Abstract
Dermal wounds, both acute and chronic, represent a significant clinical challenge and therefore the development of novel biomaterial-based skin substitutes to promote skin repair is essential. Nanofibers have garnered attention as materials to promote skin regeneration due to the similarities in morphology and dimensionality between nanofibers and native extracellular matrix proteins, which are critical in guiding cutaneous wound healing. Electrospun chitosan-poly(caprolactone) (CPCL) nanofiber scaffolds, which combine the important intrinsic biological properties of chitosan and the mechanical integrity and stability of PCL, were evaluated as skin tissue engineering scaffolds using a mouse cutaneous excisional skin defect model. Gross assessment of wound size and measurement of defect recovery over time as well as histological evaluation of wound healing showed that CPCL nanofiber scaffolds increased wound healing rate and promoted more complete wound closure as compared with Tegaderm, a commercially available occlusive dressing. CPCL nanofiber scaffolds represent a biomimetic approach to skin repair by serving as an immediately available provisional matrix to promote wound closure. These nanofiber scaffolds may have significant potential as a skin substitute or as the basis for more complex skin tissue engineering constructs involving integration with biologics.
Keywords: Nanofibers, skin, wound healing, tissue engineering, chitosan
Graphical abstract
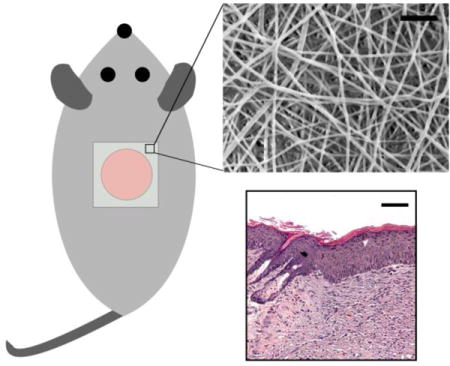
Electrospun chitosan-poly(caprolactone) (CPCL) nanofiber scaffolds showed improved wound healing rate, and overall closure, and re-epithelialization as compared with commercial Tegaderm.
Introduction
Skin is the largest organ in the human body, which functions to protect against mechanical, chemical and thermal insults and to prevent dehydration and invasion by pathogens. Because of the large surface area of skin, cutaneous wound healing, associated with acute or pathological conditions, represents a significant clinical challenge and socioeconomic burden[1–3]. Chronic wounds, such as non-healing ulcers, and wounds due to trauma, such as large burns, often cannot heal via normal mechanisms due to underlying pathology and/or severity and require the application of skin substitutes to promote repair. Autologous skin grafts, while the gold standard, are limited by tissue availability and harvest of skin grafts can result in donor site morbidity. Therefore, the development of skin substitutes for management of acute and chronic skin injuries is critical to improving patient outcomes and quality of life. Common materials utilized for skin repair include alginate, cellulose, chitosan, gelatin, hyaluronic acid, pectin, polyurethane or silicone[4–8]. These materials may take the forms of transparent films, fibers, hydrocolloids, hydrogels, sponges and/or foams[9–11]. In addition, skin tissue engineering constructs have been developed that combine these materials with biologics such as cells and/or growth factors in further attempts to promote skin regeneration[12, 13].
The first step toward development of clinically relevant skin substitutes is the design and optimization of biomaterials that engage in one or more processes associated with wound healing. An example of such a process is re-epithelialization, which is facilitated by migration of keratinocytes across the wound bed, leading to wound closure. A skin substitute that promotes migration of cells involved in skin repair may lead to faster and/or higher quality wound closure. Polymeric nanofibers, fabricated via electrospinning, have gained significant attention as clinically relevant biomaterials for skin repair because they can be fabricated with similar morphology and dimensionality to fibrillar extracellular matrix (ECM) proteins such as collagen[14–17]. Importantly, nanofibers promote cell-matrix interactions due to their high surface area-to-volume ratio that accommodates protein adsorption and presents numerous cell-binding sites. Voids between individual fibers are small and inhibit infiltration of microbes that may cause infection. In addition, the diameter of individual nanofibers and the mechanical properties and overall porosity of nanofiber scaffolds are tunable by adjusting electrospinning parameters[16]. This versatility allows for fabrication of scaffolds of different thicknesses, pore sizes or tensile strengths to accommodate different wound dimensions and/or wound properties.
Chitosan-based nanofibers have shown significant promise for a variety of tissue engineering applications due to their unique biological and physicochemical properties[18–24]. Chitosan, a naturally occurring, hydrophilic, cationic polymer derived from the exoskeleton of crustaceans, bears the proxy structure of glycosaminoglycans found in the ECM. In addition to its hydrophilicity, which promotes cell adhesion, it does not evoke a foreign body response and its degradation products are non-toxic[25]. Chitosan has demonstrated wound healing properties by activating and modulating the function of inflammatory cells such as neutrophils, macrophages, and fibroblasts as well as endothelial cells and by promoting the formation and organization of granulation tissue[26, 27]. Chitosan exhibits hemostatic and antimicrobial properties, which have been attributed to its cationic nature[28, 29]. One drawback of pure chitosan as a biomaterial is its insufficient mechanical strength and lack of structural stability in aqueous environments due to swelling. In addition, pure chitosan can be difficult to electrospin because of the high viscosity of chitosan solutions[30]. Blending chitosan with the synthetic, biocompatible polymer, polycaprolactone (PCL), yields a copolymer blend with cell affinity, enhanced strength and stability as well as spinnability[18, 31, 32]. Chitosan-PCL (CPCL) nanofibers have been utilized for applications such as nerve guides[18, 19], vascular grafts[22, 33], and muscle tissue engineering scaffolds[20]. In previous work, CPCL nanofibers, representing an epidermal scaffold, have shown to support the attachment and proliferation of keratinocytes, thereby indicating their potential as a skin substitute material[34]. CPCL nanofiber scaffolds represent a biomimetic approach to skin repair where the scaffold micro- and nanostructure recapitulates native ECM thereby presenting a provisional matrix for tissue regeneration.
Here we investigate the potential of CPCL nanofibers for promoting skin repair in vivo using a mouse cutaneous excisional skin defect model. Utilizing an in vivo model is a critical next step in corroborating our previous, in vitro results involving chitosan-polycaprolactone polymer blend nanofiber scaffolds and gaining a better understanding of cell-scaffold interactions. Full thickness skin defects covered with a commercially available wound dressing, Tegaderm™ (empty control), were compared with defects covered first with a CPCL nanofiber mat and then with Tegaderm, which is adhesive, in order to retain the CPCL over the defect. We characterized the effect of CPCL nanofibers on skin repair in terms of healing rate, extent of wound closure and quality of regenerated skin via gross appearance and histological analysis.
Experimental
Preparation of electrospun chitosan-polycaprolactone mats
Chitosan-polycaprolactone (CPCL) copolymer blend nanofibers were fabricated by electrospinning using previously optimized polymer solution concentrations with some alterations[19]. All chemicals were purchased from Sigma Aldrich (St. Louis, MO) unless otherwise stated. Stock solutions of chitosan and PCL were prepared separately and mixed to form the CPCL blend electrospinning solution. Chitosan (75–85% deacetylated, medium molecular weight) was dissolved in trifluoroacetic acid (TFA) (Fisher, Reagent >97%) to yield a 7 wt% solution and the solution was heated to 60°C under continuous mixing for 12 hours. A 12wt% polycaprolactone (PCL) (Average Mn 80,000) solution was prepared by dissolving PCL under constant stirring in 2,2,2-trifluoroethanol (TFE) (ReagentPlus®, >99%). The solutions were mixed to generate a 40 wt% chitosan and 60 wt% PCL blend and the blend solution was vortexed for two minutes to ensure homogeneity. To prevent polymer degradation, the two stock solutions were combined to make the above blend solution immediately prior to initiation of electrospinning and the electrospinning solution changed hourly.
