Abstract
Cancer has emerged as a leading cause of mortality worldwide, claiming over 8 million lives annually. Gastrointestinal (GI) cancers account for ~35% of these mortalities. Recent advances in diagnostic and treatment strategies have reduced mortality among GI cancer patients, yet a significant number of patients still develop late-stage cancer, where treatment options are inadequate. Emerging interests in ‘liquid biopsies’ have encouraged investigators to identify and develop clinically-relevant noninvasive genomic and epigenomic signatures that can be exploited as biomarkers capable of detecting premalignant and early-stage cancers. In this context, microRNAs (miRNAs), which are small non-coding RNAs that are frequently dysregulated in cancers, have emerged as promising entities for such diagnostic purposes. Albeit the future looks promising, current approaches for detecting miRNAs in blood and other biofluids remain inadequate. This review summarizes existing efforts to exploit circulating miRNAs as cancer biomarkers, evaluates their potential and challenges as liquid biopsy-based biomarkers for GI cancers.
Keywords: miRNA, biomarker, liquid biopsy, gastrointestinal cancer
Introduction
Gastrointestinal (GI) cancers primarily occur in the liver, stomach, colorectum, esophagus and pancreas, and account for ~35% of global cancer-related mortalities (1). Recent advances in surgical and endoscopic procedures have significantly improved the survival of patients with early stage disease. However, the inherently low frequency of some of these cancers, invasive nature of screening procedures, and the high costs associated with such modalities have resulted in poor compliance for current generation of screening assays. Although non-invasive screening tests such as fecal immunochemical tests (FIT) are available for screening colorectal cancer patients, their efficacy remains limited due to low sensitivity and specificity (2), and their inability to detect other types of cancers within the GI tract. Consequently, inadequate screening modalities for patients with gastrointestinal cancers highlight the imperative need for further research on this important clinically-relevant issue.
Within the context of cancer, particularly GI malignancies, ‘epigenetic’ alterations together with genetic events, have emerged as key drivers of disease development and progression (3). The term ‘epigenetic’ broadly encompasses all heritable changes in gene expression that do not involve a permanent change in the DNA sequence. In cancer, the most well-investigated epigenetic alterations include aberrant DNA methylation, histone modifications, and dysregulated expression of non-coding RNAs (4). Epigenetic alterations manifest far more frequently than genetic mutations and often appear in early stages of tumorigenesis (5). These alterations are dynamic in nature and potentially reversible, and hence have shown promise as attractive substrates for developing disease biomarkers and serve as therapeutic targets in human cancers (5). To date miRNAs are remain the most studied epigenetic alteration in circulation, both as diagnostic and prognostic cancer biomarkers. In contrast, DNA methylation has been preferentially assessed in tissues, primarily due to the limitation that significant volume of serum/plasma is needed to obtain adequate amounts of DNA for methylation analysis. Furthermore, the assessment of post-translational histone modifications in the serum is quite limited. Over the last several years, several important studies have evaluated the potential of miRNAs as liquid biopsy biomarkers, and therefore, now is perhaps the appropriate time to objectively assess their true potential as cancer biomarkers.
Among noncoding RNAs (ncRNAs), dysregulated expression of microRNAs (miRNAs) have been most widely studied over the last decade, and they appear to be promising diagnostic biomarkers for a variety of human cancers, including GI malignancies (6). A large number of these small ncRNAs have been quite well characterized for their biological function in cancer and their ability to regulate the expression of protein coding genes. From a clinical standpoint, dysregulated expression of miRNAs have been readily detected in a variety of biological fluids in cancer patients, highlighting their stability in these biofluids and providing a rationale for developing them as ‘liquid biopsy’ biomarkers. This review summarizes current efforts for implementing specific circulatory miRNAs as diagnostic biomarkers for GI cancers, and discusses how these nucleotides can incorporate into future cancer therapeutic strategies.
LIQUID BIOPSIES: NOVEL FRONTIERS IN CANCER DIAGNOSIS
The first interpretation of the term “liquid biopsy” originated in 2010 when circulating tumor cells (CTCs) were proposed as alternatives to conventional breast cancer biopsy for prognosis and evaluation of treatment responses (7). Subsequently, clinical applications of liquid biopsies have diversified from detecting early stage cancer to monitoring tumor progression, assessing tumor heterogeneity and residual disease, and potentially monitoring therapeutic response to various surgical and chemotherapeutic interventions (8). The Figure 1 depicts a theoretical progression of the clinical applicability of liquid biopsies – illustrating various types of liquid biopsy targets, the spectrum of biofluids in which these can be interrogated, and their plausible applications for improving diagnosis, prognosis, personalized therapeutics, and disease monitoring in cancer patients. These noninvasive but technologically sophisticated applications can be incorporated into existing treatment practices to decrease GI cancer-associated mortality.
Figure 1. Clinical applications of liquid biopsies.
