Abstract
A nucleos(t)ide analog (NA) is the common antiviral drug available for directly treating hepatitis B virus (HBV) infection. However, its application has led to the emergence of NA-resistant mutations mostly in a conserved region of the reverse transcriptase domain of HBV polymerase. Harboring NA-resistant mutations decreases drug effectiveness and increases the frequency of end-stage liver disease. The invention of next-generation sequencing that can generate thousands of sequences from viral complex mixtures provides opportunities to detect minor changes and early viral evolution under drug stress. The present study used ultra-deep sequencing to evaluate discrepant quasispecies in the reverse transcriptase domain of HBV including NA-resistant hotspots between seven treatment-naïve Indonesian patients infected with HBV and five at the early phase of treatment. The most common sub-genotype was HBV B3 (83.34%). The substitution rate of variants determined among amino acids with a ratio of ≥ 1% changes was higher among the population in conserved regions (23.19% vs. 4.59%, P = 0.001) and in the inter-reverse transcriptase domain (23.95% vs. 2.94%, P = 0.002) in treatment naïve, than in treated patients. Nine hotspots of antiviral resistance were identified in both groups, and the mean frequency of changes in all patients was < 1%. The known rtM204I mutation was the most frequent in both groups. The lower rate of variants in HBV quasispecies in patients undergoing treatment could be associated with virus elimination and the extinction of sensitive species by NA therapy. The present findings imply that HBV quasispecies dynamically change during treatment.
Keywords: Hepatitis B virus, Indonesia, Reverse transcriptase domain, Drug resistance, Ultra-deep sequencing
INTRODUCTION
Nucleos(t)ide analogues (NAs) must be administered indefinitely to decrease the incidence of hepatic decompensation, cirrhosis and liver cancer through the sustained suppression of hepatitis B virus (HBV) (1). This situation can lead to issues such as decreased medication compliance and the emergence of antiviral drug resistance. To date, the newer NAs tenofovir (TDF) and entecavir (ETV) are recommended as first-line therapy against naïve chronic HBV infection. Although these drugs with a high barrier against resistance have minimized the incidence and development of new mutations related to NA resistance (2–4), implementation of this clinical guideline in developing countries including Indonesia remains limited due to drug availability and the high cost of regimens (5). Appropriate treatment guidelines are particularly required in Indonesia, where the prevalence of HBV is intermediate to high and 12 million inhabitants live with chronic HBV infection (6, 7).
The NA site of action is HBV polymerase, include reverse transcriptase (RT). The RT domain comprising domains and inter-domains is used by HBV to prime protein for viral DNA synthesis. Activated NAs inhibit RT priming and/or elongation of the HBV DNA chain (8), but hotspots of NA-resistance mainly reside within RT domains. Mutations change the conformational structure of RT, which disrupts NA binding and decreases its effectiveness (9). A pool of HBV quasispecies, which consists of a highly heterogeneous viral population resulting from high replication rates and the lack of proof-reading in infected patients determine the success of HBV treatment (10). The limit of conventional sequencers to detect mutations in quasispecies is ≥ 20% of the total quasispecies population. The invention of next generation sequencers has heralded the dawn of a new era in virology because they can analyze millions of sequences that allows the detection of < 1% of minor changes and thus the early evolution of viral populations in the presence of drug stress (11, 12). Understanding the real prevalence of NA resistance in treatment-naïve patients infected with HBV and those at the early phase of treatment is important to determine rates of naturally occurring mutations in population and how they relate to treatment recommendations (10). The present study uses ultra-deep sequencing to evaluate different HBV quasispecies in the RT region including NA-resistant hotspots between Indonesian patients infected with HBV who are treatment-naïve (naïve) and those at the early phase of treatment with NA (treated).
MATERIAL AND METHODS
Subjects
Twelve patients were recruited from the outpatient clinic of the Subdivision of Gastroenterohepatology, Department of Internal Medicine, Dr. Sardjito Hospital, Yogyakarta, Indonesia. Information about clinical diagnoses and treatment status were collected from medical records and questionnaires that were completed by the attending gastroenterohepatologist. The patients provided written informed consent to participate after the study aims and protocol were explained to them. The study then proceeded according to the ethical standards established by the Ethics Committees at Kobe University, Japan and Gadjah Mada University, Indonesia (code KE/FK/194/EC).
