester cohort with valid data, 10 subjects were removed due to high SNP missing rates. Subjects were further excluded from the cohort if not of Caucasian or African-American ancestry determined by STRUCTURE, or if values for covariates were missing; this resulted in a total of 982 subjects (942 Caucasians and 40 African Americans, Table 1). On the Omni 1 genotyping array, there were 1,134,514 unique measured SNPs, of which 887,889 passed QC.
Next Generation Sequencing (mRNA-Seq)
The sequencing methods are similar or identical to those previously published. [15, 16] In summary, cryopreserved PBMCs from 30 Rochester cohort subjects (selected for an mRNA-Seq transcriptome profiling, based on their neutralizing antibody titers [15 highest and 15 lowest antibody responders] after two doses of MMR vaccine) were thawed and stimulated with live Edmonston measles virus (MV) at a multiplicity of infection (MOI) of 0.5 for 24 hours (for each subject, an aliquot of the cells was left unstimulated). The samples were randomly allocated to flow cells and lanes, balancing over the immune response and stimulation status.
Cells were stabilized with RNAprotect cell reagent (Qiagen) and total RNA was extracted using RNeasy Plus mini kit (Qiagen). Quality and quantity of RNA was determined by Nanodrop spectrophotometry (Thermo Fisher Scientific). Libraries were generated using Illumina’s mRNA TruSeq (v1) kit and sequenced (paired end sequencing) on an Illumina HiSeq 2000 (Illumina; San Diego, CA) with Illumina’s TruSeq Cluster kit (v3-cBot-HS) and 51 Cycle Illumina TruSeq SBS Sequencing Kit (v3). The sequencing reads were aligned to the human genome build 37.1 using TopHat (1.3.3) and Bowtie (0.12.7). Gene counts were performed using HTSeq (0.5.3p3), while BEDTools software (2.7.1) was used to map normalized read count to individual exons. [17–20]
RT-PCR
Total RNA was extracted from PBMCs of 30 subjects (10 CD46 rs2724374 homozygous major allele genotype subjects, 10 rs2724374 homozygous minor allele genotype subjects and 10 heterozygous subjects) using RNAprotect cell reagent and the RNAeasy Plus Mini kit (Qiagen, Valencia, CA). RT-PCR analysis of CD46 isoforms was performed with the SuperScript® One-Step RT-PCR system with Platinum Taq® DNA polymerase (Invitrogen, Carlsbad, CA), as previously described [21] using the 5′ CD46 SCR4 primer GTGGTCAAATGTCGATTTCCAGTAGTCG and the 3′ untranslated region primer CAAGCCACATTGCAATATTAGCTAAGCCACA, allowing CD46 isoform/isoforms discrimination/separation on a 3% agarose gel.
Immune Outcome Phenotyping
The immune outcome assays described herein are similar or identical to those published for our previous studies. [1–8]
Neutralizing Antibody Assay
Measles-specific neutralizing antibody titers were quantified using a high-throughput, fluorescence-based plaque reduction microneutralization assay (PRMN), using a recombinant GFP-expressing measles virus, as previously described.[3–8] The plates were scanned and counted on an automated Olympus IX71 Fluorescent microscope using the Image-Pro Plus Software Version 6.3 (MediaCybernetics). The 50% end-point titer (Neutralizing Doze, ND50) was calculated automatically using Karber’s formula, and transformed into mIU/mL (using the 3rd WHO international anti-measles antibody standard), as described previously.[3–8] The variability of the PRMN assay, calculated as a coefficient of variation (CV) based on the log-transformed ND50 values of the third WHO standard, was 5.7%.[7]
IFNγ ELISPOT Assay
Measles-specific cellular immunity was quantified using commercial Human IFNγ ELISPOT kits (R&D Systems; Minneapolis, MN) to measure the number of MV-specific IFNγ-producing cells, according to the manufacturer’s instructions and as previously described [22]. We stimulated subjects’ peripheral blood mononuclear cells/PBMCs (or, alternatively, left them unstimulated) in triplicate with the Edmonston strain of MV (MOI=0.5), and developed the reaction after 42 hours incubation at 37°C, in 5 % CO2. PHA (5 μg/mL) was used as a positive control. All plates were scanned and analyzed using the same counting parameters on an ImmunoSpot® S4 Pro Analyzer (Cellular Technology Ltd.; Cleveland, OH) using ImmunoSpot® version 4.0 software (Cellular Technology Ltd.). The ELISPOT response is presented in spot-forming units (SFUs) per 2 x 105 cells. The intraclass correlation coefficients, comparing multiple observations per sample (stimulated and unstimulated condition), was 0.94 for the stimulated values and 0.85 for the unstimulated values, indicating low assay variability. [22]
Measles-specific secreted cytokines
Secreted cytokines (IL-2, IL-6; IL-10; TNFα, IFNγ, IFNα and IFNλ1) were quantified in PBMC cultures after in vitro stimulation with live MV, as previously described. [3–5]
Statistical methods
Analysis Strategy
To achieve greatest power to detect SNPs associated with measles-specific immune response phenotypes, we pooled our data across genotyping platforms and the three cohorts. To perform the pooled analyses, we first accounted for the effects of potentially confounding factors that vary across ancestry/platform/cohort strata. After thoroughly evaluating the quality of the genotype data, we used the genetic data to define major ancestry groups. We then estimated eigenvectors within each ancestry group to account for the effects of population stratification within ancestry groups. Because the largest ancestry groups were Caucasian and African-American, we restricted our pooled analysis to these two groups. After accounting for population stratification, we evaluated the covariates that were available within each of the ancestry-platform-cohort strata to determine if the covariates were associated with the phenotype, in order to regress out the effects of potential confounding factors. This produced residuals (adjusted traits) that were then used for the GWAS analyses.
