Abstract
The telomeric single-strand DNA binding protein protection of telomeres 1 (POT1) protects telomeres from rapid degradation in Schizosaccharomyces pombe and has been implicated in positive and negative telomere length regulation in humans. Human POT1 appears to interact with telomeres both through direct binding to the 3′ overhanging G-strand DNA and through interaction with the TRF1 duplex telomere DNA binding complex. The influence of POT1 on telomerase activity has not been studied at the molecular level. We show here that POT1 negatively effects telomerase activity in vitro. We find that the DNA binding activity of POT1 is required for telomerase inhibition. Furthermore, POT1 is incapable of inhibiting telomeric repeat addition to substrate primers that are defective for POT1 binding, suggesting that in vivo, POT1 likely affects substrate access to telomerase.
Telomeres, the nucleoprotein complexes at the ends of eukaryotic linear chromosomes, serve a number of vital cellular functions. Telomere capping protects the chromosome ends from nucleolytic degradation and provides a mechanism for cells to distinguish natural from broken ends, which signal DNA damage and are substrates for DNA repair processes (reviewed in references 7 and 13). The DNA component of telomeres is composed of tandem, simple repeats (TTAGGG in mammals), terminating with a 3′-protruding single strand of the guanosine-rich strand. Due to lack of a template to replicate the 3′ overhang, most human somatic cells lose terminal DNA with each division; thus, telomeres also provide a buffer between the ends and the more internally located coding region of the genome (12, 19). In germ line cells and some cells in highly proliferative tissues, telomere loss is counterbalanced through the activities of the ribonucleoprotein telomerase. In vitro assays reveal that catalytically active telomerase is minimally composed of an RNA (TER) and a protein (TERT) subunit (5, 38, 39). Telomerase adds telomeric repeats by iteratively reverse transcribing the template portion of its RNA subunit, using the 3′ single-strand telomeric overhang as a primer (18, 42; reviewed in references 20 and 22).
Proteins that specifically bind the telomere single-strand overhang have been identified in numerous organisms. The 3′ telomeric overhang of the ciliate Oxytricha nova is bound by OnTEBP, a heterodimeric end-binding protein composed of an α subunit and a β subunit (14, 17). Crystal structures reveal that the α subunit contains three oligonucleotide/oligosaccharide binding (OB) folds: the first two OB folds bind telomeric DNA with sequence specificity, and the third participates in protein-protein interactions with the β subunit (21). A search for homologs of the OnTEBP α subunit identified the Pot1 protein in Schizosaccharomyces pombe and human based on a weak sequence similarity with the first OB fold in the α subunit (3). Genetic studies with S. pombe demonstrate a role for S. pombe POT1 in telomere end protection, since deletion of the pot1 gene results in the rapid loss of telomeric DNA and chromosome circularization. Further searching identified homologs in other organisms, including monkey, mouse, and plant (4).
Recombinant human and S. pombe POT1 have both been shown to bind with sequence specificity to the G-rich single strand of telomeric DNA in vitro but not duplex DNA. In human cells, POT1 colocalizes with telomeres (4), and results from chromatin immunoprecipitation experiments suggest that POT1 binds the telomeric 3′ overhang in vivo (25). Coimmunoprecipitation experiments also identified human POT1 as an interacting partner in the multisubunit TRF1 complex, which binds along the duplex portion of telomeric DNA through the sequence-specific DNA binding activities of the TRF1 protein (6, 8, 25). Among other proteins, known members of the TRF1 complex include TIN2, tankyrase 1, and PTOP (also known as PIP1) (23, 24, 33, 41), and recent studies suggest that POT1 interacts with the TRF1 complex through a TRF1-TIN2-PTOP/PIP1-POT1 protein bridge (24, 40, 41).
In most organisms telomere length is heterogeneous but is maintained within a defined, species-specific length range (reviewed in reference 34). In budding yeast, the duplex telomere binding protein Rap1p has been implicated in telomere length regulation, controlling telomerase activity at individual telomere ends as a function of the number of Rap1 molecules bound (28, 31). This counting mechanism involves switching of the telomere between telomerase-extendible and nonextendible states (35). In humans, TRF1 appears to regulate telomere length through a similar counting mechanism (1, 37). Two recent studies have also implicated POT1 as a modulator of telomere length. Loayza and De Lange (25) found that overexpression of N-terminally truncated POT1 (with a deletion of the DNA binding domain) leads to extreme telomere lengthening and suggested that POT1 may act as a relay of telomere length information between TRF1 and the telomere 3′ end. On the other hand, evidence has been obtained that POT1 positively regulates telomere length (2, 10).
Here we show by direct telomerase assay that recombinant human POT1 negatively regulates telomerase activity in vitro. Telomerase activity is inhibited by POT1 when it is prebound to a substrate primer and in reactions in which mutant telomeric repeats are added to POT1-bound substrates. POT1 does not affect telomerase activity on primers defective for POT1 binding, nor does a POT1 mutant with a deletion of the DNA binding domain inhibit telomerase. This suggests that POT1 negatively regulates telomerase activity at the level of substrate accessibility.
MATERIALS AND METHODS
hPOT1 expression and purification.
Full-length POT1 cDNA was generated by reverse transcription-PCR from primary human lung fibroblast RNA, using first-strand primer 5′-CAGGGAAGGTGAGTGGCAAC-3′ and subsequent amplification with primers 5′-CCGGATCCGATGTCTTTGGTTCCAGC-3′ and 5′-CGGGATCCCGTTAGATTACATCTTCTGC-3′. The plasmid pET14pot1 was constructed by cloning POT1 cDNA into the BamHI site of pET-14b (Novagen); the construct was fully sequenced and is identical to the protein coded in deposited sequence AK022580. ΔOB, encompassing amino acid residues 113 to 634, was generated by PCR and cloned into pET-14b. N-terminally His-tagged POT1 and ΔOB were expressed in Escherichia coli BL21(DE3) cells harboring plasmid pET14pot1 and pET14dOB, respectively. Cells were grown in TB supplemented with 1% glucose and 0.05 mg of carbenicillin/ml at 37°C to an optical density at 600 nm of ∼1.1. The cultures were induced to express recombinant protein with 0.005 mM isopropyl thiogalactoside for an additional 2 h at 25°C. Cells were harvested by centrifugation and stored at −70°C. Cells were lysed in one pellet volume of 20 mM NaH2PO4 (pH 8.0), 200 mM NaCl, 0.2% Tween-20, 10 mM imidazole, 20% glycerol, 5 mM β-mercaptoethanol, and complete EDTA-free protease inhibitor cocktail (Roche) and incubated with 1 mg of lysozyme/ml at 4°C for 20 min. The cell mix was lysed by sonication on ice with 12 5-s bursts and centrifuged to remove cell debris. The supernatant was adjusted to 10 mM MgCl2 and 5 mM ATP and incubated for 10 min at 37°C to release bacterial GroEL from POT1. His-POT1 and His-ΔOB were purified by fast protein liquid chromatography with sequential chromatography on Ni chelate and either blue Sepharose or MonoQ columns for full-length POT1 or ΔOB, respectively. The proteins were dialyzed in a solution containing 50 mM KCl, 20 mM NaH2PO4 (pH 8.0), 20% glycerol, 0.2% Tween 20, and 5 mM β-mercaptoethanol, concentrated with an Ultrafree-4 centrifugal filter unit, 10K MWCO (Millipore), and stored at −70°C.