Serological and biochemical tests
Automated chemiluminescent enzyme immunoassays on an Architect analyzer (Abbott Laboratories, Abbot Park, IL, USA) were used to screen hepatitis B surface antigen (HBsAg), as well as antibody to hepatitis C virus (anti-HCV) and antibody to human immunodeficiency virus (anti-HIV). Alanine aminotransferase (ALT) levels were determined using standard procedures.
HBV DNA extraction and reverse transcriptase/surface region amplification
Viral DNA was extracted from 200 mL of plasma according to the manufacturer’s instructions using QIAamp DNA Blood Mini Kits (Qiagen, Hilden, Germany). The partial RT region that partially overlaps the surface (S) region was amplified using a nested PCR approach. The primer sets were as follows: first round, HB8F nucleotides (nt) 1824 to 1843 (5′-TTCACCTCTGCCTAATCATC-3′) and HB6R nt 1421 to 1440 (5′-AACAGACCAATTTATGCCTA-3′); second round, HB2F nt 414 to 433 (5′-TGCTGCTATGCCTCATCTTC-3′) and HB3R nt 1107 to 1084 (5′-AGTTGGCGAGAAAGTGAAAGCCTG-3′). The nested PCR proceeded as described (13). The amplified products were resolved by electrophoresis through 2% agarose gels, stained with ethidium bromide and visualized by UV light.
Direct sequencing and HBV genotype analysis
The products of the second round of nested PCR were purified using ExoSAP-IT (USB Corporation, Cleveland, OH, USA) as described by the manufacturer. Products were directly sequenced using BigDye Terminator version 3.1 sequencing kits and an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems, Foster City, CA, USA). The nucleotide sequences together with reference sequences retrieved from the National Center for Biotechnology Information (NCBI) GenBank were aligned with Clustal X version 2.1 software (14). Subgenotypes of HBV were determined by constructing a phylogenetic tree of the partial S region using the neighbor-joining method with 1000 bootstrap resamplings and Molecular Evolutionary Genetics Analysis (MEGA) software.
Ultra-deep sequencing
The concentrations of second round PCR products were measured using QubitdsDNA HS Assay Kits (Q32851; Invitrogen, Carlsbad, California, USA). Meanwhile, a library of PCR products (< 500 bp) of the viral genome (50 or 0.2 ng) was prepared using Nextera DNA Sample Prep Kits (Illumina, San Diego, California, USA). The products of PCR that were uniformly sheared using kits into 500-bp fragments and 1% 8 pMPhiX (controls), were mixed with PCR product libraries, and then paired-end 151-bp sequences were analyzed on a MiSeq sequencer (Genome Analyzer; Illumina, San Diego, California, USA). Fluorescent signals were analyzed using MiSeq control software and FASTQ-formatted sequence data were obtained using reporter analysis (Illumina).
Sequence mapping and data analysis
After a quality check and data trimming, paired-end sequences with a read quality > 80% of the consensus sequence (estimated as a quality score of 30 [Q30]) were assembled using Genomic Workbench software version 6.0.1 (CLC bio, Aarhus, Denmark). Sequence reads were mapped to the reference HBV genome (NCBI GenBank accession number AB713528). Therefore, probabilistic variant detection was identified using default parameters in the mapping algorithm.
The characteristics of viral quasispecies were evaluated in terms of genetic complexity based on the number of different sequences identified in the population. This phenomenon was exposed using the ‘read conflicts’ setting in Genomic Workbench. Conflicting sequences were then annotated on the consensus sequence. Conflicts or nucleotide alterations were defined as a position where at least one of the sequence reads encoded a different nucleotide. Variants were defined as a nucleotide substitution that resulted in an amino acid change(s). The proportion of variants was calculated from the number of nucleotide substitutions that changed an amino acid at a position in the total sequence read per nucleotide position.
Statistical analysis
Data were statistically analyzed using SPSS version 21 (IBM Corporation, Armonk, NY, USA). Differences among categorical and continuous variables were compared using the χ2 test and Student’s t test, respectively. Values of P < 0.05 were considered statistically significant.
RESULTS
Characteristics of patients
Table I compares the baseline characteristics between patients (male, n = 8, age range, 18 – 77 y) who were treatment-naïve (n = 7) and treated (n = 5). Age, gender and AST levels did not differ between the two groups. The clinical diagnosis of the naïve patients ranged from asymptomatic HBV carriers (n = 3), HBV liver cirrhosis (n = 2), and HBV hepatocellular carcinoma (n = 2). Four and one of the five patients with chronic HBV infection had been administered with telbivudine (LdT) and lamivudine (LMV), respectively, for four to eight weeks.