The solution was gravity fed into the elecrospinning system through a 22-gauge needle attached to a 3 mL disposable syringe tilted 20° below the horizontal. A DC voltage of 11kV was applied to the spinneret needle tip and the spinneret was positioned 18cm from a rotating cylindrical collector covered in aluminum foil. The 8cm-diameter collector was rotated at 100RPM and the fibers were allowed to build up for 90 hours. Using a humidifier/dehumidifier, consistent environmental conditions were maintained at 40–50% humidity and 20–25°C. The nanofibrous sheet was allowed to dry for 24 hours, removed from the aluminum foil, folded in half to create 2-ply nanofiber scaffolds and cut into 1.5cm × 1.5cm squares. These CPCL nanofiber mats were neutralized in 14 wt% ammonium hydroxide for 15 minutes and washed three times with deionized water to remove residual ammonium hydroxide. The nanofiber mats were sterilized by soaking in 70% ethanol for 15 minutes followed by a change to fresh 70% ethanol for an additional 45-minute soak. The nanofibers were rinsed three times with sterile PBS and then soaked in sterile PBS overnight. PBS was changed one more time and the nanofiber mats were maintained in sterile PBS for in vivo studies.
Scanning electron microscopy and nanofiber diameter quantification
Nanofibers were sputter-coated with Au/Pd for 40s at 18mA, and imaged with a FEI Sirion XL30 SEM at an operating voltage of 3 kV.
Nanofiber diameter quantification
For fiber diameter quantification, 25 nanofibers were measured in a representative 512 × 512 pixel SEM image at 8000× magnification using ImageJ (NIH, Bethesda, Maryland, USA). This procedure was repeated for 3 separate images for a total of 75 nanofibers.
Mechanical testing of native mouse skin, C-CPL nanofiber mats and Tegaderm™
CPCL nanofiber mats, Tegaderm™, and mouse skin were tensile tested using a micro-tensile tester previously constructed in our lab to determine the elastic modulus of the materials[73]. The samples were tested with a 100N load cell and a crosshead speed of 0.4mm/min. Dry nanofiber and Tegaderm samples were cut into 20mm × 5mm strips. The mouse skin, maintained in PBS prior to testing, was tested using 30mm × 2mm strips. The elastic modulus for each condition was obtained by evaluating stress versus strain data and averaging over five samples.
Surgical procedures
All experiments conformed to the University of Washington Institutional Animal Care and Use Committee (IACUC) guidelines and approvals. Female, ~20 gram, 7 week old Balb/c mice (Charles River Laboratories, Wilmington, MA) were used for experiments. Mice were divided into two treatment groups: Tegaderm control and CPCL nanofiber. Each group contained 10 mice. Following anesthetisation (5 vol% isofluorane), the dorsal surfaces of the mice were shaved and depilated. To create full-thickness skin defects, skin was punched with a sterile 8 mm biopsy punch (Figure 1a). Empty wound sites were covered with Tegaderm™ occlusive wound dressing (3M Corporation, St. Paul, MN) and used as the empty control group. Other wound sites, representing the experimental group, were covered first with the CPCL nanofiber mats and then with Tegaderm to maintain the mats on the wounds via adhesion of Tegaderm to healthy skin surrounding the defect. Mice were sacrificed by CO2 exposure after 6 days, 10 days, or 14 days and wound area excised for histological, immunohistochemical and qRT-PCR evaluation. Three mice from each of two treatment groups were sacrificed at day 6 and at day 10. The remaining four mice from each of two treatment groups were sacrificed at day 14.
Figure 1.
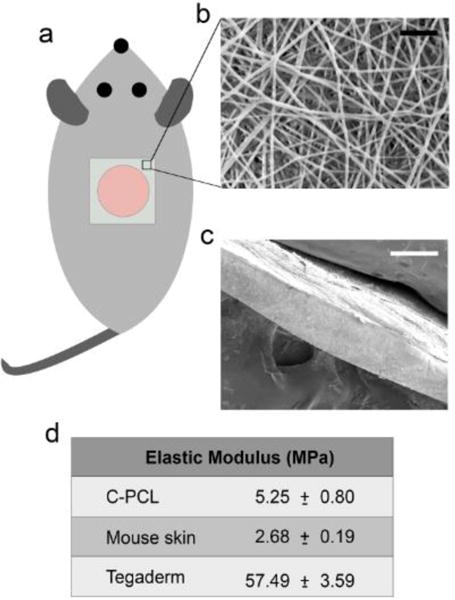
Physical properties of CPCL nanofibers. (a) Schematic of nanofiber placement on 8mm diameter cutaneous excisional skin defect. (b) Scanning electron micrograph of CPCL nanofiber scaffold en face and (c) in cross section. (d) Table summarizing elastic moduli of CPCL, mouse skin and Tegaderm. Scale bars represent (b) 2 μm and (c) 200 μm.
Quantitative evaluation of wound healing
The wound size was evaluated for both control and treatment groups every other day for 14 days by gross imaging of the site and subsequently measuring the wound area from the images using ImageJ. Wound area was measured for four defects per time point per treatment group. The extent of wound healing is expressed as percent recovery:
Histological evaluation and immunohistochemical staining
Histological and immunohistochemical staining was carried out for samples from 3 animals per time point per experimental group. Wound areas and surrounding healthy skin were fixed in 10% neutral buffered formalin for 10 minutes. Samples were dehydrated through an ethanol series, embedded in paraffin and sectioned at a thickness of 10 μm. Sections were stained with hematoxylin-eosin (H&E) or Masson’s Trichrome (Sigma Aldrich, St. Louis, MO).
For immunohistochemical staining, sections were deparaffinized and incubated in pH 6 or pH 9 antigen retrieval buffers (DAKO, Carpinteria, CA) overnight at 60°C. Paraffin sections were blocked in 0.5% normal goat serum for 30 minutes, exposed to primary antibodies against keratin 10 (1:100, Abcam, Cambridge, MA) or keratin 14 (1:100, Abcam) at 4°C overnight, incubated with biotinylated rabbit anti-mouse secondary antibody (1:200, Vector Laboratories, Burlingame, CA) at room temperature for 30 minutes, with ABC reagent (Vectastain ABC kit, Vector Laboratories) at room temperature for 30 minutes and developed with 3,3′-diaminobenzidine tetrahydrochloride (DAB) solution. The sections were counterstained with hematoxylin and mounted.
A series of 12–18 images at 10× magnification were captured for each stained tissue section using a Nikon TE300 microscope (Nikon, Japan). Tissue sections were reconstructed by stitching the images together using the Photomerge function in Adobe Photoshop.
Quantitative RT-PCR
RT-PCR was performed on samples from 4 animals per group at day 14. Tissue associated with the wound area, 14 days post-surgery, was frozen in liquid nitrogen following dissection and stored at −80°C. Samples were ground into a powder in the presence of liquid nitrogen using a dry ice-cooled mortar and pestle. RNA extraction was completed using the Qiagen RNeasy fibrous tissue kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) following manufacture’s instructions. SYBR Green PCR Master mix (Bio-Rad) was used for template amplification following the manufacture’s manual. qRT-PCR data were analyzed using the CFX Manager software (Bio-Rad). β-actin was used as the reference gene.