Genotype Quality Control and Imputation
Genotype quality control prior to imputation was conducted separately for four strata: subjects in the San Diego cohort with the Illumina 550 array genotyping data; subjects in the San Diego cohort with the Illumina 650 array genotyping data; subjects in the US cohort with the Omni 2.5 array genotyping data; and subjects in the Rochester cohort with the Omni 1M-Quad array genotyping data. SNPs on the Y chromosome and mitochondria were removed, and SNPs were eliminated if they were monomorphic or had a missing rate of 1% (5 % for the HumanHap550/650 arrays) or more. Subjects were eliminated if they had more than 5% of their SNPs missing (for the HumanHap550/650 arrays) or more than 1% missing (for the Omni 2.5 and Omni 1M-Quad arrays). The 1000 Genomes cosmopolitan samples (African, AFR; AMR; Asian, ASN; European, EUR) were used as a reference for imputation and were based on Build 37. SNPs that could not be converted to Build 37, or mapped to more than one position, or could not have their alleles verified for the forward strand, were eliminated. The reference genome was then filtered to include only those SNPs with a minor allele frequency (MAF) greater than 0.005. The data were then phased using SHAPEIT [23] and imputed via IMPUTE2. [24] SNPs with an imputation dosage allele r2 of at least 0.3, and a MAF of at least 0.01, were used in the pooled analyses.
Genetic Ancestry and Population Stratification
Genetic data was used to assign ancestry groups (African, Caucasian, or Asian) for individuals using the STRUCTURE software [25], and using the 1000 Genomes data as a reference. These estimates were done within cohort and platform (San Diego/550, San Diego/650, US/Omni 2.5, Rochester/Omni 1). Others [26] have shown that it is necessary to perform pruning of the SNPs to be used for eigenvectors in order to avoid having sample eigenvectors that are determined by small clusters of SNPs at specific locations, such as the lactose intolerance gene, or polymorphic inversion regions. Therefore, the SNPs used for STRUCTURE and for eigenvectors were selected by LD pruning from an initial pool consisting of all autosomal SNPs with the following filters: SNPs with a minor allele frequency (MAF) < 5% were excluded; influential SNPs were removed (according to the following chromosome regions: chromosome 8 [bp 1-12700000]; chromosome 2 [bp 129900001-136800000, 5700000-33500000]; chromosome 4 [bp 0900001-44900000]); correlation (r2) pruning was used to subset to uncorrelated SNPs. SNPs passing these selection criteria were input to STRUCTURE [25] to make ancestry “triangle” plots that depict the admixture proportions of ancestry groups for each subject. Subjects were classified into major ancestry groups based on the largest estimated STRUCTURE ancestry proportion.
Eigenvectors were estimated within ancestry groups and platform strata for refined control of population stratification. For this step, SNPs with a MAF < 0.01 were excluded, SNPs with a HWE p-value < 0.001 were excluded, INDELS were removed, and pruning according to variance inflation factors was used. These data were then used with smartPCA to produce a set of eigenvectors using the normalization formulas of Price et al. [27] following the procedures of EIGENSTRAT. Tracy-Widom statistics were computed to include eigenvectors as potential for adjusting covariates if they had a p-value < 0.05.