Table I.
Characteristics of patients.
| Treatment group | Samples ID | Age (y) | Sex | Clinical diagnosis | Duration of therapy | AST (U/L) | HBV geno-type | Total reads | Mapping reads | Average coverage |
|---|---|---|---|---|---|---|---|---|---|---|
| Naïve | YOGN1 | 64 | M | Cirrhosis | None | 206 | B3 | 2,676,742 | 262,044 | 9,189 |
| YOGN2 | 77 | M | HCC | None | 74 | B3 | 1,260,250 | 914,396 | 17,926 | |
| YOGN3 | 40 | M | AC | None | 74 | B3 | 1,781,062 | 679,886 | 25,336 | |
| YOGN4 | 22 | M | AC | None | 152 | B3 | 1,327,708 | 837,533 | 18,191 | |
| YOGN5 | 55 | F | HCC | None | 31 | B3 | 1,298,306 | 648,284 | 18,519 | |
| YOGN6 | 36 | M | Cirrhosis | None | 115 | B3 | 1,413,446 | 667,297 | 20,589 | |
| YOGN7 | 28 | F | AC | None | 31 | B3 | 1,638,654 | 726,531 | 23,372 | |
|
| ||||||||||
| Means ± SD | 46.00 ± 20.01 | 97.57 ± 64.55 | 1,628,024 ± 500,591 | 676,567 ± 207,242 | 19,017 ± 5,165 | |||||
|
| ||||||||||
| Under treatment | YOGT1 | 26 | F | CHB | LMV (4 w) | 72 | B7 | 4,402,758 | 1,683,940 | 60,305 |
| YOGT2 | 64 | M | CHB | LdT (4 w) | 12 | B3 | 3,785,330 | 261,026 | 9,415 | |
| YOGT3 | 34 | M | CHB | LdT (8 w) | 72 | B3 | 3,937,778 | 318,897 | 11,408 | |
| YOGT4 | 30 | F | CHB | LdT (4 w) | 69.2 | B3 | 4,134,640 | 1,587,667 | 57,346 | |
| YOGT5 | 18 | M | CHB | LdT (4 w) | 21 | C1 | 2,819,036 | 213,672 | 8,164 | |
|
| ||||||||||
| Means ± SD | 34.40 ± 17.57 | 49.24 ± 30.08 | 3,815,908 ± 603,226 | 831,040 ± 752,770 | 29,327 ± 26,972 | |||||
|
| ||||||||||
| P value | 0.314a | 0.679b | 0.117a | 0.000*a | 0.653a | 0.339a | ||||
Abbreviations: AH, asymptomatic hepatitis B carrier; AST, aspartate transaminase; CHB, chronic hepatitis B; F, female; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; ID, identity; LMV, lamivudine; LdT, telbivudine; M, male; SD, standard deviation; w: weeks; y: years.
P < 0.05, significant difference between groups.
t-test;
χ2 test.
Phylogenetic analysis revealed that 1 (6.33%), 1 (6.33%) and 10 (83.34%), patients were infected with HBV genotypes B7, C1 and B3, respectively (data not shown). Table I also compares the means of total reads, mapping reads and depth coverage of HBV DNA sequences generated by ultra-deep sequencing in the two groups.
Major variants in RT region
The major population of sequence reads that detected ≥ 20% of HBV quasispecies determined by ultra-deep sequencing were aligned (HBV rt108 – rt267) to detect major variants (Figure 1) that were most frequently detected in the A–B inter-domain. Substitutions associated with the genetic backbone of the genotype C wild-type on a sequence identified as genotype B were found at rt118, rt127, rt134, rt263, and rt267. Major variants were detected at rtH124D, rtR153Q, and rtS246C in naïve patients, and at rtI122V, rtS213T, and rtS246A only in treated patients, whereas and major variants of rtH238Q were identified in both groups.
Figure 1. Alignment of partial reverse transcriptase domain in naïve and treated patients with HBV.
The amino acids sequences of major population (sequence reads which detected ≥20% in HBV quasispecies determined by ultra-deep sequencing) were compared with consensus sequence of genotype B and C retrieved from Rhee SY et al (15). The highlighted parts show the conserved regions of RT domain.
