Abstract
Binding of the thiazolidinedione antidiabetic drug pioglitazone led to the discovery of a novel outer mitochondrial membrane protein of unknown function called mitoNEET. The protein is homodimeric and contains a uniquely ligated two iron–two sulfur cluster in each of its two cytosolic domains. Electrospray ionization mass spectrometry was employed to characterize solutions of the soluble cytosolic domain (amino acids 32–108) of the protein. Ions characteristic of dimers containing the cofactors were readily detected under native conditions. mitoNEET responded to exposure to solutions at low pH by dissociation to give monomers that retained the cofactor, followed by dissociation of the cofactor in a concerted fashion. mitoNEET formed complexes with resveratrol-3-sulfate, one of the primary metabolites of the natural product resveratrol. Resveratrol itself showed no tendency to interact with mitoNEET. The formation of complexes was evident in both electrospray ionization mass spectrometry and isothermal titration calorimetry measurements. Up to eight molecules of the compound associated with the dimeric form of the protein in a sequential fashion. Dissociation constants determined by micorcalorimetry were in the range 5–16 μM for the various binding sites. The only other known naturally occurring binding partner for mitoNEET at present is NADPH. It is very interesting that the iron–sulfur cluster containing protein interacts with two potentially redox active substances at the surface of mitochondria. These findings provide a new direction for research into two poorly understood, yet biomedically relevant, species.
Graphical Abstract
The outer mitochondrial membrane protein, mitoNEET, was discovered on the basis of its ability to bind the antidiabetic drug pioglitazone.1 The physiological role for this protein is unknown. Remarkably, it contains a redox active cofactor, an iron–sulfur cluster.2 MitoNEET is closely related in structure to the endoplasmic reticulum protein, Miner1.3 Mis-splicing of the gene for Miner1 results in Type 2 Wolfram syndrome, a disease characterized by reduced life expectancy, diabetes mellitus, and optical atrophy.4 It is not known what purpose is served by the redox cofactor being localized to the outer mitochondrial membrane by mitoNEET. It could be there to participate in electron-transfer reactions, or perhaps it plays a role in iron–sulfur cluster biogenesis or trafficking. Moreover, it is not known if mitoNEET acts biologically in concert with any binding partners, either proteins or small molecules. It was recently found that mitoNEET binds NADPH stoichiometrically, albeit weakly, and that the interaction accelerates the dissociation of the iron–sulfur cluster.5 Pioglitazone and NADPH are the only compounds reported to interact with mitoNEET so far.
MitoNEET has interesting features. It consists of a short mitochondrial membrane anchoring sequence and a cytosolic domain.2 It forms homodimers through the cytosolic domain, and each polypeptide associates with one [2Fe–2S] cluster. According to the crystallographically determined three-dimensional structure of the cytosolic domain, the protein binds the irons through three cysteines and one histidine, rather than (more commonly) four cysteines or two cysteines and two histidines.6–8
In a computer-aided docking study we recently identified potential binding sites on mitoNEET for other compounds in the thaizolidinedione family.9 The hypothesis was that an interaction with mitoNEET might account for the observed neuroprotection observed with these compounds as a consequence of an interaction at the outer mitochondrial membrane. One of the compounds predicted to bind mitoNEET altered mitochondrial function and protected neuronal cells from rotenone-induced cell death.9
In an attempt to identify additional binding partners for mitoNEET, we discovered that resveratrol-3-sulfate, but not resveratrol, forms complexes with the protein. Because there is no known bioassay for mitoNEET, direct physical methods, electrospray ionization mass spectrometry, and isothermal titration calorimetry were employed in this investigation. It was also found unexpectedly that dissociation of the mitoNEET homodimer in the absence of ligand precedes dissociation of the cofactor at low pH.
MATERIALS AND METHODS
Protein Preparation
A plasmid (pET3a) bearing the cDNA for human mitoNEET amino acids 32–108 incorporated into E. coli C43(DE3) was purchased from Entelechon (Regensburg). Additionally, the DNA coded for a thrombin cleavage site at the C-terminus (SSGLVPR/GS) of the protein followed by a hexahistidine tag. The cells were grown in 2xYT medium containing ampicilin (100 μg/mL) at 37 °C with shaking at 210 rpm to an A(595) of ~0.2. An aliquot of iron(III) chloride solution was added (1 μM final concentration), and the cells were allowed to continue to grow at 37 °C and 210 rpm to an A(595) of ~0.8. IPTG (0.5 mM) was added, and the cells were incubated at 29 °C and 210 rpm for 10 h. The cells were collected by centrifugation, washed in phosphate buffered saline, and resuspended in lysis buffer containing trypsin inhibitor (60 μg/mL), DNaseI (20 μg/mL), phenylmethylsulfonyl fluoride (100 μg/mL), β-mercaptoethanol (0.001 μL/mL), and imidazole (10 mM) in TrisHCl (0.05 M, pH 8.5, 0.3 M NaCl). The cells were stored at −80 °C. Cells were thawed, treated with lysozyme (1.6 mg/mL), and sonicated. The mixture was centrifuged (40 000 rpm, 4 °C, 1 h), and the red supernatant was combined with NiNTA Superflow (Qiagen). The red beads were washed with TrisHCl (3X, 0.05 M, pH 8.5, 0.3 M NaCl) containing 0.020 M imidazole, and the protein was released by treating the beads with TrisHCl (3X, 0.05 M, pH 8.5, 0.3 M NaCl) containing 0.250 M imidazole. The red solution was dialyzed against TrisHCl (0.05 M, pH 8.5, 0.3 M NaCl, 4 °C) and concentrated (Amicon Ultra) to 1 mg/mL. The mitoNEET solution was treated with thrombin (Enzyme Research) on ice for 30 min and was loaded onto SP-Sepharose eluted with a shallow gradient from 0.1 to 0.5 M NaCl in TrisHCl (0.05 M, pH 8.5). The protein following capture and release on NiNTA Superflow contained considerable A(260) absorbing materials, presumably nucleic acids, that were easily removed by cation exchange chromatography. Red fractions from the ion exchange column containing mitoNEET were combined and concentrated for further experimentation.
