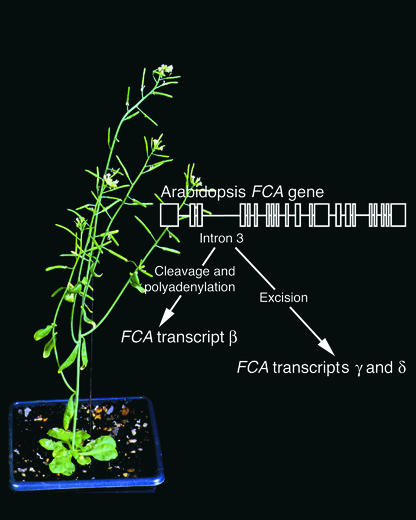
The fca mutant of Arabidopsis is one of numerous late-flowering mutants in the autonomous, or constitutive, floral promotion pathway (Koornneef et al., 1991, 1998). The autonomous pathway acts independently of the photoperiod pathway and intersects with the vernalization response pathway in the control of flowering. The FCA gene encodes a novel protein that contains RNA binding domains and a putative protein–protein interaction domain (Macknight et al., 1997). In this issue of The Plant Cell, Macknight et al. (pages 877–888) show that alternative splicing of the FCA transcript has functional significance related to its role in the promotion of the floral transition (Figure 1).
Figure 1.
Alternative Processing of FCA, a Promoter of the Floral Transition in Arabidopsis.
Cleavage and polyadenylation within intron 3 yield transcript β, which encodes a fragment of the full-length FCA protein. Transcripts γ and δ are produced from the accurate excision of intron 3. Transcript γ, produced from the accurate excision of all 21 introns, yields the only full-length functional protein. Transcript δ retains portions of intron 13, which introduces a premature translation termination codon and produces a nonfunctional protein. A fourth transcript, α, which accounts for <1% of transcripts, retains all of intron 3 and yields putatively nonfunctional products. The figure shows complementation of the fca-1 mutant phenotype of Arabidopsis via overexpression of the FCA γ transcript under the control of the 35S promoter. Overexpression of the γ transcript complements the late-flowering fca-1 mutant and causes the plant to flower significantly earlier than the wild type. The FCA β and δ transcripts were not able to complement fca-1.
STRUCTURE AND FUNCTION OF INTRONS
Most eukaryotic genes contain introns that must be spliced from pre-mRNA in the nucleus to produce functional mRNA. In Arabidopsis, 79% of nuclear genes (and 18 and 12% of plastid and mitochondrial genes, respectively) contain introns (Arabidopsis Genome Initiative, 2000), and other plant species are expected to be similar in this regard (Reddy, 2001). Thus, accurate and efficient splicing is a critical feature of the regulation of gene expression in plants as well as in other eukaryotes. Plant genes typically are shorter and contain fewer introns than vertebrate genes, but the intron-exon structure and the basic splicing mechanism are similar in all eukaryotes. Splicing takes place in a large RNA-protein complex called the spliceosome and involves two trans-esterification reactions that result in the release of the intron in a lariat form and ligation of the two exons (reviewed by Sharp, 1994; Lorković et al., 2000; Reddy, 2001). Recognition of splice sites and accurate splicing involve a large number of cis-acting elements and trans-acting factors.
The intron-exon structure of genes, which allows the duplication and rearrangement of exon units, is believed to have played an important role in the generation of new proteins during the evolution of eukaryotes (Sharp, 1994). For example, the gene for the human extracellular matrix protein fibronectin arose through tandem and dispersed duplication of exon units. The gene is composed of multiple units of three types of exons, each of which encodes a specific type of protein-folding domain. These same exon units and corresponding protein-folding domains are found in other genes as well, some of which encode cell surface receptors and blood coagulation proteins, suggesting the existence of common progenitor exon units in the evolution of numerous proteins having a variety of functions within the cell (Sharp, 1994).