Selection of Covariates to Adjust for Potential Confounders
To remove the effects of potential confounders so that the cohorts could be combined, we first screened for potential confounders relevant to each ancestry group and cohort. The immune response trait measles-specific IFNγ ELISPOT, as well as the measles-specific secreted cytokines, were transformed by normal quantiles of the difference of the mean stimulated and mean unstimulated values. Neutralizing antibody titer was transformed as the natural log of the PRMN mIU/mL value. Possible confounders were screened for their association with the trait using the following steps. Any categorical variable with a very large number of categories was binned using hierarchical clustering. This was achieved by using hierarchical clustering on the estimated regression coefficients for the different categories, binning categories with similar regression coefficients. All categorical variables were coded as dummy variables such that the most common category was used as baseline. Variables that were marginally associated with the trait with p-value < 0.1 were then included in backwards selection with a p-value threshold of 0.1. This somewhat liberal threshold achieves the goal of controlling for potential confounding covariates. Residuals from the final “covariate models” were then used as the primary adjusted traits for GWAS analyses.
GWAS Pooled Analysis
The adjusted traits for IFNγ ELISPOT and neutralizing antibody were pooled over the three cohorts and genotype platforms to perform the pooled GWAS analysis. To pool the genotypes, we used the genotypes that were imputed separately within each platform. To analyze Caucasian and African-American ancestry together, we used a linear regression model that included a cohort indicator, ancestry indicator, dose of minor allele, and interaction of ancestry with dose of minor allele. This full model was compared with a reduced model that included a cohort indicator and ancestry indicator. The full and reduced models were used to create a likelihood ratio test for the effect of a SNP, allowing for the possible interaction of the SNP with ancestry, resulting in a test with 2 degrees of freedom (df). To test for the effect of a SNP within an ancestry group, we only used cohort indicator as an adjusting factor, resulting in a test with 1 df for the SNP effect. The measles-specific secreted cytokines were only measured in the Rochester cohort, so the resulting test for SNP effects was based on a 1 df statistic. All reported p-values are two-sided. To control for multiple testing, we used the standard p-value < 5.0x10−08 [28, 29] to determine genome-wide statistical significance. Statistical analyses were performed with the R 3.2.0 statistical software and PLINK. [30]
Computer code availability
Computer code (linux shell scripts, R code) is available upon request from corresponding author.
Analysis of mRNA-Seq Data for Differential Exon Usage
We tested for evidence of differential exon usage in the CD46 and IFI44L genes using the method of Anders et al., implemented in the DEXSeq package (version 1.16.10) in the R programing language (version 3.2.3). [31, 32] This method utilizes generalized linear models (GLM) fitting per-gene negative binomial models.[33] For the full model, we included main effects for the sample, exon, and an interaction between the exon and allele state/genotype (subjects’ homozygous minor allele compared to heterozygous and homozygous major allele). The likelihood estimate from the full model is compared to the likelihood from the model with the main effects to test the hypothesis for differential exon usage between the different genotypes/allele states. A significant test for a given exon provides evidence of differential exon usage (i.e., an alternative splicing event to produce different transcripts/isoforms). The per-exon estimates of the dispersion for the negative binomial GLMs were calculated using the Cox-Reid method [31, 34, 35] applied to all of the observed exons from our experiment based on the full model. To limit the risk of false discovery, our analyses of differential exon usage were limited to the CD46 (n=14 exons) and IFI44L (n =9 exons) genes. The method of Benjamini and Hochberg was used to control the false discovery rate and the adjusted p-values are reported as q-values.[36] We used mRNA-Seq paired-end sequencing data on 28 subjects (selected based on the extremes of the distribution of the antibody response, 14 high and 14 low antibody responders) to test the hypothesis of differential exon usage (exon expression) between different genotypes of interest (for CD46 rs2724374 and for IFI44L rs1333973/rs273259).
Molecular Modeling
No full-length structure of CD46 exists, but partial structures of the extracellular domains have been solved. These cover the sequences encoded by the first six exons of the gene. Available experimental structures include the co-crystal structure between CD46 SCR1-4 and the adenovirus type 11 knob (3o8e [37]), and a co-crystal structure of CD46 SCR1-2 bound to the MV-H globular head domain dimer (3inb [38]). Computationally determined models have also been deposited in MODBASE [39]. We generated homology models of the extracellular domains (SCR1-4 and STP domains) of ABC1, BC1, and C1 isoforms of CD46 (identical to ABC2, BC2, and C2, respectively) using I-TASSER [40] and compared them to the MODBASE models and available crystal structures, in order to evaluate model quality and consistency.