Substitution rate of variants
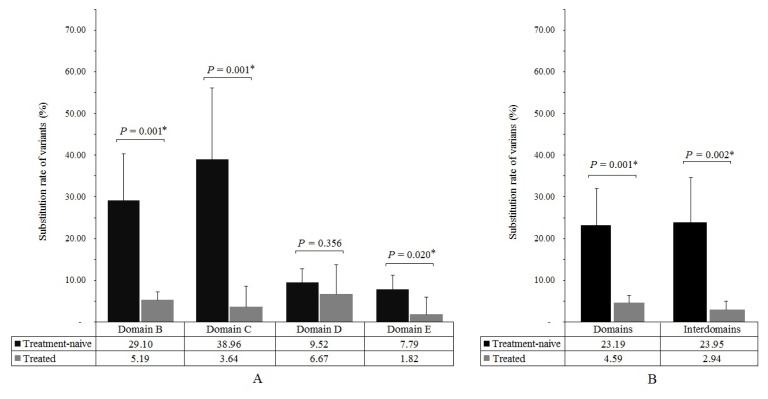
The sequencing products covered the B, C, D and E conserved regions as domains and the B–C, C–D, D–E regions as inter-domains. The substitution rate of variants was calculated as: [number of sites within region where amino acid changes with in HBV quasispecies accounted for ≥1%/number of sites within region] × 100%. The substitution rates of variants were higher at the B, C and E conserved regions of the RT domain in naïve, than in treated patients, but not in the D conserved region (Figure 2A). Overall, the substitution rates of domains and inter-domains variants were significantly higher in naïve, than in treated patients (P = 0.001 and P = 0.002, respectively; Figure 2B).
Figure 2. Substitution rate of variants in reverse transcription domain between naïve and treated Indonesian patients infected with HBV.
A variant was defined as a change in a nucleotide that caused a subsequent alteration in an amino acid. The substitution rate of variants was calculated as: [number of sites within region where the amino acid changes with population proportion in HBV quasispecies ≥ 1% / number of sites within region] × 100%. *P < 0.05, significant difference between groups. (A) Substitution rate of variants at conserved regions. (B) Substitution rate of variants at domains and inter-domains.
Proportion of NA resistance
Primary and secondary NA resistance at nine hotspots (rtI169T, rtV173L, rtL180M/I, rtA181T/V, rtT184A/S, rtS202G/I, rtM204V, rtN236T and rtM250V) was investigated using ultra-deep sequencing in treatment naïve and treated Indonesian patients infected with HBV. Mutation-related NA resistance was found in both groups. The all-proportion mean of investigated mutations in both groups was < 1%. The proportion of NA resistance in both groups was highest at rtM204I. This study found a significantly different mean proportion between the two groups at rtT184S associated with ETV resistance and with rtN236T associated with adefovir (ADV) resistance (P < 0.05 for both). The proportion of rtT184S was higher (means of proportion [means] ± standard deviation [SD]: 0.930 ± 0.038 vs. 0.567 ± 0.149), whereas that of rtN236T was lower (means ± SD: 0.335 ± 0.123 vs. 0.650 ± 0.064) in the treated, than in the naïve group (Table II).
Table II.
Proportion of primary and secondary antiviral resistance hotspots determined by ultra-deep sequencing
| Samples ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| rt169 | rt173 | rt180 | rt181 | rt184 | rt202 | rt204 | rt236 | rt250 | ||||||
|
| ||||||||||||||
| I169T | V173L | L180M | L180V | A181T | A181V | T184A | T184S | S202G | S202I | M204V | M204I | N236T | M250V | |
|
| ||||||||||||||
| ETV | LMV | LMV/LdT/ETV | LMV/LdT/ADV/ETV | ETV | ETV | LMV/LdT/ETV | ADV | ETV | ||||||
| Treatment-naïve | ||||||||||||||
| YOGN1 | 0.182 | 0.315 | 0.156 | 0.276 | 0.351 | 0.209 | 0.441 | 0.886 | 0.305 | 0.171 | 0.346 | 0.935 | 0.508 | 0.436 |
| YOGN2 | 0.190 | 0.310 | 0.120 | 0.296 | 0.310 | 0.340 | 0.960 | 0.500 | 0.510 | 0.230 | 0.630 | 1.050 | 0.670 | 0.330 |