Results and Discussion
The effect of CPCL nanofibers on wound healing was evaluated using a mouse cutaneous excisional skin defect model where 8 mm diameter defects were created on the backs of mice (Fig. 1a). Empty control defects were covered with Tegaderm to prevent infection and compared to the experimental condition where a CPCL nanofiber scaffold was placed in direct contact with the wound and held in place with a layer of adhesive Tegaderm.
Physical properties of chitosan-poly(caprolactone) nanofibers
Randomly oriented CPCL nanofibers were fabricated via electrospinning. Figure 1b shows the fiber morphology, visualized using scanning electron microscopy, where the individual nanofibers are uniform with no visible branching. The nanofiber scaffold in cross section is shown in Figure 1c and the overall scaffold thickness is ~200 μm. Although relatively thin compared with other biomaterials used for skin repair, these nanofiber scaffolds were easy to handle and apply, using forceps, for in vivo experiments. Individual nanofibers have a mean diameter of 177.6 ± 40.5 nm. Importantly, the average nanofiber diameter is similar to the diameter of mouse skin collagen fibrils (~110 nm)[35, 36].
As matrix elasticity has been shown to have a significant impact on cell phenotype and behaviors such as proliferation, differentiation and migration[37–40], the stiffness of CPCL nanofiber scaffolds was measured and compared to the stiffness of native mouse skin and Tegaderm. Based on tensile testing, the stiffness of dry CPCL nanofiber scaffolds (5.25 ± 0.80 MPa) is approximately two times that of native mouse skin (2.68 ± 0.19 MPa) whereas the stiffness of Tegaderm wound dressing (57.49 ± 3.59 MPa) is approximately one order of magnitude greater than that of CPCL nanofibers (Figure 1d). The ten-fold difference in stiffness of Tegaderm versus native mouse skin may negatively impact attachment, migration and/or proliferation of cells participating in skin repair.
Gross evaluation of wound healing
To monitor healing of empty control wounds versus wounds treated with CPCL nanofiber scaffolds over time, gross images of skin defects were captured every other day for 14 days (Figure 2). The moistened CPCL nanofiber scaffolds were semi-translucent and allowed for visualization and measurement of the defect area. No obvious signs of infection, such as redness or swelling, were present in either treatment group throughout the experimental period. Figure 2 shows that the wound diameter appears unchanged at day 2 for both experimental groups. Between day 4 and day 6, a decrease in wound diameter is visible for the CPCL group with a more significant decrease visible between day 6 and day 8. The diameter of defects in the CPCL group decreased over time and the representative defect appears closed at day 14. In the empty group, there is also a reduction in defect diameter over time with the first significant and visible decrease in wound diameter occurring between day 6 and day 8. While the diameter is significantly reduced at day 14, a scab in the very center of the defect area indicates incomplete closure.
Figure 2.
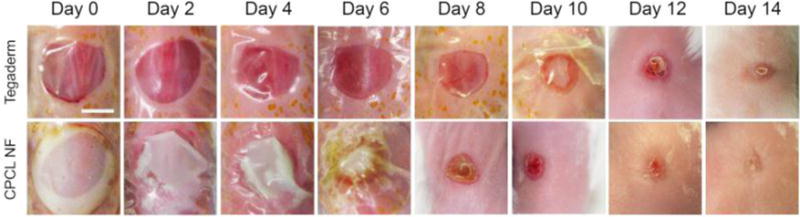
Gross imaging of defect closure over 14 days. Representative images at each time point illustrating differences in wound closure rate and quality between empty control and CPCL nanofiber-treated defects. Scale bar represents 4 mm.
Correspondingly, Figure 3 shows quantitative analysis of defect healing in terms of percent defect recovery as determined from gross images of the skin defects over time. The initial defect area at day 0 is defined treated as 0% recovery. On average, the wound size was significantly reduced between day 4 (17.6%) and day 6 (45.8%) for the CPCL treatment group, reaching 99.7% recovery by day 14. A significant change in wound size occurred between day 6 (5.9%) and day 8 (35.7%) for the control group and defects reached 94.9% closure by day 14. Based on gross observations over the two-week period, the onset of re-epithelialization was more rapid for defects in direct contact with a CPCL nanofiber scaffold, indicating a more rapid rate of recovery and more complete wound healing.
Figure 3.
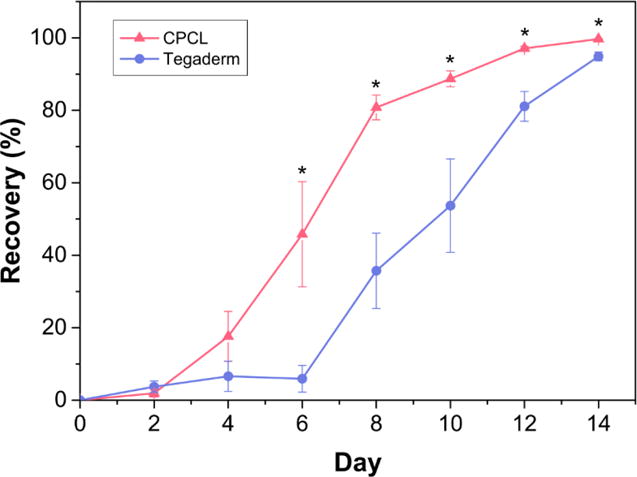
Quantitative measure of defect healing via measurement of defect area over 14 days. The initial defect area is defined as 0% recovery and data points represent mean recovery (%) of 4 defect sites +/- standard error of the mean. *(p < 0.05)
Histological analysis of wound healing
Re-epithelialization is facilitated by keratinocytes – both post-mitotic cells from the surrounding epidermis and stem cells associated with bulge regions of hair follicles[41, 42] – which are activated following cutaneous injury leading to a migratory phenotype that involves cytoskeletal changes including keratin filament rearrangement[43, 44]. Re-epithelialization begins with thickening of the epidermis near the wound margins and as keratinocytes become activated, an epidermal tongue or wedge is formed that migrates toward the center of the wound bed[45]. Migration is guided initially by a fibrin clot-associated provisional matrix, which is subsequently replaced by collagen-rich granulation tissue as fibroblasts enter the wound bed[42, 46, 47]. In addition to cell migration, re-epithelialization involves proliferation of cells located directly behind the leading edge[43, 48, 49].
H&E staining of representative skin defect sites was completed 6, 10 and 14 days post-surgery to illustrate histological differences between empty control and CPCL nanofiber scaffold-treated defects in terms of re-epithelialization and overall wound healing (Fig. 4). Masson’s trichrome staining was carried out at the same time points to highlight changes in collagen density (blue staining) over time in the regenerating dermis (Fig. 5). Both stain combinations are informative in characterizing and evaluating wound healing and the results are discussed simultaneously from one time point to the next (Fig. 4 and Fig. 5). Six days post-wounding, the wound area in the empty control group is recognizable by the interruption in epidermal continuity and the presence of a thin strip of provisional matrix within the wound bed (Figure 4a). Tegaderm is visible as a thin, transparent, wavy ribbon spanning the width of the wound above the wound bed and an epidermal tongue appears at each edge of the wound bed (Figure 4b). In the CPCL group, the eosin-stained, nanofiber scaffold spans most of the width of the wound bed (Figure 4c). A migratory epidermal tongue or migration front is also evident at the right edge of the wound bed (Figure 4d). Sample processing likely caused the healthy tissue on the left side of the tissue section to fold over such that the epidermal tongue does not appear to enter the wound bed on that side (Figure 4c). By day 6 the acute inflammatory stage has passed and there are no signs of chronic inflammation associated with the defects. A provisional matrix fills more of the wound bed in the CPCL group as compared with the control group, but the overall density of this early matrix is low with no significant collagen content as evidenced by Masson’s Trichrome staining showing little, if any, blue-stained collagen (Figure 5b).