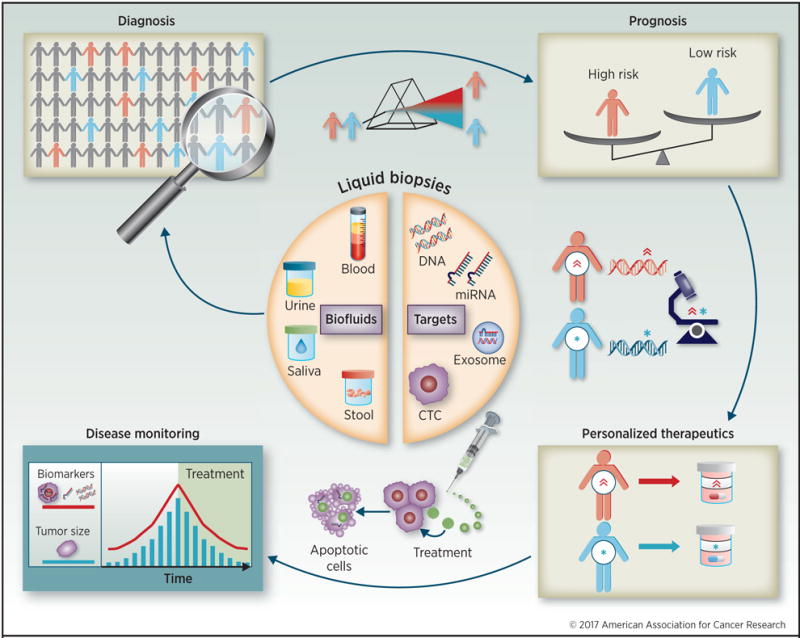
Liquid biopsies include blood, urine, saliva, and stool. These sources contain cancer-derived subcellular components, such as circulating tumor DNAs (ctDNAs), circulating microRNAs (miRNAs), circulating tumor cells (CTCs) and exosome encapsulated DNA and miRNAs. These targets circulate throughout the patient’s body and have a number of clinical applications: diagnose cancer at early stages through detection and quantification of these targets; identify aggressive phenotypes and high risk patients who necessitate intensive treatment; monitor drug efficacy to improve therapy for each patient; and monitor in real time the treatment’s effectiveness by correlating these targets with tumor size and viability. Liquid biopsy-based monitoring is potentially more sensitive at following treatment progress than computed tomography and other imaging-based strategies.
Recently, the sources of ‘liquid biopsies’ expanded beyond blood to include other body fluids including feces (9), urine (10) and saliva (10), which may directly detect cancer in associated organs. Likewise, the term “biopsy” has broadened to encompass other subcellular components including circulating tumor DNA (ctDNA) (11), ncRNAs, predominantly miRNAs (12), proteins (13) and extracellular vesicles (14) that can be used as targets for evaluated in GI cancer. In this regard, despite the initial enthusiasm for identifying a high frequency of CTCs and ctDNA in liquid biopsies from cancer patients, accumulating data indicate that although these targets offer a high degree of cancer-specificity, both entities are scarce in circulating biofluids and may be inadequate as clinically applicable diagnostic biomarkers. On average, ctDNA represents less than 1% of the total circulating free DNA found in biofluids, while in cancer patients ratio of CTCs to white blood cells is approximately 1:1 million (8). Accordingly, a study that evaluated the ability of ctDNA to identify specific mutations in individuals’ primary tumors reported success in only 73% of colorectal, 57% of gastro-esophageal, and 48% of pancreatic cancers (15). These results may be considered somewhat disappointing considering that each of these mutations was known a priori before screening (16). Consequently, other molecules derived from tumor cells, such as ncRNAs, are far more abundant than ctDNA or CTCs in biofluids, are relatively stable in a variety of biological fluids, and are frequently dysregulated even in the earliest stages of cancer. These characteristics argue in their favor for further development as noninvasive liquid biopsy biomarkers for human cancers.
Circulating miRNAs as cancer diagnostic biomarkers
In 2008, tumor-associated miRNAs (miR-155, miR-210 and miR-21) were first discovered to be upregulated in serum of lymphoma patients (17). To date hundreds more miRNAs have been identified as potential diagnostic targets in various cancers (6,18). Circulating miRNAs possess unique features making them likely candidates for development as disease-specific biomarkers. MiRNAs are generally stable in blood and other body fluids due to their small size and their ability to escape from RNase-mediated degradation, and nearly 10% of miRNAs are either secreted in membranous nano-sized vesicles called ‘exosomes’ while the remaining 90% are stabilized and packaged with other proteins, such as argonaute-2 (AGO2), high-density lipoprotein (HDL), and other RNA-binding proteins (19–21). Furthermore, both exosomal- and AGO2/HDL-attached miRNAs are actively secreted from living cells, whereas the majority of ctDNA is passively released by apoptotic or necrotic cells (8,21,22) as illustrated in Figure 2. A recent study demonstrated that miRNAs offer superior sensitivity and specificity compared to ctDNA for diagnosing colorectal cancers (23). Collectively, miRNAs appear to be promising candidates as liquid biopsy-based cancer biomarkers.
Figure 2. Screening for gastrointestinal cancers using actively or passively secreted tumor components in liquid biopsies.
