Abstract
Chemical modifications of DNA comprise epigenetic mechanisms that contribute to the maintenance of cellular activities and memory. Although the function of 5-methylcytosine (5-mC) has been extensively studied, little is known about the function(s) of relatively rarer and underappreciated cytosine modifications including 5-hydroxymethylcytosine (5-hmC). The discovery that ten-eleven translocation (Tet) proteins mediate conversion of 5-mC to 5-hmC, and other oxidation derivatives, sparked renewed interest to understand the biological role of 5-hmC. Studies examining total 5-hmC levels revealed the highly dynamic yet tissue-specific nature of this modification, implicating a role in epigenetic regulation and development. Intriguingly, 5-hmC levels are highest during early development and in the brain where abnormal patterns of 5-hmC have been observed in disease conditions. Thus, 5-hmC adds to the growing list of epigenetic modifications with potential utility in clinical applications and warrants further investigation. This review discusses the emerging functional roles of 5-hmC in normal and disease states, focusing primarily on insights provided by recent studies exploring the genome-wide distribution of this modification in mammals.
Keywords: DNA methylation, genome-wide, 5-hydroxymethylcytosine, 5-methylcytosine
Introduction
Recent advances in DNA sequencing technologies have enhanced our understanding of the functional roles of epigenetic modifications in fundamental cellular processes, including DNA repair (1, 2) and replication (3), nucleosome assembly (4), gene transcription (5), and pre-mRNA splicing (6). For studying DNA methylation, a variety of strategies have been developed that allows for extensive characterization of the genome-wide distribution of DNA methylation states at nucleosomal to single-nucleotide resolution, enabling more detailed analyses of its local activity at specific regions throughout the genome (7–11). However, the dynamic nature of DNA methylation as well as previously underappreciated variants of the modification offers additional levels of complexity to chromatin organization and function that warrants further investigation.
Although 5-methylcytosine (5-mC) is a well-described modification of DNA (12, 13), variants of DNA methylation including 5-hydroxymethylation (5-hmC) have only recently been appreciated. Nearly four decades after its discovery in mammals (14), 5-hmC was found to exist in relatively high abundance in neurons and embryonic stem cells (ESCs) (15), and produced through 5-mC oxidation catalyzed by the Tet family of proteins (16). Soon after, it was demonstrated that Tet can further oxidize 5-hmC to 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC) (17, 18). More recent studies revealed proteins that specifically bind these oxidized derivatives of methylcytosine and evidence of their involvement in facilitating DNA demethylation (19, 20). However, improvements in examining the genome-wide distribution of 5-hmC (21–23) and its reported cell- and tissue-specific dynamics (24–27) provide clues that it may exhibit additional activities.
Further investigation into the functional role(s) of 5-hmC in the genome is of considerable importance, as abnormalities in the levels and genomic distribution of 5-hmC has been described in several disease conditions (28). In this Review, we speculate on emerging functional roles of 5-hmC in normal and disease states, focusing primarily on insights provided by recent studies exploring the genome-wide distribution of this modification. Although important, we do not discuss in detail the principles and challenges of DNA methylation assays, including methods for detection of 5-hmC, and analyzing/interpreting DNA methylation data as extensively described elsewhere (29–31).
DNA methylation: a component of the epigenome
The epigenome is comprised of covalent modifications of DNA, post-translational modifications of histones that package the DNA into nucleosomal units, the dynamic deposition of histone variants, and the positioning of these nucleosomes into higher-order chromatin structure (32–34). Here, we focus on an important component of the epigenome, DNA methylation, and a renewed appreciation for its diversity.
DNA methylation, in particular 5-mC, was identified as a minor base in DNA (13, 35). The origin of this modification remained unknown until specific DNA methyltransferases (DNMTs) were isolated in bacteria (36) and then a few years later in mammals (37). Now considered the ‘writers’ of DNA methylation, DNMTs catalyze the methyl group transfer from a universal methyl donor S-adenosyl-L-methionine (SAM) to cytosine residues. In mammals, DNA methylation occurs almost exclusively within 5′-cytosine-guanine-3′ dinucleotides (CpGs), although non-CpG methylation has also been observed albeit less frequently (38, 39). DNA methylation influences gene expression by affecting the binding of methylation-sensitive DNA binding proteins and/or by interacting with various modifications of histone proteins that alter DNA accessibility (40, 41) and is generally associated with gene silencing. However, the dynamics of DNA methylation states and its tissue/cell type-specific distribution suggest additional functional roles.
