Abstract
Raccoon rabies remains a serious public health problem throughout much of the eastern seaboard of North America due to the urban nature of the reservoir host and the many challenges inherent in multi-jurisdictional efforts to administer co-ordinated and comprehensive wildlife rabies control programmes. Better understanding of the mechanisms of spread of rabies virus can play a significant role in guiding such control efforts. To facilitate a detailed molecular epidemiological study of raccoon rabies virus movements across eastern North America, we developed a methodology to efficiently determine whole genome sequences of hundreds of viral samples. The workflow combines the generation of a limited number of overlapping amplicons covering the complete viral genome and use of high throughput sequencing technology. The value of this approach is demonstrated through a retrospective phylogenetic analysis of an outbreak of raccoon rabies which occurred in the province of Ontario between 1999 and 2005. As demonstrated by the number of single nucleotide polymorphisms detected, whole genome sequence data were far more effective than single gene sequences in discriminating between samples and this facilitated the generation of more robust and informative phylogenies that yielded insights into the spatio-temporal pattern of viral spread. With minor modification this approach could be applied to other rabies virus variants thereby facilitating greatly improved phylogenetic inference and thus better understanding of the spread of this serious zoonotic disease. Such information will inform the most appropriate strategies for rabies control in wildlife reservoirs.
Keywords: raccoon rabies virus, Illumina next generation sequencing, molecular epidemiology, viral phylogeny
1. Introduction
Over the last 25 years tools to characterise RNA virus genomes have evolved considerably and the information gained from such studies is improving our understanding of the evolution of these viruses and the factors that impact their emergence and spread. Due to its zoonotic nature and its role in the etiology of a lethal encephalitis, the rabies virus (Family Rhabdoviridae, genus Lyssavirus) has been extensively investigated by molecular epidemiological tools (Nadin-Davis, 2013). Seven major phylogenetic lineages of the rabies virus (RABV) have been identified, all of which can be further subdivided into multiple variants, each maintained by a particular host reservoir occupying a certain geographical range (Bourhy et al., 2008; Nadin-Davis, 2013). In the Americas, the American indigenous RABV lineage consists of several variants circulating in chiropteran and terrestrial hosts of which the raccoon rabies virus (RRV) strain has emerged since the 1940s to become a significant public health concern with substantial economic impact (Sterner et al., 2009).
Since the initial recognition of the RRV strain in Florida in the 1940s (Jenkins et al., 1988) it has spread by both natural and anthropogenic means throughout the eastern seaboard of the USA reaching as far north as the state of Maine and causing incursions into southern parts of eastern Canada on several separate occasions (Nadin-Davis et al., 2006; Rees et al., 2011; Wandeler et al., 2000; Wandeler and Salsberg, 1999). As a result, this epizootic has come under significant scrutiny and has been the target of various control efforts (Rosatte et al., 2009; Slate et al., 2005). Studies employing partial genome characterisation of RRV have explored how landscape features influence evolution and spread of this virus over time (Szanto et al., 2011) and demonstrated how viral genetic signatures could yield information on the viral population history even in the absence of surveillance data (Biek et al., 2007).
Initial molecular epidemiological studies of RABV targeted small (<300 bases) regions of the genome (Smith et al., 1992) which were sufficient for identification of viral variants and their respective host reservoirs. However as amplification and sequencing technologies have improved the use of complete genes, multiple genes or even complete genomes has become more common (Biek et al., 2007; Bourhy et al., 2008; Delmas et al., 2008). The robustness of phylogenetic analysis improves as the amount of genetic diversity within a dataset increases; thus whole viral genome sequencing would be expected to afford the highest resolution for molecular epidemiological analysis. Moreover, complete genome characterization of RABV isolates and related viruses has had significant impact on our understanding of the taxonomy of the Lyssavirus genus (Kuzmin et al., 2005), has allowed exploration of the evolution of RABVs following a host shift (Kuzmin et al., 2012), has provided insights into the historical relationships of several attenuated viral strains used for oral vaccination of wildlife against rabies (Geue et al., 2008) and has the potential to reveal genetic differences between RABV strains which may influence host tropism and /or pathogenicity (Yu et al., 2014).
While whole genome sequencing (WGS) of RNA viruses is possible using traditional Sanger methodology, the evolution of highly parallel sequencing regimens allows much higher throughput with less technical effort (Radford et al., 2012). Methods describing the application of metagenomics RNA-seq approaches to lyssavirus WGS have been described and their application to the genome characterisation of a small number of isolates has been reported (Marston et al., 2013; Marston et al., 2015a; Marston et al., 2015b) including one study that successfully sequenced 59 isolates of African dog rabies (Brunker et al., 2015). Such an approach does not require sequence-specific primers and it is certainly preferable for detection and characterization of novel lyssaviruses. In addition, it offers the possibility of using raw sequence data not only for viral characterisation but also for parallel host species / population genetics analysis as demonstrated recently by a study on arctic fox rabies in Greenland (Hanke et al., 2016). However, a metagenomics approach presents challenges for high throughput. Due to the relatively large proportion of host RNA present in samples there is a need to remove rRNA prior to library construction and the extent of sample pooling may be limited to ensure sufficient coverage of the viral sequence target, thereby increasing the cost per sample. An alternative strategy involves amplification of the viral genome by standard reverse-transcription polymerase chain reaction (RT-PCR) and then pooling the amplicons for highly parallel sequencing (Wright et al., 2011).
This study explored the latter approach for the development of an efficient workflow for WGS of hundreds of RRV samples through optimization of the RNA extraction process, design of an efficient RT-PCR protocol for whole genome amplification and use of highly parallel sequencing technology. The value of this WGS approach to yield greatly improved insight into viral spread was explored by comparing the robustness of the phylogenies predicted using single gene and whole genome sequence data for a group of RRV specimens collected in the province of Ontario between 1999 and 2005.
2. Material and Methods
2.1 Raccoon rabies virus samples
To develop a method with broad-based coverage of the RRV strain across much of its northern range, samples of brain tissue confirmed to be infected with RABV by the direct fluorescent antibody (DFA) test (Dean et al., 1996) were obtained from several diagnostic laboratories. All samples of Canadian origin were obtained from the Centre of Expertise for Rabies of the Canadian Food Inspection Agency in Ottawa, Canada, while samples from the USA were acquired from state rabies laboratories in Albany, New York, Burlington, Vermont and Augusta, Maine. The time span of sample collection varied for each jurisdiction thus: between the early 1990s up until 2011 for New York; 1999 to 2005 in Ontario; 2005 to 2011 in Vermont; sampling was limited to select years in Maine and New Brunswick to reflect the limited incursions that occurred in that Canadian province. In areas where sympatric RABV strains associated with terrestrial hosts circulate, antigenic typing was performed as previously described (Fehlner-Gardiner et al., 2008) to confirm the presence of the RRV strain. Metadata associated with these samples, including host species, location of origin to the township level and year of collection, were also compiled (see Table S1). The sample designations employed in this report have a standard format comprised of a two letter code indicating the state or province of origin, a four digit number indicating the year of collection followed by a four digit submission number.