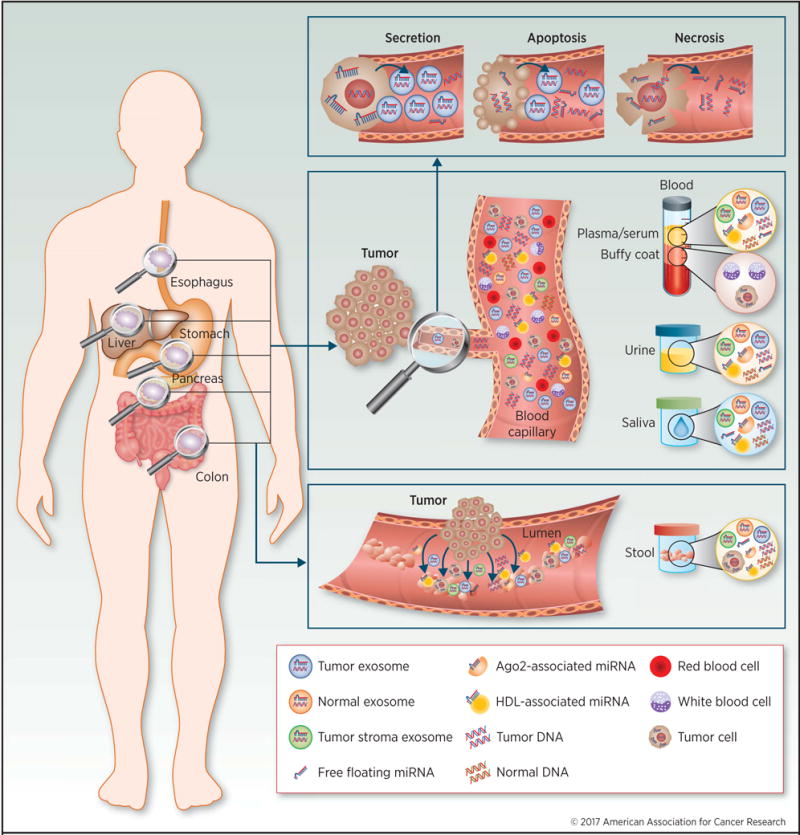
Gastrointestinal cancers, including esophageal, gastric, liver, pancreatic, and colon, shed subcellular components into the blood stream and/or intestinal lumen. These targets include circulating tumor DNAs (ctDNAs), circulating microRNAs (miRNAs), circulating tumor cells (CTCs), and exosome encapsulated DNA/miRNAs. These targets can be detected in biofluids, such as blood, urine, saliva and feces. Several morphologies of nucleotides are found in biofluids: free floating DNA/miRNA, argonaute 2 (Ago2)/high-density lipoprotein (HDL) associated miRNA, and exosome encapsulated DNA/miRNA, which are secreted from cancer cells in diverse patterns. Apoptotic or necrotic cells directly shed components extracellularly (passive secretion) as ctDNAs, while living aggressive cancer cells secrete encapsulated protein-associated miRNAs in exosomes (active secretion).
Nevertheless, there are several obstacles that must be overcome before miRNAs can be recognized and adopted as clinically relevant cancer diagnostic biomarkers. In particular, the lack of disease- and organ-specificity and uncertainty of normalization are among the most critical issues. With the significant body of literature gathered on circulating miRNAs in GI cancers, and the availability of high throughput microarray and RNA sequencing profiling results from serum and plasma samples from cancer patients, we are very likely bound to identify robust miRNAs as potential cancer diagnostic cancer markers in the near future. The diagnostic potential of many circulating miRNAs have been assessed in a variety of cancers and those within the GI tract are summarized in Table 1, and the key studies are highlighted in the following sections.
Table 1.
Circulating miRNAs as noninvasive diagnostic biomarkers in gastrointestinal cancers
| miRNA | Source | Sample size | Diagnostic value (%) | Normalizer | Year | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Cases | Controls | AUC | Sensitivity | Specificity | |||||
|
Colorectal cancer
| |||||||||
| Single marker | |||||||||
| miR-17-3p | Plasma | 90 | 50 | 0.72 | 64 | 70 | U6 | 2009 | (59) |
| miR-92 | Plasma | 90 | 50 | 0.89 | 89 | 70 | U6 | 2009 | (59) |
| miR-21 | Serum | 186 | 53 | 0.93 | 83 | 91 | cel-miR-39 | 2013 | (26) |
| miR-23a | Exosome | 88 | 11 | 0.95 | NA | NA | miR-451 | 2014 | (24) |
| miR-378 | Plasma | 29 | 29 | 0.95 | NA | NA | miR-16 | 2014 | (25) |
| miR-1246 | Exosome | 88 | 11 | 0.95 | NA | NA | miR-451 | 2014 | (24) |
| Panel | |||||||||
| miR-431, 139-3p | Plasma | 45 CRC | 26 | 0.83 | 91 | 57 | U6 | 2013 | (60) |
| miR-532-3p, 331, 195, 17, 142-3p, 15b, 532, 652 | Plasma | 16 Ad | 26 | 0.87 | 88 | 64 | U6 | 2013 | (60) |
| miR-19a-3p, 223-3p, 92a-3p, 422a | Serum | 117 CRC | 102 | 0.95 | 84 | 92 | U6 | 2014 | (27) |
| miR-19a-3p, 223-3p, 92a-3p, 422a | Serum | 73 Ad | 102 | 0.77 | NA | NA | U6 | 2014 | (27) |
| miR -21, 31, 92a, 181b, 203, let-7g | Serum | 83 | 59 | 0.92 | 96 | 88 | miR-16 | 2014 | (61) |
| miR-21, 29a, 125b | Serum | 160 Ad | 77 | 0.83 | NA | NA | cel-miR-39 | 2015 | (28) |
|
Esophageal cancer | |||||||||
| Single marker | |||||||||
| miR-18a | Plasma | 106 | 54 | 0.94 | 87 | 100 | mirVana miRNA RP | 2013 | (31) |
| miR-1246 | Serum | 101 | 46 | 0.75 | 71 | 74 | miR-16 | 2013 | (62) |
| miR-25 | Plasma | 20 | 50 | 0.86 | 85 | 86 | U6 | 2014 | (32) |
| Panel | |||||||||
| miR-10a, 22, 100, 148b, 223, 133a, 127-3p | Serum | 149 | 100 | 0.93 | 79 | 96 | Serum volume | 2010 | (33) |
| miR-21/375 (ratio) | Plasma | 50 | 20 | 0.82 | 88 | 70 | mirVana miRNA RP | 2011 | (63) |
| miR-25, 100, 193-3p, 194, 223, 337-5p, 483-5p | Serum | 63 | 63 | 0.83 | 81 | 81 | let-7d/g/i | 2014 | (64) |
|
Gastric cancer | |||||||||
| Single marker | |||||||||
| miR-16 | Serum | 50 | 47 | 0.90 | 79 | 78 | U6 | 2014 | (65) |
| miR-18a | Plasma | 82 | 65 | 0.91 | 81 | 85 | mirVana miRNA RP | 2014 | (34) |