Dynamics of DNA methylation
In mammals, DNA methylation is particularly dynamic during embryogenesis. Following fertilization, active demethylation occurs in the paternal pronucleus (42, 43), while the maternal chromosomes undergo passive demethylation that eliminates most, but not all, DNA methylation marks inherited from the gametes (44–46). After implantation, embryonic DNA methylation patterns are established through lineage-specific de novo methylation that begins in the inner cell mass of the blastocyst (44, 45, 47).
The role of these dynamic demethylation and remethylation processes during early development is not fully understood. However, it has been proposed that genome-wide demethylation during pre-implantation development may lead to chromatin decondensation and thus transcriptional activation of the zygotic genes essential for early development and/or may facilitate reprogramming of the genome through interacting with histone modifications and chromatin remodeling. In contrast, remethylation may be necessary to repress retrotransposons and establish a global gene silencing state required for embryonic development (48). Such activities appear essential as studies investigating the molecular mechanisms involved in nuclear reprogramming have suggested that correct DNA methylation patterns are required for successfully reprogramming differentiated adult cells into pluripotent embryonic stem cell like cells (49, 50). Although passive demethylation is a well-understood process, the mechanism of active demethylation, highlighted also by studies of post-mitotic neurons [reviewed in (51)], remained unclear until recently.
Role of 5-hmC in demethylation
Much excitement has been generated over the discovery that the ten-eleven translocation (TET1, TET2, and TET3) proteins facilitate the conversion of 5-mC to 5-hmC (16), as this process could potentially provide a mechanism to explain active demethylation. Tet proteins thus appear to act as ‘editors’ of DNA methylation states. Considered oxoglutarate- and iron-dependent dioxygenases, Tet proteins co-localize with 5-hmC in mouse embryonic stem cells, mammalian brain, and liver, supporting their role in converting 5-mC to 5-hmC (18). Further oxidation of 5-hmC to 5-fC and 5-caC (21, 52) have been observed and are thought to provide a possible mechanism for active removal of methyl groups from CpGs. In support of this, 5-hmC levels are increased concurrently with the loss of 5-mC in the paternal pronucleus, when global demethylation occurs actively (53). It is thought that the removal of 5-hydroxymethylated cytosines is facilitated by glycosylases (16). Indeed, mammalian glycosylases, such as thymine DNA glycosylase (TDG) and methyl-CpG-binding domain protein 4 (MBD4) exhibit strong activity on T:G mismatches that can be created through deamination of 5-mC by cytidine deaminases of the activation-induced cytidine deaminase (AID)/apolipoprotein B mRNA editing enzyme, catalytic polypeptide (APOBEC) family (54). Taken together with the discovery of 5-mC oxidation mediated by TET enzymes, these studies reveal that 5-hmC may serve as an intermediate to active DNA demethylation by a two-step process that involves deamination followed by mismatch repair [reviewed in (55)]. Interestingly, 5-hmC may also contribute to passive loss of DNA methylation as conversion of 5-mC to 5-hmC during replication inhibited DNMT1 activity (56). New data suggest a potential for DNMT-3A and -3B to directly catalyze conversion of 5-hmC, and 5-mC, to cytosine under specific conditions (57), providing another mechanism for active demethylation. Although 5-hmC may be critical to DNA demethylation, recent reports reveal there are specific proteins that ‘read’ this modification and hints at additional roles.