2.2 RNA extraction
While TRIzol™ reagent has often been employed as per the supplier’s instructions (Life Technologies Inc) for total RNA extraction from brain tissue, the resulting extract often contains impurities based on A260/280 ratios that deviate significantly from the expected value of 2.0. To improve upon this method a protocol employing a semi-automated MagMax™ instrument (Applied Biosystems) which employs magnetic beads for nucleic acid purification was assessed but this proved unsatisfactory. However use of the MagMax system to purify Trizol-extracted material generated a very clean product so a hybrid extraction method was developed. About 50 mg of tissue was homogenised in 500 μl TRIzol, 100 μl chloroform was added and samples were mixed vigorously prior to phase separation by centrifugation. The aqueous phase was recovered in 2 × 200 μl aliquots and kept frozen until further processing. An aliquot of the aqueous phase was purified on a MagMax™ instrument using an AM1830 RNA purification kit (Ambion) and the AM1830DW MagMax protocol. RNA was eluted from the magnetic beads into 50 μl elution buffer and its concentration measured spectrophotometrically using a NanoVue instrument (GE Biosciences); such preparations consistently gave an A260/280 ratio of 1.9–2.1. RNA extracts were either used immediately for cDNA preparation or stored at −80 C; freeze / thaw cycles were minimised to limit damage to the RNA template.
2.3 RT-PCR
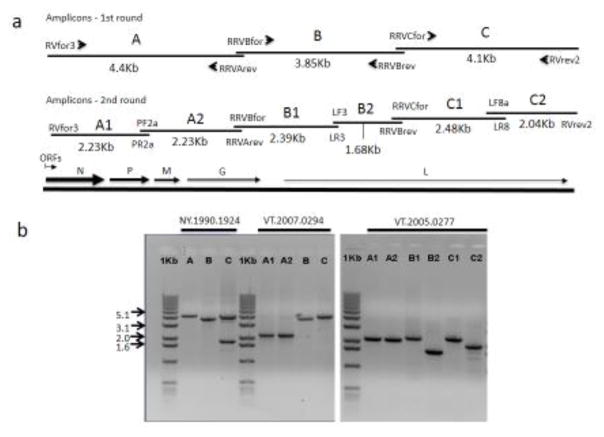
To generate amplicons covering the complete viral genome of approximately 12Kb, three primer pairs suitable for generation of overlapping PCRs were designed; in addition, two sets of primers suitable for generation of overlapping hemi-nested PCR products on each of these three amplicons were also designed (see Fig. 1a, 1b and Table 1). Primer design incorporated general PCR guidelines and targeted conserved regions of the lyssavirus genome with particular consideration of the sequence of a RRV reference genome (GenBank accession EU311738).
Fig. 1.
(a) A schematic of the RRV genome indicating the position of the five ORFs above which the locations of the 1st and 2nd round amplicons are shown. The PCR primers used are indicated at the ends of each amplicon while the size of each product is given in Kb below. (b) Gel analysis of representative purified RT-PCR products generated for sequencing. Products generated from three RRV samples are shown; NY.1990.1924 generated 1st round amplicons (A, B, C) for all targets; VT.2007.0294 generated 1st round amplicons for targets B and C but required 2nd round PCR to generate the A1 and A2 products; VT2005.0277 required 2nd round PCR for all three targets to generate six products (A1, A2, B1, B2, C1, C2). A 1Kb size marker run in parallel with all products confirms their expected sizes.
Table 1.
Primers employed for RT-PCR of RRV samples.
| Amplicon | Primer name | Primer sequence 5′ - 3′ | Location in RRV genome* | Sense | Amplicon size (bp)** |
|---|---|---|---|---|---|
| A | RVfor3 | ggACGCTTAACAACAAAATCAGAGAAGAAGTAG | 1 to 31 | +ve | 4397 |
| RRVArev | TGAGGGGATGATCTCGCTCCAAG | 4373 to 4395 | -ve | ||
| B | RRVBfor | CTGGGTTTGGGAAGGCATACACCA | 4298 to 4321 | +ve | 3842 |
| RRVBrev | TGTGAAACCTCCCAAGAGACATTCCAG | 8113 to 8139 | -ve | ||
| C | RRVCfor | TCTCTAACGACCAAATAGTCAACCTCGCTA | 7852 to 7881 | +ve | 4080 |
| RVrev2 | gcggccgcACGCTTAACAAATAAAC | 11907 to 11923 | -ve | ||
| A1 | RVfor3 | as above | 2230 | ||
| PR2a | TGACTATGTCATCAAGGTTCATCT | 2228–2205 | -ve | ||
| A2 | PF2a | CGATTTGTCTGTGGAGGCAGA | 2092–2112 | +ve | 2304 |
| RRVArev | as above | ||||
| B1 | RRVBfor | as above | 2390 | ||
| LR3 | GTGGATCTAGATACCATTTGGAGTA | 6687–6663 | -ve | ||
| B2 | LF3 | GGATCAATTCGACAAYATACA | 6464–6484 | +ve | 1676 |
| RRVBrev | as above | ||||
| C1 | RRVCfor | as above | 2485 | ||
| LR8 | GCAACCTGTTGGGCAGAGCA | 10336–10317 | -ve | ||
| C2 | LF8a | ACCACAATGAAAGAGGGCAACAGATC | 9885–9910 | +ve | 2047 |
| RVrev2 | as above |
refers to bases in uppercase based on co-ordinates of the reference RRV genome, GenBank accession number EU311738
includes the bases in lowercase which were included in the terminal primers to increase their annealing temperature
Two distinct amplification protocols were evaluated. Based on previous experience (Nadin-Davis, 1998) initial work was performed using the reverse transcription (RT) protocol RT1: 1 μg RNA was mixed with 30 pmol of each of the two amplification primers (see Table 1 where primer pairs are listed for amplicons A, B or C) in a 12 μl reaction and incubated at 70 C, 5 min, 37/45 C for 10 min. The hybridization was brought to 20 μl with the addition of 4 μl 5x RT buffer, 1 μl 100 mM DTT, 1 μl 10mM dNTPs, 1 μl RNase OUT and 1 μl Superscript III. After 2 hr at 50 C the RT was terminated by heating at 70 C for 15 min. and chilled on ice. Ten μl of the RT reaction was added to 40 μl of PCR mix that included 5X HiFi PCR buffer (provided with enzyme), 1.0 μl 10 mM dNTPs and 1.0 μl KAPA HiFi™ DNA polymerase (Kapa Biosystems). Cycling conditions were 95 C, 4 min; 40 cycles of: 98 C, 20 sec; 65 C, 15 sec; 72 C, 5 min followed by 72 C, 5 min and a 4 C hold. An alternative protocol employing random primers for the reverse transcription was also explored so as to reduce the required number of reactions. The RT2 protocol was similar to RT1 except that 30 pmol of random octamers were substituted for the sequence specific primers in the initial incubation and the PCR primers (15 pmol of each) were included in the PCR master mix prior to cycling.