Telomerase purification and reconstitution.
Affinity purification of telomerase has been previously described (32). Briefly, HeLa S3 cells (3 × 108) were resuspended in 4 ml of ice-cold buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride) supplemented with complete EDTA-free protease inhibitor cocktail (Roche) and allowed to swell on ice for 20 min. Nonidet NP-40 was added to 0.6%, the cells were vortexed vigorously for 10 s, and nuclei were pelleted by centrifugation. Nuclei were resuspended in buffer C′ (buffer A plus 0.4 M KCl) and agitated for 20 min at 4°C, and insoluble material was pelleted by centrifugation. Nuclear extracts were adjusted to 20 mM HEPES (pH 7.9), 300 mM KCl, 10% glycerol, and 0.5% Triton X-100. Yeast tRNA (50 μg/ml) and 1 μM affinity oligonucleotide [5′-biotin-CTAGACCTGTCATCA-rmeG-(rmeU)2-rmeA-(rmeG)3-(rmeU)2-rmeA-rmeG-3′ (rme = 2′O-methyl ribonucleotides)] were added, and the extract was incubated for 5 min at 30°C. Bead binding and wash steps were as described previously (32). Telomerase was eluted by incubation for 30 min at 25°C with 3 μM displacement oligonucleotide (5′-CTAACCCTAACTGATGACAGGTCTAG-3′) in WB300 (20 mM HEPES [pH 7.9], 300 mM KCl, 1 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, 0.15% Triton X-100), snap frozen, and stored under liquid nitrogen.
The expression, reconstitution, and purification of telomerase from baculovirus-infected insect cells has been previously described (39). Reconstituted telomerase was concentrated with an Ultrafree-4 centrifugal filter unit, 50K MWCO (Millipore), and protein concentrations were estimated by Western blotting, comparing the signal intensity to a known concentration of human telomerase reverse transcriptase (hTERT) C-terminal peptide fragment.
In vitro transcription and translation of hTERT were carried out with the TNT T7 Quick coupled transcription-translation system (Promega), following the manufacturer's protocol with 0.5 μg of plasmid (N-terminal Myc-tagged hTERT cloned into vector pcDNA6) per 50 μl of reaction mixture. Following 90 min of incubation at 30°C, a 2 μM concentration of in vitro-transcribed hTER was added, and incubation continued for 90 min at 30°C.
Gel shift analysis of protein-DNA complexes.
Typical binding reactions were carried out in a 10-μl solution containing 25 mM HEPES (pH 7.5), 15 to 50 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 1 to 4 nM 5′-32P-labeled DNA oligonucleotide, 200-fold molar excess nontelomeric competitor oligonucleotide (16pAT), and 1 to 2 μl of diluted POT1. Reaction mixtures were allowed to come to equilibrium at 37°C for 30 min. Products were then separated on 6% polyacrylamide-1× TGE (50 mM Tris, 380 mM glycine, 2 mM EDTA) gels supplemented with 3% glycerol in 1× TGE at 11.5 V/cm for 3 to 4 h at 4°C. Gels were dried and then visualized and quantitated by phosphorimager analysis.
For gel shift assays carried out in parallel with direct assays, the protein-DNA complexes were allowed to come to equilibrium under the conditions of the binding step preceding the direct assay: 79 mM Tris acetate (pH 8.5), 1.6 mM spermidine, 1.59 mM DTT, 1.6 mM Na2-EDTA, 19 mM KCl. Five nanomolar 5′-32P-labeled nonbiotinylated primer was included with 155 nM biotinylated primer to visualize the products. Protein-DNA complexes were then analyzed as described above.
For supershift assays, 1 nM 5′-32P-labeled tel4.0 was incubated with 5 nM POT1 and 20 pmol of mouse monoclonal horseradish peroxidase-conjugated anti-His antibody (Sigma) or mouse monoclonal anti-Hsp90 (Stressgene) in a solution containing 25 mM HEPES (pH 7.5), 15 mM NaCl, 1 mM Na2-EDTA, 5% glycerol, 1 mM DTT with 200 nM 16pAT at 25°C for 40 min. Protein-DNA complexes were then analyzed as described above.
Telomerase activity assay.
Direct telomerase assays were carried out in 10-μl reactions. Primer-POT1 (or where indicated primer-ΔOB, -vector control lysate, -E. coli SSB [USB] or -T4 gene 32 protein [gp32] [USB]) complexes were allowed to come to equilibrium in 1.9× DTA buffer (see below) with 1.6 mM EDTA at 37°C for 30 min. Extension reaction mixtures with affinity-purified telomerase contained 1× DTA buffer (50 mM Tris-acetate [pH 8.5], 50 mM KCl, 1 mM spermidine, 1 mM DTT), 1 mM MgCl2, 0.2 mM dATP, 0.2 mM dTTP, 0.006 mM dGTP, 20 μCi of [α-32P]dGTP (3,000 Ci/mmol), and 100 nM 5′-biotinylated telomeric primer. Reaction mixtures carried out with in vitro-translated hTERT reconstituted with in vitro-transcribed hTER contained 45 mM Tris-HCl (pH 8.0), 40 mM KCl, 0.9 mM MgCl2, 0.9 mM spermidine, 1 mM DTT, 0.5 mM dATP, 0.5 mM dTTP, 0.5 mM dCTP, 0.006 mM dGTP, 20 μCi of [α-32P]dGTP (3,000 Ci/mmol), and primer as described above. Extension reactions were initiated by the addition of ∼4 to 20 nM affinity-purified or 1 μl of in vitro-translated hTERT reconstituted with in vitro-transcribed hTER. Incubation was at 37 or 30°C, respectively, for 2 h, and reactions were stopped by the addition of EDTA to 17 mM. Unincorporated nucleotides were removed by binding biotinylated primers to streptavidin paramagnetic beads (Dynal), preincubated with 3′-32P-labeled B-10mer (5′-B-CAAGTCATCT-3′) as a loading control, washing twice with 2× BW (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 M NaCl) and once with TE (10 mM Tris HCl [pH 8.0], 1 mM EDTA). Bead-bound products were resuspended in 98% formamide-10 mM EDTA, heated to 98°C for 5 min, and analyzed on 10% polyacrylamide-urea gels.
For preloading assays, 1 pmol of biotinylated primer per reaction was bound to streptavidin paramagnetic beads and then washed once with 2× BW, twice with TE, and once with 1× binding buffer (25 mM HEPES [pH 7.5], 180 mM KCl, 5% glycerol, 1 mM DTT, 1 mM EDTA). Primers were preloaded by adding 3.2 pmol of POT1 (or an equivalent volume of vector control lysate) and 250 pmol of oligonucleotide 16pAT, per reaction, in 1× binding buffer and incubating for 60 min at 37°C. Unbound proteins were removed by washing twice with a solution containing 25 mM HEPES (pH 7.5), 15 mM KCl, 5% glycerol, 1 mM DTT, and 1 mM EDTA followed by one wash with a solution containing 50 mM Tris-acetate (pH 8.5), 15 mM KCl, 1 mM spermidine, and 1 mM EDTA. Extension reactions were adjusted to the direct assay conditions described above, and where appropriate, diluted POT1 or vector control lysate was added immediately prior to the addition of telomerase to start the reaction. Bead-bound products were purified and analyzed as described above.