| YOGN3 | 0.190 | 0.280 | 0.120 | 0.301 | 0.300 | 0.340 | 0.980 | 0.490 | 0.530 | 0.230 | 0.630 | 1.030 | 0.640 | 0.330 |
| YOGN4 | 0.200 | 0.290 | 0.120 | 0.283 | 0.310 | 0.360 | 0.990 | 0.510 | 0.540 | 0.250 | 0.540 | 0.590 | 0.680 | 0.370 |
| YOGN5 | 0.180 | 0.240 | 0.110 | 0.295 | 0.300 | 0.380 | 0.920 | 0.500 | 0.540 | 0.230 | 0.610 | 1.060 | 0.690 | 0.370 |
| YOGN6 | 0.190 | 0.260 | 0.120 | 0.300 | 0.270 | 0.380 | 0.910 | 0.480 | 0.520 | 0.260 | 0.570 | 1.060 | 0.680 | 0.350 |
| YOGN7 | 0.190 | 0.270 | 0.120 | 0.297 | 0.320 | 0.380 | 0.940 | 0.580 | 0.530 | 0.260 | 0.630 | 1.050 | 0.680 | 0.330 |
|
| ||||||||||||||
| Mean ± SD | 0.189 ± 0.007 | 0.281 ± 0.027 | 0.124 ± 0.015 | 0.293 ± 0.009 | 0.309 ± 0.024 | 0.341 ± 0.061 | 0.877 ± 0.195 | 0.564 ± 0.146 | 0.496 ± 0.085 | 0.233 ± 0.031 | 0.565 ± 0.103 | 0.968 ± 0.172 | 0.650 ± 0.064 | 0.359 ± 0.038 |
|
| ||||||||||||||
| Treated | ||||||||||||||
| YOGT1 | 0.259 | 0.559 | 0.209 | 0.536 | 0.389 | 0.292 | 0.827 | 0.976 | 0.494 | 0.341 | 0.451 | 1.122 | 0.493 | 0.443 |
| YOGT2 | 0.259 | 0.327 | 0.096 | 0.326 | 0.293 | 0.292 | 0.543 | 0.883 | 0.419 | 0.188 | 0.674 | 0.507 | 0.232 | 0.350 |
| YOGT3 | 0.120 | 0.272 | 0.127 | 0.386 | 0.127 | 0.386 | 0.603 | 0.906 | 0.324 | 0.219 | 0.399 | 1.472 | 0.304 | 0.449 |
| YOGT4 | 0.259 | 0.481 | 0.183 | 0.483 | 0.309 | 0.265 | 0.784 | 0.960 | 0.438 | 0.335 | 0.416 | 1.049 | 0.431 | 0.443 |
| YOGT5 | 0.123 | 0.241 | 0.182 | 0.070 | 6.748 | 0.207 | 0.723 | 0.923 | 0.305 | 0.210 | 0.444 | 0.430 | 0.213 | 0.330 |
|
| ||||||||||||||
| Mean ± SD | 0.204 ± 0.075 | 0.376 ± 0.138 | 0.159 ± 0.046 | 0.360 ± 0.181 | 1.573 ± 2.894 | 0.288 ± 0.064 | 0.696 ± 0.120 | 0.930 ± 0.038 | 0.396 ± 0.080 | 0.258 ± 0.073 | 0.477 ± 0.112 | 0.916 ± 0.440 | 0.335 ± 0.123 | 0.403 ± 0.057 |
|
| ||||||||||||||
| P valuea | 0.601 | 0.099 | 0.081 | 0.339 | 0.265 | 0.179 | 0.097 | 0.000* | 0.066 | 0.421 | 0.187 | 0.780 | 0.000* | 0.146 |
Abbreviation: ADV, adefovir; ETV, entecavir; ID, identity; LdT, telbivudine; LMV, lamivudine; SD, standard deviation.
P < 0.05, significant difference between groups.
t-test.
DISCUSSION
The present findings showed that most of the treatment-naïve patients harbored the genotype B3, whereas the treated group harbored genotypes B3, B7 and Cl. Previous studies have identified one-third of these genotypes in an Indonesian population (16, 17). Here, we used ultra-deep sequencing to identify differences in HBV quasispecies variants within RT between Indonesian patients who had never been treated (naïve) and those who had received four to eight weeks of treatment (treated). We initially found more total reads, mapping reads and higher average coverage generated by ultra-deep sequencing in the treated, than in the naïve patients. The increased average coverage might have exposed more minor sequence variants (18). As described at previous studies that the minor variants was generated from coverage only around 4 to 6 thousands reads (18, 19), we suggested that these present results were higher enough to analyze the presence of minor population.
