Abstract
We have previously demonstrated that GATA-2 and GATA-3 are expressed in adipocyte precursors and control the preadipocyte-to-adipocyte transition. Constitutive expression of both GATA-2 and GATA-3 suppressed adipocyte differentiation, partially through direct binding to the peroxisome proliferator-activated receptor γ (PPARγ) promoter and suppression of its basal activity. In the present study, we demonstrate that both GATA-2 and GATA-3 form protein complexes with CCAAT/enhancer binding protein α (C/EBPα) and C/EBPβ, members of a family of transcription factors that are integral to adipogenesis. We mapped this interaction to the basic leucine zipper domain of C/EBPα and a region adjacent to the carboxyl zinc finger of GATA-2. The interaction between GATA and C/EBP factors is critical for the ability of GATA to suppress adipocyte differentiation. Thus, these results show that in addition to its previously recognized function in suppressing PPARγ transcriptional activity, interaction of GATA factors with C/EBP is necessary for their ability to negatively regulate adipogenesis.
Adipocyte differentiation is a process controlled by multiple regulators, principally the CCAAT/enhancer binding protein (C/EBP) family of transcription factors and the peroxisome proliferator-activated receptor γ (PPARγ), a nuclear hormone receptor (12, 15). Upon hormonal stimulation, the expression of C/EBPβ and C/EBPδ temporally increases (3), followed by expression of PPARγ and C/EBPα (3, 16). A cooperative interaction between PPARγ and C/EBPα drives the expression of genes that are necessary for the generation and maintenance of the adipogenic phenotype, such as genes producing morphological changes, lipid accumulation, and insulin sensitivity (22).
The C/EBP family of transcription factors contains a highly conserved basic leucine zipper domain (bZip) that mediates homo- or heterodimerization with other isoforms in this protein family. Gain- or loss-of-function studies of preadipocyte cell lines and loss-of-function experiments in vivo all indicate that C/EBPα is a key regulator of adipogenesis (6, 20, 23, 25). Furthermore, two other members of this family, C/EBPβ and C/EBPδ, are also highly expressed in adipose tissue and have been demonstrated to be vital components of the signaling cascade which initiates adipocyte differentiation (13). Clearly, the coordinated activity of these three members of the C/EBP family makes key contributions to adipogenesis.
We have previously demonstrated that the zinc finger transcription factors GATA-2 and GATA-3 are expressed predominantly in white and not brown adipose tissue in vivo and that their expression is restricted to preadipocytes and down-regulated upon adipocyte differentiation. Constitutive expression of both GATA-2 and GATA-3 suppressed adipocyte differentiation and trapped cells at the preadipocyte stage, as found when morphology and gene expression were assessed. This effect was mediated, at least in part, through direct binding of PPARγ and inhibition of its basal promoter activity. However, coexpression of PPARγ and GATA can only partially rescue the suppression of adipogenesis by GATA (14), which suggests the existence of additional mechanisms by which GATA regulates adipocyte differentiation.
In this study, we demonstrated that both GATA-2 and GATA-3 can form protein complexes with C/EBPα or C/EBPβ and that this interaction is critical for the suppression of adipocyte differentiation by GATA. We mapped the interaction domain on GATA-2 and discovered that mutations within this region abolish protein interaction between GATA-2 and C/EBP and subsequently diminish the ability of GATA-2 to suppress adipocyte differentiation.
MATERIALS AND METHODS
Cell culture and RNA isolation.
3T3-F442A preadipocytes, NIH 3T3 fibroblasts, and COS-7 monkey kidney cells were maintained in Dulbecco's modified Eagle medium (Gibco, Carlsbad, Calif.) containing 10% calf serum (HyClone, Logan, Utah). 3T3-F442A cells were differentiated in 10% cosmic calf serum (CCS; HyClone) containing 5 μg of insulin/ml (Sigma, St. Louis, Mo.) for 7 days. Similarly, NIH 3T3 cells were differentiated in 10% CCS containing 5 μg of insulin/ml, 0.5 mM isobutylmethylxanthine (Sigma), and 1 μM dexamethasone (Sigma) in either the presence or the absence of the thiazoladinedione BRL49653 (0.1 μM; Calbiochem, San Diego, Calif.) for the first 3 days and subsequently fed 10% CCS containing 5 μg of insulin/ml every other day for another week. Total RNA was harvested from differentiated cells using TRIzol (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. Retroviral transduction was used to ectopically express full-length GATA or deletion mutants in 3T3-F442A and NIH 3T3 cells as previously described (10). Briefly, BOSC23 viral packaging cells were transfected with various GATA mutants in a retroviral vector by calcium phosphate transfection (Amersham Pharmacia, Piscataway, N.J.). Forty-eight hours after transfection, the supernatant containing viral particles was harvested, passed through a filter (pore size, 0.45 μm), and added to 20 to 30% confluent 3T3-F442A cells. Positive infectants were selected with 2 μg of puromycin/ml (Sigma) for 2 to 4 days or 50 μg of hygromycin/ml (Sigma) for 7 days before experiments were performed.