Figure 4.
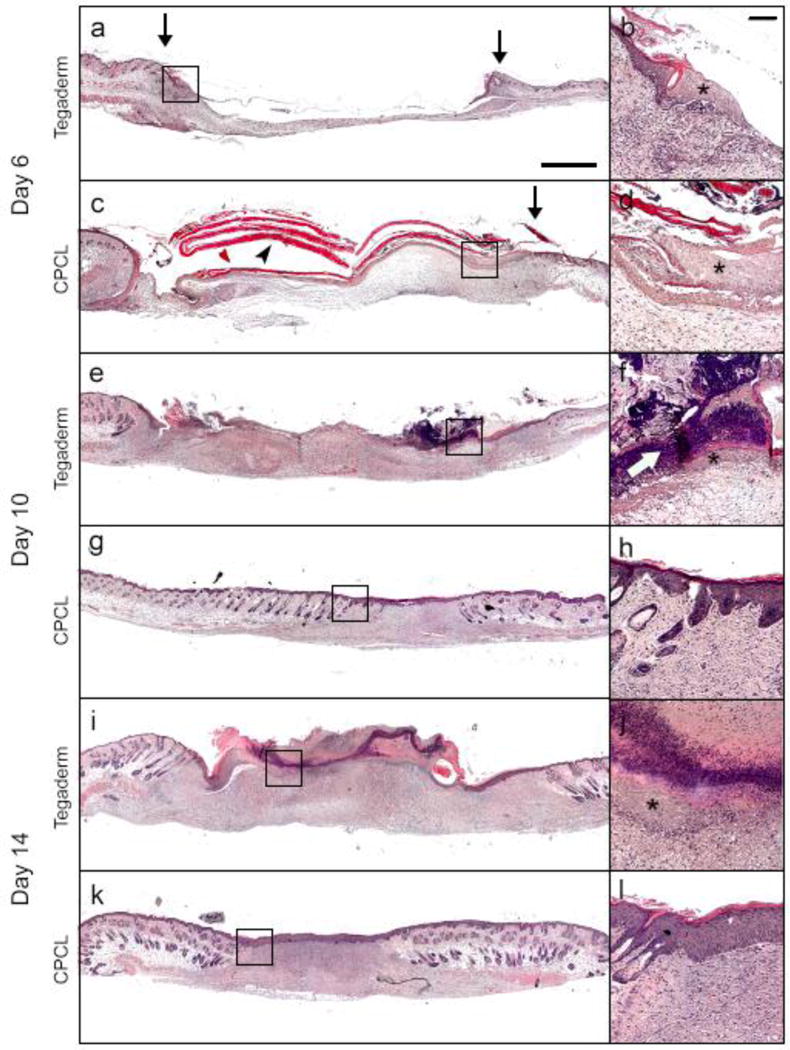
H&E staining of representative skin defect sites. Each panel (a, c, e, g, i, k) is a composition of 12–18 images to provide high resolution over a large area with accompanying inset (b, d, f, h, j, l) providing a higher magnification view near the wound edge. (↓) defect margin; (➤) CPCL; (✱) epidermal tongue; (⇨) scab. Scale bar represents 1 mm, inset scale bar represents 200 μm.
Figure 5.
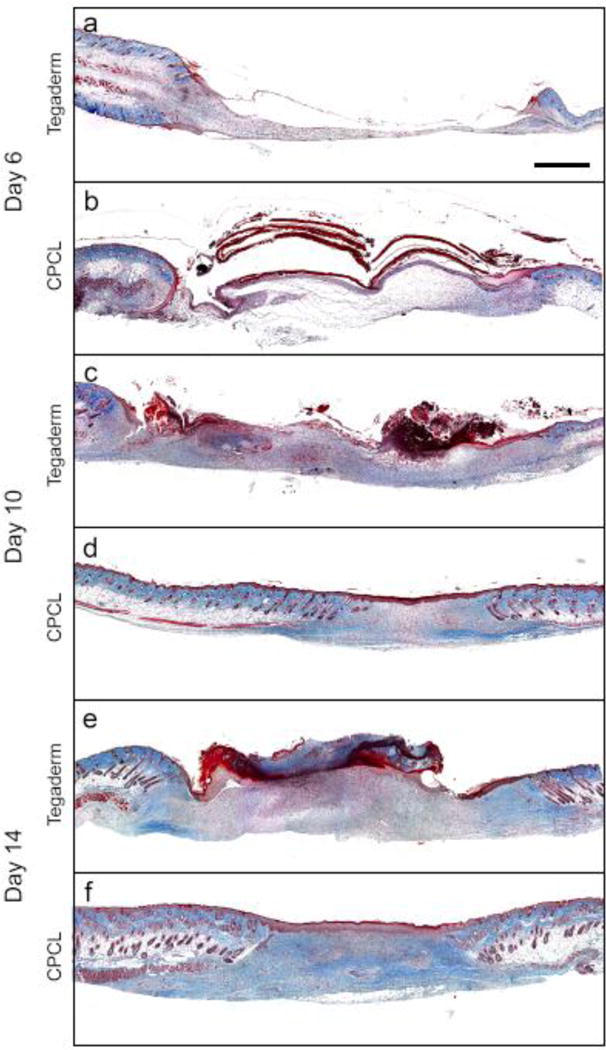
Masson’s Trichrome staining of representative skin defect sites at 6 (a–b), 10 (c–d) and 14 (e–f) days post-surgery to illustrate histological differences in collagen deposition between Tegaderm treated and CPCL nanofiber scaffold treated defects. Scale bar represents 1 mm.
Ten days post-wounding, eschar or scab formation has initiated above the empty control wound bed (Figure 4f). The migratory epidermal tongue advanced from the wound edge toward the center of the defect and was visible between the scab and provisional matrix. The thickness and density of the provisional matrix has increased significantly including a minimal amount of collagen (Figure 5c). In the CPCL group, re-epithelialization of the wound bed is complete by day 10 (Figure 4g) and the density of the neo-dermal tissue has increased significantly due to collagen deposition by invading fibroblasts (Figure 5d).
At day 14, a scab covers the width of the wound bed in the empty control group (Figure 4i). Hypertrophic epidermis is present between the scab and the neo-dermal tissue, but re-epithelialization remains incomplete whereas more collagen is present in the neodermis (Figure 5e). A more mature, stratified neo-epidermis is present in the CPCL group (Figure 4k), which is thicker than that found in normal epidermal tissue surrounding the wound. The epidermis can be identified by polarized basal keratinocytes, a spinous layer composed of cuboidal, differentiating keratinocytes, a granular cell layer and finally the outermost stratum corneum composed of flattened, dead cells (Figure 4l). Additionally, collagen deposition in the neo-dermis is more uniform in density than at the previous time point (Figure 5f) and no hair follicles or sebaceous glands are present within the regenerating wound area, which is consistent with this stage of normal wound healing[41]. Finally, there is no evidence of the CPCL nanofiber scaffold remaining in the wound bed.
