Abstract
Purpose of review
Myelodysplastic syndromes (MDS) have remarkably diverse somatic mutation patterns that are challenging to interpret clinically. Yet, genetic information is increasingly available to physicians. This review will examine several implications of genetic diversity in MDS.
Recent findings
Somatic mutations can serve as clinically relevant biomarkers in MDS. Molecular subtypes may exist that share clinical features including risk of progression to acute myeloid leukemia (AML), response to treatment, and overall survival. Several mutated genes are known to have prognostic value that is independent of common risk stratification tools. Mutations of several genes identify low-blast percentage patients with greater than predicted disease risk while only SF3B1 mutations predict lower disease risk than expected. Mutations of TP53 are associated with adverse features, yet demonstrate inferior outcomes than predicted by these risk factors. SF3B1 and TP53 mutations may identify clinically relevant subtypes of MDS and allow for better refinement of risk within these groups. Using somatic mutations to diagnose MDS is more challenging since they can occur in healthy individuals. Yet, patients with unexplained cytopenias have a high rate of clonal hematopoiesis that may be an important risk factor to identify clinically.
Summary
Patterns of somatic mutations are diverse in MDS, but can inform the prediction of prognosis and aid in its diagnosis.
Keywords: Myelodysplastic syndromes, somatic mutations, clinical genetics
Introduction
Advances in cancer genetics have made a tremendous impact in how clinicians evaluate and treat patients with a wide variety of tumor types. In the myeloid malignancies mutation testing is often considered standard of care as it provides information critical for making the diagnosis, selecting therapies, and predicting outcomes. Several myeloproliferative neoplasms (MPN) are defined by recurrent mutations in genes like JAK2, CALR, MPL, and KIT or gene rearrangements like BCR-ABL.(1) Acute myeloid leukemias (AML) are also classified by chromosomal rearrangements and several are further subdivided by point mutations in genes like DNMT3A, FLT3, CEBPA, and NPM1.(1)
Establishing similar practices for patients with myelodysplastic syndromes (MDS) has been more challenging. This is largely due to the oft cited clinical and genetic heterogeneity of these disorders in which particular mutations may be less common and less specific for important disease phenotypes. Despite this diversity, we have long considered chromosomal abnormalities to be important clinical biomarkers in MDS; primarily for risk stratification, but also for diagnosis and prediction of response in the case of MDS with isolated del(5q).(1) Somatic mutations of individual genes are now beginning to play similar roles. As driver events responsible for the development and progression of MDS, somatic mutations may be more closely tied to disease phenotypes and therefore, may be more reliable biomarkers.(2)
It is estimated that one or more mutated genes can be identified in over 90% of cases.(3, 4) Several mutations are tightly associated with clinical features such as bone marrow blast percentage, severity of certain cytopenias, chromosomal instability, or the presence of dysplasia.(2) These lesions often carry prognostic significance that is independent of existing risk stratification tools and to a lesser extent, can help predict response to certain therapies.(5–8) In some cases, the presence of a somatic mutation in cytopenic patients may also aid in establishing a diagnosis. This brief review will give examples of how somatic mutations detected in MDS patients might impact their clinical care and reshape how we classify these disorders.