Analytical Methods
Amino acid analyses were conducted by AAA Service Laboratory (Damascus, OR). Iron determinations were carried out on a Perkin-Elmer Model 5100PC atomic absorption spectrophotometer operating in the flame ionization mode using a certified atomic absorption iron standard (Fisher).
Crystallization
Red crystals formed in hanging drop vapor diffusion experiments when solutions of mitoNEET (23.3 mg/mL, TrisHCl, 0.05 M, pH 8.5, 0.3 M NaCl) were combined (1:1) with 1.7 M ammonium sulfate, TrisHCl, 0.1 M, pH 8.5. Cryoprotection was afforded by a short soak in the mother liquor containing 20% xylitol. Crystal soaking experiments were conducted by transferring mitoNEET crystals into a 12 mM solution of resveratrol-3-sulfate in mother liquor.
ESI-MS Experiments
Mass spectrometry measurements were obtained with a Q-Tof Micro (Waters) mass spectrometer with electrospray ionization interface operating in the positive ion mode. The ESI capillary voltage was set at 3 kV, sample cone voltage was 30 V, and extraction cone voltage was 2.5 V. The collision energy applied to all MS lenses preceding the collision cell was varied between 2 and 10 V, and it was found that the optimal value for the transmission and detection of intact protein ions was 5 V. The desolvation temperature was 100 °C while the source temperature was 90 °C. The samples were infused into the mass spectrometer at a flow rate of 5 μL/min using a syringe pump (Fisher). Nitrogen gas was used for desolvation and nebulization of the samples. The data were acquired and analyzed using MassLynx software (Waters). Protein solutions (10 μM) were prepared in 10 mM ammonium acetate buffer adjusted to the appropriate pH with ammonium hydroxide or acetic acid. A concentrated stock solution of each compound tested was prepared in ammonium acetate, 10 mM, pH 8.5, and combined with appropriate dilutions of the protein in the same buffer to achieve the reported molar ratios.
Microcalorimetry
Isothermal titration calorimetry was performed in a VP-ITC MicroCalorimeter (MicroCal). A thoroughly degassed solution of mitoNEET (TrisHCl, 0.05 M, pH 8.5, 0.3 M NaCl) was transferred to the sample cell and combined with 0.010 mL aliquots of the compounds dissolved in the same buffer at 30 °C. The buffers for the protein and the titrant were carefully matched. The protein was dialyzed extensively against the buffer employed in the titrations, and the final dialysis buffer was used to dissolve the resveratrol-3-sulfate. The concentrations were mitoNEET, 0.203 mM, and resveratrol-3-sulfate, 2.6 mM; mitoNEET, 0.044 mM, and resveratrol, 0.412 mM. The integration of the observed heats was carried out in Origin 7.0 for the VP-ITC (Microcal).
Molecular Modeling and Docking Studies
Molecular modeling was carried out as previously reported, with minor modification.9 Briefly, resveratrol-3-sulfate was drawn using MarvinSketch (ChemAxon), after which it was minimized in MOE (Chemical Computing Group) with the MMFF94s force field and a termination criterion of 0.01 kcal/mol, for a total of 1000 iterations. The compound was also protonated to correspond to pH 7.4. Docking studies with mitoNEET were done with the protein file 2Q7H from the Protein Data Bank. To prepare the structure, hydrogens were added and amino acids were protonated for pH 7.4. The SiteFinder module in MOE identified seven binding sites, which were then used in the docking studies. After using MOE-Dock, the top-scoring pose was used for analysis such as interactions in the binding pocket with amino acids.