| miR-222 | Plasma | 114 | 56 | 0.85 | 66 | 88 | U6 | 2014 | (66) |
| miR-21 | Serum | 50 | 50 | 0.91 | 88 | 80 | U6 | 2015 | (35) |
| Panel | |||||||||
| miR-106a/let-7a ratio | Plasma | 69 | 30 | 0.88 | 86 | 80 | U6 | 2010 | (67) |
| miR-120a, 27a, 34, 423-5p | Serum | 142 | 105 | 0.88 | 80 | 81 | miR-16 | 2011 | (36) |
| miR-223, 21, 218 | Plasma | 70 | 70 | 0.95 | 84 | 93 | cel-miR-39 | 2012 | (68) |
|
Hepatocellular carcinoma | |||||||||
| Single marker | |||||||||
| miR-21 | Serum | 101 | 89 | 0.87 | 84 | 74 | miR-181a, 181c | 2011 | (69) |
| miR-122 | Serum | 101 | 89 | 0.79 | 71 | 69 | miR-181a, 181c | 2011 | (69) |
| miR-223 | Serum | 101 | 89 | 0.86 | 80 | 77 | miR-181a, 181c | 2011 | (69) |
| miR-18a | Serum | 86 | 45 | 0.88 | 86 | 75 | U6 | 2012 | (70) |
| Panel | |||||||||
| miR-122, 192, 21, 223, 26a, 27a, 801 | Plasma | 196 | 66 | 0.94 | 83 | 94 | miR-1228 | 2011 | (37) |
| miR-375, 25, let-7f | Serum | 55 | 100 | 0.99 | 98 | 99 | plant miR-168 | 2011 | (71) |
| miR-23b, 423, 375, 23a, 342-3p | Serum | 55 | 100 | 0.99 | 97 | 99 | plant miR-168 | 2011 | (71) |
| miR-29a, 29c, 133a, 143, 145, 192, 505 | Serum | 229 | 108 | 0.82 | 75 | 89 | cel-miR-67 | 2015 | (38) |
|
Pancreatic cancer | |||||||||
| Single marker | |||||||||
| miR-200a | Serum | 45 | 32 | 0.86 | 84 | 88 | miR-16 | 2010 | (72) |
| miR-200b | Serum | 45 | 32 | 0.85 | 71 | 97 | miR-16 | 2010 | (72) |
| miR-27a-3p | Whole blood | 129 | 60 | 0.86 | 82 | 79 | U6 | 2013 | (73) |
| miR-1290 | Serum | 41 | 19 | 0.96 | 88 | 84 | miR-16 | 2013 | (42) |
| Panel | |||||||||
| miR-16, 196a (with CA19-9) | Plasma | 140 | 68 | 0.98 | 92 | 96 | cel-miR-39 | 2012 | (74) |
| miR-20a, 21, 24, 25, 99a, 185, 191 | Serum | 95 | 81 | 0.99 | 94 | 93 | Serum volume | 2012 | (43) |
| miR-145, 150, 223, 636 | Whole blood | 86 | 44 | 0.83 | 85 | 48 | ath-miR159a | 2014 | (75) |
| miR-26b, 34a, 122, 126*, 145, 150, 223, 505, 636, 885-5p | Whole blood | 86 | 44 | 0.82 | 85 | 55 | ath-miR159a | 2014 | (75) |
|
Biliary cancer | |||||||||
| Single marker | |||||||||
| miR-21 | Plasma | 94 | 50 | 0.93 | 85 | 100 | miR-16 | 2013 | (40) |
| miR-126 | Serum | 31 | 40 | 0.87 | 68 | 93 | cel-miR-39 | 2015 | (76) |
| miR-1281 | Serum | 31 | 40 | 0.83 | 55 | 90 | cel-miR-39 | 2015 | (76) |
| Panel | |||||||||
| miR-6075, 4294, 6880-5p, 6799-5p, 125a-3p, 4530, 6836-3p, 4476 | Serum | 98 | 150 | 0.95 | 80 | 98 | microarray-based normalization | 2015 | (77) |
NA, not available; Ad, adenoma; CRC, colorectal cancer; mirVana miRNA RP, Thermo Fisher Scientific mirVana™ miRNA reference panel
Colorectal cancer
Among all GI cancers, miRNA-based diagnostic biomarkers largely have been studied in colorectal cancer (CRC) patients (18). It is beyond the scope of this article to discuss all reports on this topic, but the more promising miRNA-based diagnostic markers in CRC have been miR-21, miR-23a, miR-378, and miR-1246 based upon reported AUC values (24–26). More recently, another panel of miRNAs (miR-19a-3p, miR-223-3p, miR-92a-3p, and miR-422a) was derived from pooled serum samples obtained from CRC patients and healthy subjects using next generation sequencing (NGS); its robustness was confirmed in a collection of 219 specimens, in which these markers successfully distinguished both cancers and adenomas from healthy controls (27). Recent efforts have attempted to translate these findings to liquid biopsy markers for detection of early colorectal neoplasia. In a cohort of 237 patients, circulating levels of miR-21, miR-29a and miR-125b independently differentiated colorectal neoplasms from healthy controls. However, when combined into a panel, the accuracy of detection improved significantly (28). Collectively, these studies highlight the ability of miRNA biomarkers to identify patients with CRC, and more importantly, screen for and detect patients with advanced polyps and early stage cancer.