Readers of DNA methylation states are comprised of proteins that preferentially bind methylated DNA and recruit other chromatin-associated protein complexes. They include, but are not limited to, methyl-binding domain proteins (MBD) such as MeCP2 and its family members (MBD1-4). Much of the functional role for DNA methylation and what reader it recruits depends on its distribution within the genome. At promoters, DNA methylation generally precludes transcription by blocking the binding of transcriptional activators directly or indirectly through the recruitment of methyl-binding proteins and co-repressor complexes containing histone deacetylases that cooperatively facilitates the formation of heterochromatin (58). However, genome-wide studies have revealed that DNA methylation within gene bodies is far more frequent than at promoters (5, 59–61), playing a role in tissue-specific alternative promoter usage (5). Preferential positioning of DNA methylation over exons compared with introns (62–65) prompted speculation for its potential role in pre-mRNA splicing, which has been confirmed by further studies (6, 66, 67). Interestingly, readers of DNA methylation including MeCP2 and methylation-sensitive proteins such as CTCF play roles in exon recognition and alternative splicing (6, 66).
In addition to proteins that associate with methylcytosine, recent studies have shown a number of potential readers of oxidized derivatives of methylcytosine (19, 20). RPL26, PRP8 and the DNA mismatch repair protein MHS6 seem to have a preference for 5-hmC, whereas transcriptional regulators (FOXK1, FOXK2, FOXP1, FOXP4 and FOXI3), DNA repair factors (TDG and MPG) and chromatin regulators (EHMT1, L3MBTL2 and all components of the NuRD complex) have a strong preference for 5-fC (19), a stable modification with highest levels observed in brain (68). Furthermore, some of these interactions are dynamic during differentiation including the binding of Uhrf2 to 5-hmC containing DNA (20). That the oxidized derivatives of methylcytosine recruit distinct transcription regulators as well as a large number of DNA repair proteins implicate DNA damage response as a major player in active DNA demethylation and chromatin remodeling.
Tissue specificity of 5-hmC
Studies examining the total chromatin abundance of 5-hmC across mammalian tissue/cell types revealed early clues into its dynamics and potential role. One such study observed that 5-hmC comprises 0.6% of total nucleotides in cerebellar Purkinje cells and 0.2% in granule cells, with 5-hmC being ~40% as abundant as 5-mC in Purkinje cells (15). Following this discovery, quantification of bulk levels of 5-hmC found that it is relatively rare, with levels varying by tissue from < 0.1% to 0.7% of all cytosines globally (70) (Figure 1). Additionally, 5-hmC levels appeared to be dynamic: levels in embryonic stem (ES) cells decreased by ~40% upon differentiation, while levels increased in cerebellum and hippocampus tissues (16, 22). Localization of 5-hmC in mature neuronal cells, but not immature neurons, was also reported (22). Further evidence of the connection between 5-hmC and neurodevelopment was discovered in autism cerebellum, where 5-hmC DNA hypermethylation was associated with increased engrailed-2 (EN-2) expression, a gene normally downregulated in Purkinje cell maturation (71). Taken together, these findings indicate a crucial role of 5-hmC in development, particularly in the central nervous system where the modification is most concentrated (70). Although, the precise molecular function of 5-hmC remains incompletely understood, new technologies enabling the genome-wide mapping of 5-hmC states offer insight into the potential functional activities of 5-hmC in normal and disease conditions beyond its role as an intermediate in active demethylation.
Figure 1.
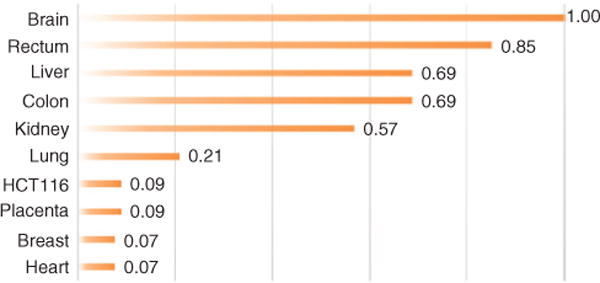
5-Hydroxymethylcytosine levels relative to brain tissue. Fold-change was calculated by dividing tissue total percent 5-hmC by brain total percent 5-hmC.
HCT116 (a colon cancer cell line) showed ~9-fold decrease in 5-hmC levels compared to normal colon tissue healthy colon. Data adapted from Ref. (69).