In order to evaluate the effect of differential splicing on the dynamics of the CD46 structure, we generated Anisotropic Network Models (ANMs)[41, 42] using a distance-dependent interaction strength [43]. Models were evaluated using computed Mean Squared Fluctuations as a per-residue measure of mobility, commute time [44] as a measure of efficiency with which information passes through the structure, and angle monitors to evaluate overall domain-domain orientations. The motions computed by ANM are theoretically local (small-scale), but often correlate strongly with large-scale functional motions. Thus, the functional magnitudes of ANM motions are not intrinsically defined. Therefore, we deformed each structure to 2Å RMSD in each direction of a given mode as a consistent and realistic extent of motion.
The crystal structure of the MV-H head domain dimer and its interactions with CD46 were taken from PDB 3INB [38] and we modeled its dynamics using ANM. Motions of bound CD46 molecules induced by these intrinsic motions of MV-H were inferred by rigidly tethering SCR1 and SCR2 to their interacting residues in the crystal structure.
Diagram mModels of MV-F were produced by Gaussian smoothing of the molecular surface of 1G5G [45] – the fusion protein of Newcastle disease virus/NDV which, like MV, is a member of the Paramyxoviridae family.
Supplementary Material
Top 20 genome-wide associations of SNPs with measles-specific IFNγ ELISPOT response after MMR vaccination (Combined analysis, n=2872a)
Associations between chromosome 1 SNPs and measles virus-specific secreted IFNα (Caucasians, n=625 subjects with available measles virus-specific cytokine data)
Manhattan and Q-Q plots summary of GWAS associations between SNPs and measles-specific neutralizing antibody titers.
A. Manhattan (left panel) and Q-Q (right panel) plots for the combined cohort analysis (n=2872, reduced to 2818 after excluding subjects with neutralizing antibody data, that failed QC).
B. Manhattan (left panel) and Q-Q (right panel) plots for the subset analysis of subjects of Caucasian ancestry (n=2555, reduced to 2506 after excluding subjects with neutralizing antibody data, that failed QC).
Haplotype block structure of the significant IFI44L and CD46 SNPs associated with measles-specific antibody response (combined sample).
The schematic representation and LD block structure of IFI44L (left) and CD46 (right) genetic regions are depicted (significantly associated SNPs only). The LD block structure was analyzed using Haploview software, version 4.2. The r2 color scheme is: white (r2=0), shades of grey (0< r2<1), black (r2=1). The numbers report the r2 value multiplied by 100.
Manhattan plot summary of GWAS associations between SNPs and measles-specific IFNγ ELISPOT response in the combined cohort (n=2872a)
aReduced to 2618 after excluding subjects with immune outcome data that failed QC
CD46 isoforms exhibit different flexibilities, specifically about the hinge between the SCR4 and STP domains.
A. Molecular structure of the full length CD46, zoomed in to emphasize the differentially spliced exons.
B. Using the first 3 modes of an ANM model (see Methods), we compute the mobility of each residue. There is increased mobility for the C1 isoform. (inset) The normal mode frequencies are plotted on a log-log scale and indicate a dramatically lower collectivity for the C1 isoform.
C. Commute times are computed for each structure and show a decrease in C1 relative to BC1. (inset) Example matrix of commute times from the BC1 isoform with the N-terminus at the top left and C-terminus bottom right.
D. and E. We choose representative Cα atoms to define the hinge angle between the exon 6 subdomain and the subdomain comprised of the isoform-specific sequences. D. for BC1, relatively low mobility about this hinge region is observed (ANM mode 2), while greater flexibility is observed in C1 (ANM mode 2) in E. We show representative structures from the ANM modes, deformed to 2Å RMSD in both directions and superimposed about the sequences encoded by the variable exons.
F. Across the first 5 low-frequency ANM modes, we indicate the change in this angle observed when deforming each structure to 2Å RMSD in each direction.