Diverse Mutational Spectrum
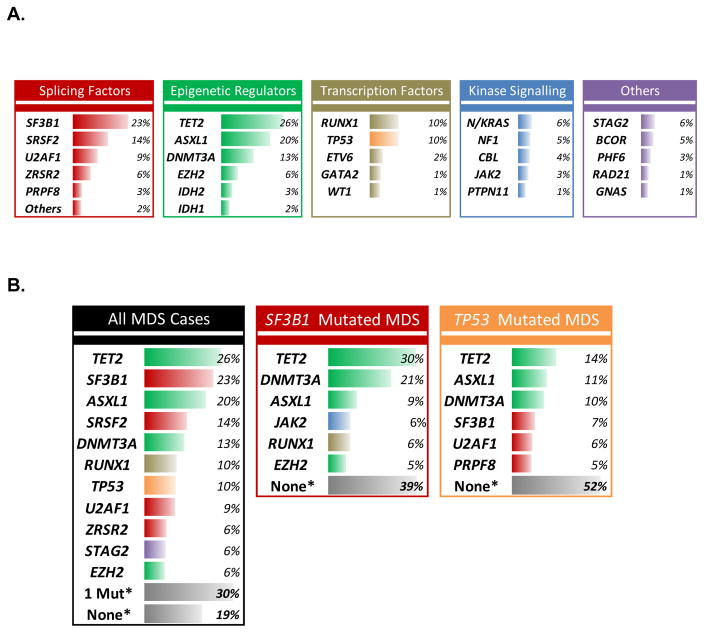
The overall pattern of mutations observed in MDS is similar to that seen in many solid tumors. A few genes are mutated with relatively high frequency while many more are found mutated only in a small minority of patients. This long tail of rarely mutated genes makes it difficult to discern their clinical implications. As a consequence, the mutated genes reported to have clinical utility tend to be the few that are either more frequent or that have very strong effects when present. The most frequently mutated MDS genes are SF3B1, TET2, ASXL1, SRSF2, DNMT3A, RUNX1, U2AF1, ZRSR2, STAG2, TP53, NRAS, and EZH2 in rough order of descending frequency (Figure 1).(3, 4) No single gene is mutated in the majority of cases and the most mutated genes are found in fewer than 5% of cases.(2–4)
Figure 1. Somatic mutation frequencies by class and mutation group.
A) Frequency of gene mutations organized by functional groups in all cases of MDS. B) Comparison of mutation rates in all MDS cases vs. those with either SF3B1 mutation or TP53 mutation. * - This refers to the percentage of patients with one (1 Mut) or no (None) mutation of the 17 most frequently mutated MDS genes – mutations in more rarely mutated genes may still be present in these cases.
Looking at the frequently mutated genes it is apparent that MDS are diseases of disordered spliceosome function and epigenetic regulation. Nearly two-thirds of MDS patients carry a splicing factor mutation and more than half have a mutation in an epigenetic regulator. Several additional pathways are frequently affected including hematopoietic transcription factors, tyrosine kinase signaling, cohesin genes, and DNA damage response and repair enzymes. However, clinical phenotypes are much more closely tied to the individual genes mutated that the pathway they belong to.(9) For example, MDS patients with SF3B1 mutations very often have ring sideroblasts which is a rare feature in patients with other mutated splicing factors. Similarly, patients with NRAS or CBL mutations tend to have higher risk disease while those with JAK2 mutations do not.
Finally, it is important to note that genetic variability in MDS refers to more than the vast array of combinations in which genes can be co-mutated. There is also a diverse clonal architecture to these disorders.(10) In some patients, a mutated gene may exist in a small subclone while in another it might represent an ancestral mutation present in every tumor cell. Furthermore, this clonal architecture evolves over time and with treatment.(11) The clinical implication of mutations must be considered in the context of other mutations as well as their associated clone size.(12) This degree of genetic variability might make it seem unlikely that we would find reliable clinical correlates of genetic mutations, but several important observations have been validated and can clearly impact care.
Prognostic Value
The greatest evidence for the role of somatic mutations in the clinical care of MDS patients comes from studies on disease risk and prognosis. Several mutated genes are strongly associated with known clinical risk factors. Mutations of TP53, RUNX1, ASXL1, SRSF2, and NRAS are enriched in patients with elevated bone marrow blast proportion, for example.(2, 4) The same genes are associated with thrombocytopenia while TP53 mutant patients are more likely to have a low neutrophil count and a complex karyotype. SF3B1 mutations, on the other hand, are inversely associated with these adverse risk factors.(3) Given these associations, it is not surprising that somatic mutations carry prognostic information and can be used to risk stratify patients.