Relative telomerase activity was quantified by using the AIDA software (Raytest), measuring the integrated activity of each major band minus the background level and normalizing for the number of 32P-labeled dGTPs incorporated. The normalized activity between 4 and 43 nucleotides (nt) was summed and expressed as a percentage of the activity compared with the water control.
RESULTS
Bacterially expressed POT1 binds single-stranded telomeric DNA in vitro.
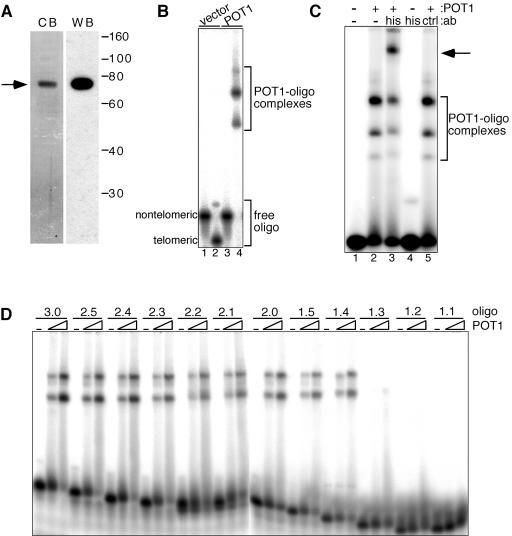
Human POT1 was expressed in E. coli and purified by two-step affinity chromatography over a nickel chelate column and a blue Sepharose column. The purified N-terminally His-tagged POT1 migrated as a single band on a sodium dodecyl sulfate-polyacrylamide gel that was stained with Coomassie brilliant blue (Fig. 1A, left panel). The apparent mobility of the tagged protein corresponded to the expected size of 74 kDa. Recombinant POT1 was also recognized by a monoclonal anti-His antibody on a Western blot, confirming its identity (Fig. 1A, right panel).
FIG. 1.
POT1 binds telomeric DNA in vitro. (A) Purification of His-POT1. The protein fraction was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane following purification. Transferred protein was revealed by immunoblotting with an anti-His antibody (right), after which the membrane was stained with Coomassie brilliant blue (left). (B) Gel shift analysis of purified POT1. Reaction mixtures containing 1 μM unlabeled oligonucleotide (oligo) 16pAT as a nonspecific competitor and 4 nM 5′-labeled oligonucleotide tel3.0 (lanes 2 and 4) or nontelomeric oligonucleotide NTermTA- (lanes 1 and 3) were incubated with 50 nM POT1 (lanes 3 and 4) or an equivalent volume of purified lysate from cells expressing the empty vector (lanes 1 and 2). (C) Supershift of POT-DNA complex. Reactions contained 1 nM oligonucleotide tel4.0, 5 nM POT1 (lanes 2, 3, and 5), and 20 pmol of anti-His antibody (lanes 3 and 4) or the nonspecific control Hsp90 antibody (lane 5). Arrow, POT1-oligonucleotide-antibody supershift complex. (D) Minimum telomeric sequence allowing POT1 binding. two-nanomolar 5′-labeled telomeric oligonucleotides with between 7 and 18 nt of telomeric repeat sequence (see Table 1) were incubated with 0, 2.6 nM, or 10 nM POT1 for 30 min at 37°C. Reactions were carried out at 200 nM NaCl and included 500 nM oligonucleotide 16pAT as a nonspecific competitor. POT1-oligonucleotide complexes were separated as in panel B.
To determine the DNA binding activity and specificity of recombinant POT1, an electrophoretic mobility shift assay (EMSA) was carried out with either purified POT1 or lysate from E. coli cells purified in parallel as a negative control. Shown in Table 1 is a list of oligonucleotides used in these studies. Similar to results from others (3, 26), we found that POT1 efficiently bound a DNA oligonucleotide consisting of 11 nt of nontelomeric sequence at the 5′ end followed by three telomeric repeats (Fig. 1B, lane 4) but showed no binding activity for a 30-nt nontelomeric oligonucleotide (NTermTA-) (Fig. 1B, lane 3). Purified lysate from vector control cells showed no binding activity with either oligonucleotide (Fig. 1B, lanes 1 and 2).
TABLE 1.
Oligonucleotides used in gel shift and direct assay studies
| Name | Sequence (5′→3′) |
|---|---|
| tel1.1 | CGCTCTAGAGCGGGTTAG |
| tel1.2 | CGCTCTAGAGCGGGTTAGG |
| tel1.3 | CGCTCTAGAGCGGGTTAGGG |
| tel1.4 | CGCTCTAGAGCGGGTTAGGGT |
| tel1.5 | CGCTCTAGAGCGGGTTAGGGTT |
| tel2.0 | CGCTCTAGAGCGGGTTAGGGTTA |
| tel2.1 | CGCTCTAGAGCGGGTTAGGGTTAG |
| tel2.2 | CGCTCTAGAGCGGGTTAGGGTTAGG |
| tel2.3 | CGCTCTAGAGCGGGTTAGGGTTAGGG |
| tel2.4 | CGCTCTAGAGCGGGTTAGGGTTAGGGT |
| tel2.5 | CGCTCTAGAGCGGGTTAGGGTTAGGGTT |
| tel3.0 | CGCTCTAGAGCGGGTTAGGGTTAGGGTTA |
| tel4.0 | CGCTCTAGAGCGGGTTAGGGTTAGGGTTAGGGTTA |
| TS-C | AATCCGTCGAGCAGGTTAG |
| GGGTTA3 | GCTGGTGATACCCTGAGTCTCCGGGTTAGGGTTAGGGTTA |
| 16pAT | CGACTCTTCCATTCCCCTTGACAGTGCATC |
| NtermTA- | GCTGGTGATACCCTGAGTCTCCTTTTTATATACTTGAAT |
We observed several POT1-oligonucleotide complexes with different mobilities by EMSA. The ratio of two predominant species (Fig. 1B) did not change when increasing concentrations of POT1 were added (Fig. 2C; also data not shown), suggesting that they are not due to the binding of additional POT1 molecules at higher protein concentrations. The complexes have been observed with all telomeric oligonucleotides tested. Additionally, we found that only one POT1 molecule stably binds the tel3.0 oligonucleotide (data to be published elsewhere). We verified that the shift species observed by EMSA contained POT1 by supershift analysis in which the POT1-oligonucleotide complex incubated with mouse monoclonal anti-His antibody prior to electrophoresis gave rise to a new, slower-mobility band and a decrease in the intensity of the bands corresponding to the POT1-oligonucleotide complexes (Fig. 1C, lane 3). The anti-His antibody did not bind the oligonucleotide in the absence of POT1 (lane 4), and no supershift was observed when the anti-His antibody was replaced by a mouse monoclonal antibody to an unrelated protein (lane 5).
FIG.2.