When the expected amplicon was either faint or not visible upon gel electrophoresis, two hemi-nested PCRs were performed using the first round product. Reactions were performed as for first round PCRs except that slightly modified cycling conditions were employed. When using first round PCRs with no visible product the cycling profile was 95 C, 3 min; 35 cycles of: 98 C, 20 sec; 60 C, 30 sec; 72 C, 2 min followed by 72 C, 5 min and a 4 C hold. When first round amplicons produced faint bands the number of PCR cycles was reduced to 20–25 cycles. Samples that failed to generate hemi-nested products were abandoned. PCR products were purified using either a Wizard DNA clean up kit (Promega) or a Gene Jet kit (ThermoFisher) and stored at −20 C.
2.4 Nucleotide sequencing
Initially, standard Sanger sequencing was performed on individual purified amplicons using internal sequencing primers and a BDTv3.1 cycle sequencing kit (Applied Biosystems) followed by purification with a BD XTerminator kit (Applied Biosystems) and electrophoresis with a 3500xl Genetic Analyser (Applied Biosystems). Complete coverage of the genome in both directions for each sample required use of 36 sequencing primers (see Table S2). Reads were assembled using either Variant Reporter software (ABI) or the SeqMan Pro programme of the DNASTAR Lasergene software package (v. 11).
Alternatively high throughput parallel sequencing was adopted for processing of many samples during the course of these studies. The amplicons generated from a single sample (total of 3–6) were quantified individually using a Qubit instrument (ThermoFisher) and then pooled in equimolar amounts. Pooled amplicons representing either 24 or 96 samples were used to prepare indexed libraries using Nextera XT and index kits as directed (Illumina) and 200 or 250 base paired end reads were generated on a MiSeq instrument. Reference-based assembly of the paired fastq files was achieved using the NGen programme of the DNASTAR Lasergene software (v. 11) using either a reference RRV genome (GenBank accession EU311738) or more closely related sequences obtained as the study progressed. Due to amplicon overlap all internal sequence could be unequivocally determined; however the terminal 5′ and 3′ sequences corresponding to the terminal PCR primer targets were manually adjusted to conform to the primer sequences as these regions could not be verified by this approach. The inability to determine the sequences of the genome termini was not a major concern as it is known that, even between rabies virus strains, variation at the termini is very limited (Delmas et al., 2008) and unlikely to impact phylogenetic analysis. Complete consensus sequences were exported as individual fasta files.
2.5 Phylogenetic analysis
Phylogenetic analysis was performed using WGS data and single gene sequences extracted from the WGS information for 62 RRV isolates, details of which are provided in the Supplementary Table S1. A publicly available reference sequence of the RABV Pasteur Virus (PV) strain (NCBI accession number NC_001542) (Tordo et al., 1986, 1988) was also included as an outgroup. Sequences were aligned using MUSCLE (Edgar, 2004) and the GTR+I model was identified as the best fitting nucleotide substitution model for these sequences using jModelTest v2.1.7 (Darriba et al., 2012). Maximum Likelihood (ML) phylogenies were generated under this model in PhyML (Guindon and Gascuel, 2003) for the WGS alignment, and for the complete genes encoding the N, P and G proteins. Node support was evaluated for each phylogeny based on 1000 bootstrap replicates. All trees were imported into R v3.3.1 and the ggtree package (Yu et al., 2016) was used for annotation and generation of quality graphical illustrations. Bayesian molecular clock analysis was also conducted for the WGS alignment using BEAST v1.8.2 (Drummond et al., 2012) under a relaxed molecular clock model with branch rates drawn from a log normal distribution (Drummond et al., 2006). The clock prior was specified as a normal distribution with mean of 1.44 × 10−4 nucleotide substitutions per site based on the results of Brunker et al. (2015), with a wide standard deviation of 0.0144 nucleotide substitutions per site, and truncated to range between 0 and 0.15. The Bayesian skyline model (Drummond et al., 2005) was used as a flexible demographic prior, and two independent MCMC chains were run for 107 iterations, with the first 10% removed as burn-in. Convergence between runs was confirmed using Tracer.
2.6 Generation of map
The township of sample origin was used to determine latitude and longitude co-ordinates for mapping using a stamen map in the ggmap package in R, v. 3.0, (Kahle and Wicklam, 2013; R_Core_Team, 2016)..
3. Results and Discussion
3.1 Complete RRV genome amplification by RT-PCR
The RRV WGS workflow described in this report incorporated a number of technical modifications to previously published amplification methods (Nadin-Davis, 1998). Preliminary studies indicated that RT-PCR products up to 4 Kb in size could be produced fairly consistently provided the RNA template was very pure and a high fidelity DNA polymerase was employed. Accordingly, an amplification scheme in which the RRV genome is copied as three overlapping amplicons (A, B and C) as illustrated in Fig. 1a was designed; details of the primers employed as well as additional primers for hemi-nested PCR of each of these three fragments are provided (Table 1). While extracts generated using the standard TRIzol reagent protocol were sometimes sufficient to support such amplification, use of the hybrid TRIzol/MagMax system consistently generated RNA preparations suitable for efficient RT-PCR with infrequent need for hemi-nested PCR. While other means of RNA purification were not explored alternative protocols may yield extracts appropriate for this purpose.
Amplicons A and B were amplified well using either sequence-specific primers (RT1 protocol) or random primers (RT2 protocol); accordingly a single cDNA generated by the RT2 protocol was employed for generation of these PCRs so as to minimise the number and hence cost of reverse transcription reactions. However, this approach performed poorly for generation of amplicon C, probably because of the shorter sequence match of primer RVrev2 to the template RNA that precluded efficient binding to the target at an annealing temperature appropriate for PCR, so this amplicon was generated by the RT1 protocol. Typical RT-PCR products generated by this protocol are illustrated in Fig. 1b. Hemi-nested PCR was required to generate products in some instances; this was particularly true for amplicon A perhaps because as the largest of the three fragments the length of this amplicon was at the size limit for routine amplification.
3.2 Sequence platform evaluation
While Sanger sequencing is a feasible option for small numbers of samples, much higher throughput with less technical effort was achieved through use of Illumina technology. Two initial trials employing amplicons from 24 samples per sequencing run were followed by runs comprised of amplicons from 96 samples. Each sample in the 24 sample libraries generated between 250,000 to 1.6 million sequence reads of which 90–96% assembled into a single contig representing the complete viral genome. For samples in the 96 sample libraries sequence reads generally ranged between 60,000 to 800,000 with a very similar proportion incorporated into the genome assembly. Occasional sequencing failures for individual samples were observed for the 96 sample libraries (range of zero to 2 samples per library); repeats of these failed samples using the same amplicons in subsequent libraries or by Sanger sequencing were usually successful, suggesting that the initial failures were due to technical error. Sequence coverage along the genome was often somewhat uneven but was virtually always > 200 except for the terminal one to five bases at both ends of the genome which often had a coverage of <10 but which, in any event, were defined by amplification primers. Average sequence coverage across the entire genome was usually > 1,000x but ranged as high as 25,000x and 15,000x for samples in the 24 and 96 sample libraries respectively; Consequently, assemblies consistently generated consensus sequences having quality values of 34–36 along the entire length of the genome.