RESULTS
Purification and Characterization of the Human MitoNEET Employed in the Study
The protein was obtained from an E. coli expression procedure. Briefly, the cDNA for the presumed cytosolic domain of human mitoNEET (32–108) was purchased as an insert in the pET3a vector. The construct included a C-terminal six-histidine sequence beyond a thrombin cleavage site. Cells (C43/DE3) were grown in 2xYT supplemented with iron chloride. Induction with IPTG led to the overexpression of a red protein that was collected on nickel–NTA beads. The thrombin cleaved product was purified to homogeneity by ion exchange chromatography. A solution of the protein was subjected to iron determination (triplicate), amino acid analysis (two samples, each duplicated), and UV–vis spectrophotometry. According to the amino acid analysis, the concentration of the solution was 47.8 ±1.7 μM in monomer. Atomic absorbance spectrophotometry conducted on the same solution provided an iron concentration of 94.2 ± 1.4 μM or 1.98 iron atoms per monomer. From the UV–vis absorbance spectrum for this solution, the molar absorptivity at 457 nm was 8286 L/(mol cm) for the monomer of this soluble domain of human mitoNEET (hereafter mitoNEET). A bona fide molar absorptivity value made it possible to determine concentrations of solutions of the protein for all subsequent measurements. Concentrated solutions of the purified protein yielded red crystals from vapor diffusion hanging drop experiments with ammonium sulfate in Tris-HCl buffer at pH 8.5. The crystals diffracted X-rays (Life Sciences-Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory) to 1.76 Å resolution and contained mitoNEET dimers in the I4122 space group. The three-dimensional structure of the protein (PDB 3REE) solved by the molecular replacement method (PDB 2R13 as model) was not significantly different from earlier reported crystal structures.6–8 These observations simply verify that we are investigating the same protein as other investigators in this area and that there is a sound basis for the determination of the concentrations of mitoNEET solutions.
Solution Characteristics of Human MitoNEET Using ESI-MS
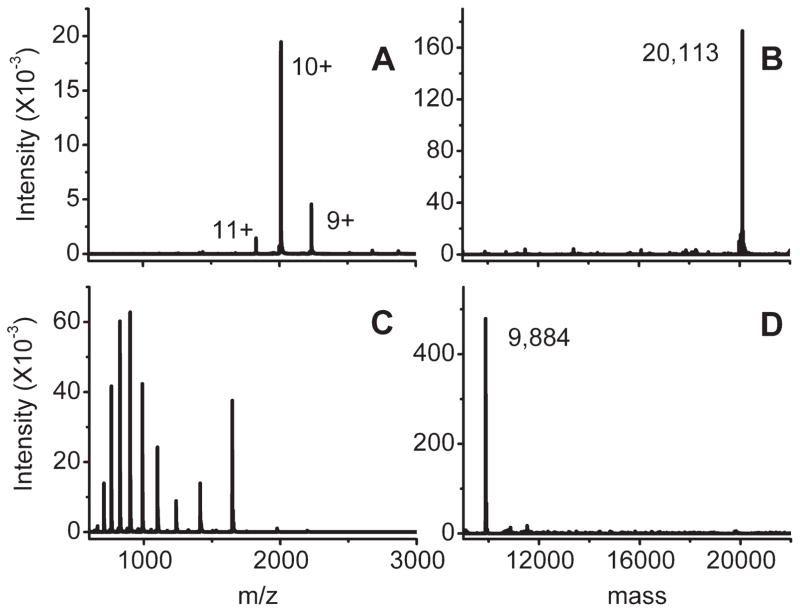
Native and denatured solutions of mitoNEET were subjected to ESI-MS (Figure 1). Solutions of the protein under native conditions (pH 8.5, ammonium acetate buffer) afforded ions characteristic exclusively of dimers (Figure 1A,B). The calculated mass of the dimer was 20 112 (vs 20 113 observed), which included the mass of four iron atoms and four sulfur atoms. When the protein solution was combined with acetonitrile (1:1) and formic acid (1%) and allowed to stand at room temperature for several hours, the mass spectra in Figure 1C,D were obtained. All of the ions in the spectra were characteristic of monomers that contained no iron or sulfur (calculated MW 9884).
Figure 1.
Electrospray ionization mass spectra, panels A and C, and deconvolutions, panels B and D: (A, B) 10 μM mitoNEET, 10 mM ammonium acetate, pH 8.5; (C, D) 10 μM mitoNEET treated with formic acid and acetonitrile for 3 h at room temperature.