Acknowledgments
We thank the Mayo Clinic Vaccine Research Group staff and the study participants. We wish to recognize Julie M. Cunningham and the Mayo Advanced Genomic Technology Center for the genotyping and next generation sequencing efforts, and Nathaniel D. Warner (Division of Biomedical Statistics and Informatics, Mayo Clinic Department of Health Science Research) for his programming assistance and contribution to statistical analysis. We thank Caroline L. Vitse for her editorial assistance with this manuscript. Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under Award Number R01AI033144 and R37AI048793. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Presented in part: Ovsyannikova IG, Haralambieva IH, Kennedy RB, Larrabee BR, Schaid DJ, Poland GA. A Genome-Wide Association Study Identifies Major Loci Associated with Measles Vaccine-Specific Immune Responses. The 19th Annual Conference on Vaccine Research, Baltimore, MD, April 18–20, 2016
Competing Interests: Dr. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. Dr. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Dynavax, Novartis Vaccines and Therapeutics, Emergent Biosolutions, Adjuvance, Microdermis, Seqirus, NewLink, Protein Sciences, GSK Vaccines, and Sanofi Pasteur. Drs. Poland and Ovsyannikova hold three patents related to measles and vaccinia peptide research. Dr. Kennedy has received funding from Merck Research Laboratories to study waning immunity to measles and mumps after immunization with MMR-II®. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
References
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome research. 2012;22:2008–17. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophysical journal. 2001;80:505–15. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarczyk RA, Orenstein WA, Omer SB. Estimating the Number of Measles-Susceptible Children and Adolescents in the United States Using Data From the National Immunization Survey-Teen (NIS-Teen) American Journal of Epidemiology. 2016;184:148–56. doi: 10.1093/aje/kwv320. [DOI] [PubMed] [Google Scholar]
- Buchholz CJ, Gerlier D, Hu A, Cathomen T, Liszewski MK, Atkinson JP, Cattaneo R. Selective expression of a subset of measles virus receptor-competent CD46 isoforms in human brain. Virology. 1996a;217:349–355. doi: 10.1006/viro.1996.0122. [DOI] [PubMed] [Google Scholar]
- Buchholz CJ, Schneider U, Devaux P, Gerlier D, Cattaneo R. Cell entry by measles virus: Long hybrid receptors uncouple binding from membrane fusion. Virology. 1996b;70:3716–3723. doi: 10.1128/jvi.70.6.3716-3723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol. 2004;78:4385–4388. doi: 10.1128/JVI.78.9.4385-4388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennubhotla C, Bahar I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS computational biology. 2007;3:1716–26. doi: 10.1371/journal.pcbi.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford HD, Hayden CM, Khoo SK, Zhang G, Le Souef PN, Richmond P. CD46 measles virus receptor polymorphisms influence receptor protein expression and primary measles vaccine responses in naive Australian children. Clin Vaccine Immunol. 2012;19:704–710. doi: 10.1128/CVI.05652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Lam KC, Dias C, Majewski J. Fine-scale variation and genetic determinants of alternative splicing across individuals. PLoS genetics. 2009;5:e1000766. doi: 10.1371/journal.pgen.1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman N, Cunningham JM, Jacobson RM, Vierkant RA, Wu Y, Ovsyannikova IG, Pankratz VS, Poland GA. Variations in measles vaccine-specific humoral immunity by polymorphisms in SLAM and CD46 measles virus receptors. Journal of Allergy and Clinical Immunology. 2007;120:666–672. doi: 10.1016/j.jaci.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Feenstra B, Pasternak B, Geller F, Carstensen L, Wang T, Huang F, Eitson JL, Hollegaard MV, Svanstrom H, Vestergaard M, Hougaard DM, Schoggins JW, Jan LY, Melbye M, Hviid A. Common variants associated with general and MMR vaccine-related febrile seizures. Nature Genetics. 2014;46:1274–82. doi: 10.1038/ng.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Xie X. Common polymorphic transcript variation in human disease. Genome research. 2009;19:567–75. doi: 10.1101/gr.083477.108. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Atkinson JP, Fremeaux-Bacchi V, Kemper C. CD46-induced human Treg enhance B-cell responses. European Journal of Immunology. 2009;39:3097–109. doi: 10.1002/eji.200939392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Kennedy RB, Ovsyannikova IG, Whitaker JA, Poland GA. Variability in Humoral Immunity to Measles Vaccine: New Developments. Trends in molecular medicine. 2015;21:789–801. doi: 10.1016/j.molmed.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz SV, Jacobson RM, Poland GA. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011a;29:7883–7895. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Ovsyannikova IG, O’Byrne M, Pankratz VS, Jacobson RM, Poland GA. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine. 2011b;29:4485–4491. doi: 10.1016/j.vaccine.2011.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Ovsyannikova IG, Pankratz VS, Kennedy RB, Jacobson RM, Poland GA. The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert Review of Vaccines. 2013;12:57–70. doi: 10.1586/erv.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Ovsyannikova IG, Umlauf BJ, Vierkant RA, Pankratz SV, Jacobson RM, Poland GA. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011c;29:8988–8997. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralambieva IH, Zimmermann MT, Ovsyannikova IG, Grill DE, Oberg AL, Kennedy RB, Poland GA. Whole Transcriptome Profiling Identifies CD93 and Other Plasma Cell Survival Factor Genes Associated with Measles-Specific Antibody Response after Vaccination. PLos ONE. 2016;11:e0160970. doi: 10.1371/journal.pone.0160970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, Kwiatkowski D. Identification of common genetic variation that modulates alternative splicing. PLoS genetics. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata K, Seya T, Ueda S, Ariga H, Nagasawa S. Modulation of complement regulatory function and measles virus receptor function by the serine-threonine-rich domains of membrane cofactor protein (CD46) The Biochemical journal. 1994;304(Pt 1):169–75. doi: 10.1042/bj3040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Ovsyannikova IG, Haralambieva IH, O’Byrne MM, Jacobson RM, Pankratz VS, Poland GA. Multigenic control of measles vaccine immunity mediated by polymorphisms in measles receptor, innate pathway, and cytokine genes. Vaccine. 2012a;30:2159–2167. doi: 10.1016/j.vaccine.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Ovsyannikova IG, Pankratz VS, Haralambieva IH, Vierkant RA, Jacobson RM, Poland GA. Genome-wide genetic associations with IFNgamma response to smallpox vaccine. Human Genetics. 2012b;131:1433–1451. doi: 10.1007/s00439-012-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RB, Ovsyannikova IG, Shane PV, Haralambieva IH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Human Genetics. 2012c;131:1403–1421. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert ND, Haralambieva IH, Kennedy RB, Ovsyannikova IG, Pankrantz VS, Poland GA. Polymorphisms in HLA-DPB1 are associated with differences in rubella-specific humoral immunity after vaccination. Journal of Infectious Diseases. 2015;211:898–905. doi: 10.1093/infdis/jiu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlof J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HP, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Hasler R, Syvanen AC, van Ommen GJ, Brazma A, Meitinger T, Rosenstiel P, Guigo R, Gut IG, Estivill X, Dermitzakis ET. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Tedja I, Atkinson JP. Membrane cofactor protein (CD46) of complement. Processing differences related to alternatively spliced cytoplasmic domains. The Journal of Biological Chemistry. 1994;269:10776–9. [PubMed] [Google Scholar]
- Manchester M, Liszewski MK, Atkinson JP, Oldstone MBA. Multiple isoforms of CD46 (membrane cofactor protein) serve as receptors for measles virus. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:2161–2165. doi: 10.1073/pnas.91.6.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie JC, Astier AL, Rivailler P, Rabourdin-Combe C, Wild TF, Horvat B. Linking innate and acquired immunity: divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nature immunology. 2002;3:659–66. doi: 10.1038/ni810. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--a replication study and examination of novel polymorphisms. Human Heredity. 2011a;72:206–223. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Haralambieva IH, Vierkant RA, Pankratz VS, Poland GA. The role of polymorphisms in toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Human Genetics. 2011b;130:547–561. doi: 10.1007/s00439-011-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30:2146–2152. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BD, Schmitz NB, Santiago C, Zocher G, Larvie M, Scheu U, Casasnovas JM, Stehle T. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathogens. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Jacobson RM. The re-emergence of measles in developed countries: time to develop the next-generation measles vaccines? Vaccine. 2012;30:103–104. doi: 10.1016/j.vaccine.2011.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post TW, Liszewski MK, Adams EM, Tedja I, Miller EA, Atkinson JP. Membrane cofactor protein of the complement system: alternative splicing of serine/threonine/proline-rich exons and cytoplasmic tails produces multiple isoforms that correlate with protein phenotype. J Exp Med. 1991;174:93–102. doi: 10.1084/jem.174.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention CfDCa. [Accessed February 12, 2015 2015];Measles Cases and Outbreaks. 2015 http://www.cdc.gov/measles/cases-outbreaks.html. http://www.cdc.gov/measles/cases-outbreaks.html.