In both Sanger and Illumina methods, some samples yielded a few ambiguous base calls (range 0–3 per genome). Manual review of Sanger sequencing trace profiles usually identified a predominant base which was assigned to the position but in a few cases where the traces were virtually superimposed (<60% of reads were a majority) the base call could not be resolved and remained ambiguous (usually Y or R). The default consensus base call threshold employed by the DNASTAR software for the Illumina data was 75%. As a result, the consensus sequences generated from Illumina data contain a slightly higher proportion of ambiguous bases which may better reflect the true genetic heterogeneity of these samples.
To establish whether there were any significant differences between the consensus sequences being generated by Sanger and Illumina protocols, amplicons from six isolates representative of the viral populations under study were sequenced using both methods. Alignments of the two consensus sequences for the six genomes (representing a total of 71,543 bases analyzed) identified just three positions for which consistent unequivocal base reads were not obtained (see data for group 1, Table 2). Notably in all three cases there was an ambiguous base call in at least one of the sequence runs. Thus in no case was there a clear sequence disagreement between the two methods, thereby substantiating the interchangeable nature of the sequence data generated by both platforms. For these six samples two indels were consistently identified by both methods: an additional A was present in ON.2005.4941 at position 5355 (which added an extra A to the polyadenylation signal of the G gene) while a T was deleted from both Vermont samples at position 11899 just before the cDNA 3′ terminus. These two indels determine the observed variation in genome length (11923–11925 nucleotides). Overall pairwise distance values for this set of six isolates ranged from 0.0034 to 0.0141.
Table 2.
Evaluation of the effects of sequencing platform and RT-PCR on whole genome sequences
| Sample designation | Total length (nucleotides) | Number and nature of non-conserved bases | Position in sequence alignment | Sequencing platform used: on PCR1 | Sequencing platform used: on PCR2 |
|---|---|---|---|---|---|
| Group 1 | |||||
| ME.2013.0131 | 11924 | 0 | Both1 | - | |
| NB.2014.0941 | 11924 | 0 | Both1 | - | |
| NY.2004.2473 | 11924 | 0 | Both1 | - | |
| ON.2005.4941 | 11925 | 1, R by both reads | 6880 | Both2 | - |
| VT.2006.0225 | 11923 | 1, R by Illumina, G by Sanger | 4302 | Both2 | - |
| VT.2011.0122 | 11923 | 1, G by Illumina, K by Sanger | 3780 | Both1 | - |
| Group 2 | |||||
| ON.2003.1519 | 11925 | 0 | Illumina2 | Sanger1 | |
| ON.2003.2760 | 11925 | 0 | Illumina1 | Sanger1 | |
| VT.2006.0259 | 11924 | 0 | Sanger1 | Sanger4 | |
| VT.2007.0522 | 11925 | 1, G (PCR1), R (PCR2) | 4308 | Illumina3 | Sanger4 |
| VT.2008.0209 | 11925 | 0 | Sanger1 | Sanger4 | |
| VT.2008.0237 | 11923 | 1, R (PCR1), G (PCR2) | 4894 | Sanger1 | Sanger4 |
| VT.2011.0122 | 11923 | 0 | Illumina1 | Sanger4 | |
amplified as three fragments A, B, C
amplified as four fragments A1, A2, B, C
amplified as four fragments A, B C1 and C2
amplified as six fragments A1, A2, B1, B2, C1, C2
The potential for incorporation of experimentally-induced errors had been a significant concern given the extensive amplification that samples undergo before and during DNA library construction and this was further examined by sequencing seven isolates in duplicate using independently generated PCR products. This analysis included five samples from Vermont since it was found that these samples, which were less well preserved due to delays during inter-laboratory shipment, more often required nested PCR to generate sufficient products for sequencing compared to samples from other jurisdictions. Comparison of the duplicate sequences for these seven genomes identified two inconsistent base calls, one each in samples VT.2007.0522 and VT.2008.0237, both involving G/A mutations (Group 2, Table 2). In each case one set of Sanger sequence reads was ambiguous. It is unclear if these differences represent poor quality sequencing by the Sanger method at these positions or if indeed the samples exhibit genetic heterogeneity that was differentially amplified during independent PCRs. If it is assumed that these differences were experimentally induced then a PCR error rate of 2 mutations over 83,470 bases sequenced (= 2.39 × 10−5 mutations per site) was estimated; however given the uncertainty in interpreting these data this value may be an overestimate.
The KAPA HiFi™ enzyme used for PCR has a reported error rate of 1 in 2.8 × 107 nucleotides incorporated (KAPA HiFi™ Technical data sheet version 4.1, Kapa Biosystems) which is equivalent to 3.57 × 10−8 errors per site. The estimated error rate observed in these studies (2.39 × 10−5 mutations per site) was much higher, perhaps in part due to the effects of the reverse transcription by Superscript III as well as the amplification during Nextera XT library generation. However this is significantly lower than the estimated viral mutation rate (2.7 × 10−4 per site), confirming that method-induced errors will have minimal impact on the accuracy of these sequences. Notably one sample, VT.2011.0122, included in both groups in Table 2, was sequenced three times and only one ambiguous base call was noted in one sequence run.
Since the consensus sequences generated by Sanger sequencing and Illumina HTS were comparable (Table 2) data from both methods has been used interchangeably in subsequent phylogenetic analyses. It should be noted however, that we estimate that switching from Sanger sequencing to processing of 96 samples by Illumina technology reduced the technical effort required for the sequencing portion of the workflow by 6–8 fold and this was pivotal to our ability to process large numbers of viral isolates. Moreover as the number of samples that can be pooled in a single MiSeq run is dependent only on the number of indices available for discrimination of each isolate, the availability of additional indices for future studies could further streamline this process as the sequence coverage obtained with 96 sample libraries was far more than adequate to generate consensus sequences.
3.3 Phylogenetic analysis of the RRV outbreak in Ontario 1999–2005
The WGS methodology described here has been applied to a re-evaluation of an outbreak of RRV in Ontario that began in 1999 and resulted in a total of 132 reported cases before the outbreak was eliminated in 2005 (Rosatte et al., 2009; Wandeler and Salsberg, 1999). In this study attempts were made to amplify the RRV genome from 59 samples representative of the temporal and spatial characteristics of the outbreak. Fifty-six samples were successfully amplified (overall sample success rate of 94.8%) and sequenced. Of the three samples that were ultimately abandoned, one failed to amplify any products. This translates to a rate of 1.7% (one in 59) of samples that failed amplification for all three amplicons; a similar failure rate of 1.5% (7 of 462 samples) was observed in a parallel study of RRV in New York state (data not shown). We suspect that failure was due either to very limited amounts of tissue, extremely low levels of viral infection or poor sample condition rather than primer bias, given the successful amplification of RRV samples from several jurisdictions. These same factors can preclude amplification of shorter single gene fragments although failure rates are likely lower for shorter targets. Of the other two samples that were eventually abandoned one gave only weak hemi-nested PCR products for the A fragment and another failed to generate a C1 product. This yields an overall amplicon failure rate of 2.8% (five failures out of 177 attempts).