Oil red O lipid staining.
Differentiated adipocytes were fixed in 0.5% glutaraldehyde and stained for 30 to 120 min in a 4:6 dilution in water of 0.25% oil red O (Sigma) in isopropanol. Cells were visualized by phase-contrast microscopy at a magnification of ×40.
Plasmids.
For retroviral transfection, full-length human GATA-2 (hGATA-2) or murine GATA-3 (mGATA-3) in the pBabePuro retroviral vector were used as described previously (14). To generate pBabeHygro-hGATA2Δ377-415 (mutant A), full-length pBabeHygro-hGATA2 was digested with HpaI/SalI to eliminate the entire carboxyl terminus following the C-terminal zinc finger. Then, a fragment consisting only of residues 416 to 475 was excised from full-length hGATA-2 by digestion with SphI (mung bean nuclease treated) and SalI and reinserted by blunt-end ligation into HpaI/SalI-digested pBabeHygro-hGATA2 in order to result in a mutant with only 39 amino acid (aa) residues missing from the region immediately following the carboxy-terminal zinc finger. To generate pBabeHygro-hGATA2Δ415-475 (mutant B), a fragment of hGATA-2 not including the region coding for the last 61 aa residues was generated by EcoRI/SphI excision, treated with mung bean nuclease, and inserted into the SnaBI cloning site of the pBabeHygro vector.
To create FLAG-tagged GATA constructs, an EcoRI site was inserted into hGATA-2 or mGATA-3 by PCR, just following the ATG translational initiation codon. The GATA-2 or GATA-3 coding sequences were then excised with EcoRI, treated with mung bean nuclease, and ligated into the EcoRV cloning site of pFLAG-CMV2 (Sigma). pFLAG-GATA2Δ377-415 (mutant A) was generated by digesting pFLAG-GATA2 with HpaI/XbaI and reinserting an SphI (mung bean nuclease-treated)/XbaI-digested insert by blunt-end ligation. pFLAG-GATA2Δ415-475 (mutant B) was generated by HindIII/SphI (mung bean nuclease) digestion, followed by blunt-end ligation into HindIII/EcoRV sites on pFLAG-CMV2.
Missense mutations were generated in the region adjacent to the C-terminal zinc finger to create G2Ala381-385 (GATA2 with Ala at aa 381 to 385 [mutant C]) and G2Ala388-394 (mutant D). A substantial region following the C-terminal zinc finger was first deleted (and named G2ΔC1B) and then replaced with PCR fragments containing alanine substitutions between residues 381 and 384 or 388 and 394. G2ΔC1B was generated by PCR amplification of the last 243 nucleotides of the hGATA-2 coding region with primers AGTTAACTCCAACAAGTCCAAGAAGAG and GCTGGTAAGGGTTTGGTC . The fragment was digested with HpaI and then ligated to the HpaI site of hGATA-2 to replace the last 294 bp of the hGATA-2 coding sequence, resulting in a deletion of 51 bp. This construct was digested with HpaI and reinserted with PCR-synthesized fragments containing the missense mutations (G2Ala381-385 or G2Ala388-394). G2Ala381-385 (mutant C) was generated by annealing the partially complementary oligomers AATAGGCCACTGGCAGCAGCTGCTGCAGGGATC and CTTCCGGTTCCGAGTCTGGATCCCTGCAGCAGC, which were then filled in with Pwo polymerase (Roche, Indianapolis, Ind.) to form a double-stranded fragment. This synthesized sequence was phosphorylated and then inserted into the HpaI site of G2ΔC1B. Likewise, G2Ala388-394(mutant D) was generated by annealing the partially complementary oligomers AATAGGCCACTGACCATGAAGAAGGAAGGGATC and TGCTGCAGCAGCTGCTGCGATCCCTTCCTTCTTC and subsequently filling in with Pwo polymerase to form a double-stranded fragment, which then was inserted into the HpaI site of G2ΔC1B.
To generate pGEX-(p12)C/EBPα, a 0.6-kb NotI/HindIII fragment was excised from rat C/EBPα DNA, treated with mung bean nuclease, and blunt-end ligated into the SmaI site of the pGEX-2T vector (Amersham Pharmacia).