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SM, Sparrow RL, McKenzie IFC, Purcell DFJ. Tissue-specific and allelic expression of the complement regulator CD46 is controlled by alternative splicing. European Journal of Immunology. 1992;22:1513–1518. doi: 10.1002/eji.1830220625. [DOI] [PubMed] [Google Scholar]
- Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nature structural & molecular biology. 2010;17:124–9. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature Genetics. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Tan PL, Jacobson RM, Poland GA, Jacobsen SJ, Pankratz SV. Twin studies of immunogenicity - determining the genetic contribution to vaccine failure. Vaccine. 2001;19:2434–2439. doi: 10.1016/s0264-410x(00)00468-0. [DOI] [PubMed] [Google Scholar]
- Team RDC. R: a language for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
- Wang G, Liszewski MK, Chan AC, Atkinson JP. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J Immunol. 2000;164:1839–1846. doi: 10.4049/jimmunol.164.4.1839. [DOI] [PubMed] [Google Scholar]
- Whitaker JA, Poland GA. Measles and mumps outbreaks in the United States: Think globally, vaccinate locally. Vaccine. 2014;32:4703–4. doi: 10.1016/j.vaccine.2014.06.088. [DOI] [PubMed] [Google Scholar]
- Wiltshire SA, Leiva-Torres GA, Vidal SM. Quantitative trait locus analysis, pathway analysis, and consomic mapping show genetic variants of Tnni3k, Fpgt, or H28 control susceptibility to viral myocarditis. Journal of Immunology. 2011;186:6398–405. doi: 10.4049/jimmunol.1100159. [DOI] [PubMed] [Google Scholar]
- Yang L, Song G, Jernigan RL. Protein elastic network models and the ranges of cooperativity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12347–52. doi: 10.1073/pnas.0902159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Lu ZX, Park JW, Zhou Q, Xing Y. GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome biology. 2013;14:R74. doi: 10.1186/gb-2013-14-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Detailed Methods
- 1.Kennedy RB, Ovsyannikova IG, Haralambieva IH, et al. Multigenic control of measles vaccine immunity mediated by polymorphisms in measles receptor, innate pathway, and cytokine genes. Vaccine. 2012;30:2159–2167. doi: 10.1016/j.vaccine.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ovsyannikova IG, Haralambieva IH, Vierkant RA, Pankratz VS, Poland GA. The role of polymorphisms in toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Human Genetics. 2011;130:547–561. doi: 10.1007/s00439-011-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--a replication study and examination of novel polymorphisms. Human Heredity. 2011;72:206–223. doi: 10.1159/000331585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haralambieva IH, Ovsyannikova IG, Umlauf BJ, et al. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29:8988–8997. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haralambieva IH, Ovsyannikova IG, Kennedy RB, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011;29:7883–7895. doi: 10.1016/j.vaccine.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ovsyannikova IG, Kennedy RB, O’Byrne M, Jacobson RM, Pankratz VS, Poland GA. Genome-wide association study of antibody response to smallpox vaccine. Vaccine. 2012;30:4182–4189. doi: 10.1016/j.vaccine.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haralambieva IH, Ovsyannikova IG, O’Byrne M, Pankratz VS, Jacobson RM, Poland GA. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine. 2011;29:4485–4491. doi: 10.1016/j.vaccine.2011.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ovsyannikova IG, Pankrantz VS, Salk HM, Kennedy RB, Poland GA. HLA alleles associated with the adaptive immune response to smallpox vaccine: a replication study. Human Genetics. 2014;133:1083–92. doi: 10.1007/s00439-014-1449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy RB, Ovsyannikova IG, Pankratz VS, et al. Genome-wide genetic associations with IFNgamma response to smallpox vaccine. Human Genetics. 2012;131:1433–1451. doi: 10.1007/s00439-012-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy RB, Ovsyannikova IG, Shane PV, Haralambieva IH, Vierkant RA, Poland GA. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Human Genetics. 2012;131:1403–1421. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert ND, Haralambieva IH, Kennedy RB, Ovsyannikova IG, Pankrantz VS, Poland GA. Polymorphisms in HLA-DPB1 are associated with differences in rubella-specific humoral immunity after vaccination. Journal of Infectious Diseases. 2015;211:898–905. doi: 10.1093/infdis/jiu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haralambieva IH, Zimmermann MT, Ovsyannikova IG, et al. Whole Transcriptome Profiling Identifies CD93 and Other Plasma Cell Survival Factor Genes Associated with Measles-Specific Antibody Response after Vaccination. PLos ONE. 2016;11:e0160970. doi: 10.1371/journal.pone.0160970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30:2146–2152. doi: 10.1016/j.vaccine.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Poland GA. Associations between polymorphisms in the antiviral TRIM genes and measles vaccine immunity. Human Immunology. 2013;74:768–74. doi: 10.1016/j.humimm.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haralambieva IH, Oberg AL, Ovsyannikova IG, et al. Genome-wide characterization of transcriptional patterns in high and low antibody responders to rubella vaccination. PLos ONE. 2013;8:e62149. doi: 10.1371/journal.pone.0062149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy RB, Oberg AL, Ovsyannikova IG, Haralambieva IH, Grill DE, Poland GA. Transcriptomic profiles of high and low antibody responders to smallpox vaccine. Genes and Immunity. 2013;14:277–285. doi: 10.1038/gene.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Liszewski MK, Chan AC, Atkinson JP. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J Immunol. 2000;164:1839–1846. doi: 10.4049/jimmunol.164.4.1839. [DOI] [PubMed] [Google Scholar]