Genomic localization of 5-hmC and potential functional roles
Prior challenges in distinguishing 5-hmC from 5-mC have prevented fully elucidating its function. In particular, widely used methods to probe 5-mC, such as bisulfite sequencing and methylation-sensitive restriction digestion, cannot discriminate between 5-hmC and 5-mC (72, 73). Emerging approaches have largely overcome such challenges [reviewed in Ref. (74)]. In addition, advances in next generation sequencing (NGS) platforms have made genomic sequencing affordable and applicable, even leading to the first FDA approved NGS applications for the clinic in 2013 (75). Coupled with NGS, data derived from such innovative methods allow further insight into 5-hmC. While there are other methods of measuring 5-hmC, here we focus only on those studies that apply NGS to specifically and directly examine the distribution of 5-hmC genome-wide (Table 1).
Table 1.
Genome-wide sequencing methods targeting 5-hmC modifications.
| ‘Seq’-assays | DNA amount (ng) | Enrichment | Resolution | Sequencing platform | References |
|---|---|---|---|---|---|
| OxBS-Seq | 100–1000 | No | 1 bp | Illumina | (76) |
| TAB-Seq | 500 | No | 1 bp | Illumina | (77) |
| JBP1-Seq | 50 | Yes | 36–50 bp | Illumina | (78) |
| MeDIP-Seq | 4000 | Yes | 200–400 bp | Illumina | (79) |
| MeDIP-Seq | 5000 | Yes | 100 bp | Illumina | (80) |
| MeDIP-Seq | 1000 | Yes | 50 bp | Ion Torrent | (67) |
The distribution of 5-hmC throughout the genome appears specific. Although the studies listed here examine different tissue/cell types and samples, a few common features of the distribution of 5-hmC can be observed that suggest that this modification may play a structural role in the organization of the genome. Studies in mouse ES cells reported 5-hmC enrichment in gene-rich regions, especially in and near transcriptional start and end sites, gene bodies, and distal regions (31, 81, 82). Wu et al. reported a majority of 5-hmC in intragenic and distal intergenic regions (58% and 30%, respectively) (82), and in the same year Song et al. reported concentrated 5-hmC in intragenic regions linked to neurodegenerative disease (31). Additionally, L1 long interspersed nuclear element (LINE1) subgroup displayed enriched levels of 5-hmC, whereas intracisternal A-particle (IAP) regions returned low amounts of hydroxymethylation (83). Booth et al. posited that LINE1 and IAP trends could implicate 5-hmC in the demethylation process of specific repeat classes, as LINE1 elements are reprogrammed during preimplantation and IAPs remain unchanged (83). Genetic localization in these elements and recent studies of 5-hmC patterns in neuronal development support the notion that this modification plays a structural role in vivo (26, 84). Indeed, it was recently observed that 5-hmC concentrated at exon-intron boundaries (85). We observed a similar pattern (Figure 2). Related to its role in genome organization, a recent study reported the enrichment of 5-hmC at bivalent domains, a specific chromatin configuration thought to regulate gene activity (86), implicating this epigenetic mark in gene regulation by potentially inducing ‘poised’ chromatin states that dynamically modulate the levels and timing of gene expression (87).
Figure 2.
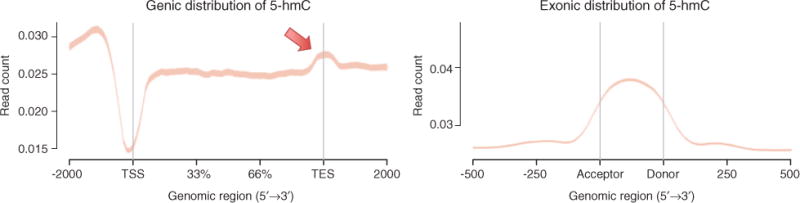
Genic and exonic distribution of 5-hmC in brain.
Using antibodies specific for 5-hmC combined with a MeDIP-Seq method optimized for Ion Torrent sequencing, we measured the distribution of 5-hmC in postmortem human brain tissue. Reads from the sequenced library were mapped to the hg19 reference genome and read count (per million mapped reads) is displayed ±2 kb over TSS, gene bodies, and TES (left panel) and over canonical exons (right panel). Arrow highlights area of 5-hmC enrichment at TES. Adapted from Ref. (67).