Some mutated genes, like CBL, EZH2, PTPN11, and PRPF8, are not strongly linked to clinical risk factors yet still predict a shorter overall survival.(13) Mutations in these genes may have prognostic value that is independent of the IPSS-R. In actuality, even mutated genes like TP53 that are strongly tied to clinical risk factors have independent prognostic significance.(14) This may be because mutations associated with an adverse prognosis often arise in small subclones that can be detected by sensitive sequencing techniques.(3) These small clones may be harbingers of disease progression, but not yet large enough to alter clinical risk factors like blast proportion or cytopenias. For adverse mutations, it has been shown that their presence in a small subclone carries the same risk as when they are present in the dominant clone.(3)
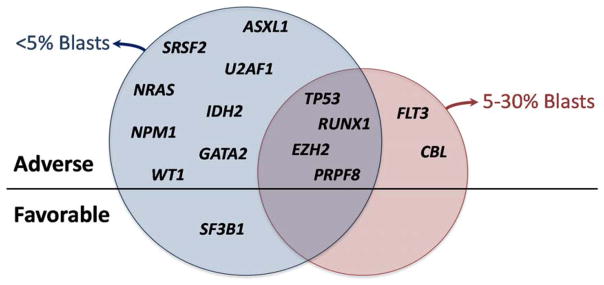
Mutations of TP53, EZH2, RUNX1, ETV6, and ASXL1 have been associated with greater risk that predicted by the IPSS and later, the IPSS-R.(2) Subsequent studies have found additional mutated genes with adverse prognostic associations independent of the IPSS-R, including NRAS, CBL, PRPF8, PTPN11, and NF1 to name a few (Figure 2).(3, 4) Surprisingly, one or more of these adverse mutations can be found in over one-third of patients, indicating that we routinely underestimate disease risk using conventional assessment tools.(13) In contrast, we may be over estimating risk in patients with SF3B1 mutation even though the majority of this group is considered to have lower risk disease by the IPSS-R.
Figure 2. Mutated genes with independent prognostic significance by MDS bone marrow blast proportion.
Mutated genes associated with overall survival in MDS patients after adjustment for IPSS-R risk groups are listed below. Those in the blue circle are significant in patients with less than 5% blasts in their bone marrow. Those in the red circle are significant in patients with 5–30% blasts. Gene listed above the straight line are prognostically adverse. Only mutations of SF3B1, listed below the line, are independently favorable. Data based on IWG-PM presentation at ASH 2015.(13)
Similarly, patients with a complex karyotype are considered to have high disease risk, but in practice, are a heterogeneous group. Complex karyotype MDS patients have fewer mutations in other genes, but roughly half will carry a TP53 mutation and have the worst overall outcomes, even after treatment.(6, 8, 15, 16) Complex karyotype patients without TP53 mutations appear to do as well as MDS patients without a complex karyotype, highlighting the importance of TP53 mutation status in this group.(12)
Why is this clinically relevant? Treatment for MDS is highly risk stratified. Patients perceived to have lower risk disease are either observed or treated with supportive measures such as erythropoiesis stimulating agents. MDS patients at greater risk are treated more aggressively with hypomethylating agents or considered for stem cell transplantation. Misassigning risk could lead to over or under treatment. In addition, there may be uncertainty about how best to treat patients with intermediate risk according to tools like the IPSS and IPSS-R. Additional information about somatic mutations can refine risk in these cases and help with the selection of appropriate therapies.
How best to incorporate mutational information into clinical risk assessments is not yet clear as expert consensus guidelines for this have not been published. Ideally, a molecularly integrated risk assessment tool would improve accuracy without adding undue complexity or barriers to use. A straightforward method would be to use a two-step process in which patients are scored using conventional prognostic models like the IPSS or IPSS-R, and then have their risk group adjusted based on somatic mutations.(2) For example, an MDS patient with IPSS Intermediate-1 risk or IPSS-R Intermediate risk would be considered to have “higher” risk disease if they carried an ASXL1 mutation or another adverse gene mutation. Such a patient might be referred to stem cell transplantation or afforded hypomethylating agent sooner than they otherwise might. This approach leverages existing scoring systems without requiring physicians to adopt a completely novel one. The disadvantage could be some loss of accuracy in exchange for simplicity as mutations in different genes would not be assigned different weights nor could the relative contribution of clinical measures like blast count be changed.
Alternatively, a new model that reweights the contribution of genetic and clinical measures could be created.(4, 17) The disadvantage to this approach is its complexity, which could represent a barrier to adoption. It also may not consider subtleties that might limit its prognostic accuracy. For example, somatic mutations that predict increased disease risk might do so only in certain clinical situations and some gene mutations may not necessarily have additive risk when they are found to co-occur.