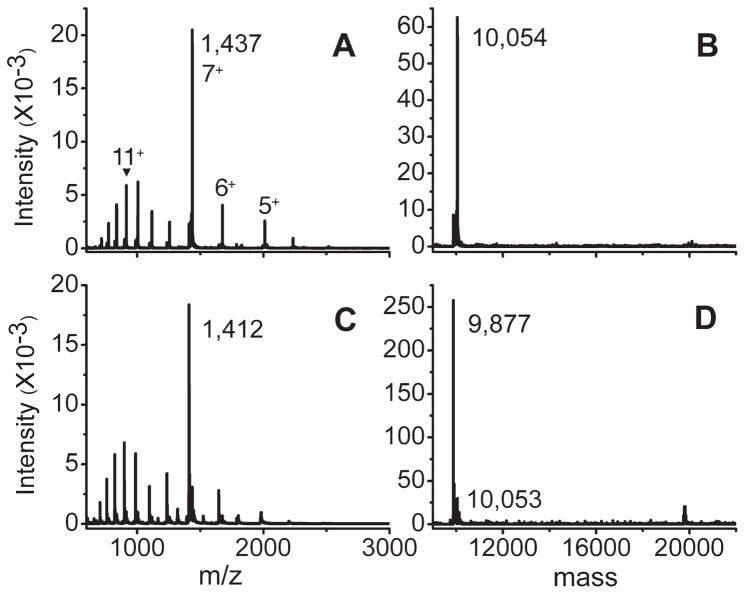
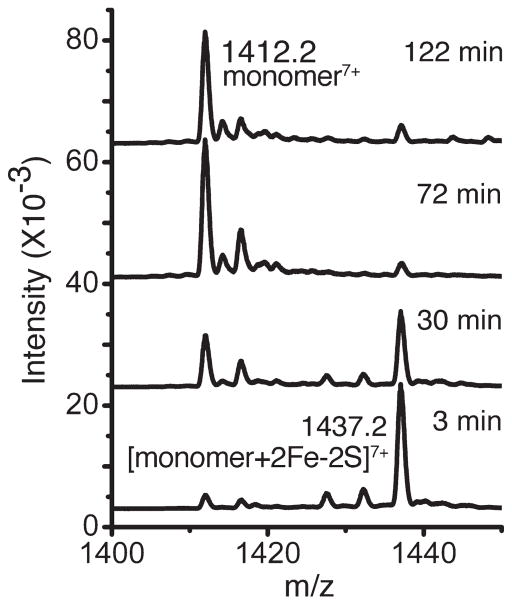
When the pH of the mitoNEET solution was reduced to 5.1 in ammonium acetate buffer for ESI-MS, two sets of ions, m/z 700–1200 and m/z 1250–2500, were present in the spectrum (Figure 2A). The existence of a bimodal distribution of charge states is commonplace in protein ESI-MS and indicates the existence of two conformational states that are more and less compact.10 Rather unexpectedly, however, in this experiment both sets of ions were from monomers that retained the iron–sulfur clusters (Figure 2B, calculated MW 10 056 vs 10 054 observed). Spectra recorded at subsequent time points reflected a smooth conversion to the apo form of each set of ions simultaneously. For example, after 2 h on ice (Figure 2C,D), both sets of ions were still present, but both were then devoid of the iron sulfur clusters. This is further illustrated in Figure 3. There was not a discernible stepwise process involved in the loss of the iron–sulfur clusters. Rather, the ions shifted to the lower mass values in a more or less concerted fashion. In response to a reduced pH, it was clear from this experiment that mitoNEET dissociated to monomers first and subsequently lost the iron sulfur clusters more slowly. This contrasts with the dimeric protein containing iron sulfur clusters that was stable for studies by ESI-MS when maintained at pH > 8.
Figure 2.
Electrospray ionization mass spectra, panels A and C, and deconvolutions, panels B and D: (A, B) 10 μM mitoNEET, 10 mM ammonium acetate, pH 5.1, measured immediately; (C, D) solution from (A, B) measured after 2 h on ice.
Figure 3.
Electrospray ionization mass spectra, 10 μM mitoNEET, 10 mM ammonium acetate, pH 5.1, incubated for the indicated times on ice.
Resveratrol-3-sulfate Forms Complexes with MitoNEET
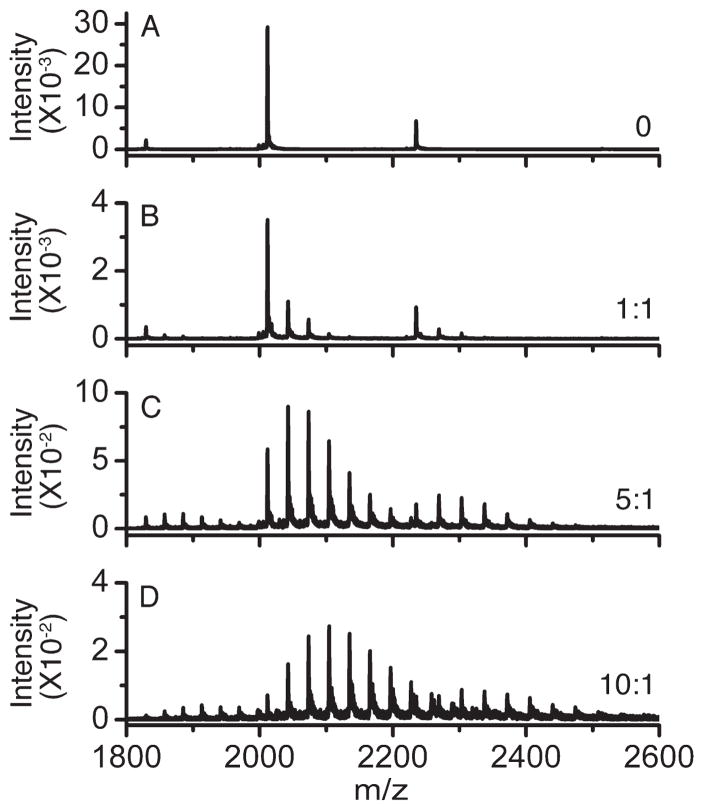
MitoNEET was combined with resveratrol-3-sulfate in various molar ratios in ammonium acetate buffer at pH 8.5 and subjected to ESI-MS. Ions with masses characteristic of complexes between the protein and the compound were readily evident in the spectra (Figure 4). Remarkably, ions for complexes of as many as eight molecules per mitoNEET dimer were present. Only ions of masses attributable to the intact dimer were observed. That is, there was no evidence for dimer dissociation upon ligand binding. Spectra for solutions of mitoNEET treated with resveratrol in the same way contained no ions that could be attributed to a complex of any stoichiometry (data not shown).