Given the general chromatin landscape of 5-hmC, we speculate additional potentially novel functional roles for the modification. In particular, the abundance of 5-hmC at transcription end sites (TES) is substantially higher than neighboring regions within genes (67). MeDIP-Seq results of 5-hmC levels in brain tissue from our lab clearly show depletion of the mark in transcription start sites (TSS) and enrichment precisely over exons (Figure 2), similarly to 5-mC distribution (5, 6). As has been described for 5-mC (6), such exonic patterns of 5-hmC appears to be a common observation (84, 85) and suggest it may play a role in pre-mRNA splicing. In contrast to 5-mC where depletion is typically observed (5, 29, 67), 5-hmC appears significantly enriched at TES regions (67, 81–83) (Figure 2). Although the functional role of 5-hmC at TES has not been explicitly examined, integrating this distribution with studies of RNA revealing tissue-specific transcription initiation near TES (88, 89) and novel post-transcriptional RNA methylation at 3′-UTRs (90) implicate additional roles for 5-hmC in transcription and RNA methylation, respectively. Additional biochemical and genetic assays targeting 5-hmC and other variants are needed to substantiate these potential roles.
Implications of 5-hmC in disease
Although the role of 5-hmC remains poorly understood, alterations to the epigenome have been well studied in the progression of cancer with the focus on 5-mC modifications. Global DNA demethylation (loss of 5-mC or ‘hypomethylation’) is a hallmark of most cancer types (91, 92). DNA hypomethylation affects chromosome stability, promoting translocations and deletions (93). Another characteristic of cancer is DNA ‘hypermethylation’, typically occurring at tumor suppressor genes and is generally associated with gene silencing (94). Over 700 mutations in writers, editors, and readers of DNA methylation, histone modifications, and chromatin remodeling were found across multiple tumor types, highlighting the disturbance of the epigenetic machinery in cancer (95). The advances in sequencing technologies coupled with new methods to examine different variants of DNA methylation genome-wide has revealed insights into potential consequences to changes in 5-hmC in cancer. Global 5-hmC levels were first observed to be significantly decreased in solid tumors of the colon and rectum (69) (compare colon with HCT116, a colon cancer cell line in Figure 1). Later, this phenomenon was also observed in melanomas and myeloid leukemia as a result of TET2 mutations (96, 97). Significantly, expression of active TET2 in animal models re-establishes 5-hmC landscapes and suppresses melanoma growth in animal models (96). The consequences of such extensive changes in 5-hmC levels remain largely unknown, though dysregulation of gene expression and genome stability are suspected (24). Genome-wide methods are just beginning to reveal that 5-hmC is redistributed in cancer, including its aberrant enrichment at oncogenic promoters such as GATA6 (98), warranting further studies.
In addition to cancer (95), the role of 5-hmC in gene regulation and neurodevelopment has led to the modification being implicated in a wide range of diseases including Rett syndrome and Fragile X syndrome (28), among others (Table 2). Investigators have also observed epigenetic perturbations in neurodegenerative diseases such as Huntington’s and amyotrophic lateral sclerosis (ALS) [reviewed in Ref. (109)] and neurodevelopmental diseases such as autism (99). Given that 5-hmC is enriched and dynamic in neural tissue, abnormal differential hydroxymethylation in neurological diseases is perhaps not surprising. Indeed, abnormal hydroxymethylation and Tet1 expression was also observed in genes related to drug addiction after cocaine use in mouse nucleus accumbens tissue, implicating epigenetic mechanisms involving 5-hmC in related pathologies (110). In support of this, a recent study observed a link between increased levels of 5-hmC at the EN-2 promoter and autism in postnatal cerebellum (71). EN-2 expression represses transcription during fetal and early postnatal development, while EN-2 downregulation in later development is central to normal brain structure (71). James et al. also reported increased TET1 and TET3 expression coupled with decreased MeCP2 binding. The investigators hypothesized decreased binding of repressive MeCP2, presumably facilitated by increased 5-hmC, contributed to EN-2 overexpression and abnormal development. Recently, we reported that a mutation affecting the activity of Tet1 on establishing 5-hmC resulted in dysregulation of genes involved in neural tube closure and contributed to craniofacial malformations in tuft mice (111). Altogether, these studies implicate a role for 5-hmC in disease and suggest that 5-hmC may be a useful clinical marker of diseases depending on its genomic localization. We anticipate further, more precise mechanistic insight into the role of 5-hmC as improvements in evaluating DNA methylation states genome-wide are currently being investigated (112).