Preliminary results presented at ASH by the International Working Group for MDS (IWG) molecular prognosis committee demonstrated how some mutated genes have independent prognostic significance only in MDS patients with < 5% bone marrow blasts (Figure 2).(13) In patients with excess blasts, genes like U2AF1, SRSF2, SF3B1, and ASXL1 lost their independent prognostic significance. This may represent a more nuanced approach that subdivides MDS patients based on prognostic clinical features, such as blast count, and then performs molecularly based risk stratification. The result could be greater accuracy that retains the simplicity of a two-step model without adding burdensome complexity. Ongoing efforts of large collaborative groups like the IWG will provide guidance about how best to utilize MDS mutation data in practice.
Diagnostic Value
As with MPNs, cases of MDS can often be difficult to distinguish from reactive processes. Many patients with unexplained cytopenias who lack excess blasts or sufficient dysplasia in their bone marrow are left without a diagnosis and little clarity about their prognosis. It is tempting to think that the identification of a somatic mutation can be used as a surrogate marker of disease in these patients who do not demonstrate the morphologic criteria required for MDS. However, the reality is much more challenging. No mutation is present in the majority of cases and no mutated gene is highly specific for MDS. In fact, somatic mutations indicative of clonal hematopoiesis are strikingly more common in persons without hematologic abnormalities. Several studies have demonstrated that somatic mutations can be found in the blood of unselected individuals with a frequency that increases sharply with age.(18–20) For persons aged 75, the prevalence of clonal hematopoiesis approaches 15%. The somatically mutated genes are the same as those found in MDS, but at different frequencies. Mutation of DNMT3A, TET2, and ASXL1 are the most common, but also include TP53, SF3B1, SRSF2, JAK2, and CBL, typically at low abundance.
The relative risk of developing a hematologic malignancy is increased in these individuals with clonal hematopoiesis, but the absolute risk remains very low. Since the vast majority will never develop a hematologic disorder they are described as having clonal hematopoiesis of indeterminate potential (CHIP).(19–21) If a person with CHIP develops cytopenias caused by a vitamin deficiency, alcohol use, a viral infection, or another non-malignant cause, the identification of a somatic mutation could be misleading and should not be used to indicate that MDS is present.(22)
With that caveat in place, there is a role for mutation sequencing in patients with unexplained cytopenias. Patients who do not meet the diagnostic criteria for MDS are often described as having idiopathic cytopenias of undetermined significance (ICUS). The prognosis in ICUS patients remains largely unknown. A recent study of cytopenic patients suspected of having MDS and who had a bone marrow biopsy performed showed that the incidence of ICUS is high.(23) Of 144 patients prospectively screened, only 24 were diagnosed with MDS. Another 21 had evidence of dysplasia, but did not meet diagnostic criteria for MDS and 99 had no dysplasia whatsoever. Karyotype analysis and sequencing of 22 MDS genes found evidence of clonal hematopoiesis in 71% of patients with some dysplasia and in 27% of those with none. These rates are many times higher than the background rate of CHIP in similarly aged individuals. Furthermore, the size of the mutant clones in these cytopenic patients was much greater than that seen in CHIP. Overall, 40% of patients with ICUS had evidence of clonal hematopoiesis and were described as having clonal cytopenias of undetermined significance (CCUS). Eventual outcomes for CCUS patients versus those with non-clonal ICUS were not examined and remain unclear.(22)
A complementary study examined 69 patients with MDS or AML who had a prior bone marrow biopsy that failed to meet diagnostic criteria for any hematologic malignancy.(24) When DNA samples from these earlier non-diagnostic biopsies were sequenced, 91% carried at least one somatic mutation. Only 43% of the cohort developed an additional mutation by the time that they were diagnosed with MDS or AML. These studies indicate that somatic mutations in cytopenic patients are likely risk factors for the development of myeloid malignancies (Table). Studies are ongoing to better characterize this risk and to determine which features are most predictive. In the meantime, identification of CCUS patients is useful, as it will allow us to learn more about their outcomes.
Table.