Figure 4.
Electrospray ionization mass spectra: resveratrol-3-sulfate: mitoNEET at the indicated molar ratios and 10 μM mitoNEET in each, 10 mM ammonium acetate, pH 8.5.
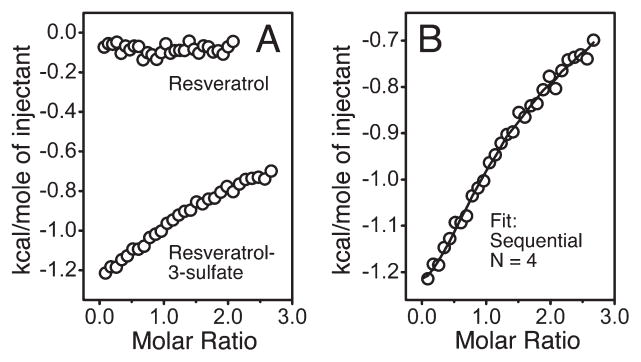
The formation of complexes between mitoNEET and resveratrol-3-sulfate in solution was confirmed by isothermal titration calorimetry. Injections of resveratrol-3-sulfate, but not resveratrol, into solutions of mitoNEET in the microcalorimeter produced heat (Figure 5A). The data were fit with a stoichiometry of four molecules per monomer (Figure 5B) using a sequential binding model. This was consistent with the mass spectrometric findings. The four dissociation constants for the fit were in the range of 5–16 μM (Table 1). In each case, the enthalpy and entropy changes associated with the fit were small but favorable.
Figure 5.
Isothermal titration calorimetry (30 °C) for mitoNEET titrated with resveratrol and resveratrol-3-sulfate: (A) comparison of resveratrol and resveratrol-3-sulfate; (B) resveratrol-3-sulfate data were fit with a sequential model and four binding sites.
Table 1.
Thermodynamic Parameters for the Titration of MitoNEET with Resveratrol-3-sulfate Using a Sequential Binding Sites Model
| site | K (M), 1/K (μM) | ΔH (cal/mol) | ΔS (cal/(deg mol)) |
|---|---|---|---|
| 1 | (1.88 ± 0.20) × 105, 5.3 | −1246 ± 13.1 | 20.0 |
| 2 | (1.04 ± 0.11) × 104, 9.6 | −1143 ± 83.0 | 19.2 |
| 3 | (6.12 ± 0.69) × 104, 16.3 | −152.7 ± 213 | 21.4 |
| 4 | (6.44 ± 1.1) × 104, 15.5 | −1077 ± 225 | 18.4 |
Molecular modeling of resveratrol-3-sulfate docking with mitoNEET identified a number of potential binding sites. Figure 6 illustrates some of the predicted binding sites from the perspective of the presumed cytosolic side of mitoNEET. In some of the sites, the ligand was accommodated in more than one orientation (sites 1/3 and 2/4). Sites were predicted both at the interface between the monomers and exclusively to one of the molecules. When crystals of mitoNEET were soaked briefly in solutions containing resveratrol-3-sulfate, no new density was evident in electron density difference maps upon X-ray diffraction analysis. Longer soaking experiments led to catastrophic degradation of the mitoNEET crystals.
Figure 6.