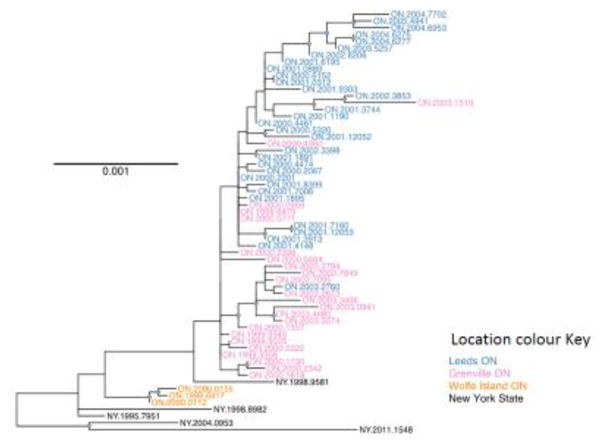
The 56 samples that were successfully amplified were sequenced either by Sanger or Illumina technologies. These WGS data, together with the sequence of one additional Ontario RRV sample characterised previously (Szanto et al., 2008) (GenBank Accession EU311738), were compared to five samples from areas of New York state that border the province. A phylogeny based on these 62 sequences is presented in Fig. 2 and the locations of origin of all samples are illustrated in Fig. 3.
Fig. 2.
A ML tree of 62 RRV genomes (57 from Ontario and five from New York) using the PV reference strain (NC_001542) as outgroup (not shown). Nodes with greater than 75% bootstrap support are indicated by grey diamonds. Scale bar gives the branch length in nucleotide substitutions per site.
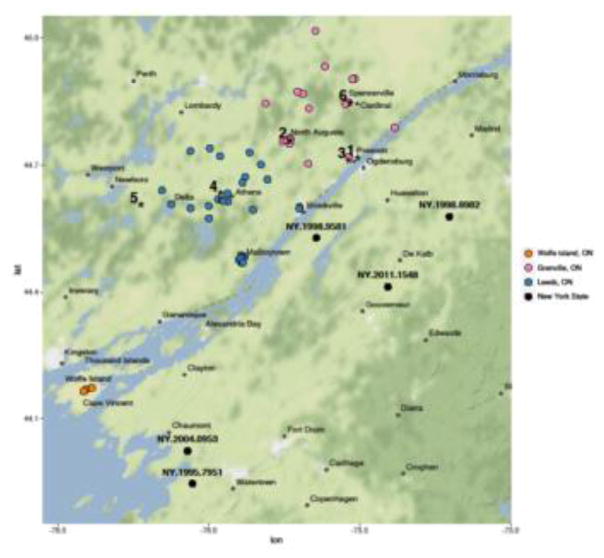
Fig. 3.
A map showing the study area of eastern Ontario and northern New York state. Locations from which sequenced isolates were recovered are shown using the colour coding as detailed in the key except for specimens of special note which are indicated by stars and identified numerically as follows: 1, ON.1999.3306 (index case); 2, ON.1999.3545, (2nd case); 3, ON.2003.1519 (outlier); 4, ON.2003.2760 (outlier); 5, ON.2002.3853 (outlier related); 6, ON.2003.2673, (outlier related).
The ML tree generated using WGS data clearly identified two distinct clades of viruses within the Ontario samples. Three isolates representative of a group of six recovered from Wolfe Island in Lake Ontario southwest of the main study area between December 1999 and January 2000 (Rosatte et al., 2007), formed a separate branch. The other 54 samples originating from the Ontario mainland appeared to form a large monophyletic clade, with the sample NY.1998.9581 identified as an outgroup. The other four NY samples recovered from the border area were more distantly related to the virus responsible for this outbreak. The phylogeny appears to respect the spatial distribution of the samples to a large degree, with most cases from the two counties affected by the outbreak appearing to segregate to separate groups.
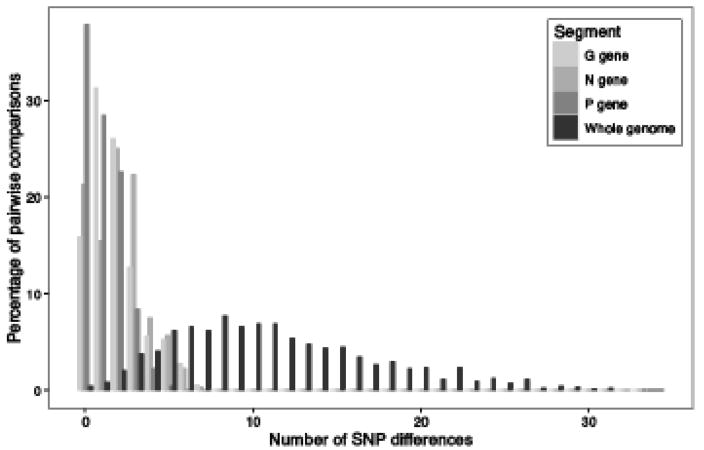
To compare the resolution of individual isolates afforded by WGS compared to single gene sequencing, plots of the proportion of pairwise sequence comparisons as a function of number of SNP differences for the 54 Ontario samples comprising the mainland outbreak are shown (Fig. 4). Based on single gene sequences a high proportion of the sequence pairs have either no SNP differences, and are thus indistinguishable, or differ by just one to three SNPs with a maximum number of SNP differences per gene of seven. In contrast, based on WGS data the vast majority of sequence pairs exhibited at least one SNP difference (range of 0 to 34), thereby providing much enhanced differentiation of individual samples.
Fig. 4.
Histogram showing the proportion of pairwise sequence comparisons from the mainland Ontario RRV outbreak that differ in their number of SNPs. Comparisons were carried out for the G gene, the N gene, the P gene, and for whole genome sequences.
To further illustrate the advantage of WGS for following the dynamics of the outbreak, phylogenetic trees generated from WGS data (Fig 2) were compared with trees generated using datasets corresponding to single gene (N, P and G) sequences (Supplemental Fig S1). While 16 internal nodes of the tree produced using the WGS dataset had bootstrap values > 75%, the trees generated using just N, P and G genes contained only two, zero and one node respectively that reached this value. These analyses clearly illustrate the improved phylogenetic resolution achieved with the WGS dataset. Indeed a prior study of 127 isolates from the Ontario RRV outbreak, in which a highly variable 550 bp portion of the P gene had been targeted (Nadin-Davis et al., 2002; Nadin-Davis et al., 2006), identified a number of SNPs, many of which were confirmed in this smaller-scale study, but most of the isolates were identical to the index case. The limited genetic variation observed for that large collection of RRV isolates was insufficient to support the performance of any meaningful phylogenetic analysis as confirmed by the tree generated from whole P gene sequence in this study.