Esophageal cancer
The primary causes of esophageal cancers include excessive alcohol consumption, tobacco use, and chronic gastro-esophageal reflux disease (1). Currently, esophageal cancer is difficult to resect, highly aggressive, and has a low 5-year survival rate of 17–19% (29). The flat morphology of early-stage esophageal cancers makes their diagnosis challenging even with endoscopy, emphasizing the need for markers that can facilitate detection of the earliest stages of disease and improve patient survival (30). Based on extensive interrogation of miRNAs upregulated in primary esophageal cancers, miR-18a and miR-25 appear as promising diagnostic markers (31,32). NGS on pooled serum specimens from patients with advanced esophageal cancer identified a panel of miRNAs comprising of miR-10a, miR-22, miR-100, miR-148b, miR-223, miR-133a, and miR-127-3p (33). Subsequent validation of this diagnostic panel in two large independent clinical cohorts yielded an impressive AUC value of 0.93 (33).
Gastric cancer
Historically, H. pylori infections were considered one of the major causes of gastric cancer, but cancer-associated mortality has declined significantly ever since effective antibiotic regimens were implemented to eradicate this pathogen (1). A thorough review of literature on the diagnostic accuracy of miRNA biomarkers for gastric cancer revealed that miR-18a and miR-21 are among the leading candidates that deserve further interrogation and validation (34,35). Comprehensive RNA sequencing on 20 gastric cancers and healthy controls revealed a panel of miRNAs (miR-1, miR-20a, miR-27a, miR-34, and miR-423-5p) that can differentiate patients with gastric cancer from healthy controls (36). Since diffused-type gastric cancers are difficult to detect using endoscopy, miRNA-based liquid biopsy approaches may provide an attractive, noninvasive and inexpensive option for improved detection of such lesions.
Hepatocellular and biliary cancer
Hepatitis viruses B and C are major contributors to hepatocellular carcinoma development, while other risk factors include cirrhosis, obesity, aflatoxin exposure and high alcohol consumption (1). A recent large-scale clinical trial developed a plasma-based miRNA panel comprising of miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801 (37), which robustly differentiated between in two large independent cohorts of 407 and 390 specimens of hepatocellular carcinomas and healthy controls. However, these results could not be replicated in another study, which identified miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192 and miR-505 as diagnostic markers (38). This discrepancy emphasized the need for more carefully designed discovery and validation cohorts for liquid biopsy biomarker discovery.
Biliary cancer is a rare disease and affects 2,000.3,000 people each year in the U.S. (39). Similar to other GI cancers, miR-21 appears to be the most promising circulating miRNA-based diagnostic marker for biliary cancer (40). However, a recent study which compared microarray expression profiles in serum from healthy subjects and cancer patients identified dysregulation of several previously unreported miRNAs including: miR-6075, miR-4294, miR-6880-5p, miR-6799, miR-4530, miR-6836-3p and miR-4476. It is interesting to notice that most of these biliary cancer-associated miRNAs are somewhat unique, and are not frequently altered in other GI cancers.
Pancreatic cancer
Pancreatic cancer has the lowest 5-year survival rate of ~7% among all GI cancers because of the basic biology of the disease, which is further compounded by a dearth of optimal detection methods (41). It cannot be screened by endoscopy while imaging methods include abdominal ultrasonography – the gold standard for detection. These methods are limited detection rates due to the anatomical location of pancreas, particularly for smaller lesions. High-throughput PCR arrays identified serum miR-1290 as a robust circulating diagnostic marker for pancreatic cancer (42). NGS identified a miRNA panel (miR-20a, miR-21, miR-24, miR-25, miR-99a, miR-185 and miR-191) that remarkably differentiated pancreatic cancer patients from healthy controls with an AUC of 0.99 (43). However, there remains a need for noninvasive liquid biopsy based biomarkers that will improve survival of patients by detecting precancerous or early-stage pancreatic cancers.
MiRNA Diagnostics: A panel based-approach
In spite of recent discoveries that promote circulating miRNAs to diagnose GI cancers, none have led to the implementation of markers for clinical use due to the inadequacy of solitary miRNA biomarkers in clinical testing. A growing interest to combine biomarker into panels (44) confronts the issue of tumor heterogeneity and low specificity and sensitivity of solitary miRNAs to detect a particular cancer. In this regard, several mathematical models were utilized to evaluate the performance of combinations of miRNAs as cancer diagnostic markers. These strategies include threshold-based methods, decision trees, logistic regression and support vector machine (45). Although combining markers clearly improved the diagnostic potential of miRNAs, unfortunately one of the limitations is that most miRNA panels reported to date were derived using insufficient sample sizes and validations were performed inadequately in clinical applications. Furthermore, these biomarker panels failed to exploit the statistical leverage associated with combining multiple markers, and instead contributed to vast discrepancies and noise across various studies that selected miRNAs. However, such inadequacies are expected and, considering the wealth of knowledge gathered on this discipline, future studies will address these concerns and hopefully yield liquid biopsy biomarker panels that can routinely detect GI cancers.