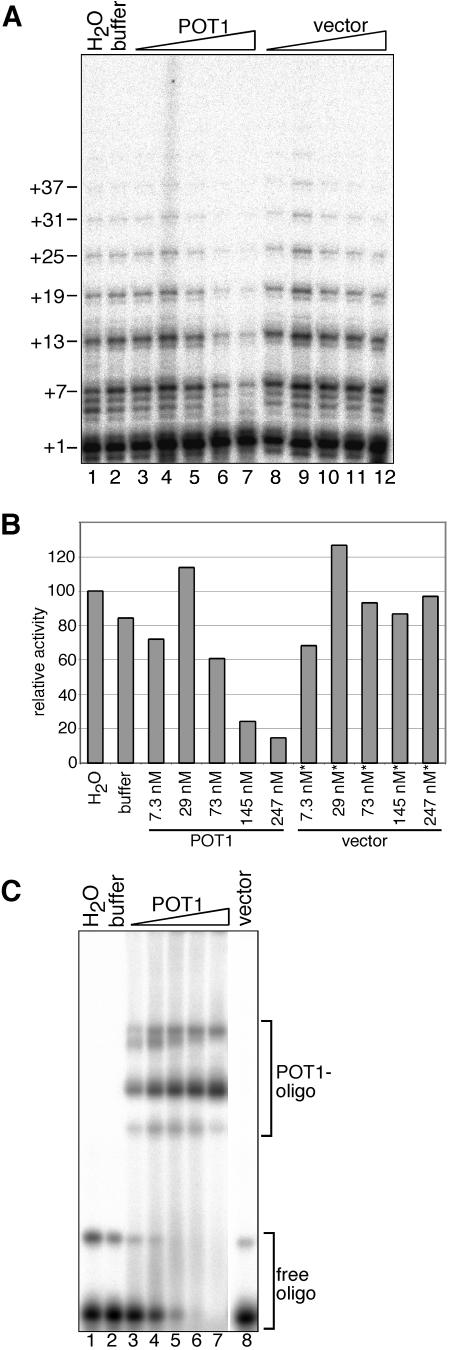
POT1 reduces telomerase activity. (A) Direct telomerase assay. tel3.0 was incubated with affinity-purified recombinant telomerase expressed in insect cells and H2O (lane 1), POT1 dialysis buffer (lane 2), or POT1 (diluted 1:34 [7.3 nM], 1:8.5 [29 nM], 1:3.4 [73 nM], or 1:1.7 [145 nM] or undiluted [247 nM; lanes 3 to 7]) or purified lysate from cells expressing empty vector (diluted 1:34, 1:8.5, 1:3.4, or 1:1.7 or undiluted; lanes 8 to 12). Telomerase products were separated on a sequencing gel. Numbers (left) show the number of nucleotides added to the primer at each pause site. (B) Plot of the sum of the relative activities of the products of telomerase extended 4 nt and more, compared to the water control. Asterisks in panel B indicate that a dilution of the vector control lysate equivalent to the dilution used to obtain the POT1 concentration shown was added. (C) Gel shift analysis of POT1-DNA complexes under conditions of the direct assay binding step (see Materials and Methods). H2O (lane 1), POT1 dialysis buffer (lane 2), POT1 (7.3, 29, 73, 145, or 247 nM; lanes 3 to 7) or purified vector control lysate (undiluted; lane 8) was incubated with biotinylated tel3.0 primer and trace amounts of 5′-labeled nonbiotinylated tel3.0 (to visualize the protein-oligonucleotide (oligo) complexes).
To determine what minimum length of telomeric sequence was required for a stable POT1-primer interaction to be formed, we tested a series of oligonucleotides (see Table 1) by EMSA, each shorter by one base at the 3′ end. We found that POT1 efficiently bound oligonucleotides with as few as 10 nt of telomeric sequence (Fig. 1D, oligonucleotide tel1.4) but bound oligonucleotides with fewer bases of telomeric sequence poorly or not at all (Fig. 1D, oligonucleotides tel1.3, tel1.2, and tel1.1; also data not shown).
POT1 inhibits the activity of reconstituted telomerase in a direct telomerase assay.
We tested whether POT1 affects telomerase activity using the direct telomerase assay. hTERT was expressed from a baculovirus vector in insect cells and reconstituted with in vitro-transcribed hTER. The ribonucleoprotein was purified by affinity chromatography as described previously (39). Prior to initiation of the extension reaction, primer containing three telomeric repeats was incubated for 30 min with POT1 or lysate from vector control cells that was purified in parallel, allowing binding to come to an equilibrium. Telomerase was then added, and the reaction mixture was incubated for an additional 2 h. In control reactions in which water or POT1 buffer alone was added, extension products consisting of up to six telomeric repeats were generated by telomerase (Fig. 2A, lanes 1 and 2). By contrast, although the amount of activity migrating at the +1 position did not decrease upon POT1 addition, there was a clear decrease in the total amount of activity corresponding to synthesis products greater than four nucleotides (Fig. 2A, compare lane 7 to lanes 3 to 5, bands +7 and +13) such that at the highest POT1 concentration, products corresponding to more than three repeats (+19 and larger) were virtually eliminated. Addition of equivalent dilutions of vector control lysate resulted in a very modest decrease in the number of telomeric repeats detected (Fig. 2A, lanes 8 to 12).
Quantification of these data revealed that at the highest POT1 concentration, the amount of telomerase extension products longer than four nucleotides was reduced more than fivefold compared to the water control (Fig. 2B), while the percentage of the total products extended by either one or two nucleotides slightly increased. Control assays carried out under identical conditions, with the highest POT1 concentration but in the absence of telomerase, revealed a minor nucleotide addition activity in the POT1 and vector control preparations. When RNase-treated telomerase was added to the control reaction, a signal migrating at the +1 position was observed (data not shown). We deduce that an interaction between the biotinylated telomeric primer and the displacement oligonucleotide used in the telomerase affinity purification generates a primer template for telomerase-independent addition of one nucleotide to the primer. Therefore, we excluded product migrating at the +1 position from calculations of telomerase activity.
In parallel with the direct assay described above, a gel shift assay was carried out to assess the extent to which the telomeric oligonucleotide primer was bound by POT1 under the conditions of the direct assay (Fig. 2C). As expected, no shifted species were detected in the presence of water, buffer, or the highest concentration of vector control lysate (Fig. 2C, lanes 1, 2, and 8), whereas a dose-dependent decrease in the free oligonucleotide and an increase in POT1-oligonucleotide complexes were seen as the POT1 concentration increased (Fig. 2C, lanes 3 to 7). Importantly, POT1 concentrations that significantly inhibited telomerase activity corresponded to essentially 100% binding of the telomeric primer by POT1 (Fig. 2A and 2C, lanes 6 and 7).
Efficient telomerase inhibition requires saturation of the binding sites on the telomeric primer by POT1.
As noted above, there appeared to be a slight decrease in the mean length of the telomerase extension products in the direct assay carried out in the presence of the highest concentration of vector control lysate. To try to ensure that the inhibition seen in the presence of POT1 was indeed due to the POT1 protein rather than a trace contaminant copurified from the lysate, we developed an assay that permitted the total concentration of POT1 added to the reaction to be increased while significantly decreasing the amount of any potentially contaminating material added. Shown schematically in Fig. 3A, biotinylated primer tel3.0 bound to streptavidin beads was incubated with a molar excess of POT1 and allowed to come to equilibrium in the presence of a large excess of unbiotinylated nontelomeric oligonucleotide to aid the removal of any nonspecific DNA binding factors. Unbound proteins were removed by several washes. As unbound POT1 was also removed during the washes, a substoichiometric amount of POT1 (with respect to the primer) was added at the start of the reaction to replace POT1 that may have transiently dissociated and been lost during the wash steps. Primer extension was then initiated by the addition of telomerase.