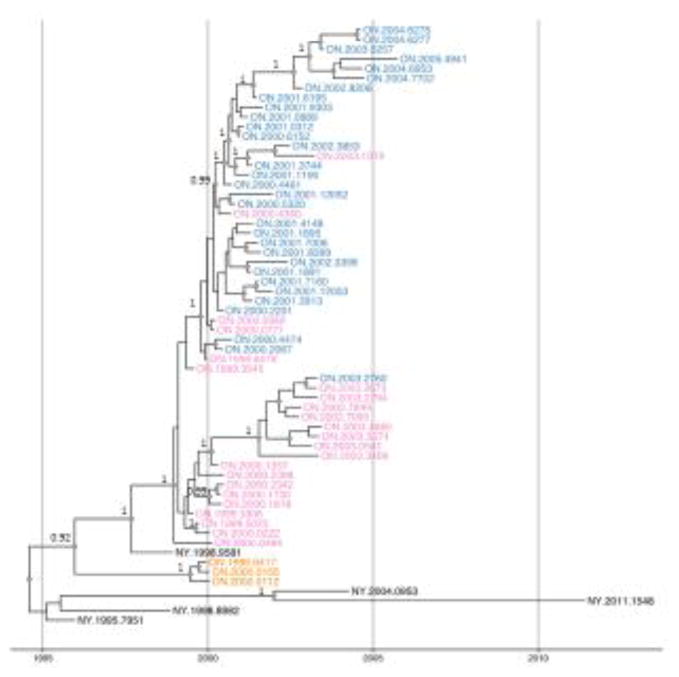
A time-calibrated phylogeny (maximum clade credibility tree) generated from the WGS dataset (Fig. 5) had a very similar topology to that generated by ML analysis (Fig. 2) and identified separate clades for the Wolfe Island and mainland isolates. Furthermore this tree more robustly supports the independent emergence of two distinct clades in the two counties of the main Ontario outbreak area. An alignment of all mainland Ontario isolate sequences identified two SNPs that were closely correlated with these two viral groups. At alignment position 131, viruses in the Grenville county (GC) clade all encoded A while the Leeds county (LC) viruses encoded a G; at position 287 the GC clade encoded T and the LC clade encoded C. The only exception was ON.1999.3545 which is assigned to clade LC but which in fact is essentially indistinguishable from the index case and one isolate in the LC clade (ON.2001.3744) that exhibited a reversion to A131.
Fig. 5.
Time-scaled BEAST phylogeny of RRV whole genome sequences from the Ontario outbreak between 1999 and 2005, and from New York State. Tip labels are colored by location using the key shown in Fig. 2. All nodes with posterior support values > 0.9 are indicated by grey diamonds and the actual posterior support values for several of these major clades are indicated in black to the left of the node.
Using the estimated molecular clock rate of 2.7×10−4 nucleotide substitutions per site for these sequences (95% Highest Posterior Density (HPD): 2.1×10−4 – 3.4×10−4), the time of the most recent common ancestor (tMRCA) of the mainland Ontario outbreak (blue and pink clades, Fig. 5) was December 1998 (95% HPD: April 1998 – April 1999), a date consistent with the first reported case in July 1999. The tMRCA for the Wolfe Island outbreak (orange clade, Fig. 5) was May 1999 (95% HPD: September 1998–October 1999). By contrast, the tMRCA of the most recent ancestor of both Ontario outbreaks was estimated to be November 1995 (95% HPD: March 1994–March 1997). This latter date is consistent with the independence of these two incursions given the unlikely scenario that RRV was circulating in the province several years before detection. The independence of the Wolfe island and mainland outbreaks had been suggested previously based on the geographic separation of the two events and a SNP difference in the partial P gene sequences between the mainland index case and the Wolfe Island cases (Nadin-Davis et al., 2006) and the current study substantiates this conclusion.
All of the Ontario samples recovered from the mainland emerged from a single progenitor which was shared with a sample from New York, NY.1998.9581, recovered from the northwestern part of the state close to the St. Lawrence seaway that forms the international border with Canada and in proximity to where the Ontario index case was recovered. The first RRV case in mainland Ontario (ON.1999.3306) was reported on July 14, 1999, in the small community of Prescott, across from Ogdensburg, NY, on the St. Lawrence seaway (Wandeler and Salsberg, 1999). Less than two weeks later a second case (ON.1999.3545) was reported in the community of North Augusta approximately 20 km northwest of the index case. These two isolates were virtually identical in sequence but appear to have resulted in the emergence of two distinct groups of viruses that tended to be restricted to distinct geographical areas. While the outbreak began in Grenville county (colour-coded as pink, Figs. 3,5) one viral branch (GC) spread northwards and circulated almost exclusively in that county while the other branch (LC) which initially straddled the border of the two counties spread westwards and southwards to invade Leeds county (colour-coded as blue, Figs. 3,5).
These data address some long-standing questions about whether later RRV cases (2002–2005) were in fact a continuation of the Ontario outbreak or a result of new incursions from New York given the proximity of communities such as Mallorytown to the USA. Previous partial genetic characterization of these viruses was insufficiently sensitive to discriminate between these two alternatives (Nadin-Davis et al., 2006). The current analysis clearly shows that the cases in clade GC from 2002 and 2003 share a progenitor with ON.2000.1357 and are a continuation of this outbreak in Grenville county. Similarly, the group of samples in the LC clade collected between 2003 and 2005 from Mallorytown have a common lineage with isolate ON.2002.8206, also collected from this community, and thus represent continued circulation of this variant. This latter information suggests that rabies cases had eluded the extensive provincial control efforts that were ongoing from 1999 until 2005 when the last case of this incursion was recorded (Rosatte et al., 2009). These types of insight regarding patterns of continued disease persistence and differentiation of programme failure vs re-introductions could be highly informative to control efforts especially if this information could be provided in real-time.
While the segregation of the two viral clades between the two Ontario counties holds well for the most part, with some overlap of variants at the county borders, there are two samples which appear to be out of place: ON.2003.1519 (from Prescott) and ON.2003.2760 (from Athens) (see Fig. 3). The variants found in these submissions are more closely related to viruses circulating in different parts of the study area. In particular, the sample from Athens (#4, Fig. 3) clusters with another 2003 sample from the town of Spencerville (#6, Fig. 3) which is at the easternmost edge of the mainland outbreak area and a considerable distance (about 40 kms) from Athens. Considering that these two RRV samples are very closely related and raccoons in Ontario tend to range over an area not exceeding 4.5 kms (Rosatte et al., 2006), the location of the Athens sample is unexpected. Similar arguments can be applied to the sample from Prescott (#3, Fig. 3) although that sample was genetically divergent from its most closely related isolate (ON.2002.3853, #5, Fig. 3) collected nine months earlier. Samples ON.2003.1519 and ON.2003.2760 were submitted to repeat testing from original infected brain tissue to ascertain if there could have been a mix-up during specimen processing; however, sequencing of the re-extracted RNA generated results identical to those produced initially (Group 2, Table 2). While it remains possible that the tissues from one or both of these two submissions has been misidentified for long term storage, other possible explanations for these results were considered. Either these represent infected animals which undertook unusually long-range dispersion or there was human-mediated transfer of diseased animals during the course of the outbreak. The role of humans in inadvertently mediating the expansion of the raccoon rabies epizootic from Florida to other US states through inter-state transportation of diseased animals has been frequently cited (Jenkins et al., 1988). Moreover, long distance spread of raccoon rabies has been inferred from surveillance data within the state of Connecticut previously (Smith et al., 2005). Unfortunately, given the retrospective nature of this study, we are unable to resolve whether the spatial placement of these two samples is accurate or not but this observation does demonstrate the potential insights into the mechanisms of viral spread that WGS analysis can provide.