The presence of CPCL nanofibers in direct contact with the wound bed may expedite re-epithelialization by presenting an additional provisional matrix to which activated keratinocytes can attach and migrate. Whereas early provisional matrix (fibrin clot) formation takes time, the CPCL nanofiber scaffold is immediately in contact with the wound bed post-wounding. Importantly, we have previously shown that keratinocytes attach to CPCL nanofibers and proliferate in vitro indicating scaffold tissue compatibility with respect to skin regeneration[34].
CPCL nanofiber scaffolds also appear to promote the formation and maturation of neo-dermal tissue within the wound bed. Early granulation tissue, formed by invading fibroblasts contains significant amounts of fibronectin and type III collagen that is later remodelled to include mostly type I collagen as neo-dermis is formed. Masson’s trichrome staining shows minimal collagen (blue staining) in the wound bed at all time points for empty control defects despite an overall increase in density of the neo-dermal tissue in that volume (Figure 5a, c, e). Collagen density increases over time in the CPCL treatment group with no collagen visible at day 6 (Figure 5b) and significant collagen deposition by day 14 (Figure 5f).
In the empty control group, the Tegaderm is in direct contact with the wound bed, but does not appear to significantly promote keratinocyte or fibroblast migration in support of wound closure. Crosstalk between keratinocytes and fibroblasts is crucial for timely and complete wound closure[50] and therefore improvement in rate and quality of both neo-epidermis and neo-dermis formation in the CPCL treatment group demonstrates how CPCL nanofibers engage in multiple processes associated with wound healing. Tegaderm, a polyurethane based occlusive dressing with an adhesive acrylic layer[51], has been shown to support attachment and proliferation of keratinocytes in vitro[52], but our results and those of others demonstrate that it is a suboptimal material for expediting skin repair and hence its utilization as a control material in studies of biomaterials for wound healing.
Immunohistochemical evaluation of wound healing
Keratins are intermediate filaments found within the cytoskeleton of epithelial cells. The temporal keratin expression profile of keratinocytes changes as they undergo differentiation due to changes in cell morphology and function and therefore characterization of key keratins provides some insights into keratinocyte activity during wound healing[53]. We examined the expression of keratin 14 (Krt14) by mitotically active, basal keratinocytes and keratin 10 (Krt10) by post-mitotic, differentiating, suprabasal keratinocytes within the regenerating epidermis[54–56]. Keratin 14 expression at day 6 for the CPCL group, is present throughout the migratory epidermal tongue from the apical to basal surface with highest intensity staining of basal cells (Figure 6a). A similar pattern of expression throughout the migratory tongue is observed for the control group at all time points. At days 10 and 14, when re-epithelialization is complete in the CPCL group, keratin 14 staining remains in the basal layer with no keratin 14 expression in the apical stratum corneum layer. While keratin 14 expression is generally downregulated in differentiating, suprabasal keratinocytes, the relative stability of epidermal keratins means that keratin 14 expression is often still observed in the suprabasal spinous layers of the epidermis as seen here[57]. Reversion to basal cell-specificity of keratin 14 expression occurs later in wound healing[53].
Figure 6.
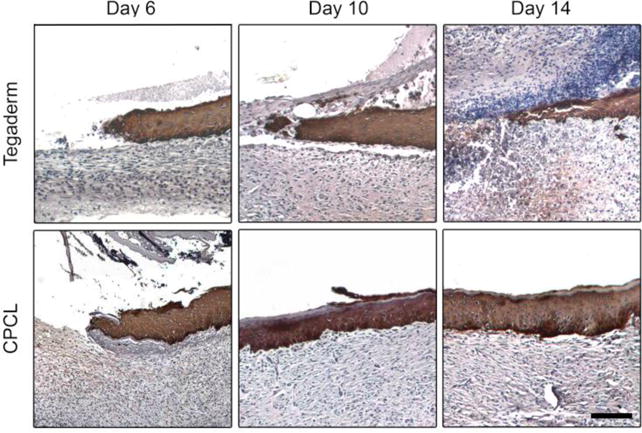
Keratin 14 immunohistochemical staining (brown/black) with haematoxylin counterstain (blue) of skin defect sites 6, 10 and 14 days post surgery to illustrate proliferating keratinocyte population in Tegaderm treated and CPCL nanofiber scaffold treated defects. The scale bar represents 100 μm.
At day 6, keratin 10 is not expressed by keratinocytes at the very tip of the migratory tongue, in either treatment group, with diffuse expression in adjacent regions that transitions into more intense and uniform expression away from the tip and toward established epidermal tissue in the surrounding native skin (Figure 7a–d). Keratin 10 is a marker of keratinocyte differentiation and maturation, so lack of expression at the tip of a migratory epidermal tongue is indicative of the dedifferentiated state of cells at the leading edge[58]. The same pattern of keratin 10 expression (no expression at the leading edge of the epidermal tongue) is observed in the empty control group at days 10 and 14 as the migratory front advances across the wound bed under the scab (Figure 7e, i). Keratin 10 expression becomes relatively uniform throughout the neo-epidermis, from the basal to apical surface, by day 10 in the CPCL treatment group (Figure 7g, h) and is then limited to suprabasal cells by day 14 (Figure 7l). Here keratin 10 expression occurs uniformly in the spinous layer, but not the basal layer or apical stratum corneum. This indicates that keratinocytes in the neo-epidermis have returned to a normal program of terminal differentiation by day 14 in the CPCL group while further maturation and remodelling is required to attain native epidermal tissue characteristics such as thickness and organization.
Figure 7.
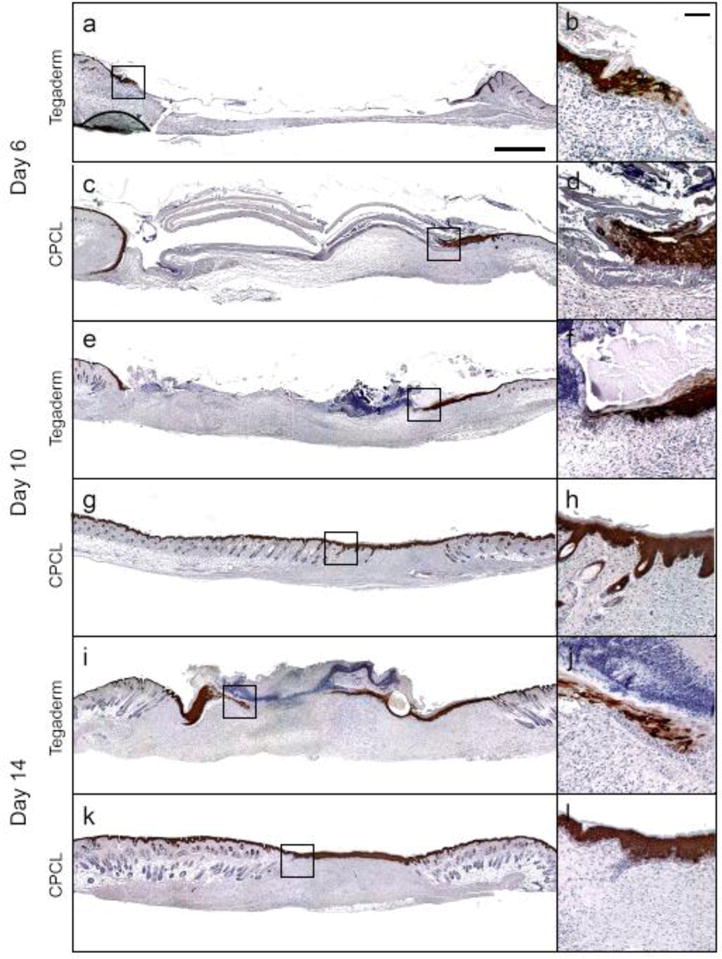
Keratin 10 immunohistochemical staining (brown/black) with haematoxylin counterstain (blue) of skin defect sites 6, 10 and 14 days post-surgery to illustrate localization of differentiating keratinocyte population in Tegaderm treated and CPCL nanofiber scaffold treated defects. The scale bar represents 1 mm, and the inset scale bar represents 200 μm.