- 22.Ovsyannikova IG, Haralambieva IH, Vierkant RA, O’Byrne MM, Jacobson RM, Poland GA. Effects of vitamin A and D receptor gene polymorphisms/haplotypes on immune responses to measles vaccine. Pharmacogenetics and Genomics. 2012;22:20–31. doi: 10.1097/FPC.0b013e32834df186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England Journal of Medicine. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 29.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic Epidemiology. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. AmJ HumGenet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome research. 2012;22:2008–17. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Team RDC. R: a language for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 33.McCullagh Nelder. Generalized Linear Models. Chapman and Hall Lt; Boca Raton, FL: 1989. [Google Scholar]
- 34.Cox DR, Reid N. Parameter orthogonality and approximate conditional inference. J R Stat Soc Ser B Methodol. 1987;49:1–39. [Google Scholar]
- 35.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research. 2012;40:4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 37.Persson BD, Schmitz NB, Santiago C, et al. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathogens. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–9. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 39.Pieper U, Eswar N, Braberg H, et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Research. 2004;32:D217–22. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmermann MT, Kloczkowski A, Jernigan RL. MAVENs: motion analysis and visualization of elastic networks and structural ensembles. BMC Bioinformatics. 2011;12:264. doi: 10.1186/1471-2105-12-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–15. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Song G, Jernigan RL. Protein elastic network models and the ranges of cooperativity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12347–52. doi: 10.1073/pnas.0902159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chennubhotla C, Bahar I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS Comput Biol. 2007;3:1716–26. doi: 10.1371/journal.pcbi.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Gorman JJ, McKimm-Breschkin J, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–66. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top 20 genome-wide associations of SNPs with measles-specific IFNγ ELISPOT response after MMR vaccination (Combined analysis, n=2872a)
Associations between chromosome 1 SNPs and measles virus-specific secreted IFNα (Caucasians, n=625 subjects with available measles virus-specific cytokine data)
Manhattan and Q-Q plots summary of GWAS associations between SNPs and measles-specific neutralizing antibody titers.
A. Manhattan (left panel) and Q-Q (right panel) plots for the combined cohort analysis (n=2872, reduced to 2818 after excluding subjects with neutralizing antibody data, that failed QC).
B. Manhattan (left panel) and Q-Q (right panel) plots for the subset analysis of subjects of Caucasian ancestry (n=2555, reduced to 2506 after excluding subjects with neutralizing antibody data, that failed QC).
Haplotype block structure of the significant IFI44L and CD46 SNPs associated with measles-specific antibody response (combined sample).
The schematic representation and LD block structure of IFI44L (left) and CD46 (right) genetic regions are depicted (significantly associated SNPs only). The LD block structure was analyzed using Haploview software, version 4.2. The r2 color scheme is: white (r2=0), shades of grey (0< r2<1), black (r2=1). The numbers report the r2 value multiplied by 100.
Manhattan plot summary of GWAS associations between SNPs and measles-specific IFNγ ELISPOT response in the combined cohort (n=2872a)
aReduced to 2618 after excluding subjects with immune outcome data that failed QC
CD46 isoforms exhibit different flexibilities, specifically about the hinge between the SCR4 and STP domains.
A. Molecular structure of the full length CD46, zoomed in to emphasize the differentially spliced exons.
B. Using the first 3 modes of an ANM model (see Methods), we compute the mobility of each residue. There is increased mobility for the C1 isoform. (inset) The normal mode frequencies are plotted on a log-log scale and indicate a dramatically lower collectivity for the C1 isoform.
C. Commute times are computed for each structure and show a decrease in C1 relative to BC1. (inset) Example matrix of commute times from the BC1 isoform with the N-terminus at the top left and C-terminus bottom right.
D. and E. We choose representative Cα atoms to define the hinge angle between the exon 6 subdomain and the subdomain comprised of the isoform-specific sequences. D. for BC1, relatively low mobility about this hinge region is observed (ANM mode 2), while greater flexibility is observed in C1 (ANM mode 2) in E. We show representative structures from the ANM modes, deformed to 2Å RMSD in both directions and superimposed about the sequences encoded by the variable exons.
F. Across the first 5 low-frequency ANM modes, we indicate the change in this angle observed when deforming each structure to 2Å RMSD in each direction.