Immunoprecipitation and immunoblot analysis.
For protein expression, COS-7 cells were transfected with 10 μg of the FLAG-tagged GATA-2 DNA constructs and 10 μg of pcDNA.3-C/EBPα by calcium phosphate precipitation. Cell lysates were prepared using radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% Na deoxycholate, 1 mM polymethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, 1 mM NaF, 1 mM activated Na3VO4) and immunoprecipitated with M2 anti-FLAG antibody conjugated to agarose beads (Sigma), and the eluate was separated by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride (PVDF) membrane, and immunoblotted with an antibody against C/EBPα or C/EBPβ (Santa Cruz Biotechnology, Santa Cruz, Calif.). The endogenous interaction between C/EBPα and GATA was similarly examined in whole-cell lysates from 3T3-L1 cells induced to differentiate for 24 h. Immune complexes were immunoprecipitated with anti-GATA-2 antibody (Santa Cruz Biotechnology) and subsequently subjected to SDS-PAGE and immunoblot analysis using anti-C/EBPα antibody.
GST pull-down assay.
The carboxy-terminal 12-kDa portion of rat C/EBPα was expressed as a fusion protein with glutathione S-transferase (GST) in the pGEX-2T plasmid. After expression was induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma) for 2 h, the fusion protein was affinity purified with glutathione Sepharose (Sigma). The bound recombinant protein was incubated with lysates of COS-7 cells expressing FLAG-tagged GATA-2, resolved by denaturing SDS-PAGE, and transferred to a PVDF membrane. Finally, GATA-2 proteins were detected by immunoblotting with an anti-FLAG antibody.
Gel shift assay.
The electrophoretic mobility shift assays were performed as described previously (14). Briefly, in a 20-μl reaction volume, 2 μl of nuclear extract of COS-7 cells expressing GATA or C/EBP proteins was incubated with 0.5 μg of poly(dI-dC) and 4 × 104 cpm of DNA probe that was end labeled with 32P in the presence or absence of 1 μg of specific or nonspecific competitors at room temperature for 20 min. Total protein expression was examined by SDS-PAGE of nuclear extracts and subsequent immunoblot analysis using an anti-FLAG antibody.
Luciferase reporter assays.
A luciferase reporter construct under the direction of a 1,575-bp 5′-flanking sequence of the C/EBPα gene was provided by M. Daniel Lane (Johns Hopkins University, Baltimore, Md.). One microgram of the reporter construct was transfected into NIH 3T3 cells along with GATA-2 or GATA-3 (0.3 μg) expression plasmids by calcium phosphate precipitation. The 0.6-kb PPARγ2 promoter fragment (nucleotides −603 to +62) in the pXP2 luciferase reporter construct (14) (1.0 μg) was cotransfected into NIH 3T3 or 3T3-F442A cells with various GATA-2 deletion mutants (0.2 μg) and, when appropriate, with an expression plasmid for C/EBPα (1.0 μg) by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. One microgram of a reporter gene construct driven by the 5′-end flanking region of the human C3 complement factor gene (nucleotides −127 to −199) (gift of Gretchen Darlington, Baylor College of Medicine, Houston, Tex.) was transfected into 3T3-F442A cells along with 0.5 μg of expression plasmids for C/EBPα and either GATA-2 or mutant C by use of Lipofectamine 2000. One microgram of a reporter gene construct driven by the 5′-end flanking region of the interleukin-5 gene (gift of I-Cheng Ho, Harvard Medical School, Boston, Mass.) was transfected into HEK293 cells along with 3.5 μg of expression plasmids for GATA-2, mutant B, or mutant C by use of Lipofectamine 2000. In all experiments, Renilla luciferase reporter (0.1 μg) was used as an internal control for transfection efficiency. Two days after transfection, cells were lysed, and luciferase activity was measured with the dual luciferase kit (Promega, Madison Wis.) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Subcellular fractionation.
Nuclear and cytosolic fractions were prepared from COS-7 cells 2 days after the transfection of DNA constructs expressing various GATA-2 mutants with the NE-PER fractionation kit (Pierce, Rockford, Ill.) according to the manufacturer's instructions. Lysates were separated by denaturing SDS-PAGE and transferred to a PVDF membrane. The presence of GATA-2 in subcellular fractions was detected with anti-FLAG antibody (Sigma). Nuclear fractions were identified by immunoblotting with an antihistone antibody (Santa Cruz Biotechnology), and cytosolic fractions were identified likewise with an anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Ambion, Austin, Tex.).