qRT-PCR analysis of genes associated with skin wound healing
In addition to histological and immunohistochemical staining to evaluate overall wound healing and protein expression, qRT-PCR was utilized to evaluate expression of genes associated with cutaneous wound healing (Krt10, Krt14, Col1a, Col3a). As previously mentioned, the keratin expression profile of keratinocytes is dynamic and changes upon cell differentiation. Keratin 10 and keratin 14 protein expression at days 6, 10 and 14, as seen in Figures 6 and 7, was informative and show differences in protein expression between the two groups and over time. Evaluation of keratin mRNA expression at day 14 provides supporting information regarding extent of and continuation of wound healing.
Keratin 10 (Krt10) and keratin 14 (Krt14) mRNA expression was compared between the empty control and experimental nanofiber conditions at day 14 using qRT-PCR (Fig. 8). Expression of Krt10 and Krt14, relative to expression of the same gene in empty control wounds, was down-regulated in CPCL-treated wounds. This result is consistent with the overall rate and extent of wound healing reported above where healing is more complete in the CPCL treatment group therefore representing a different keratinocyte activity/differentiation profile from empty control wounds. There are likely more proliferating or differentiating keratinocytes expressing Krt14 and Krt10 in empty control wounds concomitant to re-epithelialization, which remains incomplete at day 14. In addition, it is well understood that keratin 14/keratin 5 expression switches to keratin 10/keratin 1 expression as keratinocytes commit to and enter terminal differentiation[59, 60], which occurs more quickly in CPCL-treated wounds. The stability of keratin proteins correlates with a long half-life[61, 62] meaning that high keratin 14 and keratin 10 protein expression in regenerated epidermis of CPCL-treated wounds at day 14 (Fig. 6 and 7) can reasonably correlate with down-regulation of gene expression at the same time point. Overall, the presence of CPCL nanofiber scaffolds not only increases the rate of re-epithelialization, but also appears to expedite maturation of the neo-epidermal tissue.
Figure 8.
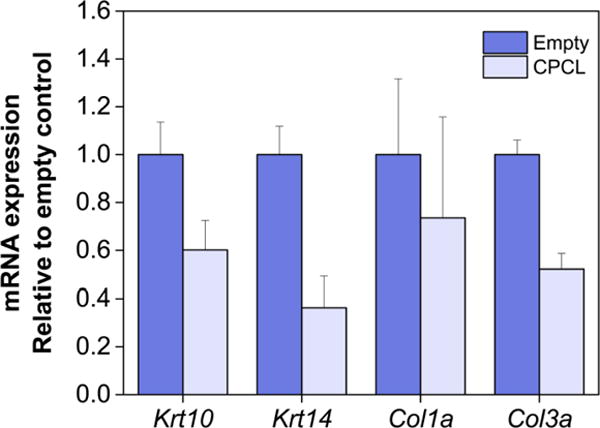
Keratin 10 (Krt10), keratin 14 (Krt14), collagen 1A (Col1a), collagen 3A (Col3a) mRNA qRT-PCR analysis of empty control vs. CPCL nanofiber-treated skin wounds 14 days post-wounding. Error bars represent standard error of the mean.
Type I collagen is the predominant form of collagen in skin, which replaces early type III collagen during later stages of wound healing, but is also found to be co-localized with type III collagen[1, 63, 64]. qRT-PCR analysis of collagen mRNA expression 14 days post-wounding (Fig. 8) shows down-regulated expression of Col3a in CPCL-treated wounds relative to empty control wounds. This is indicative of the improved rate of wound closure in the CPCL treatment group as type III collagen is generally produced during early phases of wound healing[65, 66]. Col1a expression does not differ between the treatment and control conditions (Fig. 8) 14 days post-wounding likely because type I collagen production remains at high levels at day 14 in both groups. Production may be in an earlier phase in the empty control group and a later phase related to remodeling in the CPCL group.
Possible mechanisms associated with CPCL nanofiber facilitation of wound healing
Some intrinsic properties of chitosan have been linked to biological responses such as promotion of hemostasis and wound healing as well as antimicrobial effects. Yet the overall mechanisms by which CPCL nanofiber scaffolds promote skin repair remain to be elucidated. These mechanisms are likely intimately related to scaffold architecture and physical properties. The large surface area presented by nanofibers promotes protein adsorption thereby supporting cell adhesion, migration and proliferation and allows the scaffolds to act as a provisional matrix in support of re-epithelialization and neo-dermal tissue formation. Similar to the role of the fibrin clot during early stages of wound healing associated with inflammation, CPCL nanofibers may also serve to bind growth factors and/or cytokines critical to wound healing and act as a reservoir for these factors. Keratinocyte activation, migration and proliferation is a result of a plethora of coordinated chemical and mechanical signals initiated during the inflammatory stage of the wound healing cascade[41, 67]. Growth factors and cytokines released by platelets and inflammatory cells include PDGF, EGF, FGFs, TGF-βs, VEGF, interleukins and colony stimulating factors, to name a few[1, 68].
Finally, the stiffness of the CPCL scaffolds, which we reported as approximately two times that of native mouse skin (Fig. 1), likely influences keratinocyte phenotype due to mechanosensing and mechanotransduction. It has been well-established that ECM stiffness significantly effects cell behaviour due to cell-matrix interactions whereby cell-generated forces inform intracellular signalling and changes to cell phenotype[37, 69]. The elastic modulus of Tegaderm is an order of magnitude greater than that of mouse skin and may be too stiff to promote normal keratinocyte phenotype during would healing. Some evidence has been reported that matrix elasticity has a significant effect on keratinocyte activity during wound healing such as migration velocity, proliferation, colony formation and differentiation, but a consistent correlation has yet to be established[70–72]. Regardless, targeting the elasticity of biomaterials within the range of native skin is likely critical to development of novel and effective skin substitutes.
Conclusions
CPCL nanofiber scaffolds increased wound healing rate and overall closure, re-epithelialization, maturity of neo-epidermis, and collagen deposition when compared with (Tegaderm only) empty control. These results indicate that CPCL nanofiber scaffolds possess significant potential in promoting skin repair and this is likely due to inherent biological properties of chitosan and physical properties of nanofibers such as fiber diameter and surface area as well as scaffold porosity and stiffness. Importantly, the architecture of the scaffold is biomimetic for native extracellular matrix in terms of fiber morphology and dimensionality and likely serves as an immediate provisional matrix for keratinocyte and fibroblast migration in support of wound closure. CPCL nanofibers are effective in promoting skin repair when utilized alone and can be utilized in the future as a component of more complex tissue engineering constructs involving delivery of cells and/or proteins to further enhance cutaneous wound healing.
Statistical analysis
The results are presented as mean values ± standard error of the mean. Statistical differences were determined by one-way analysis of variance (ANOVA). Values were considered statistically significant at p < 0.05.