Although this article primarily focused on describing the diagnostic potential of circulating miRNAs, the clinical usefulness of these biomarkers also extends their ability to serve as prognostic and predictive biomarkers for response to chemotherapy as summarized in Supplementary Tables 1 and 2.
A current perspective on miRNA-based diagnostic cancer biomarkers
Several well recognized obstacles must be overcome before miRNA biomarkers can realistically transition to clinic. First, qRT-PCR-based quantification of miRNAs is imperfect due to lack of a consensus endogenous normalizer. Currently, the expression of miRNAs in serum or plasma is commonly normalized using either endogenous internal controls (house-keeping genes) or synthetic spiked-in controls (i.e. cel-miR-39 or ath-miR159a) in a standardized sample volume. While synthetic spike-in controls are simple and an accurate way to quantify miRNAs, standardization of expression values across multiple-cohorts remains challenging. Considering that differences in extraction procedures and storage conditions can affect RNA quality and subsequently influence the outcome of spike-in control normalized data, spike-in controls may not be suitable for clinical circumstances. Furthermore, the use of spike-in controls is not adequate for analyzing expression of circulating miRNAs contained in exosomes as it requires an additional step of ultracentrifugation-based purification. Therefore, the current practice remains the use of endogenous controls, such as U6, miR-451, and miR-16, even though several studies have found the expression of these markers to be dysregulated in cancers, making them unsuitable for normalization purposes (46). Alternatively, as the cost associated with RNA-sequencing becomes more affordable, RNA-seq based global normalization procedures such as RPKM (reads per kilo-base per million mapped reads) could be used eliminate the biases associated with endogenous and spike-in controls. Second, the low disease and organ specificity of circulating miRNAs hampers miRNA-based cancer biomarker research. There is a school of thought that changes in circulating miRNAs in cancer patients often occur holistically and may not truly reflect alterations present in the tumor itself. For example, several cancer-associated circulating miRNAs are also elevated in inflammatory diseases such as colitis and rheumatoid arthritis (47). Well-established oncogenic miRNAs such as miR-21 and miR-155 have been linked to inflammation and, despite extensive research, the question remains whether the overexpression of these miRNAs are causally linked to cancer or are a consequence of inflammation (48). Similarly, certain oncogenic miRNAs are upregulated in multiple cancer types and thus are not organ-specific. A significant step to overcome this problem was addressed in a recent NGS-based study where multiple cancer types were compared and 71 organ-specific iso-miRNAs (iso-miRs) were identified (49). A follow-up study developed a panel of iso-miRs that adequately identified patients with triple-negative breast cancers (50), highlighting the potential of iso-miRs to identify the organ of origin. In addition, several miRNAs have been identified to undergo RNA-editing in cancers and these miRNAs with edited sequences appear to acquire new biological functions (51). In melanoma, edited miR-445 enhanced tumor growth and metastasis. Likewise, high throughput sequencing profiling identified a small population of edited miRNAs in colorectal cancer (51,52). Collectively, discovery of both iso-miRNAs and edited-miRNAs broadens potential candidates for miRNA-based biomarkers and highlights the functional complexity of miRNAs.
Moreover, recent research identified exosomal miRNA populations that are organ and cancer-specific (53). For instance, A33 is an epithelial cell-specific antigen found exclusively on the surface of exosomes released from the colon (54). Similarly, a recent report demonstrated that exosomes expressing Glypican-1, a cell surface proteoglycan, are released exclusively from pancreatic cancer cells and not from normal cells (53). Microarray based comparison between plasma and exosomal miRNA showed significant difference in miRNA contents indicating that exosomal miRNAs could improve the biomarker potential of conventional serum based circulating miRNA markers (55). Furthermore, recent technological advancements could make an enormous impact on the development of new screening methods for detection of cancer exosomes. Recently, a modification was made to the conventional fluorescence activated cell sorting (FACS) methodology which allows detection and sorting of a specific population of exosomes by labelling surface proteins with fluorescence antibodies (56). This study assessed surface EGFR and CD9 in exosomes isolated from a colorectal cancer cell line as well as plasma-derived human exosomes. Similarly, surface plasmon resonance spectroscopy was used to assess the levels of CD44, CD24 and Epcam on the breast cancer cell line-derived exosomes (57). Not only these methodologies will be utilized for biomarker research in the near future, they will also clarify the physiological and mechanistic roles of cancer exosomes. Adding to our fundamental understanding of exosomal miRNAs, we have now recognized that such miRNAs are frequently taken up by neighboring or distant cells and subsequently functionally modulate recipient cells (58). Collectively, comprehensive characterization of epigenome and proteome in cancer derived-exosomes appear to be the focal points of miRNA-based biomarker field and could transform the conventional school of blood-based molecular cancer diagnostic markers.