FIG. 3.
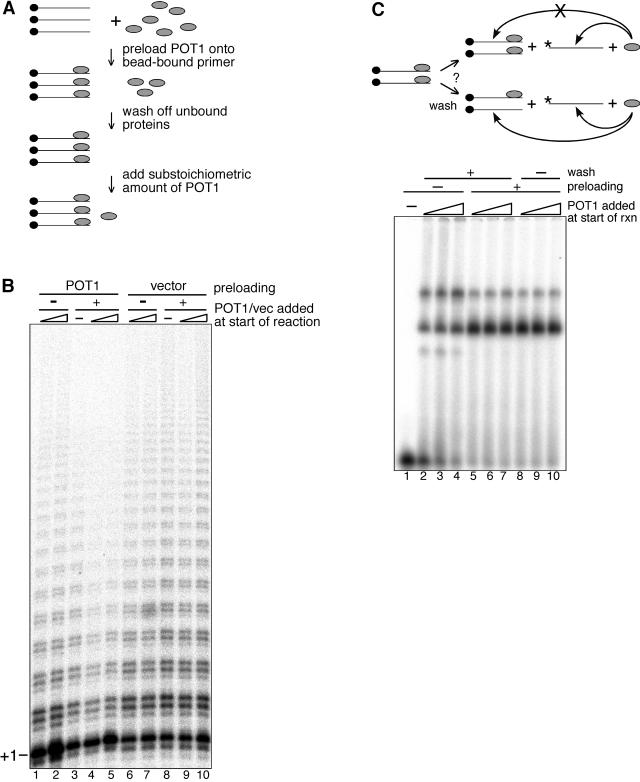
Direct telomerase assay with POT1 preloaded primers. (A) Schematic of the experimental design to preload substrate primers with POT1 (see the text). (B) Direct telomerase activity assay. Biotinylated primer tel3.0 bound to streptavidin beads was incubated with POT1 dialysis buffer (lanes 1, 2, 6, and 7), a molar excess of POT1 (lanes 3 to 5), or an equivalent volume of vector control lysate (lanes 8 to 10). Following washes to remove unbound proteins, extension reactions were initiated by the addition of affinity-purified, baculovirus-expressed recombinant telomerase and POT1 dialysis buffer (lanes 3 and 8), 0.032 pmol (3.2 nM) of POT1 (lanes 1 and 4), 0.13 pmol (13 nM) of POT1 (lanes 2 and 5), vector lysate diluted 1:100 (lanes 6 and 9), or vector lysate diluted 1:25 (lanes 7 and 10). (C) The schematic of the EMSA control is indicated in the upper panel. Biotinylated primer tel3.0 bound to streptavidin beads was incubated with POT1. To test if the wash steps also removed some transiently dissociated POT1, trace amounts of 5′-labeled tel3.0 were added following the wash steps, and competition by preloaded biotinylated primer tel3.0 was assessed by EMSA (lower panel). Biotinylated primer tel3.0 bound to streptavidin beads was incubated with POT1 dialysis buffer (lanes 1 to 4) or a molar excess of POT1 (lanes 5 to 10). Protein-DNA complexes were washed (lanes 2 to 7) or not washed (lanes 1 and 8 to 10), and trace 5′-labeled tel3.0 was added. Dialysis buffer (lane 1) or POT1 at 3.2 nM (lanes 2, 5, and 8), 6.4 nM (lanes 3, 6, and 9), or 13 nM (lanes 4, 7, and 10) was added, and reactions (rxn) were incubated for 30 min at 37°C. Reaction products were separated on a 6% polyacrylamide TGE gel.
When substoichiometric amounts of POT1 were added at the start of reactions in which the primer was first preloaded with POT1, a striking decrease in longer extension products was observed (Fig. 3B, lanes 4 and 5) compared with control reactions in which the same substoichiometric amounts of POT1 were added to primers not preloaded with POT1 (Fig. 3B, lanes 1 and 2). Preloading the primer with purified vector lysate in control reactions had no effect on the telomerase extension reaction (lane 8), and likewise, no change in telomerase activity was observed when additional vector lysate was added at the start of the reaction (lanes 6, 7, 9, and 10). The finding that synthesis of longer extension products was reduced only in those cases where the primer was preloaded with POT1 (Fig. 3B, lanes 3 to 5) suggests that the inhibition effected by POT1 requires the primer to be fully occupied by POT1.
A gel shift reaction carried out in parallel with the above direct assay was used to assess the fraction of the telomeric oligonucleotide that remained POT1 bound following the steps designed to wash away nonspecific unbound proteins. Since binding to the bead-bound biotinylated tel3.0 primer could not be measured directly, a competition experiment was designed in which trace amounts of 5′-labeled unbiotinylated tel3.0 were added to the binding reactions after the preloading and wash steps (Fig. 3C, schematic). In control reactions in which no POT1 was added during the preloading step, 41 to 81% (10 to 40 nM POT1) of the labeled primer was shifted (Fig. 3C, lanes 2 to 4). As expected, when the biotinylated primer was first preloaded with POT1, significantly more (86 to 90%) labeled primer was shifted, since the POT1 binding sites on the biotinylated oligonucleotide were occupied by the preloaded POT1 and therefore could not compete for binding with the end-labeled primer (Fig. 3C, lanes 5 to 7). Little difference in the fraction of labeled primer shifted (89 to 92%) was observed when the wash step was eliminated following preloading (Fig. 3C, lanes 8 to 10), suggesting that the majority of the bound POT1 remains stably associated with the primer throughout the washes.
DNA binding domain mutant POT1 does not inhibit telomerase activity.
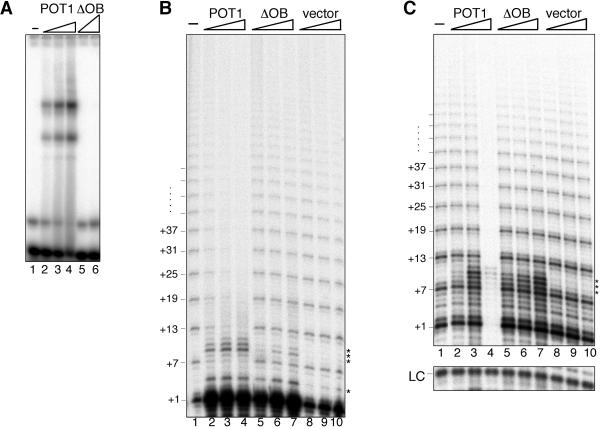
To assess whether binding of POT1 to the telomeric substrate primer was required for the negative regulation of telomerase activity, we expressed and purified from E. coli an N-terminal truncation mutant of POT1 (ΔOB) lacking the DNA binding domain (25). By gel shift analysis, purified ΔOB showed no DNA binding activity when incubated with the tel3.0 telomeric oligonucleotide (Fig. 4A, lanes 5 and 6) even when present at a concentration 60 times greater than that required for full-length POT1 to shift 50% of the labeled oligonucleotide (Fig. 4A, compare lanes 4 and 6).