Table 1.
Sequences of primers used in qRT-PCR analysis
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Keratin 10 | 5′-CAGATAGGCCAGCTCTTCAGT-3′ | 5′-GACATCAACGGCCTGCGTA-3′ |
| Keratin 14 | 5′-AGACCAAAGGTCGCTACTGC-3′ | 5′-TGGAGGAGGTCACATCTCTGG-3′ |
| Collagen 1A | 5′-ACATGTTCAGCTTTGTGGACC-3′ | 5′-CATGGTACCTGAGGGCGTTC-3′ |
| Collagen 3A | 5′-ATGTTGTGCAGTTTGCCCAC-3′ | 5′-TCGTCCGGGTCTACCTGATT-3′ |
Acknowledgments
This work is supported in part by Kyocera Endowment to Miqin Zhang. Sheeny Lan Levengood acknowledges support from the Ruth L. Kirschstein NIH Training grant T32CA138312. Ariane E. Erickson acknowledges support from the National Science Foundation Graduate Research Fellowship Program. The authors acknowledge the use of resources at the Molecular Analysis Facility at the University of Washington.
References
- 1.Sun BK, Siprashvili Z, Khavari PA. Science. 2014;346:941. doi: 10.1126/science.1253836. [DOI] [PubMed] [Google Scholar]
- 2.Gomathysankar S, Halim AS, Yaacob NS. Arch Plast Surg. 2014;41:452. doi: 10.5999/aps.2014.41.5.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard D, Laverdet B, Buhe V, Trouillas M, Ghazi K, Alexaline M, Egles C, Misery L, Coulomb B, Lataillade JJ, Berthod F, Desmouliere A. Tissue Eng Part B Rev. 2016 doi: 10.1089/ten.TEB.2016.0195. [DOI] [PubMed] [Google Scholar]
- 4.Mayet N, Choonara YE, Kumar P, Tomar LK, Tyagi C, Du Toit LC, Pillay V. J Pharm Sci. 2014;103:2211. doi: 10.1002/jps.24068. [DOI] [PubMed] [Google Scholar]
- 5.Mogosanu GD, Grumezescu AM. Int J Pharm. 2014;463:127. doi: 10.1016/j.ijpharm.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Cam C, Zhu S, Truong NF, Scumpia PO, Segura T. J Mater Chem B Mater Biol Med. 2015;3:7986. doi: 10.1039/C5TB00807G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adolph EJ, Guo R, Pollins AC, Zienkiewicz K, Cardwell N, Davidson JM, Guelcher SA, Nanney LB. J Biomed Mater Res B Appl Biomater. 2015 doi: 10.1002/jbm.b.33515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caetano GF, Frade MA, Andrade TA, Leite MN, Bueno CZ, Moraes AM, Ribeiro-Paes JT. J Biomed Mater Res B Appl Biomater. 2015;103:1013. doi: 10.1002/jbm.b.33277. [DOI] [PubMed] [Google Scholar]
- 9.Moura LI, Dias AM, Carvalho E, de Sousa HC. Acta Biomater. 2013;9:7093. doi: 10.1016/j.actbio.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 10.Yin H, Ding G, Shi X, Guo W, Ni Z, Fu H, Fu Z. Colloids Surf B Biointerfaces. 2016;145:226. doi: 10.1016/j.colsurfb.2016.04.056. [DOI] [PubMed] [Google Scholar]
- 11.Griffin DR, Weaver WM, Scumpia PO, Di Carlo D, Segura T. Nat Mater. 2015;14:737. doi: 10.1038/nmat4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi SK, Park JK, Kim JH, Lee KM, Kim E, Jeong KS, Jeon WB. J Control Release. 2016;237:89. doi: 10.1016/j.jconrel.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Ye J, Zhu J, Xiao Z, He C, Shi H, Wang Y, Lin C, Zhang H, Zhao Y, Fu X, Chen H, Li X, Li L, Zheng J, Xiao J. Biomacromolecules. 2016;17:2168. doi: 10.1021/acs.biomac.6b00398. [DOI] [PubMed] [Google Scholar]
- 14.Kumbar SG, James R, Nukavarapu SP, Laurencin CT. Biomed Mater. 2008;3:034002. doi: 10.1088/1748-6041/3/3/034002. [DOI] [PubMed] [Google Scholar]
- 15.Vasita R, Katti DS. Int J Nanomedicine. 2006;1:15. doi: 10.2147/nano.2006.1.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy VJ, Radhakrishnan S, Ravichandran R, Mukherjee S, Balamurugan R, Sundarrajan S, Ramakrishna S. Wound Rep Reg. 2013;21:1. doi: 10.1111/j.1524-475X.2012.00861.x. [DOI] [PubMed] [Google Scholar]
- 17.Uitto J, Olsen DR, Fazio MJ. J Invest Dermatol. 1989;92:61S. doi: 10.1111/1523-1747.ep13075039. [DOI] [PubMed] [Google Scholar]
- 18.Bhattaria N, Li Z, Gunn J, Leung M, Cooper A, Edmondson D, Veiseh O, Chen MH, Zhang Y, Ellenbogen RC, Zhang M. Adv Mater. 2009;21:2792. [Google Scholar]
- 19.Cooper A, Bhattaria N, Zhang M. Carbohydr Polym. 2011;85:149. [Google Scholar]
- 20.Jana S, Leung M, Chang J, Zhang M. Biofabrication. 2014;6:035012. doi: 10.1088/1758-5082/6/3/035012. [DOI] [PubMed] [Google Scholar]
- 21.Shalumon KT, Anulekha KH, Chennazhi KP, Tamura H, Nair SV, Jayakumar R. Int J Biol Macromol. 2011;48:571. doi: 10.1016/j.ijbiomac.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Qiao W, Liu Z, Shang T, Qiao T, Mao C, Liu C. Tissue Eng Part A. 2014;20:79. doi: 10.1089/ten.tea.2013.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen SH, Chen CH, Fong YT, Chen JP. Acta Biomater. 2014;10:4971. doi: 10.1016/j.actbio.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Peng H, Wu Y, Zhang C, Cai Y, Xu G, Li Q, Chen X, Ji J, Zhang Y, OuYang HW. Biomaterials. 2013;34:4404. doi: 10.1016/j.biomaterials.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 25.Jana S, Florczyk SJ, Leung M, Zhang M. J Mater Chem. 2012;22:6291. [Google Scholar]
- 26.Senel S, McClure SJ. Adv Drug Deliv Rev. 2004;56:1467. doi: 10.1016/j.addr.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Ueno H, Mori T, Fujinaga T. Adv Drug Deliv Rev. 2001;52:105. doi: 10.1016/s0169-409x(01)00189-2. [DOI] [PubMed] [Google Scholar]
- 28.Dai T, Tanaka M, Huang YY, Hamblin MR. Expert Rev Anti Infect Ther. 2011;9:857. doi: 10.1586/eri.11.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aranaz I, Mengibar M, Harris R, Panos I, Miralles B, Acosta N, Galed G, Heras A. Curr Chem Biol. 2009;3:203. [Google Scholar]
- 30.Levengood SL, Zhang M. J Mater Chem B Mater Biol Med. 2014;2:3161. doi: 10.1039/C4TB00027G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing X, Mi HY, Wang XC, Peng XF, Turng LS. ACS Appl Mater Interfaces. 2015;7:6955. doi: 10.1021/acsami.5b00900. [DOI] [PubMed] [Google Scholar]
- 32.Prasad T, Shabeena EA, Vinod D, Kumary TV, Anil Kumar PR. J Mater Sci Mater Med. 2015;26:5352. doi: 10.1007/s10856-014-5352-8. [DOI] [PubMed] [Google Scholar]