Table 2.
Human diseases and observed aberrant 5-hydroxymethylcytosine levels.a
| Disease | Observation | References |
|---|---|---|
| Autism spectrum disorders | Enrichment in autism related genes in cerebellar cortex | (84, 99, 100) |
| Fragile X syndrome (FXS) | Enrichment in FXS related genes in cortex | (84) |
| Alzheimer’s disease | Decrease or increase in the genome in hippocampus, cerebellum and cortex | (101–105) |
| Huntington’s disease | Decrease in 5′ UTR region of ADORA2A in putamen | (103, 106) |
| Amyotrophic lateral sclerosis | Global increase in spinal cord | (107) |
| Friedreich’s ataxia | Increase in 5′ GAA repeat region of FXN in cerebellum and heart | (108) |
Conclusion
Relative to 5-mC, the 5-hmC variant of DNA methylation remains poorly understood. To date, studies have indicated that 5-hmC not only serves as a DNA demethylation intermediate but functions as a stable epigenetic mark that is read by specific chromatin-associated proteins. Indeed, 5-hmC, as well as other oxidative derivatives of 5-mC, can be stable (68). The enrichment of 5-hmC over gene bodies, certain promoters, and transcription factor binding sites integrated with transcriptomic data offer suggestive, multiple roles for 5-hmC in regulating gene expression potentially via influencing transcription initiation, pre-mRNA splicing, and/or RNA methylation. As current evidence reveal correlative relationships, the precise mechanism by which 5-hmC may play a role in these critical cellular activities remains under investigation. Meanwhile, abnormalities in the levels and distribution of 5-hmC in the context of disease conditions are only now being recognized and described, briefly reviewed here. Although we have speculated on a few potential roles, we have yet to understand the consequences of perturbations to 5-hmC in the genome. We anticipate more precise mechanistic insight into the role(s) of 5-hmC as single-cell analysis of DNA methylation states genome-wide advance. Single-cell genome sequencing has been reviewed recently (113), with the promise of new knowledge informing future diagnostic and therapeutic strategies of epigenetic-based diseases.
Acknowledgments
This work was supported in part by National Institutes of Health grants P30GM103341, U54MD007584, G12MD007601, P20GM103457, and K01HL125504 (A.K.M.) as well as by the Queen’s Health Systems Native Hawaiian Health Initiative, Queen’s Medical Center (A.K.M.).
List of abbreviations
- 5-hmC
5-hydroxymethylcytosine
- 5-mC
5-methylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- TET
ten-eleven translocation methylcytosine dioxygenase
- DNMT
DNA methyltransferase
- SAM
S-adenosyl-L-methionine
- NGS
next generation sequencing
- CpG
cytosine-phosphate-guanine
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
References
- 1.Rossetto D, Truman AW, Kron SJ, Cote J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res. 2010;16:4543–52. doi: 10.1158/1078-0432.CCR-10-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore SP, Toomire KJ, Strauss PR. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst) 2013;12:1152–8. doi: 10.1016/j.dnarep.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genetics. 2013;9:e1003542. doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3. 3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–69. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maunakea AK, Chepelev I, Zhao K. Epigenome mapping in normal and disease States. Cir Res. 2010;107:327–39. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–19. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 9.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–66. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 11.Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, Jäger N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 13.Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–8. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 14.Penn NW, Suwalski R, O’Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–90. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ, Andrews S, Balasubramanian S, Reik W. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Müller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C. Genomewide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–91. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature Neurosci. 2011;14:1607–16. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–77. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 28.Sun W, Zang L, Shu Q, Li X. From development to diseases: the role of 5hmC in brain. Genomics. 2014;104:347–51. doi: 10.1016/j.ygeno.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–51. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 30.Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr Opin Cell Biol. 2013;25:289–96. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–16. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 33.Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 34.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 35.Hotchkiss RD. The mode of action of chemotherapeutic agents. Annu Rev Microbiol. 1948;2:183–214. doi: 10.1146/annurev.mi.02.100148.001151. [DOI] [PubMed] [Google Scholar]