FIG. 4.
ΔOB mutant of POT1 does not inhibit telomerase activity, while POT1 inhibits recombinant and native telomerase. (A) Gel shift analysis of ΔOB DNA binding activity. POT1 dialysis buffer (lane 1), POT1 (0.5, 1.5, or 5 nM; lanes 2 to 4), or ΔOB (0.5 or 300 nM; lanes 5 and 6) were incubated with labeled tel3.0 primer and a 250-fold molar excess of 16pAT nontelomeric competitor. POT1-oligonucleotide complexes were resolved on a native TGE polyacrylamide gel. Direct telomerase assay with telomerase purified from HeLa cell extract (B) or in vitro-transcribed and -translated telomerase (C) is also shown. Primer tel3.0 and a 240-fold excess of 16pAT nontelomeric competitor oligonucleotide were incubated with telomerase and POT1 dialysis buffer (lane 1) or POT1 (124, 247, or 425 nM; lanes 2 to 4), ΔOB (124, 247, or 425 nM; lanes 5 to 7), or purified vector control lysate (equivalent dilutions as POT1; lanes 8 to 10). Telomerase products were separated on a sequencing gel. Numbers (left) indicate nucleotide position of telomerase pause sites. Asterisks indicate RNase-insensitive non-telomerase-directed products. Lower panel, B-10mer loading control (LC).
We investigated the effect of adding increasing concentrations of full-length or ΔOB mutant POT1 on telomerase activity by direct assay. In addition, to test whether other components of the telomerase holoenzyme or potential posttranslational modification of telomerase might modulate the POT1 inhibition of telomerase, we assayed telomerase activity using telomerase isolated and affinity purified from HeLa cell nuclear extracts in these assays. As with hTERT purified from insect cells, the products of direct assays with HeLa-derived telomerase showed a characteristic six-base repeat pattern when separated on a sequencing gel (Fig. 4B, lane 1), whose intensity diminished when increasing concentrations of purified POT1 were added (Fig. 4B, lanes 2 to 4). Importantly, addition of equivalent concentrations of ΔOB or equivalent dilutions of vector control lysate did not negatively affect telomerase activity (Fig. 4B, lanes 5 to 10).
Since an RNase-insensitive accumulation of primer +1 nt product (see above) prevented the clear determination of whether the presence of POT1 or ΔOB influenced addition of the first nucleotide by telomerase (Fig. 4B, lanes 2 to 7), we repeated the direct assay reactions with telomerase from a third source. hTERT was in vitro transcribed and translated in rabbit reticulocyte lysate, reconstituted with in vitro-transcribed hTER, and used in the direct assay without prior purification. Strikingly, at the highest concentration of POT1 added to the reaction (4.25× molar excess over the primer), telomerase activity was completely eliminated (Fig. 4C, lane 4), whereas little or no change in the product intensity was detected when lower POT1 concentrations were used (Fig. 4C, compare lanes 2 and 3 with lane 1). As with telomerase from HeLa cell extracts (Fig. 4B), neither ΔOB nor vector control lysate negatively affected telomerase activity (Fig. 4C, lanes 5 to 10). Furthermore, although the signal corresponding to primer +1 nt increased slightly when 125 nM POT1 was added to the reaction mixture (lane 3), similar or greater accumulations at this position were observed when ΔOB or vector lysate were added (lanes 5 to 10), suggesting that this is not a specific effect of the added POT1.
Prebinding of POT1 to the primer is required for telomerase inhibition.
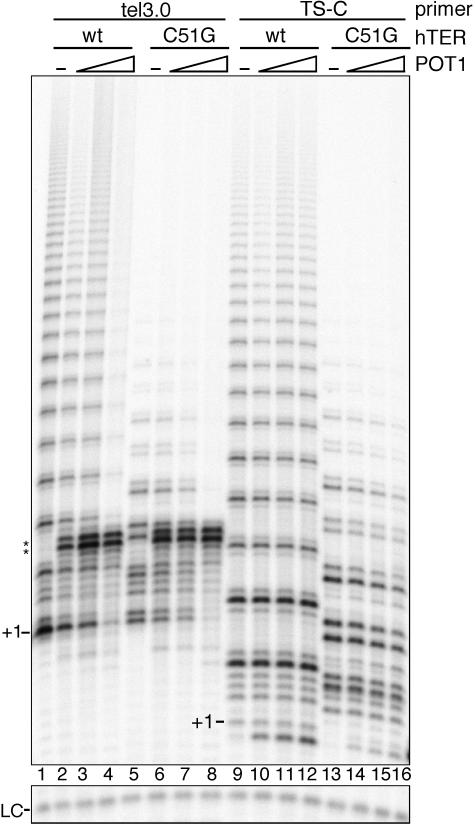
The data presented above suggest that telomerase activity is inhibited on POT1-bound substrate primers. We nonetheless wondered whether the inhibition was due strictly to POT1-oligonucleotide binding or whether potential POT1-telomerase interactions may also play a role. To address this question, we tested the effect of adding POT1 to direct assay reactions in which in vitro-translated hTERT was reconstituted with mutant hTER. The hTER mutant used contains a single nucleotide substitution in the template region (C51G), resulting in the synthesis of (TTAGCG)n telomeric repeats by the mutant telomerase. By gel shift assay we verified that POT1 had no detectable DNA binding activity with an oligonucleotide ending with three repeats of the mutant telomeric sequence -TTAGCG-3′ (data not shown). As primers for the direct assay, we used either the tel3.0 oligonucleotide, which binds POT1 with high affinity, or oligonucleotide TS-C (see Table 1), which is not detectably bound by POT1.
We observed that the telomerase activity and processivity of the C51G template mutant were reduced on both primer substrates compared with wild-type (wt) telomerase (Fig. 5, compare lanes 5 and 13 with lanes 1 and 9). Nonetheless, a six-base repeat pattern, which was dependent on the addition of dCTP to the reaction (data not shown), was generated by the mutant telomerase, confirming that it adds mutant telomeric repeats in the direct assay. In reactions with primer tel3.0 and mutant hTER, telomerase activity was inhibited when POT1 was added at the highest concentrations (Fig. 5, lane 8), as was the case with primer tel3.0 and wt hTER (Fig. 5, lane 4). POT1 did not inhibit a contaminating terminal transferase activity that was detected in reactions with tel3.0 primer (Fig. 5, lanes 2 to 4 and 6 to 8, bands +10 and +11; also data not shown). By contrast, the addition of equivalent concentrations of POT1 to reactions with either wt or C51G hTER, but in which synthesis was primed by oligonucleotide TS-C, had no negative effect on telomerase activity (Fig. 5, lanes 10 to 12 and 14 to 16).
FIG. 5.
Direct telomerase assay with hTER template mutant. Telomerase extension was primed by oligonucleotide tel3.0 (lanes 1 to 8) or oligonucleotide TS-C (lanes 9 to 16) after incubation of the primer with POT1 dialysis buffer (lanes 1, 5, 9, and 13), 124 nM POT1 (lanes 2, 6, 10, and 14), 247 nM POT1 (lanes 3, 7, 11, and 15), or 425 nM POT1 (lanes 4, 8, 12, and 16). Reactions were initiated by addition of in vitro-translated hTERT reconstituted with wt hTER (lanes 1 to 4 and 9 to 12) or C51G template mutant hTER (lanes 5 to 8 and 13 to 16). +1 indicates the position of the first incorporated nucleotide. Asterisks indicates RNase A-insensitive bands likely stemming from non-telomerase-directed nucleotide addition. Lower panel, B-10mer loading control (LC).