To our knowledge, this is the first study to compare quasispecies variants in RT between naïve and treated Indonesian patients infected with HBV. DNA polymerase of HBV consists of a terminal protein, a spacer, RT and an RNase domain. Variations that affect or are affected by NA are located in the RT region, which comprises conserved G, F, A, B, C, D and E domains as well as F–A, A–B, B–C, C–D and D–E inter-domains that play roles in HBV replication. Furthermore, mutations in these domains affect RT function even in the absence of NA therapy (20–22). However, RT variants are found mostly, but not exclusively, in the A–B inter-domain that is structurally distant from the functional domain. Moreover, mutations in the A–B inter-domain that overlap the α-determinant and major hydrophilic region (MHR) of HBsAg would reduce binding affinity for neutralizing hepatitis B surface antibodies (anti-HBs) (23, 24).
Ultra-deep sequencing found that ≥ 20% of variants in the total population of variants were located in inter-domains and that variants in the major population were most frequently located in the A–B inter-domain. We found that that 6 (85.71%) of 7 naïve patients harbored the rtR153Q mutation, which is putatively associated with LMV therapy. Moreover, this amino acid change in RT was concomitant with a sG145R mutation in the surface region. Importantly, the sG145R mutation caused vaccination or immunoglobulin (HBIg) therapy to fail (25–27). The present findings together with those of previous studies detected major variants at rtH214D and rtS246C in naïve patients (22, 28, 29). Meanwhile major variants at rtI122V, rtS213T, rtS246A were detected only in treated patients. A study reported the emergence of mutation rtS213T in LMV-treated patients (30). The rtI122V and rtS246A were identified as new mutations, which might be involved in the evolution of NAs-resistance. An rtH238Q mutation in domain D was identified in both naïve and treated patients. Previous study reported that rtH238Q was identified among 18 of 35 naïve Indonesian patients using direct sequencing (31). In studies by Li et al. (32) and Zhong et al. (33), rtH238 was frequently found in HBV genotype B and there was a tendency of increasing mutation in NA-treated patients. However, an in-vitro study failed to demonstrate that rtH238 affected viral replication or NAs-drug resistance (33). Furthermore, the correlation rtH238Q in clinical relevancies requires further investigation in future studies.
We used ultra-deep sequencing to select a population of variants from total reads per sequence with a ≥ 1% cutoff and then compared the substitution rates of RT variants in different domains between naïve and treated patients. The results showed that the substitution rates of variants in domain B, C and E and in the inter-domains of RT were significantly higher in the naïve, than in the treated group. These results indicated a greater population diversity in the naïve, than in the treated patients, suggesting that a decrease in variation within the RT domain is influenced by NA therapy. Thus, we considered that the clonal selection in the present study have been lower during the first month of NA therapy. In addition, HBV with variations that are susceptible to NA would be lost during NA therapy. Genetic complexity decreases during the first two weeks of interferon therapy (11). It was recently reported that lower diversity in early phase of treatment was found in NAs-responders than those of nonresponders (34). Future studies of cohort samples before and after therapy with NA are required to clarify the dynamics and diversity of viral populations
This study continues an investigation conducted by Widasari et al. (35) who applied deep sequencing to detect several known drug-resistant mutations such as I169L/M, S202R, M204I/L or N236S with a proportion > 1% among Indonesian treatment-naïve patients. We compared HBV quasispecies of RT between Indonesian patients with HBV infection undergoing early NA therapy and treatment-naïve patients. The treated group comprised patients who were administered with LMV or LdT for four to eight weeks. Mutations that are known to confer NA drug resistance were identified in < 2% of the total population of mutations in both groups. Importantly, the average proportion of rtM204I was the highest among other known mutations that cause drug resistance in the naïve and treated patients (0.968% vs. 0.916%), although the prevalence did not significantly differ between the groups. The primary rtM204I mutation confers resistance to LMV or LdT (9, 23). We also found that the prevalence of mutations such as rtT184S associated with ETV resistance was significantly higher in the treated, than in the naïve group, whereas that of rt236T, which confers resistance to ADV was significantly more prevalent among the naïve patients. The present findings showed that treatment for less than eight weeks is insufficient to detect increases in the proportion of known mutations in a minor population. In addition, because of the limitation of sample collection, naïve and treated patients of present study were completely different. It will be better when the diversity before and after NA therapy in the same patient are analyzed and compared in the future. We suggest that further analysis in a cohort study should confirm the effect of prolonged LMV/LdT therapy in Indonesian patients who harbor minor variants of rtM204I before starting therapy, because LMV/LdT remains the most common strategy for treating patients infected with HBV.