Nonetheless, a careful review of literature still supports the notion that a small panel of miRNAs is consistently upregulated in various cancers and detected in the blood of the cancer patients (Table-2). Since these miRNA biomarkers have shown validation in multiple, independent studies, there is a growing enthusiasm that some of these may likely be ready for clinical applications in the near future.
Table 2.
Most promising miRNA biomarkers with diagnostic significance
| miRNA | Supporting evidence | Limitations | Source |
|---|---|---|---|
|
Colorectal cancer
| |||
| miR-21 | • One of the most abundant miRNAs | • Not cancer specific | Serum, Plasma |
| • Highly upregulated miRNAs in solid tumors | • Upregulated by inflammation | ||
| • One of the most studied diagnostic circulating miRNAs | • Affected by hemolysis | ||
| • Suitable for early diagnosis | |||
| miR-29a | • Unaffected by hemolysis | Serum, Plasma | |
| • Suitable for early diagnosis | |||
| miR-92a | • One of the most abundant miRNAs | • Influenced by hemolysis | Serum, Plasma |
|
Gastric cancer | |||
| miR-21 | • Same as above | • Upregulated by H. pylori infection | Serum, Plasma |
| miR-27a | • Well-established oncogene | • Upregulated by H. pylori infection | Serum, Plasma |
| • Unaffected by hemolysis | |||
|
Hepatocellular carcinoma | |||
| miR-21 | • Same as above | • Upregulated by hepatitis virus infection | Serum, Plasma, Exosome |
| miR-192 | • Suitable for early diagnosis | • Upregulated by hepatitis virus infection | Serum, Plasma |
|
Pancreatic cancer | |||
| miR-21 | • Same as above | • Same as above | Serum, Plasma, Exosome |
| miR-223 | • Unaffected by hemolysis | • Influenced by aspirin | Plasma, Whole blood |
| • Overexpressed in early stage pancreatic cancer | |||
CONCLUSIONS
The field of miRNA-based cancer research has witnessed a remarkable evolution over the last two decades. While much effort to date has been to identify specific miRNAs and their role in cancer, interest has grown to evaluate their potential as disease biomarkers, as well as recent attempts at exploiting their significance as therapeutic targets. Their small size and stability in a variety of body fluids makes them attractive substrates for biomarker development. As this field continues to mature with identification of more specific subtypes of miRNAs and increasing focus on large-scale multi-center comprehensive studies miRNA-based diagnostic approaches are likely to usher in a new era of personalized medicine for cancer patients.
Supplementary Material
Acknowledgments
Funding: The present work was supported by grants R01 CA72851, CA18172, CA184792 and U01 CA187956 from the National Cancer Institute, National Institutes of Health, funds from the Baylor Research Institute and a pilot grant from Charles A Sammons Cancer Center. This work was also supported by grant from Uehara Memorial Foundation.
Footnotes
Conflicts of interest: The authors have no conflict of interest directly relevant to the content of this article.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570–95. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27(9):858–63. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–56. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 7.Lianidou ES, Mavroudis D, Sotiropoulou G, Agelaki S, Pantel K. What’s new on circulating tumor cells? A meeting report Breast Cancer Res. 2010;12(4):307. doi: 10.1186/bcr2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Janku F, Zhan Q, Fan JB. Accessing Genetic Information with Liquid Biopsies. Trends in Genetics. 2015;31(10):564–75. doi: 10.1016/j.tig.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Lin CC, Huang WL, Wei F, Su WC, Wong DT. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev Mol Diagn. 2015;15(11):1427–40. doi: 10.1586/14737159.2015.1094379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 12.Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61(1):56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 13.Shimada H. Is “liquid biopsy” useful for assessing HER2 status in gastric cancer? J Gastroenterol. 2015;50(1):119–20. doi: 10.1007/s00535-014-0967-6. [DOI] [PubMed] [Google Scholar]
- 14.Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Translational Cancer Research. 2015;4(3):280–90. doi: 10.3978/j.issn.2218-676X.2015.06.05. [DOI] [Google Scholar]
- 15.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141(5):672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 18.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149(5):1204–25 e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Ganepola GA, Nizin J, Rutledge JR, Chang DH. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J Gastrointest Oncol. 2014;6(4):83–97. doi: 10.4251/wjgo.v6.i4.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. Plos One. 2014;9(4):e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, Perrone F, et al. Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer. 2014;110(4):1001–7. doi: 10.1038/bjc.2013.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111(10):1985–92. doi: 10.1038/bjc.2014.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res. 2015;21(18):4234–42. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry MF. Esophageal cancer: staging system and guidelines for staging and treatment. J Thorac Dis. 2014;6(Suppl 3):S289–97. doi: 10.3978/j.issn.2072-1439.2014.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturm MB, Wang TD. Emerging optical methods for surveillance of Barrett’s oesophagus. Gut. 2015;64(11):1816–23. doi: 10.1136/gutjnl-2013-306706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A, et al. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108(9):1822–9. doi: 10.1038/bjc.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komatsu S, Ichikawa D, Hirajima S, Kawaguchi T, Miyamae M, Okajima W, et al. Plasma microRNA profiles: identification of miR-25 as a novel diagnostic and monitoring biomarker in oesophageal squamous cell carcinoma. Br J Cancer. 2014;111(8):1614–24. doi: 10.1038/bjc.2014.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang CN, Wang C, Chen X, Yang CH, Li K, Wang JJ, et al. Expression Profile of MicroRNAs in Serum: A Fingerprint for Esophageal Squamous Cell Carcinoma. Clin Chem. 2010;56(12):1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 34.Su ZX, Zhao J, Rong ZH, Wu YG, Geng WM, Qin CK. Diagnostic and prognostic value of circulating miR-18a in the plasma of patients with gastric cancer. Tumour Biol. 2014;35(12):12119–25. doi: 10.1007/s13277-014-2516-6. [DOI] [PubMed] [Google Scholar]
- 35.Wu JH, Li GX, Wang ZY, Yao YL, Chen R, Pu XY, et al. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Disease Markers. 2015 doi: 10.1155/2015/435656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47(5):784–91. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–8. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 38.Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16(7):804–15. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 39.Yalcin S. Diagnosis and management of cholangiocarcinomas: a comprehensive review. Hepato-gastroenterology. 2004;51(55):43–50. [PubMed] [Google Scholar]
- 40.Kane JM, Kishimoto T, Correll CU. Assessing the comparative effectiveness of long-acting injectable vs. oral antipsychotic medications in the prevention of relapse provides a case study in comparative effectiveness research in psychiatry. J Clin Epidemiol. 2013;66(8 Suppl):S37–41. doi: 10.1016/j.jclinepi.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NIH. http://seer.cancer.gov/statfacts/. Accessed in December, 2015.