Telomerase activity is blocked by noncognate single-strand binding proteins, and terminal transferase activity is reduced by POT1.
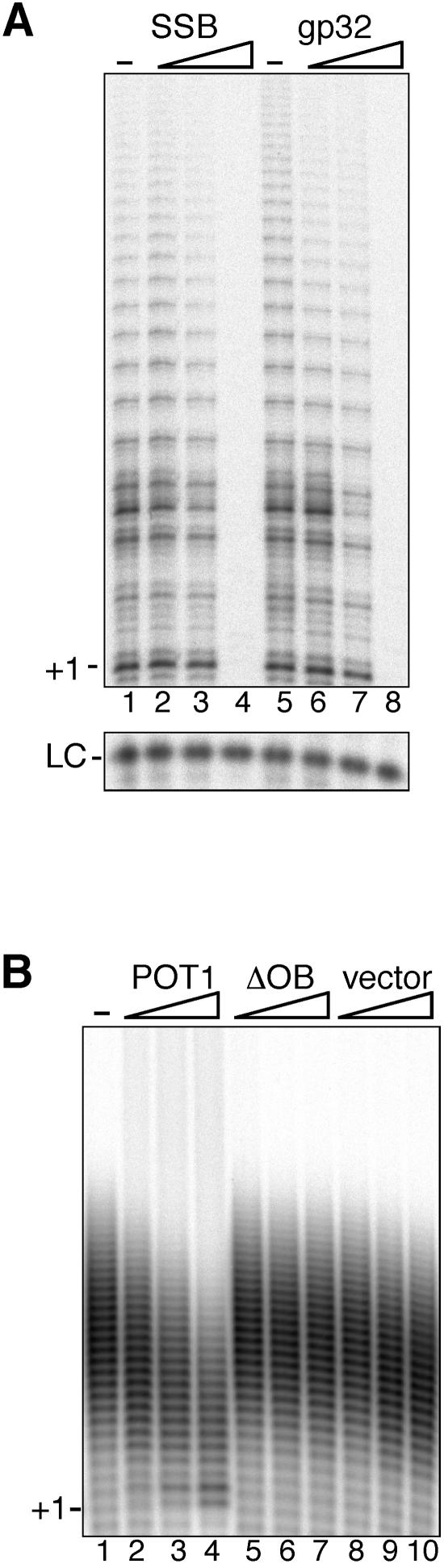
The results described above suggest that POT1 inhibits telomerase activity by blocking the access of telomerase to the primer. To determine whether this activity was specific to POT1, we tested whether telomerase could extend primers bound by either E. coli single-strand binding protein (SSB) or phage T4 gp32. Similar to the results with POT1, both SSB and gp32 effectively blocked telomerase activity when added at high enough concentrations (Fig. 6A, lanes 4 and 8, respectively). As was the case with POT1, inhibition of telomerase activity by SSB or gp32 occurred at concentrations corresponding to those at which ∼100% of the primer was shifted by EMSA (data not shown). These data are consistent with a recent report showing that Tetrahymena telomerase activity was inhibited by E. coli SSB at concentrations sufficient to bind all primer (9). Also, as reported for Tetrahymena telomerase, a very modest enhancement of human telomerase activity was detected at lower SSB concentrations (lane 2) (9).
FIG. 6.
The effect of noncognate single-strand binding proteins on telomerase activity and of POT1 on terminal transferase activity. (A) Direct telomerase assays with single-strand binding protein bound primers. Primer GGGTTA3 was incubated with dilution buffer (lanes 1 and 5), E. coli SSB (50, 100, or 500 nM; lanes 2 to 4), or T4 gene 32 protein (0.1, 1, or 10 μM; lanes 6 to 8), following which the extension reactions were initiated by the addition of in vitro-transcribed and -translated telomerase. Lower panel, B-10mer loading control (LC). (B) Terminal transferase activity assay. Primer GGGTTA3 was incubated with dilution buffer (lanes 1), POT1 (124, 247, or 425 nM; lanes 2 to 4), ΔOB (124, 247, or 425 nM; lanes 5 to 7), or purified vector control lysate (dilutions equivalent to those of POT1; lanes 8 to 10) Extension reactions were initiated by the addition of terminal transferase (0.4 U), and incubation continued for 1 h. Products were separated on a 10% sequencing gel. +1 indicates the position of the first incorporated nucleotide.
We also investigated the specificity of the effect of POT1 on telomerase activity by testing whether POT1-bound oligonucleotides could still serve as primers for terminal transferase. A dose-dependent decrease in terminal transferase activity and decrease in mean product length was observed as increasing concentrations of POT1 were added to the reactions (Fig. 6B, lanes 2 to 4). As expected, neither the addition of ΔOB nor that of vector lysate significantly affected the terminal transferase activity.
DISCUSSION
The role of human POT1 in telomere length regulation has been examined previously with cell culture. The expression of an N-terminal truncation mutant of POT1, with a deletion in the DNA binding domain, was shown to cause telomere elongation (24, 25), leading to speculation that the mutant acts as a dominant negative, disrupting signaling between the duplex telomere and the 3′ end (24, 25). Consistent with this conclusion, it was shown that reduction in endogenous POT1 levels by RNA interference led to telomere lengthening, implying that POT1 negatively regulates telomere length (41). Increased tankyrase activity, which removes TRF1 from telomeres, or decreases in TIN2, which protects TRF1 from tankyrase, also leads to telomere elongation (11, 33, 40), as does partial knockdown of PTOP/PIP1 or overexpression of a small peptide of PTOP/PIP1 encompassing the POT1 interaction domain (24, 41). Thus, disruption of the TRF1 complex, and in particular its interaction with POT1, leads to unregulated telomere elongation, further supporting the notion that POT1, responding to signaling conveyed through the TRF1 complex, negatively regulates telomere length. On the other hand, Colgin et al. (10) overexpressed full-length POT1 in telomerase-positive cells and, after long passage in culture, observed substantially increased telomere lengths in a fraction of their cloned cells, leading to the conclusion that POT1 acts as a telomerase-dependent positive regulator of telomere length.
In this study we took a biochemical approach, using purified components under defined reaction conditions, to determine whether human POT1 directly influences telomerase activity. We began by characterizing the DNA binding activity of the bacterially expressed, purified POT1. Although the oligonucleotide tel1.4 has the shortest telomeric repeat sequence we found that was efficiently bound by POT1, this sequence does not contain the minimum binding site for POT1 of 5′-TAGGGTTAG-3′, as recently reported by Loayza and colleagues (26). These authors did not test POT1 binding to a substrate containing the tel1.4 telomeric sequence of 5′-GGGTTAGGGT-3′; thus, the results cannot be directly compared. Nonetheless, since both short sequences appear to be efficiently bound by POT1, but neither contains within it a permutation of the other, these findings may suggest that POT1 is capable of carrying out nucleotide shuffling, as has been demonstrated by crystallographic studies on the Oxytricha α subunit complexed with different permutations of the O. nova telomeric sequence (36).