In conclusion, our results showed that the substitution rates of variants with a prevalence of ≥ 1% in RT domains B, C and E and inter-domains detected using ultra-deep sequencing significantly differed between naïve- and treated patients. We also found that the proportion of rtM204I associated with LMV/LdT resistance was the highest in a minor population even among treatment-naïve patients. Therefore, the existence of drug resistance variants in HBV minor population could be considered by clinicians to select therapeutic strategies for Indonesian patients infected with HBV. Our concern, in the future research, a large number of clinical patients are needed to get the reproducible data and assure the results, although the limited size of samples in the present study had significant meaning and provide insight about the pattern of quasispecies population of RT domain in HBV isolated from Indonesian population.
ACKLOWLEDGMENTS
This study was supported by a grand-in-aid from the Japan Initiative for Global Research Network on Infectious Disease (J-GRID) Program of the Ministry of Education, Culture, Sport, Science and Technology; the Japan International Cooperation Agency; and by the Ministry of Health, Labor and Welfare of Japan (grant H-25-general-008).
REFERENCES
- 1.Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, et al. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int. 2012;6:531–61. doi: 10.1007/s12072-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 2.Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–2. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 3.European Association For The Study Of The Liver. EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol. 2012;57:167–85. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Ayoub WS, Keeffe EB. Review article: current antiviral therapy of chronic hepatitis B. Aliment Pharmacol Ther. 2011;34:1145–58. doi: 10.1111/j.1365-2036.2011.04869.x. [DOI] [PubMed] [Google Scholar]
- 5.Gish R, Jia JD, Locarnini S, Zoulim F. Selection of chronic hepatitis B therapy with high barrier to resistance. Lancet Infect Dis. 2012;12:341–53. doi: 10.1016/S1473-3099(11)70314-0. [DOI] [PubMed] [Google Scholar]
- 6.Yano Y, Utsumi T, Lusida MI, Hayashi Y. Hepatitis B virus infection in Indonesia. World J Gastroenterol. 2015;21:10714–20. doi: 10.3748/wjg.v21.i38.10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niederau C. Chronic hepatitis B in 2014: great therapeutic progress, large diagnostic deficit. World J Gastroenterol. 2014;20:11595–617. doi: 10.3748/wjg.v20.i33.11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones SA, Hu J. Hepatitis B virus reverse transcriptase: diverse functions as classical and emerging targets for antiviral intervention. Emerg Microbes Infect. 2013;2:e56. doi: 10.1038/emi.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strasfeld L, Chou S. Antiviral drug resistance: mechanisms and clinical implications. Infect Dis Clin North Am. 2010;24:809–33. doi: 10.1016/j.idc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, Dong H, He Y, Sun J, Jin W, Xie Q, et al. Composition and interactions of hepatitis B virus quasispecies defined the virological response during telbivudine therapy. Sci Rep. 2015;5:17123. doi: 10.1038/srep17123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasu A, Marusawa H, Ueda Y, Nishijima N, Takahashi K, Osaki Y, et al. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PLoS One. 2011;6:e24907. doi: 10.1371/journal.pone.0024907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012;142:1303–1313 e1. doi: 10.1053/j.gastro.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugauchi F, Mizokami M, Orito E, Ohno T, Kato H, Suzuki S, et al. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001;82:883–92. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Rhee SY, Margeridon-Thermet S, Nguyen MH, Liu TF, Kagan RM, Beggel B, et al. Hepatitis B virus reverse transcriptase sequence variant database for sequence analysis and mutation discovery. Antiviral Res. 2010;88:269–75. doi: 10.1016/j.antiviral.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nurainy N, Muljono DH, Sudoyo H, Marzuki S. Genetic study of hepatitis B virus in Indonesia reveals a new subgenotype of genotype B in east Nusa Tenggara. Arch Virol. 2008;153:1057–65. doi: 10.1007/s00705-008-0092-z. [DOI] [PubMed] [Google Scholar]