3.4 Conclusions and future studies
While single gene sequencing still has an important role to play in RABV strain identification for routine viral typing purposes, WGS clearly has vastly superior capability to reveal the detailed phylogeny of a selected viral strain. Obtaining accurate data regarding wildlife disease spread from passive surveillance is known to be problematic since many cases go unrecognised and submissions are biased towards wildlife with human contact. Given these limitations inherent in wildlife disease surveillance, viral phylogenetic analysis provides the best available means of exploring the spatio-temporal characteristics of disease spread. Moreover, while in this report analysis of the WGS data has focused on use of consensus sequences to explore the detailed molecular epidemiology of RRV, the availability of deep sequencing data for many of these samples would provide an opportunity to explore the role of rare mutations in the evolution of the virus as it spreads across the landscape as reported by a study examining a host shift of skunk–associated rabies viruses to foxes in California (Borucki et al., 2013).
The WGS methodology described in this report has enabled a reassessment of the incursion of RRV into Ontario between 1999 and 2005. In particular the ability of WGS phylogenies to discriminate between isolates that are very closely related both genetically and geographically distinguishes this approach from those that employ only single gene targets as clearly supported by the data in Figs. 2, 4 and 5. These analyses unequivocally demonstrate that the mainland outbreak was due to a single incursion from New York state and that, contrary to the belief at the time, cases of RRV continued to spread within Ontario during 2002 to 2004 despite the extensive control activities undertaken to eradicate the outbreak. Such information could not be deduced with more limited viral characterisation (Nadin-Davis et al., 2006). The availability of such a genetic tool to investigate future outbreaks of RRV will be an invaluable asset for informing better wildlife rabies control strategies. Further studies on RRV in additional jurisdictions will expand the knowledge developed in this study. Furthermore, with minimal changes in primer design this same methodology is being applied to other RABV strains of interest, including Canadian isolates of the arctic strain (Hanke et al., 2016).
Supplementary Material
Highlights.
A protocol for high throughput whole genome sequencing (WGS) of raccoon rabies virus
WGS vastly improves phylogenetic resolution of virus isolates
New insights from a re-examination of the spread of raccoon rabies in Ontario
Methodology has broad application to many rabies virus variants
Acknowledgments
We are most grateful to the following for provision of raccoon rabies virus infected samples together with related metadata in support of the molecular epidemiological analysis presented in this work: Laura Kamhi, Rabies Unit, Vermont Public Health Laboratory, Burlington, Vermont; Robert Rudd, Rabies Laboratory, New York State Department of Health, Albany, New York. The contributions of staff of the Ontario Ministry of Natural Resources and Forestry in providing survey samples and metadata in support of these studies is gratefully acknowledged. We also thank Mary Sheen, Sarah Kamm and Qigao Fu for excellent technical assistance and staff of the diagnostic unit of the CFIA’s Rabies Centre of Expertise at Ottawa Laboratory Fallowfield for logistical support. This research was supported by NIH grant RO1 AI047498 to L.A. Real.
Abbreviations
- RABV
rabies virus
- RRV
raccoon strain rabies virus
- WGS
whole genome sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Biek R, Henderson CI, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceedings of the National Academy of Sciences USA. 2007;104:7993–7998. doi: 10.1073/pnas.0700741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki MK, Chen-Harris H, Lao V, Vanier G, Wadford DA, Messenger S, Allen JE. Ultra-deep sequencing of intra-host rabies virus populations during cross-species transmission. PLoS Neglected Tropical Diseases. 2013;7(11):e2555. doi: 10.1371/journal.pntd.0002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H, Reynes JM, Dunham EJ, Dacheux L, Larrous F, Thi Que Huong V, Xu G, Yan J, Miranda MEG, Holmes EC. The origin and phylogeography of dog rabies virus. Journal of General Virology. 2008;89:2673–2681. doi: 10.1099/vir.0.2008/003913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker K, Marston DA, Horton DL, Cleaveland S, Fooks AR, Kazwala R, Ngeleja C, Lembo T, Sambo M, Mtema ZJ, Sikana L, Wilkie G, Biek R, Hampson K. Elucidating the phylodynamics of endemic rabies virus in eastern Africa using whole-genome sequencing. Virus Evolution. 2015;1(1):1–11. doi: 10.1093/ve/vev011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModeltest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas O, Holmes EC, Talbi C, Larrous F, Dacheux L, Bouchier C, Bourhy H. Genomic diversity and evolution of the Lyssaviruses. PLoS ONE. 2008;3(4):e2057. doi: 10.1371/journal.pone.0002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLos Biology. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular and Biological Evolution. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geue L, Schares S, Schnick C, Kliemt J, Beckert A, Freuling C, Conraths FJ, Hoffmann B, Zanoni R, Marston D, McElhinney L, Johnson N, Fooks AR, Tordo N, Mueller T. Genetic characterisation of attenuated SAD rabies virus strains used for oral vaccintion of wildlife. Vaccine. 2008;26:3227–3235. doi: 10.1016/j.vaccine.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hanke D, Freuling CM, Fischer S, Hueffer K, Hundertmark K, Nadin-Davis S, Marston D, Fooks AR, Botner A, Mettenleiter TC, Beer M, Rasmussen TB, Mueller TF, Hoper D. Saptio-temporal analysis of the genetic diversity of Arctic rabies viruses and their reservoir hosts in Greenland. PLoS Neglected Tropical Diseases. 2016;10(7):e0004779. doi: 10.1371/journal.pntd.0004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins SR, Perry BD, Winkler WG. Ecology and epidemiology of raccoon rabies. Reviews of Infectious Diseases. 1988;10(Supplement 4):S620–S625. doi: 10.1093/clinids/10.supplement_4.s620. [DOI] [PubMed] [Google Scholar]
- Kahle D, Wicklam H. ggmap : Spatial visualization with ggplot2. R J. 2013;5:144–161. [Google Scholar]