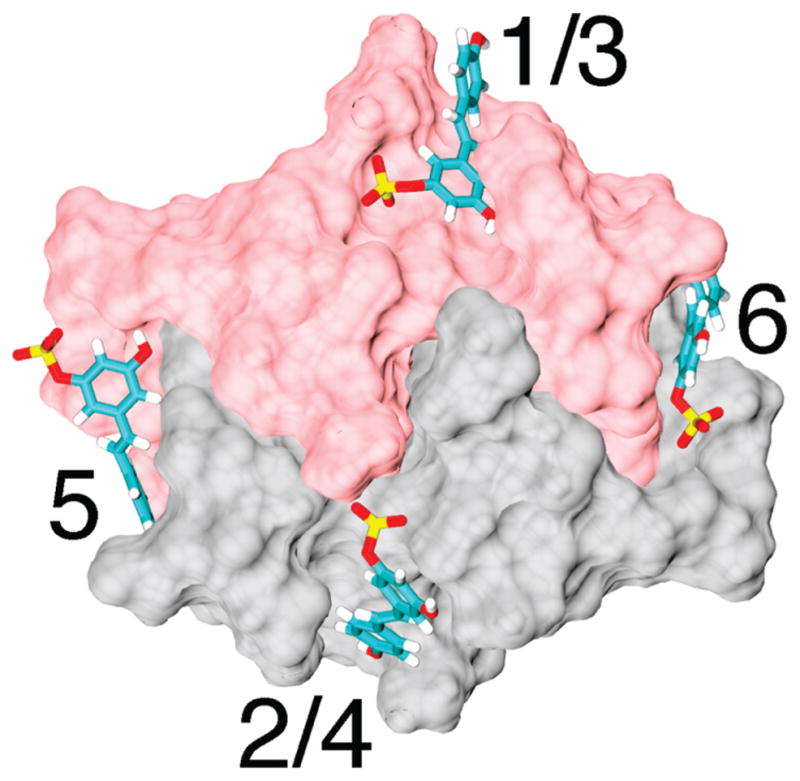
Structural model from the presumed cytoplasmic perspective for mitoNEET indicating predicted binding sites for resveratrol-3-sulfate.
DISCUSSION
Dimers of mitoNEET containing iron–sulfur clusters were readily detected in the ESI-MS experiments conducted under native conditions, pH 8.5 in ammonium acetate buffer. The electrospray ionization of the mitoNEET samples under native conditions preserved the quaternary structure of the protein ions. Such analysis is called native mass spectrometry and is readily performed on the Q-Tof mass spectrometers.11 Because the charge of the mitoNEET ions was lower under native conditions, the Q-Tof mass spectrometer used in this study was optimized in order to increase transmission of the ions with with high m/z values. The optimization included the minimization of the collision energy applied to the protein ions in order to prevent their fragmentation prior to MS detection. Acidification and exposure to organic solvent led to dissociation of the dimers as well as loss of the iron sulfur clusters. Interestingly, there were contributions from two sets of ions in the spectra even under harsh denaturing conditions. The ions reflect the existence of two conformations of the protein with different degrees of compactness. While the ions at the lowest m/z values (700–1200) in acidified solutions containing organic solvent were likely from the fully denatured protein, ions with m/z values (1250–2500) intermediate between fully denatured and native solution structures were also present. Because of the acid labile nature of the sulfur in iron–sulfur clusters,12 loss of all or part of the prosthetic group, e.g., one or more sulfur atoms, from mitoNEET prior to unfolding could have been a reasonable expectation. Quite to the contrary, the iron–sulfur cluster was retained in the monomer in two conformations of the monomeric protein at least initially at pH 5.1 in aqueous solution. When the cluster did dissociate from the protein, it appeared to do so all at once.
Proteins containing [2Fe–2S] clusters, e.g., ferredoxins, were characterized by ESI-MS previously. Some of these proteins had acidic pIs and ions from the holoprotein could best be observed in the negative-ion mode.13 MitoNEET is a basic protein, so this was not necessary even at pH > 8. ESI with FT-ICR mass spectrometry was used to assign both stoichiometry and oxidation state to a variety of iron–sulfur cluster proteins.14 For example, spectra for the [2Fe–2S] cluster in iron hydrogenase from T. maritima, contained ions for the holoprotein as well as ions corresponding to the loss of one or two sulfur atoms from the cluster.15 Two enzymes containing [2Fe–2S] clusters were also previously studied by ESI-MS, APS reductase, and biotin synthase. Under nondenaturing conditions, the spectrum of APS reductase contained ions that were attributed to the apoprotein, an intermediate containing the [2Fe–2S] cluster, and the holoprotein with a [4Fe–4S] cluster.16 Biotin synthase is a homodimer that contains one [2Fe–2S] cluster at the interface. ESI spectra for the synthase were populated with ions from the apomonomer, the monomer containing the [2Fe–2S] cluster, and the dimeric holoprotein.17 None of the previous reports included an investigation of the effect of the solution conditions, e.g., solution pH, on the ESI-MS characteristics of the [2Fe–2S] cluster proteins.