- 17.Mulyanto Depamede SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, et al. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154:1047–59. doi: 10.1007/s00705-009-0406-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher R, van Zyl GU, Travers SA, Pond SLK, Engelbrech S, Murrell B, et al. Deep sequencing reveals minor protease resistance mutations in patients failing a protease inhibitor regimen. J Virol. 2012;86:6231–7. doi: 10.1128/JVI.06541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuyver LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, et al. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33:751–7. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]
- 21.Warner N, Locarnini S, Kuiper M, Bartholomeusz A, Ayres A, Yuen L, et al. The L80I substitution in the reverse transcriptase domain of the hepatitis B virus polymerase is associated with lamivudine resistance and enhanced viral replication in vitro. Antimicrob Agents Chemother. 2007;51:2285–92. doi: 10.1128/AAC.01499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu BM, Li T, Xu J, Li XG, Dong JP, Yan P, et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naive Chinese patients. Antiviral Res. 2010;85:512–9. doi: 10.1016/j.antiviral.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Locarnini S. Primary resistance, multidrug resistance, and cross-resistance pathways in HBV as a consequence of treatment failure. Hepatol Int. 2008;2:147–51. doi: 10.1007/s12072-008-9048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locarnini S. Transmission of antiviral drug resistant hepatitis B virus: implications for public health and patient management. J Gastroenterol Hepatol. 2010;25:649–51. doi: 10.1111/j.1440-1746.2010.06255.x. [DOI] [PubMed] [Google Scholar]
- 25.Torresi J, Earnest-Silveira L, Civitico G, Walters TE, Lewin SR, Fyfe J, et al. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology. 2002;299:88–99. doi: 10.1006/viro.2002.1448. [DOI] [PubMed] [Google Scholar]
- 26.Sheldon J, Rodes B, Zoulim F, Bartholomeusz A, Soriano V. Mutations affecting the replication capacity of the hepatitis B virus. J Viral Hepat. 2006;13:427–34. doi: 10.1111/j.1365-2893.2005.00713.x. [DOI] [PubMed] [Google Scholar]
- 27.Carman WF. The clinical significance of surface antigen variants of hepatitis B virus. J Viral Hepat. 1997;4(Suppl 1):11–20. doi: 10.1111/j.1365-2893.1997.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Wu B, Wang JH, Huang L, Wang DY, Zhao L, et al. Pre-existing mutations in reverse transcriptase of hepatitis B virus in treatment-naive Chinese patients with chronic hepatitis B. PLoS One. 2015;10:e0117429. doi: 10.1371/journal.pone.0117429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colonno RJ, Rose R, Baldick CJ, Levine S, Pokornowski K, Yu CF, et al. Entecavir resistance is rare in nucleoside naive patients with hepatitis B. Hepatology. 2006;44:1656–65. doi: 10.1002/hep.21422. [DOI] [PubMed] [Google Scholar]
- 30.Pallier C, Rodriguez C, Brillet R, Nordmann P, Hezode C, Pawlotsky JM. Complex dynamics of hepatitis B virus resistance to adefovir. Hepatology. 2009;49:50–9. doi: 10.1002/hep.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pribadi J, Gani RA, Syam AF, Lesmana CR, Budiman Bela B, Setiati S, et al. Pola Mutasi Resistensi terhadap Analog Nukleosida Virus Hepatitis B pada Pengidap Kronis yang Belum Mendapatkan Terapi. Research Series. 2013. Abstract in Indonesian. Available from: http://pphi-online.org/alpha/?p=579 [cited 28th November 2015]
- 32.Li XG, Liu BM, Xu J, Liu XE, Ding H, Li T. Discrepancy of potential antiviral resistance mutation profiles within the HBV reverse transcriptase between nucleos(t)ide analogue-untreated and -treated patients with chronic hepatitis B in a hospital in China. J Med Virol. 2012;84:207–16. doi: 10.1002/jmv.23182. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Y, Lv J, Li J, Xing X, Zhu H, Su H, et al. Prevalence, virology and antiviral drugs susceptibility of hepatitis B virus rtN238H polymerase mutation from 1865 Chinese patients with chronic hepatitis B. Antiviral Res. 2012;93:185–90. doi: 10.1016/j.antiviral.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Peveling-Oberhag J, Herrmann E, Kronenberger B, Farnik H, Susser S, Sarrazin C, et al. Dynamics of hepatitis B virus quasispecies heterogeneity and virologic response in patients receiving low-to-moderate genetic barrier nucleoside analogs. J Viral Hepat. 2013;20:234–9. doi: 10.1111/jvh.12013. [DOI] [PubMed] [Google Scholar]
- 35.Widasari DI, Yano Y, Heriyanto DS, Utsumi T, Yamani LN, Rinonce HT, et al. A deep-sequencing method detects drug-resistant mutations in the hepatitis B virus in Indonesians. Intervirology. 2014;57:384–92. doi: 10.1159/000366420. [DOI] [PubMed] [Google Scholar]