- 33.Fukunishi T, Best CA, Sugiura T, Shoji T, Yi T, Udelsman B, Ohst D, Ong CS, Zhang H, Shinoka T, Breuer CK, Johnson J, Hibino N. PLoS One. 2016;11:e0158555. doi: 10.1371/journal.pone.0158555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lou T, Leung M, Wang X, Chang JY, Tsao CT, Sham JG, Edmondson D, Zhang M. J Biomed Nanotechnol. 2014;10:1105. doi: 10.1166/jbn.2014.1793. [DOI] [PubMed] [Google Scholar]
- 35.Kounadi E, Tzaphlidou M, Fountos G, Glaros D. Micron. 1995;26:113. doi: 10.1016/0968-4328(94)00056-v. [DOI] [PubMed] [Google Scholar]
- 36.Tzaphlidou M, Kounadi E, Leontiou I, Matthopoulos DP, Glaros D. Int J Radiat Biol. 1997;71:109. doi: 10.1080/095530097144481. [DOI] [PubMed] [Google Scholar]
- 37.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. J Cell Sci. 2012;125:5974. doi: 10.1242/jcs.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Curr Biol. 2009;19:1511. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raab M, Swift J, Dingal PC, Shah P, Shin JW, Discher DE. J Cell Biol. 2012;199:669. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw TJ, Martin P. J Cell Sci. 2009;122:3209. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solanas G, Benitah SA. Nat Rev Mol Cell Biol. 2013;14:737. doi: 10.1038/nrm3675. [DOI] [PubMed] [Google Scholar]
- 43.Viticchie G, Lena AM, Cianfarani F, Odorisio T, Annicchiarico-Petruzzelli M, Melino G, Candi E. Cell Death Dis. 2012;3:e435. doi: 10.1038/cddis.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Usui ML, Mansbridge JN, Carter WG, Fujita M, Olerud JE. J Histochem Cytochem. 2008;56:687. doi: 10.1369/jhc.2008.951194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Escamez MJ, Garcia M, Larcher F, Meana A, Munoz E, Jorcano JL, Del Rio M. J Invest Dermatol. 2004;123:1182. doi: 10.1111/j.0022-202X.2004.23473.x. [DOI] [PubMed] [Google Scholar]
- 46.Clark RA, Folkvord JM, Wertz RL. J Invest Dermatol. 1985;84:378. doi: 10.1111/1523-1747.ep12265466. [DOI] [PubMed] [Google Scholar]
- 47.Braiman-Wiksman L, Solomonik I, Spira R, Tennenbaum T. Toxicol Pathol. 2007;35:767. doi: 10.1080/01926230701584189. [DOI] [PubMed] [Google Scholar]
- 48.Martin P. Science. 1997;276:75. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 49.Santoro MM, Gaudino G. Exp Cell Res. 2005;304:274. doi: 10.1016/j.yexcr.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 50.El Ghalbzouri A, Hensbergen P, Gibbs S, Kempenaar J, van der Schors R, Ponec M. Lab Invest. 2004;84:102. doi: 10.1038/labinvest.3700014. [DOI] [PubMed] [Google Scholar]
- 51.Kirker KR, Luo Y, Nielson JH, Shelby J, Prestwich GD. Biomaterials. 2002;23:3661. doi: 10.1016/s0142-9612(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 52.Chua AW, Ma DR, Song IC, Phan TT, Lee ST, Song C. Burns. 2008;34:175. doi: 10.1016/j.burns.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe S, Osumi M, Ohnishi T, Ichikawa E, Takahashi H. Histochem Cell Biol. 1995;103:425. doi: 10.1007/BF01457542. [DOI] [PubMed] [Google Scholar]
- 54.Liang X, Bhattacharya S, Bajaj G, Guha G, Wang Z, Jang HS, Leid M, Indra AK, Ganguli-Indra G. PLoS One. 2012;7:e29999. doi: 10.1371/journal.pone.0029999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Cheng X, Merched-Sauvage M, Caulin C, Roop DR, Koch PJ. J Cell Sci. 2006;119:5067. doi: 10.1242/jcs.03298. [DOI] [PubMed] [Google Scholar]
- 56.DiTommaso T, Cottle DL, Pearson HB, Schluter H, Kaur P, Humbert PO, Smyth IM. PLoS Genet. 2014;10:e1004706. doi: 10.1371/journal.pgen.1004706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coulombe PA, Kopan R, Fuchs E. J Cell Biol. 1989;109:2295. doi: 10.1083/jcb.109.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo S, Yufit T, Carson P, Fiore D, Falanga J, Lin X, Mamakos L, Falanga V. Int J Low Extrem Wounds. 2011;10:122. doi: 10.1177/1534734611418157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Byrne C, Tainsky M, Fuchs E. Development. 1994;120:2369. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 60.Kartasova T, Roop DR, Holbrook KA, Yuspa SH. J Cell Biol. 1993;120:1251. doi: 10.1083/jcb.120.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haines RL, Lane EB. J Cell Sci. 2012;125:3923. doi: 10.1242/jcs.099655. [DOI] [PubMed] [Google Scholar]
- 62.Toftgard R, Yuspa SH, Roop DR. Cancer Res. 1985;45:5845. [PubMed] [Google Scholar]
- 63.Gay S, Vijanto J, Raekallio J, Penttinen R. Acta Chir Scand. 1978;144:205. [PubMed] [Google Scholar]
- 64.Liu X, Wu H, Byrne M, Krane S, Jaenisch R. Proc Natl Acad Sci U S A. 1997;94:1852. doi: 10.1073/pnas.94.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fathke C, Wilson L, Hutter J, Kapoor V, Smith A, Hocking A, Isik F. Stem Cells. 2004;22:812. doi: 10.1634/stemcells.22-5-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Volk SW, Wang Y, Mauldin EA, Liechty KW, Adams SL. Cells Tissues Organs. 2011;194:25. doi: 10.1159/000322399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kippenberger S, Bernd A, Loitsch S, Guschel M, Muller J, Bereiter-Hahn J, Kaufmann R. J Invest Dermatol. 2000;114:408. doi: 10.1046/j.1523-1747.2000.00915.x. [DOI] [PubMed] [Google Scholar]
- 68.Bahou WF, Gnatenko DV. Semin Thromb Hemost. 2004;30:473. doi: 10.1055/s-2004-833482. [DOI] [PubMed] [Google Scholar]
- 69.Evans ND, Oreffo RO, Healy E, Thurner PJ, Man YH. J Mech Behav Biomed Mater. 2013;28:397. doi: 10.1016/j.jmbbm.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Wang G, Luo X, Qiu J, Tang C. Burns. 2012;38:414. doi: 10.1016/j.burns.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Zarkoob H, Bodduluri S, Ponnaluri SV, Selby JC, Sander EA. Cell Mol Bioeng. 2015;8:32. doi: 10.1007/s12195-015-0377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anon E, Serra-Picamal X, Hersen P, Gauthier NC, Sheetz MP, Trepat X, Ladoux B. Proc Natl Acad Sci U S A. 2012;109:10891. doi: 10.1073/pnas.1117814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edmondson D, Bhattaria N, Jana S, Kim A, Zhang M. Appl Phys Lett. 2009;94:103101. [Google Scholar]