It is clear from the concentration dependence of the ESI-MS data for mitoNEET in the presence of resveratrol-3-sulfate that multiple molecules associated with the protein before saturation occurred for the first binding molecule, suggesting a sequential and potentially cooperative binding effect. It seems extremely likely that electrostatics play an important role in the interactions. MitoNEET bears a calculated pI of 9.41, making it substantially positive even at the pH of 8.5 used for the ESI-MS experiments. That it would attract and interact with the negatively charged resveratrol-3-sulfate is entirely logical. Nonspecific interactions based solely on electrostatics tend not to be cooperative in nature.18 Ion intensities for proteins bearing nonspecifically bound ligands decline monotonically as the number of ligands increases. A recent example where it was possible to separate the specific from the nonspecific effects mathematically was the association of ADP with creatine kinase.19 In that case, a lot was already known about the stoichiometry and thermodynamics of the interaction. The concentration dependence of the ESI-MS for mitoNEET and resveratrol-3-sulfate clearly resembles the data for creatine kinase and ADP. The other molecules known to interact with mitoNEET are pioglitazone and NADPH.5,8 While NADPH had a destabilizing effect on the protein and its cofactor, the effect of the thizolidinedione was a stabilizing one. The interaction of NADPH with mitoNEET was roughly 3 orders of magnitude weaker than was found for resveratrol-3-sulfate in the present study. Furthermore, in contrast to resveratrol-3-sulfate, the stoichiometry of the NADPH interaction was 1:1 with respect to the protein monomer.5 It is very interesting that mitoNEET interacts with two potentially redox active molecules located as it is on the outer surface of the mitochondrion.
The molecular modeling predicts five discrete and a total of seven potential binding sites for resveratrol-3-sulfate on the holoprotein, less than the value actually found by the physical measurements, ESI-MS, and isothermal titration calorimetry, four per monomer or potentially eight altogether. The docking calculations presuppose independent binding sites and a fixed structure for the protein. Since sequential models provide the best fit for the ligand binding experiments, it is entirely possible that conformational changes accompanying the initial interactions account for binding of additional resveratrol-3-sulfate molecules. Initial attempts to produce crystals containing complexes between mitoNEET and resveratrol-3-sulfate for structure determination by crystal soaking experiments were not successful. The native crystals were quite sensitive to the presence of ligand. These observations were not too surprising since multiple molecules bind, the predicted binding sites are surface oriented, and conformational changes induced by ligand binding seem likely.
Resveratrol is a natural product that has attracted a lot or research interest for its numerous favorable biological activities. For example, it extended the lifespan of S. cerevisiae, C. elegans, and D. melanogaster.20–22 It was found to have both chemopreventive activity for cancer23 and cardioprotective effects.24 Mechanisms that account for some of these effects include activation of sirtuins25 and stimulation of AMP-activated protein kinase.26 Resveratrol undergoes conversion in vivo such that only the primary metabolites, resveratrol-3-O-glucuronide and resveratrol-3-sulfate, were detected in serum or urine from experimental animals treated with oral or intravenous doses.27 Similarly, in humans, orally administered resveratrol could only be detected at trace levels in serum, whereas metabolites accounted for at least 70% of the administered dose.28 Resveratrol-3-sulfate has many of the same biological effects that have been attributed to resveratrol.29 This leads to the supposition that the metabolites of resveratrol may account for the some of the favorable biological effects ascribed to the natural product. The discovery that resveratrol-3-sulfate binds to mitoNEET provides an intriguing new lead in this line of inquiry.
Acknowledgments
The superb technical assistance of Dr. Panee Burkel in conducting the atomic absorption spectrophotometry measurements is gratefully acknowledged.
Funding Sources
This research was supported by funding from the University of Toledo, the Stark Community Foundation, and the National Heart, Lung, and Blood Institute of the National Institutes of Health (HL091482).
ABBREVIATIONS
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- 2Fe–2S
two iron–two sulfur cluster
- ESI-MS
electrospray ionization mass spectrometry
- APS
adenosine 5′-phosphosulfate
- AMP
adenosine-5′-monophosphate
References
- 1.Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR. Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am J Physiol Endocrinol Metab. 2004;286:E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 2.Wiley SE, Rardin MJ, Dixon JE. Localization and function of the 2Fe-2S outer mitochondrial membrane protein mitoNEET. Met Enzymol. 2009;456:233–246. doi: 10.1016/S0076-6879(08)04413-3. [DOI] [PubMed] [Google Scholar]