ALTERNATIVE SPLICING AND CONTROL OF GENE EXPRESSION
The possibility of alternative splicing is another consequence of intron-exon gene structure and is in fact a widespread mechanism for regulating gene expression and generating isoform diversity. Alternative splicing can cause the deletion or modification of protein activity, generate completely different protein activities, alter protein localization within the cell, and/or affect RNA stability and translational efficiency (Smith et al., 1989). There are a number of well-known examples in vertebrates. The fibronectin gene, mentioned above, is alternatively spliced into mRNAs that encode 20 different proteins, some of which have different patterns of localization and slightly different functions in human cells (Sharp, 1994). A remarkable gene for a K+ channel in birds has 576 possible alternatively spliced forms, and a subset of these is expressed in a specific gradient along the sensory receptor cells of the inner ear, contributing to the highly accurate perception of different sound frequencies in birds (Black, 1998). Until relatively recently, there were very few examples of alternatively spliced genes in higher plants and still fewer examples known to generate functionally distinct proteins. However, the number of known alternatively spliced plant genes is expanding rapidly, and it is likely that alternative splicing plays a major role in the regulation of gene expression in plants and in other eukaryotes.
ALTERNATIVE SPLICING IN HIGHER PLANTS
Ribulose-1,5-bisphosphate carboxylase/ oxygenase (Rubisco) activase represents one of the best-known examples of alternative splicing in higher plants. Alternative splicing in most plant species yields two polypeptides of different size that are identical except for an additional number of amino acids at the C terminus of the larger isoform (Werneke et al., 1989). In plants that contain both isoforms, Rubisco activase regulates the activity of Rubisco in response to light-induced changes in the redox potential, via thioredoxin f, as well as to changes in the ADP/ATP ratio. Although both isoforms are capable of regulating Rubisco activity, only the larger isoform is redox regulated via thioredoxin as a result of the presence of two Cys residues in the additional portion of the C terminus, and the larger isoform is capable of interacting with and altering the activity of the smaller isoform (Zhang and Portis, 1999). Thus, alternative splicing appears to have created a mechanism for fine-tuning of the light regulation of Rubisco activase.
Other examples include diacylglycerol kinase in tomato, which is alternatively spliced to remove or retain the last exon containing the coding region for calmodulin binding (Snedden and Blumwald, 2000); the N resistance gene of tobacco, which is alternately spliced to produce short and long transcripts, both of which are required for complete resistance against Tobacco mosaic virus (Dinesh-Kumar and Baker, 2000); and chloroplast ascorbate peroxidase in spinach, which is alternately spliced to produce four isoforms, one of which is a thylakoid-bound enzyme and the other three of which are found in the stroma (Yoshimura et al., 1999). Reddy (2001) presents a list of 29 plant pre-mRNAs that undergo alternative splicing, many of which are known to encode proteins with different functions and/or different cellular or subcellular localization patterns. As with vertebrates and other eukaryotes (Smith et al., 1989), the list soon may become so large that it is no longer useful to compile comprehensive catalogs of alternatively spliced plant genes.
ALTERNATIVE SPLICING OF FCA
The FCA floral promotion gene contains 20 introns, and alternative splicing of introns 3 and 13 produces four different transcripts, designated α, β, δ, and γ, which account for <1, 55, 10, and 35%, respectively, of FCA transcripts (Figure 1) (Macknight et al., 1997). Transcript γ, in which all of the introns are accurately spliced and removed, encodes the full-length protein, including two RNA binding motifs and a WW domain that is known to play a role in protein–protein interactions. The most abundant transcript, β, is produced from cleavage and polyadenylation within intron 3, resulting in a truncated transcript that encodes a protein containing little more than the N terminus and lacking the RNA binding and WW domains. Transcript δ, which accounts for ∼10% of transcripts, is alternatively spliced in intron 13 to produce an early in-frame translation termination codon that would yield a protein containing the RNA binding domains but lacking the WW domain and 63 amino acids downstream of this domain. Transcript α, which accounts for <1% of transcripts, is alternatively spliced to retain intron 3, which introduces an early translation termination codon (encoding a protein identical to that of transcript β) and includes a second downstream open reading frame. RNase protection assays on RNA isolated from different tissues and under different growth conditions did not reveal any differences in the relative ratios of the four transcripts (Macknight et al., 1997).