- Kuzmin IV, Hughes GJ, Botvinkin AD, Orciari LA, Rupprecht CE. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Research. 2005;111:28–43. doi: 10.1016/j.virusres.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Kuzmin IV, Shi M, Orciari LA, Yager PA, Velasco-Villa A, Kuzmina NA, Streicker DG, Bergman DL, Rupprecht CE. Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarniovores in Arizona during 2001–2009. PLOS Pathogens. 2012;8(6):e1002786. doi: 10.1371/journal.ppat.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DA, McElhinney LM, Ellis RJ, Horton DL, Wise EL, Leech SL, David D, de Lamballerie X, Fooks AR. Next generation sequencing of viral RNA genomes. BMC Genomics. 2013;14:444. doi: 10.1186/1471-2164-14-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DA, Vazquez-Moron S, Ellis RJ, Wise EL, McElhinney LM, De Lamballerie X, Fooks AR, Echevarria JE. Complete genomic sequence of European bat lyssavirus 1, isolated from Eptesicus isabellinus in Spain. Genome Announcements. 2015a;3(1):e01518–01514. doi: 10.1128/genomeA.01518-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DA, Wise EL, Ellis RJ, McElhinney LM, Banyard AC, Johnson N, Deressa A, Regassa F, de Lamballerie X, Fooks AR, Sillero-Zubiri C. Complete genomic sequence of rabies virus from an Ethiopian wolf. Genome Announcements. 2015b;3(2):e00157–00115. doi: 10.1128/genomeA.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadin-Davis SA. Polymerase chain reaction protocols for rabies virus discrimination. Virol Methods. 1998;75:1–8. doi: 10.1016/s0166-0934(98)00106-2. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA. Molecular epidemiology. In: Jackson AC, editor. Rabies: Scientific Basis of the Disease and its Management. 3. Academic Press; Oxford, UK: 2013. pp. 123–177. [Google Scholar]
- Nadin-Davis SA, Abdel-Malik M, Armstrong J, Wandeler AI. Lyssavirus P gene characterisation provides insights into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology. 2002;298:286–305. doi: 10.1006/viro.2002.1492. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Muldoon F, Wandeler AI. A molecular epidemiological analysis of the incursion of the raccoon strain of rabies virus into Canada. Epidemiology and Infection. 2006;134:534–547. doi: 10.1017/S0950268805005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next-generation sequencing technologies in virology. Journal of General Virology. 2012;93:1853–1868. doi: 10.1099/vir.0.043182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees EE, Belanger D, Lelievre F, Cote N, Lambert L. Targeted surveillance of raccoon rabies in Quebec, Canada. Journal of Wildlife Management. 2011;75(6):1406–1416. [Google Scholar]
- Rosatte RC, Donovan D, Allan M, Bruce L, Buchanan T, Sobey K, Stevenson B, Gibson M, MacDonald T, Whalen M, Davies JC, Muldoon F, Wandeler A. The control of raccoon rabies in Ontario, Canada: proactive and reactive tactics, 1994–2007. Journal of Wildlife Diseases. 2009;45:772–784. doi: 10.7589/0090-3558-45.3.772. [DOI] [PubMed] [Google Scholar]
- Rosatte R, MacDonald E, Sobey K, Donovan D, Bruce L, Allan M, Silver A, Bennett K, Brown L, MacDonald K, Gibson M, Buchanan T, Stevenson B, Davies C, Wandeler A, Muldoon F. The elimination of raccoon rabies from Wolfe Island, Ontario: Animal density and movements. Journal of Wildlife Diseases. 2007;43(2):242–250. doi: 10.7589/0090-3558-43.2.242. [DOI] [PubMed] [Google Scholar]
- Rosatte RC, Sobey K, Donovan D, Bruce L, Allan M, Silver A, Bennett K, Gibson M, Simpson H, Davies C, Wandeler A, Muldoon F. Behavior, movements, and demographics of rabid raccoons in Ontario, Canada: Management implications. Journal of Wildlife Diseases. 2006;42(3):589–605. doi: 10.7589/0090-3558-42.3.589. [DOI] [PubMed] [Google Scholar]
- Slate D, Rupprecht CE, Rooney JA, Donovan D, Lein DH, Chipman RB. Status of oral rabies vaccination in wild carnivores in the United States. Virus Research. 2005;111:68–76. doi: 10.1016/j.virusres.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Smith DL, Waller LA, Russell CA, Childs JE, Real LA. Assessing the role of long-distance translocation and spatial heterogeneity in the raccoon rabies epidemic in Connecticut. Preventive Veterinary Medicine. 2005;71:225–240. doi: 10.1016/j.prevetmed.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Orciari LA, Yager PA, Seidel HD, Warner CK. Epidemiologic and historical relationships among 87 rabies virus isolates as determined by limited sequence analysis. The Journal of Infectious Diseases. 1992;166:296–307. doi: 10.1093/infdis/166.2.296. [DOI] [PubMed] [Google Scholar]
- Sterner RT, Meltzer MI, Shwiff SA, Slate D. Tactics and economics of wildlife oral rabies vaccination, Canada and the United States. Emerging Infectious Diseases. 2009;15:1176–1184. doi: 10.3201/eid1508.081061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto AG, Nadin-Davis SA, Rosatte RC, White BN. Genetic tracking of the raccoon variant of rabies virus in eastern North America. pidemics. 2011;3:76–87. doi: 10.1016/j.epidem.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Szanto AG, Nadin-Davis SA, White BN. Complete genome sequence of a raccoon rabies virus isolate. Virus Research. 2008;136:130–139. doi: 10.1016/j.virusres.2008.04.029. [DOI] [PubMed] [Google Scholar]
- Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proceedings of the National Academy of Sciences USA. 1986;83:3914–3918. doi: 10.1073/pnas.83.11.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Completion of the rabies virus genome sequence determination: Highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology. 1988;165:565–576. doi: 10.1016/0042-6822(88)90600-9. [DOI] [PubMed] [Google Scholar]
- Wandeler AI, Rosatte RC, Williams D, Lee TK, Gensheimer KF, Montero JT, Trimarchi CV, Morse DL, Eidson M, Smith PF, Hunter JL, Smith KA, Johnson RH, Jenkins SR, Berryman C. Update: Raccoon rabies epizootic -- United States and Canada, 1999. Morbidity and Mortality Weekly Review. 2000;49(2):31–35. [PubMed] [Google Scholar]
- Wandeler AI, Salsberg E. Raccoon rabies in eastern Ontario. Canadian Veterinary Journal. 1999;40:731. [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Morelli MJ, Thébaud G, Knowles NJ, Herzyk P, Paton DJ, Haydon DT, King DP. Beyond the consensus: dissecting within-host viral population diversity of Foot-and-Mouth disease virus by using next-generation genome sequencing. Journal of Virology. 2011;85(5):2266–2275. doi: 10.1128/JVI.01396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2016;1:28–36. [Google Scholar]
- Yu F, Zhang G, Zhong X, Han N, Song Y, Zhao L, Cui M, Rayner S, Fu ZF. Comparison of complete genome sequences of dog rabies viruses isolated from China and Mexico reveals key amino acid changes that may be associated with virus replication and virulence. Archives of Virology. 2014;159:1593–1601. doi: 10.1007/s00705-013-1966-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.