Comparison of genetic characteristics between CHIP, CCUS, and MDS.
| CHIP Unselected Population |
CCUS At Diagnosis |
CCUS Prior to MDS/AML Progression |
MDS All Risk Groups |
|
|---|---|---|---|---|
| Commonly Mutated Genes | DNMT3A, TET2,ASXL1, PPM1D, JAK2, TP53 | TET2, DNMT3A,ASXL1, SRSF2, TP53 | TET2, SRSF2, ASXL1, U2AF1, DNMT3A | SF3B1, TET2, ASXL1, SRSF2, DNMT3A |
| Mean # of Mutations | ~1 | ~1.6 | ~2 | ~2.6 |
| Typical VAF | 9–12% | 30–40% | ~40% | 30–50% |
| Incidence | About 10–15% of 70 year-olds | About 35% of ICUS | About 90% of ICUS | May be <50% of cytopenic patients |
Most recently, the World Health Organization (WHO) committee on the diagnosis of myeloid neoplasms revised the diagnostic criteria for MDS to include a single gene mutation.(25) Cytopenic patients with as few as 5% ring sideroblasts (RS) are considered to have MDS if they carry at typical SF3B1 mutation. This represents a relaxation of the criteria that MDS-RS patients have at least 15% ring sideroblasts and is the first example of a genetic mutation included in the diagnostic criteria for MDS.
Conclusion
Recurrent somatic mutations are the molecular drivers responsible for the development and progression of MDS. They are associated with clinical manifestations, prognosis, and in some cases, how patients respond to therapy. The genetic variability associated with MDS has made it challenging to establish simple rules governing the clinical interpretation of somatic mutations. Despite these challenges, somatic mutation testing for MDS is useful as an aid to clinical decision making today as long as they are interpreted in light of the clinical context and not used as the sole means of diagnosing, risk stratifying, or selecting therapy for patients. Consensus guidelines have begun to describe the role of somatic mutations in the risk assessment of patients with MDS. Novel prognostic models are being developed that will incorporate mutational information. Molecular subgroups, like SF3B1 or TP53 mutant MDS, are being explored as has recently been done with AML.(9, 26) Further investigation into the risk associated with somatic mutations in CCUS patients will likely redefine the diagnostic boundaries of MDS. Together, these advances will leverage the genetic diversity of MDS to create clinical tools that improve the personalization of MDS patient care.
Key Points.
Somatic mutations are common in MDS and occur in highly variable patterns
The most common class of mutations involve splicing factors and epigenetic regulators
Several mutations are associated with clinical features and may identify genetic subtypes of MDS with more homogenous disease phenotypes, responses to treatment, and predicted prognosis.
Somatic mutations should not be used to diagnose cytopenic patients with MDS in the absence of morphologic or cytogenetic criteria, but can identify patients with clonal cytopenias that are predicted to have increased risk disease risk.
Acknowledgments
Financial support and sponsorship
This work was supported by a MDS Foundation grant to the International Working Group for MDS and the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant K08DK091360.
Footnotes
Conflicts of interest
Dr. Bejar has received honoraria from Genoptix, Celgene, Illumina, Foundation Medicine, and Alexion. RB has IP licensed to Genoptix and serves on a data safety monitoring board for a clinical trial sponsored by Celgene.
References
- 1.Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–51. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 2.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, Lord A, Stevenson K, Bar-Natan M, Perez-Ladaga A, Zaneveld J, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124(17):2705–12. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejar R, Stevenson KE, Caughey B, Lindsley RC, Mar BG, Stojanov P, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–8. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25(7):1147–52. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 8.Della Porta MG, Galli A, Bacigalupo A, Zibellini S, Bernardi M, Rizzo E, et al. Clinical Effects of Driver Somatic Mutations on the Outcomes of Patients With Myelodysplastic Syndromes Treated With Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.67.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **9.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015 doi: 10.1182/blood-2015-03-633537. An excellent example of genetic clustering used to identify molecular subgroups of MDS patients with shared genetic and clinical features, particularly SF3B1 mutant MDS as a distinct entity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter MJ, Shen D, Shao J, Ding L, White BS, Kandoth C, et al. Clonal diversity of recurrently mutated genes in myelodysplastic syndromes. Leukemia. 2013;27(6):1275–82. doi: 10.1038/leu.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Mossner M, Jann JC, Wittig J, Nolte F, Fey S, Nowak V, et al. Mutational hierarchies in myelodysplastic syndromes dynamically adapt and evolve upon therapy response and failure. Blood. 2016;128(9):1246–59. doi: 10.1182/blood-2015-11-679167. A fascinating study on clonal dynamics in serially sampled MDS patients before and after therapy. [DOI] [PubMed] [Google Scholar]