Macknight et al. (2002) confirm that the γ transcript encodes the only protein that functions in the control of flowering time, and using a series of translational FCA:β-glucuronidase (GUS) fusions, they show that expression of the γ transcript appears to be restricted predominantly to shoot and root apices and young flower buds. They also found that alternative processing of intron 3 is conserved in Brassica napus and in pea, which is consistent with a functional role of alternative splicing in the control of FCA gene expression. The functionality of the γ transcript was confirmed by the overexpression of various FCA constructs under the control of the 35S promoter in the late-flowering fca-1 mutant (Figure 1). Overexpression of the γ transcript complemented the late-flowering phenotype of the mutant and caused the plants to flower earlier than the wild type, whereas overexpression of the β or δ transcript had no effect on the mutant phenotype.
The expression of the γ transcript relative to total FCA transcripts was examined with the use of fusion constructs with the GUS reporter gene. GUS fused to an FCA construct designed to show GUS activity in all tissues having an active FCA promoter (i.e., all transcripts) showed high activity in new cotyledons within 2 days after germination and increasing activity in many tissues throughout development, including vascular tissue and all parts of the flower. In contrast, GUS fused to a construct designed to produce only transcript γ did not show high GUS activity until 6 days after germination, and the activity was restricted to the shoot and root apices and lateral root primordia. In the shoot apex, GUS activity associated with the γ transcript was confined to the meristematic region of the shoot, flower buds, and new leaf primordia and decreased below the level of detection in leaves after they reached >1 mm in length.
FCA FUNCTION IN THE FLORAL TRANSITION
FCA appears to function by repressing the activity of FLOWERING LOCUS C (FLC), which is a key regulator of flowering in photoperiod-independent floral induction pathways. FLC encodes a MADS box transcription factor that acts as a repressor of floral induction (Michaels and Amasino, 1999a; Sheldon et al., 1999, 2000). FLC expression is undetectable in wild-type early-flowering accessions, but it is enhanced in late-flowering autonomous pathway mutants such as fca. The fca mutant is late flowering because of an increased level of FLC expression; the late-flowering mutant phenotype can be rescued by a loss-of-function FLC allele (Michaels and Amasino, 2001) or by a reduction in FLC transcripts brought about by antisense inhibition (Sheldon et al., 2000). The late-flowering phenotype of fca and other autonomous pathway mutants also can be rescued by prolonged exposure to cold (i.e., they show a vernalization response) via cold-induced repression of FLC, demonstrating that the autonomous and vernalization pathways converge on FLC.
A third pathway converging on FLC, identified through the analysis of natural variation in flowering time, is mediated by FRIGIDA (FRI), which encodes a novel protein that functions to enhance FLC expression (Johanson et al., 2000). Many laboratory strains of Arabidopsis are rapid-cycling, early-flowering accessions that have little or no vernalization response, but a number of naturally occurring ecotypes are late-flowering winter annuals that show a vernalization response. This variation in the vernalization response can be accounted for by the activities of FLC and FRI. Winter annual ecotypes of Arabidopsis carry a dominant FRI allele, which is associated with increased FLC expression. Thus, FRI functions as a repressor of the floral transition by enhancing the expression of the floral repressor FLC. In the winter annual ecotypes, vernalization overrides FRI and causes the repression of FLC, which then leads to promotion of the flowering response. Early-flowering laboratory accessions carry loss-of-function mutations at FRI (Johanson et al., 2000). FLC expression in these accessions is normally repressed, presumably as a result of the activity of FCA and other components of the autonomous pathway. Thus, a “winter annualism” phenotype is conferred by the presence of a dominant FRI allele or by recessive mutations in FCA and other components of the autonomous pathway via effects on FLC (reviewed by Michaels and Amasino, 1999b).