- 3.Conlan AR, Axelrod HL, Cohen AE, Abresch EC, Zuris J, Yee D, Nechustai R, Jennings PA, Paddock ML. Crystal Structure of Miner1: the redox active 2Fe-2S protein causative in Wolfram Syndrome. J Mol Biol. 2009;392:143–153. doi: 10.1016/j.jmb.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amr S, Heisey C, Zhang M, Xia XJ, Shows KH, Ajlouni K, Pandya A, Satin LS, El-Shanti H, Shiang R. A homozygous mutation in a novel zinc finger protein, ERIS, is responsible for Wolfram Syndromg 2. Am J Hum Genet. 2007;81:673–683. doi: 10.1086/520961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T, Lin J, Feng Y, Wang J. Binding of reduced nicotinamide adenine dinucleotide phosphate destabilizes the iron-sulfur clusters of human mitoNEET. Biochemistry. 2010;49:9604–9612. doi: 10.1021/bi101168c. [DOI] [PubMed] [Google Scholar]
- 6.Hou X, Liu R, Ross S, Smart EJ, Zhu H, Gong W. Crystallographic studies of human mitoNEET. J Biol Chem. 2007;282:33242–33246. doi: 10.1074/jbc.C700172200. [DOI] [PubMed] [Google Scholar]
- 7.Lin J, Zhou T, Ye K, Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron-sulfur proteins. Proc Natl Acad Sci USA. 2007;104:14640–14645. doi: 10.1073/pnas.0702426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings PA. MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci USA. 2007;104:14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geldenhuys WJ, Funk MO, Barnes KF, Carroll RT. Structure based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg Med Chem Lett. 2010;20:819–823. doi: 10.1016/j.bmcl.2009.12.088. [DOI] [PubMed] [Google Scholar]
- 10.Natalello A, Benetti F, Doglia SM, Legname G, Grandori R. Compact conformations of alpha-synuclein induced by alcohols and copper. Prot Struct Funct Bioinform. 2011;79:611–621. doi: 10.1002/prot.22909. [DOI] [PubMed] [Google Scholar]
- 11.Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nature Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 12.Tsibris JCM, Tsai RL, Gunsalus IC, Orme-Johnson WH, Hansen RE, Beinert H. The number of iron atoms in the paramagnetic center (G=1.94) of reduced putidaredoxin, a nonheme iron protein. Proc Natl Acad Soc USA. 1968;59:959–965. doi: 10.1073/pnas.59.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petillot Y, Forest E, Meyer J, Moulis JM. Observation of holoprotein molecular ions of several ferredoxins by electrospray ionization mass spectrometry. Anal Biochem. 1995;228:56–63. doi: 10.1006/abio.1995.1314. [DOI] [PubMed] [Google Scholar]
- 14.Taylor PK, Kurtz DM, Amster IJ. Electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry of multimeric metalloproteins. Int J Mass Spectrom. 2001;210/211:651–663. [Google Scholar]
- 15.Johnson KA, Verhagen MFJM, Brereton PS, Adams MWW, Amster IJ. Probing the stoichiometry and oxidation states of metal centers in iron-sulfur proteins using electrospray FTICR mass spectrometry. Anal Chem. 2000;72:1410–1418. doi: 10.1021/ac991183e. [DOI] [PubMed] [Google Scholar]
- 16.Carroll KS, Gao H, Chen H, Leary JA, Bertozzi CR. Investigation of the iron-sulfur cluster in Mycobacterium tuberculosis APS reductase: implications for substrate binding and catalysis. Biochemistry. 2005;44:14647–14657. doi: 10.1021/bi051344a. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez H, Hewitson KS, Roach P, Shaw NM, Baldwin JE, Robinson CV. Observations of the iron-sulfur cluster in Escherichia coli biotin synthase by nanoflow electrospray mass spectrometry. Anal Chem. 2001;73:4154–4161. doi: 10.1021/ac0102664. [DOI] [PubMed] [Google Scholar]
- 18.Sun N, Soya N, Kitova EN, Klassen JS. Nonspecific interactions between proteins and charged biomolecules in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2010;21:472–481. doi: 10.1016/j.jasms.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Shimon L, Sharon M, Horovitz A. A method for removing effects of nonspecific binding on the distribution of binding stoichiometries: application to mass spectroscopy data. Biophys J. 2010;99:1645–1649. doi: 10.1016/j.bpj.2010.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 21.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 22.Bauer JH, Goupil SG, Garber GB, Helfand SL. An accelerated assay for the identification of lifespan-extending interventions in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:12980–12985. doi: 10.1073/pnas.0403493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang MS, Cai EN, Udeani GO, Slowing KV, Thomas CF, Beecher CWW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 24.Hung LM, Chen JK, Huang SS, Lee RS, Su MJ. Cardioprotective effect of resveratrol, a natural antioxidant derived from grapes. Cardiovasc Res. 2000;47:549–555. doi: 10.1016/s0008-6363(00)00102-4. [DOI] [PubMed] [Google Scholar]
- 25.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliot P, Geny B, Laasko M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci USA. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C, Shin YG, Chow A, Li Y, Kosmeder JW, Lee YS, Hirschelman WH, Pezzuto JM, Mehta RG, van Breemen RB. Human, rat, and mouse metabolism of resveratrol. Pharm Res. 2002;19:1907–1914. doi: 10.1023/a:1021414129280. [DOI] [PubMed] [Google Scholar]
- 28.Meng XF, Maliakal P, Lu H, Lee MJ, Yang CS. Urinary and plasma levels of resveratrol and quercetin in humans, mice, and rats after ingestion of pure compounds and grape juice. J Agric Food Chem. 2004;52:935–942. doi: 10.1021/jf030582e. [DOI] [PubMed] [Google Scholar]
- 29.Hoshino J, Park EJ, Kondratyuk TP, Marler L, Pezzuto JM, van Breemen RB, Mo S, Li Y, Cushman M. Selective synthesis and biological evaluation of sulfate-conjugated resveratrol metabolites. J Med Chem. 2010;53:5033–5043. doi: 10.1021/jm100274c. [DOI] [PMC free article] [PubMed] [Google Scholar]