- *12.Sallman DA, Komrokji R, Vaupel C, Cluzeau T, Geyer SM, McGraw KL, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia. 2016;30(3):666–73. doi: 10.1038/leu.2015.304. A nice example of the adverse impact of TP53 mutations at various mutant allele frequencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **13.Bejar R, Papaemmanuil E, Haferlach T, Garcia-Manero G, Maciejewski JP, Sekeres MA, et al. Somatic Mutations in MDS Patients Are Associated with Clinical Features and Predict Prognosis Independent of the IPSS-R: Analysis of Combined Datasets from the International Working Group for Prognosis in MDS-Molecular Committee. Blood. 2015;126(23):907. An early look at the prognostic significance of mutations in MDS patients studied retrospectively from the worldwide collaboration of the IWG-PM. [Google Scholar]
- 14.Bejar R, Papaemmanuil E, Haferlach T, Garcia-Manero G, Maciejewski JP, Sekeres MA, et al. TP53 Mutation Status Divides MDS Patients with Complex Karyotypes into Distinct Prognostic Risk Groups: Analysis of Combined Datasets from the International Working Group for MDS-Molecular Prognosis Committee. Blood. 2014;124(21):532. [Google Scholar]
- *15.Takahashi K, Patel K, Bueso-Ramos C, Zhang J, Gumbs C, Jabbour E, et al. Clinical implications of TP53 mutations in myelodysplastic syndromes treated with hypomethylating agents. Oncotarget. 2016;7(12):14172–87. doi: 10.18632/oncotarget.7290. A nice study elucidating the adverse impact of TP53 mutations in MDS patients treated with hypomethylating agents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Mossner M, Jann JC, Nowak D, Platzbecker U, Giagounidis A, Gotze K, et al. Prevalence, clonal dynamics and clinical impact of TP53 mutations in patients with myelodysplastic syndrome with isolated deletion (5q) treated with lenalidomide: results from a prospective multicenter study of the german MDS study group (GMDS) Leukemia. 2016;30(9):1956–9. doi: 10.1038/leu.2016.111. A recent look at the clinical impact of TP53 mutations in MDS patients traditionally considered to have favorable risk and good responses to lenalidomide. [DOI] [PubMed] [Google Scholar]
- *17.Nazha A, Narkhede M, Radivoyevitch T, Seastone DJ, Patel BJ, Gerds AT, et al. Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia. 2016 doi: 10.1038/leu.2016.138. An example of how somatic mutations can be incorporated into a clinical scoring system for MDS. [DOI] [PubMed] [Google Scholar]
- 18.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **19.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. An excellent perspective on CHIP as a premalignant lesion and how somatic mutations should not be considered synonymous with MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Link DC, Walter MJ. ‘CHIP’ping away at clonal hematopoiesis. Leukemia. 2016;30(8):1633–5. doi: 10.1038/leu.2016.130. An excellent commentary on the intepretation of clonal hematopoiesis in various clinical contexts. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Pol S, Ma L, Ohgami RS, Arber DA. Significance of myelodysplastic syndrome-associated somatic variants in the evaluation of patients with pancytopenia and idiopathic cytopenias of undetermined significance. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2016 doi: 10.1038/modpathol.2016.100. [DOI] [PubMed] [Google Scholar]
- **23.Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126(21):2355–61. doi: 10.1182/blood-2015-08-667063. Prospective and restrospective studies on clonal hematopoiesis defined by somatic mutations in cytopenic patients suspected of having MDS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Cargo CA, Rowbotham N, Evans PA, Barrans SL, Bowen DT, Crouch S, et al. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126(21):2362–5. doi: 10.1182/blood-2015-08-663237. A retrospective look at the high rate of clonal hematopoiesis in cytopenic patients with non-diagnostic bone marrow studies. [DOI] [PubMed] [Google Scholar]
- **25.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. A summary of proposed changes to the WHO classification of myeloid disorders ighlighting the diagnostic value of SF3B1 mutations in MDS. [DOI] [PubMed] [Google Scholar]
- *26.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–21. doi: 10.1056/NEJMoa1516192. An example in AML of how genetic clustering can be used to identify subgroups of patients with more homogenouse clinical phenotypes and response to treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]