We find that POT1 inhibits telomerase activity, apparently by obstructing access to the primer 3′ end. A reduction in telomerase activity was observed in assays with POT1-bound telomeric primer and recombinant human telomerase expressed either in insect cells or in vitro, in rabbit reticulocyte lysate, and with native telomerase, affinity-purified from HeLa cell nuclear extracts. On the other hand, ΔOB POT1 did not negatively affect telomerase activity, consistent with the model that binding of POT1 to the single-strand telomere 3′ end is necessary for the negative regulation of telomerase in vivo (25). We found that telomerase activity was unaffected by the addition of POT1 when oligonucleotide TS-C (which does not support POT1 binding) was used as a primer, arguing that POT1 may modulate telomerase activity by regulating the access of telomerase to the primer but not during extension. Since terminal transferase access was not entirely blocked on POT1-bound primers, it appears that the 3′ end is not completely occluded by POT1 binding, but the interaction with telomerase was prohibited through steric hindrance by the POT1 protein, preventing base pairing between the DNA primer and the telomerase RNA template.
The O. nova TEBP α/β heterodimer, when complexed with telomeric DNA, blocks telomerase access to the substrate primer (15). Attenuation of telomerase activity by the TEBP α subunit alone, however, became evident only when the concentration of α exceeded that of the telomeric primer by 2,000-fold and was not eliminated even when the protein concentration was 10,000 times greater than that of the primer. By comparison, we observed that POT1 negatively affected telomerase activity when the POT1 concentration exceeded that of the DNA primer by fivefold or less, likely due to the very strong binding affinity of POT1 for the tel3.0 primer (our unpublished data). Our results nonetheless suggest that effective obstruction of the 3′ end by POT1 in vivo would require POT1 levels that are, at a minimum, stoichiometrically equivalent or in excess of telomeric ends. Evidence that an approximate 50% loss of single-stranded overhang through overexpression of the TRF2(ΔBΔM) dominant-negative allele of TRF2 resulted in a roughly twofold reduction in POT1 associated with telomeric DNA (25) suggests that such stoichiometric binding may occur in vivo. It remains to be determined whether POT1 binding activity varies throughout the cell cycle.
Our results are interesting in light of recent findings with Saccharomyces cerevisiae showing that telomeres switch between telomerase-extendible and nonextendible states in a length-dependent manner, whereas the number of nucleotides added during one cell cycle is independent of the length of the telomere (35). The molecular nature of the extendible and nonextendible telomeric states is unknown, but it is conceivable that POT1 bound to telomeric ends corresponds to the nonextendible telomeric state, as previously proposed (28). But how is the inhibitory role of POT1 relieved, in vivo, to allow telomerase-mediated extension? It is conceivable that binding of POT1 to the telomeric 3′ overhang occurs in a telomere length-dependent fashion (28). On the other hand, it is possible that POT1 loses its inhibitory role when modified or assembled with other polypeptides. The high affinity and specificity of POT1 for the telomeric 3′ overhang makes this protein ideally situated to recruit other complexes involved in telomere maintenance. In S. cerevisiae, Cdc13p appears to play such a multivalent role, being described as a landing platform for complexes involved in telomere maintenance (27, 30); perturbation of these functions can lead to either gain or loss of telomeric sequence (16, 29). Further studies will be necessary to determine if POT1 interacts with factors in addition to TRF1-PTOP/PIP1 complex and whether, like Cdc13p, human POT1 participates in both telomere protection and length regulation pathways.
Acknowledgments
We thank Gael Cristofari for comments on the manuscript.
This work was supported by grants from the Swiss Cancer League and the Swiss National Science Foundation.
REFERENCES
- 1.Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22:3474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armbruster, B. N., C. M. Linardic, T. Veldman, N. P. Bansal, D. L. Downie, and C. M. Counter. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, P., and T. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, P., E. Podell, and T. R. Cech. 2002. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi, A., R. M. Stansel, L. Fairall, J. D. Griffith, D. Rhodes, and T. de Lange. 1999. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18:5735-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, S. W., and E. H. Blackburn. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21:553-563. [DOI] [PubMed] [Google Scholar]
- 8.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, S., E. Jacob, and H. Manor. 2004. Effects of single-stranded DNA binding proteins on primer extension by telomerase. Biochim. Biophys. Acta 1679:129-140. [DOI] [PubMed] [Google Scholar]
- 10.Colgin, L. M., K. Baran, P. Baumann, T. R. Cech, and R. R. Reddel. 2003. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13:942-946. [DOI] [PubMed] [Google Scholar]
- 11.Cook, B. D., J. N. Dynek, W. Chang, G. Shostak, and S. Smith. 2002. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 14.Fang, G., and T. R. Cech. 1993. Oxytricha telomere-binding protein: DNA-dependent dimerization of the alpha and beta subunits. Proc. Natl. Acad. Sci. USA 90:6056-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froelich-Ammon, S. J., B. A. Dickinson, J. M. Bevilacqua, S. C. Schultz, and T. R. Cech. 1998. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 12:1504-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschling, D. E., and V. A. Zakian. 1986. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47:195-205. [DOI] [PubMed] [Google Scholar]
- 18.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 19.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 20.Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194:139-154. [DOI] [PubMed] [Google Scholar]
- 21.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 22.Kelleher, C., M. T. Teixeira, K. Forstemann, and J. Lingner. 2002. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27:572-579. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. H., P. Kaminker, and J. Campisi. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, D., A. Safari, M. S. O'Connor, D. W. Chan, A. Laegeler, J. Qin, and Z. Songyang. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6:673-680. [DOI] [PubMed] [Google Scholar]
- 25.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013-1018. [DOI] [PubMed] [Google Scholar]
- 26.Loayza, D., H. Parsons, J. Donigian, K. Hoke, and T. de Lange. 2004. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279:13241-13248. [DOI] [PubMed] [Google Scholar]
- 27.Lustig, A. J. 2001. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 8:297-299. [DOI] [PubMed] [Google Scholar]
- 28.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 29.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 30.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 31.Ray, A., and K. W. Runge. 1999. The yeast telomere length counting machinery is sensitive to sequences at the telomere-nontelomere junction. Mol. Cell. Biol. 19:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnapp, G., H. P. Rodi, W. J. Rettig, A. Schnapp, and K. Damm. 1998. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 26:3311-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484-1487. [DOI] [PubMed] [Google Scholar]
- 34.Smogorzewska, A., and T. de Lange. 2004. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73:177-208. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira, M. T., M. Arneric, P. Sperisen, and J. Lingner. 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117:323-335. [DOI] [PubMed] [Google Scholar]
- 36.Theobald, D. L., and S. C. Schultz. 2003. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 22:4314-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 38.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-502. [DOI] [PubMed] [Google Scholar]
- 39.Wenz, C., B. Enenkel, M. Amacker, C. Kelleher, K. Damm, and J. Lingner. 2001. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20:3526-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, J. Z., and T. de Lange. 2004. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 36:618-623. [DOI] [PubMed] [Google Scholar]
- 41.Ye, J. Z., D. Hockemeyer, A. N. Krutchinsky, D. Loayza, S. M. Hooper, B. T. Chait, and T. de Lange. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, G. L., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344:126-132. [DOI] [PubMed] [Google Scholar]