- 42.Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–10. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–8. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 44.Baker SG, Kramer BS, Srivastava S. Markers for early detection of cancer: statistical guidelines for nested case-control studies. BMC Med Res Methodol. 2002;2:4. doi: 10.1186/1471-2288-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robin X, Turck N, Hainard A, Lisacek F, Sanchez JC, Muller M. Bioinformatics for protein biomarker panel classification: what is needed to bring biomarker panels into in vitro diagnostics? Expert Rev Proteomics. 2009;6(6):675–89. doi: 10.1586/epr.09.83. [DOI] [PubMed] [Google Scholar]
- 46.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210–4. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 47.Churov AV, Oleinik EK, Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev. 2015;14(11):1029–37. doi: 10.1016/j.autrev.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Yang S, Guo L, Zhao Y, Shao F, Chen F. Comparisons of isomiR patterns and classification performance using the rank-based MANOVA and 10-fold cross-validation. Gene. 2015;569(1):21–6. doi: 10.1016/j.gene.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 50.Telonis AG, Loher P, Jing Y, Londin E, Rigoutsos I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic acids research. 2015;43(19):9158–75. doi: 10.1093/nar/gkv922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 2015;17(3):311–21. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng Y, Li T, Ren R, Shi D, Wang S. Revealing editing and SNPs of microRNAs in colon tissues by analyzing high-throughput sequencing profiles of small RNAs. BMC Genomics. 2014;15(Suppl 9):S11. doi: 10.1186/1471-2164-15-S9-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9(2):197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higginbotham JN, Zhang Q, Jeppesen DK, Scott AM, Manning HC, Ochieng J, et al. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J Extracell Vesicles. 2016;5:29254. doi: 10.3402/jev.v5.29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grasso L, Wyss R, Weidenauer L, Thampi A, Demurtas D, Prudent M, et al. Molecular screening of cancer-derived exosomes by surface plasmon resonance spectroscopy. Analytical and bioanalytical chemistry. 2015;407(18):5425–32. doi: 10.1007/s00216-015-8711-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta. 2012;1826(1):103–11. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 60.Kanaan Z, Roberts H, Eichenberger MR, Billeter A, Ocheretner G, Pan J, et al. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258(3):400–8. doi: 10.1097/SLA.0b013e3182a15bcc. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, et al. Identification of a circulating microRNA signature for colorectal cancer detection. Plos One. 2014;9(4):e87451. doi: 10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108(3):644–52. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105(1):104–11. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. Plos One. 2014;9(3):e92292. doi: 10.1371/journal.pone.0092292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31(12):298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- 66.Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31(9):164. doi: 10.1007/s12032-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 67.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102(7):1174–9. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. Plos One. 2012;7(7):e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50(2):136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 70.Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 2012;57(11):2910–6. doi: 10.1007/s10620-012-2317-y. [DOI] [PubMed] [Google Scholar]
- 71.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF. Serum microRNA Profiles Serve as Novel Biomarkers for HBV Infection and Diagnosis of HBV-Positive Hepatocarcinoma (vol 70, pg 9798, 2010) Cancer Res. 2011;71(5):2022. doi: 10.1158/0008-5472.CAN-11-0110. [DOI] [PubMed] [Google Scholar]
- 72.Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70(13):5226–37. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang WS, Liu LX, Li GP, Chen Y, Li CY, Jin DY, et al. Combined serum CA19-9 and miR-27a-3p in peripheral blood mononuclear cells to diagnose pancreatic cancer. Cancer Prev Res (Phila) 2013;6(4):331–8. doi: 10.1158/1940-6207.CAPR-12-0307. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer. 2012;131(3):683–91. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 75.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311(4):392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 76.Voigtlander T, Gupta SK, Thum S, Fendrich J, Manns MP, Lankisch TO, et al. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. Plos One. 2015;10(10) doi: 10.1371/journal.pone.0139305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kojima M, Sudo H, Kawauchi J, Takizawa S, Kondou S, Nobumasa H, et al. MicroRNA Markers for the Diagnosis of Pancreatic and Biliary-Tract Cancers. Plos One. 2015;10(2) doi: 10.1371/journal.pone.0118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