- 36.Gold M, Hurwitz J, Anders M. The Enzymatic Methylation of Rna and DNA, Ii. On the Species Specificity of the Methylation Enzymes. Proc Natl Acad Sci USA. 1963;50:164–9. doi: 10.1073/pnas.50.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheid B, Srinivasan PR, Borek E. Deoxyribonucleic acid methylase of mammalian tissues. Biochemistry. 1968;7:280–5. doi: 10.1021/bi00841a034. [DOI] [PubMed] [Google Scholar]
- 38.Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, Walter J. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark SJ, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–7. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 40.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 41.Lande-Diner L, Cedar H. Silence of the genes–mechanisms of long-term repression. Nat Rev Genet. 2005;6:648–54. doi: 10.1038/nrg1639. [DOI] [PubMed] [Google Scholar]
- 42.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 43.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 44.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–27. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 45.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–14. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 46.Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Pàldi A, Viegas-Péquignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–13. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 48.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 49.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 50.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma RP, Gavin DP, Grayson DR. CpG methylation in neurons: message, memory, or mask? Neuropsychopharmacology. 2010;35:2009–20. doi: 10.1038/npp.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–12. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 53.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Schöler H, Walter J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–88. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–66. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 56.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 57.van der Wijst MG, Venkiteswaran M, Chen H, Xu GL, Plosch T, Rots MG. Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase. Epigenetics. 2015;10:671–6. doi: 10.1080/15592294.2015.1062204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–86. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, Humphray S, Cox T, Lang-ford C, Bird A. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang P, Song F, Ghosh S, Morien E, Qin M, Mahmood S, Fujiwara K, Igarashi J, Nagase H, Held WA. Genome-wide survey reveals dynamic widespread tissue-specific changes in DNA methylation during development. BMC Genomics. 2011;12:231. doi: 10.1186/1471-2164-12-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol. 2011;30:267–75. doi: 10.1089/dna.2010.1094. [DOI] [PubMed] [Google Scholar]
- 63.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Krämer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corley MJ, Zhang W, Zheng X, Lum-Jones A, Maunakea AK. Semiconductor-based sequencing of genome-wide DNA methylation states. Epigenetics. 2015;10:153–66. doi: 10.1080/15592294.2014.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, Murrell A, Balasubramanian S. 5-Formylcytosine can be a stable DNA modification in mammals. Nat Chem Biol. 2015;11:555–7. doi: 10.1038/nchembio.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Elevated 5-hydroxymethylcytosine in the Engrailed-2 (EN-2) promoter is associated with increased gene expression and decreased MeCP2 binding in autism cerebellum. Transl Psychiatry. 2014;4:e460. doi: 10.1038/tp.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet. 2014;15:647–61. doi: 10.1038/nrg3772. [DOI] [PubMed] [Google Scholar]
- 75.Bijwaard K, Dickey JS, Kelm K, Tezak Z. The first FDA marketing authorizations of next-generation sequencing technology and tests: challenges, solutions and impact for future assays. Expert Rev Mol Diagn. 2015;15:33–40. doi: 10.1586/14737159.2015.979795. [DOI] [PubMed] [Google Scholar]
- 76.Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8:1841–51. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu M, Hon GC, Szulwach KE, Song CX, Jin P, Ren B, He C. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 2012;7:2159–70. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui L, Chung TH, Tan D, Sun X, Jia XY. JBP1-seq: a fast and efficient method for genome-wide profiling of 5hmC. Genomics. 2014;104:368–75. doi: 10.1016/j.ygeno.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 79.Tan L, Xiong L, Xu W, Wu F, Huang N, Xu Y, Kong L, Zheng L, Schwartz L, Shi Y, Shi YG. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 2013;41:e84. doi: 10.1093/nar/gkt091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–7. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 84.Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, Tice R, Tochigi M, Moréra S, Nazarians A, Belsham D, Wong AH, Blencowe BJ, Wang SC, Kapranov P, Kustra R, Labrie V, Klimasauskas S, Petronis A. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nature Struct Mol Biol. 2012;19:1037–43. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104:341–6. doi: 10.1016/j.ygeno.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 86.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 87.Gao F, Xia Y, Wang J, Luo H, Gao Z, Han X, Zhang J, Huang X, Yao Y, Lu H, Yi N, Zhou B, Lin Z, Wen B, Zhang X, Yang H, Wang J. Integrated detection of both 5-mC and 5-hmC by high-throughput tag sequencing technology highlights methylation reprogramming of bivalent genes during cellular differentiation. Epigenetics. 2013;8:421–30. doi: 10.4161/epi.24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776–81. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haberle V, Forrest AR, Hayashizaki Y, Carninci P, Lenhard B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res. 2015;43:e51. doi: 10.1093/nar/gkv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 93.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 94.Baylin SB, Jones PA. A decade of exploring the cancer epigenome–biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–80. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 96.Lian CG1, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Jr, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martínez JA, Pape UJ, Jacobsen SE, Peters B, Rao A. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:1361–6. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhattacharyya S, Yu Y, Suzuki M, Campbell N, Mazdo J, Vasanthakumar A, Bhagat TD, Nischal S, Christopeit M, Parekh S, Steidl U, Godley L, Maitra A, Greally JM, Verma A. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Res. 2013;41:e157. doi: 10.1093/nar/gkt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, Wu H, Warren ST, Jin P, Duan R, Li X. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet. 2012;21:5500–10. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR, Steinbusch HW, Coleman PD, Rutten BP, van den Hove DL. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol Aging. 2013;34:2091–9. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Condliffe D, Wong A, Troakes C, Proitsi P, Patel Y, Chouliaras L, Fernandes C, Cooper J, Lovestone S, Schalkwyk L, Mill J, Lunnon K. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiol Aging. 2014;35:1850–4. doi: 10.1016/j.neurobiolaging.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Villar-Menéndez I, Blanch M, Tyebji S, Pereira-Veiga T, Albasanz JL, Martín M, Ferrer I, Pérez-Navarro E, Barrachina M. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A2AR levels in Huntington’s disease. Neuromolecular Med. 2013;15:295–309. doi: 10.1007/s12017-013-8219-0. [DOI] [PubMed] [Google Scholar]
- 104.Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging. 2014;35:1334–44. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 105.Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer’s disease. Mech Ageing Dev. 2013;134:486–95. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Luo HR, Guo C, Han C, Tang TS. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington’s disease. Hum Mol Genet. 2013;22:3641–53. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- 107.Figueroa-Romero C, Hur J, Bender DE, Delaney CE, Cataldo MD, Smith AL, Yung R, Ruden DM, Callaghan BC, Feldman EL. Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS One. 2012;7:e52672. doi: 10.1371/journal.pone.0052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Al-Mahdawi S, Sandi C, Mouro Pinto R, Pook MA. Friedreich ataxia patient tissues exhibit increased 5-hydroxymethylcytosine modification and decreased CTCF binding at the FXN locus. PLoS One. 2013;8:e74956. doi: 10.1371/journal.pone.0074956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Al-Mahdawi S, Virmouni SA, Pook MA. The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front Neurosci. 2014;8:397. doi: 10.3389/fnins.2014.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, Koo JW, Ribeiro E, Labonte B, Laitman BM, Estey D, Stockman V, Kennedy P, Couroussé T, Mensah I, Turecki G, Faull KF, Ming GL, Song H, Fan G, Casaccia P, Shen L, Jin P, Nestler EJ. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18:536–44. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fong KS, Hufnagel RB, Khadka VS, Corley MJ, Maunakea AK, Fogelgren B, Ahmed ZM, LozanoffS A mutation in the tuft mouse disrupts TET1 activity and alters the expression of genes that are crucial for neural tube closure. Dis Model Mech. 2016;9:585–96. doi: 10.1242/dmm.024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marx V. Genetics: profiling DNA methylation and beyond. Nat Methods. 2016;13:119–22. doi: 10.1038/nmeth.3736. [DOI] [PubMed] [Google Scholar]
- 113.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–88. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]