ALTERNATE SPLICING AS A DEVELOPMENTAL SWITCH
Macknight et al. (2002) suggest that the expression of the functional FCA γ transcript in meristematic regions of roots as well as leaf and floral primordia may indicate that FCA and the autonomous pathway play a role in regulating a more general function of meristems, of which timing of the floral transition is just one aspect. Consistent with this idea, the authors note that other components of the autonomous pathway also are expressed in roots as well as shoots.
Alternative splicing plays a major role as an on/off switch for the function of numerous vertebrate genes. Where such genes encode regulatory proteins or transcription factors, alternative splicing emerges as a major developmental control mechanism. One of the best-documented examples is Sex-lethal (Sxl) and associated genes in the sex determination pathway of Drosophila. Alternative splicing of these genes, resulting from a signal related to the ratio of X chromosomes to autosomes, leads to gene products that in males repress female differentiation genes but in females repress male differentiation genes (Smith et al., 1989).
Alternative splicing of FCA transcripts bears resemblance to alternative splicing of Sxl in Drosophila. The male-specific Sxl transcript retains an intron (or “male-specific exon”) that introduces an in-frame termination codon, resulting in early truncation of the major open reading frame and production of a transcript of 42 to 48 codons that encodes a nonfunctional product. In females, this intron is spliced out, and the resulting transcript encodes a 354–amino acid protein that includes RNA binding motifs. Sxl splicing is autoregulated in that the functional Sxl product promotes continued female-specific splicing and the further production of full-length Sxl protein. Sxl also regulates alternative splicing of other genes in the sex determination pathway, ultimately leading to the repression of male differentiation genes in females (Smith et al., 1989).
The region encompassing the two RNA binding motifs in FCA shows similarity to the RNA binding region in Sxl, and Macknight et al. (1997) raised the possibility that FCA could play a similar role in the regulation of alternative splicing. The presence and functional requirement of the WW box—a putative protein–protein interaction domain—suggest that FCA interacts with another protein or proteins. It will be important to determine the RNA binding specificities and protein interactions of FCA.
The FCA gene provides an important example of alternative splicing in developmental control in higher plants. Further investigations into FCA function and mechanism of action promise to reveal more information regarding the role(s) and components of the autonomous pathway and the role of alternative splicing in the regulation of gene expression, meristem function, and the transition to flowering.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Black, D.L. (1998). Splicing in the inner ear: A familiar tune, but what are the instruments? Neuron 20, 165–168. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., and Baker, B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular variation of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Peeters, A.J.M. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković, Z.J., Wieczorek Kirk, D.A., Lambermon, M.H.L., and Filipowicz, W. (2000). Pre-mRNA splicing in higher plants. Trends Plant Sci. 5, 160–166. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999. a). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999. b). Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 23, 1145–1153. [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001). Nuclear pre-mRNA splicing in plants. Crit. Rev. Plant Sci. 20, 523–571. [Google Scholar]
- Sharp, P.A. (1994). Split genes and RNA splicing. Cell 77, 805–815. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W.J., Patton, J.G., and Nadal-Ginard, B. (1989). Alternative splicing in the control of gene expression. Annu. Rev. Genet. 23, 527–577. [DOI] [PubMed] [Google Scholar]
- Snedden, W.A., and Blumwald, E. (2000). Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J. 24, 317–326. [DOI] [PubMed] [Google Scholar]
- Werneke, J.M., Chatfield, J.M., and Ogren, W.L. (1989). Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1, 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, K., Yabuta, Y., Tamoi, M., Ishikawa, T., and Shigeoka, S. (1999). Alternatively spliced mRNA variants of chloroplast ascorbate peroxidase isoenzymes in spinach leaves. Biochem. J. 338, 41–48. [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., and Portis, A.R., Jr. (1999). Mechanism of light regulation of Rubisco: A specific role for the larger Rubisco activase isoform involving reductive activation by thioredoxin-f. Proc. Natl. Acad. Sci. USA 96, 9438–9443. [DOI] [PMC free article] [PubMed] [Google Scholar]