Quantitative PCR.
One microgram of total RNA extracted from 3T3-F442A cells overexpressing wild-type GATA-2 or various mutants was reverse transcribed with ThermoScript RT (Invitrogen) according to the manufacturer's instructions. Expression levels of GATA-2 were measured by quantitative reverse transcription (RT)-PCR with SYBR Core mix (Applied Biosystems, Foster City, Calif.) on an iCycler machine (Bio-Rad, Hercules, Calif.) at a melting temperature of 56°C. The sequences for forward and reverse primers (Integrated DNA Technologies, Coralville, Iowa) are as follows: for GATA-2, the forward primer is ACGGCGTCAAGTACCAGGTG and the reverse primer is TCCGCTGCTGTAGTCGTGG, and for β-actin, the forward primer is GCTGTGCTATGTTGCTCTAG and the reverse primer is CGCTCGTTGCCAATAGTG.
RESULTS
Regulation of the PPARγ2 promoter by GATA.
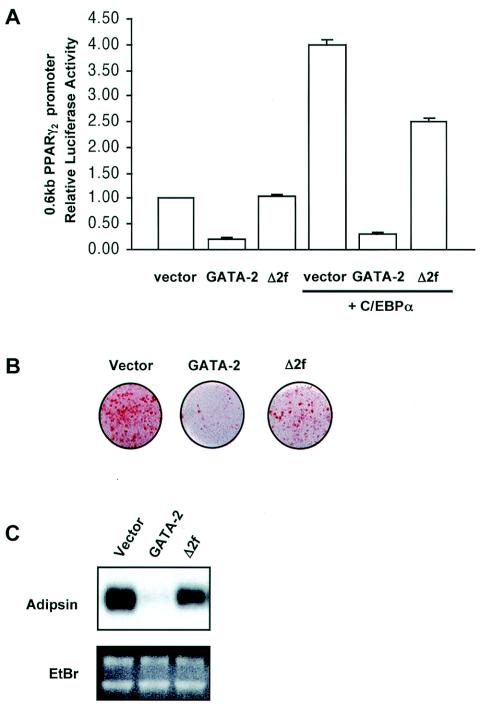
We have previously shown that either GATA-2 or GATA-3 can impair basal promoter activity of a 0.6-kb PPARγ promoter fragment and that deletion of both GATA zinc fingers can completely restore this activity (14). Additional cotransfection of an expression plasmid for C/EBPα activates the basal PPARγ promoter activity fourfold. While the C/EBPα-activated PPARγ promoter activity is also strongly suppressed with wild-type GATA-2, deletion of both zinc fingers of GATA-2 was unable to fully recapitulate C/EBP-mediated activation of the PPARγ2 promoter (Fig. 1A). This observation correlates well with adipocyte differentiation of 3T3-F442A cells expressing a GATA-2 deletion mutant lacking both zinc fingers. Whereas cells ectopically expressing wild-type GATA-2 fail to differentiate, the deletion of both zinc fingers prevents GATA-2 from suppressing adipogenesis to the same extent. However, this recovery was incomplete, as reflected by oil red O staining of intracellular lipids and adipsin gene expression (Fig. 1B and C). The failure to completely restore adipogenesis in cells expressing GATA-2 devoid of DNA binding capacity indicates that the disruption of DNA binding is insufficient for the complete restoration of normal adipocyte differentiation and therefore that additional mechanisms might be involved in GATA-mediated suppression of differentiation, such as interaction with C/EBP factors.
FIG. 1.
GATA-mediated suppression of adipogenesis is not fully reversed by disruption of DNA binding. Renilla luciferase vector was included as an internal control to account for transfection efficiency. (A) Relative luciferase activity of a PPARγ promoter construct (PPARγ2) driving a luciferase reporter was examined in NIH 3T3 cells expressing GATA-2 or a GATA-2 mutant in which both zinc fingers have been deleted (Δ2f) in the presence (+) or absence of C/EBPα. The values reported here represent the means ± standard errors of the means (SEMs) of relative luciferase activities. Error bars indicate SEMs. (B) Oil red O staining of 3T3-F442A cells constitutively expressing either wild-type GATA-2 or a mutant lacking both zinc fingers (Δ2f) and stimulated for adipocyte differentiation for 8 days with insulin. (C) Northern blot analysis for gene expression of adipsin, which is expressed in a differentiation-dependent manner, in differentiated 3T3-F442A cells. Ethidium bromide (EtBr) staining demonstrates equal loading in each lane as well as the integrity of mRNA.
GATA and C/EBP factors are in the same protein complexes.
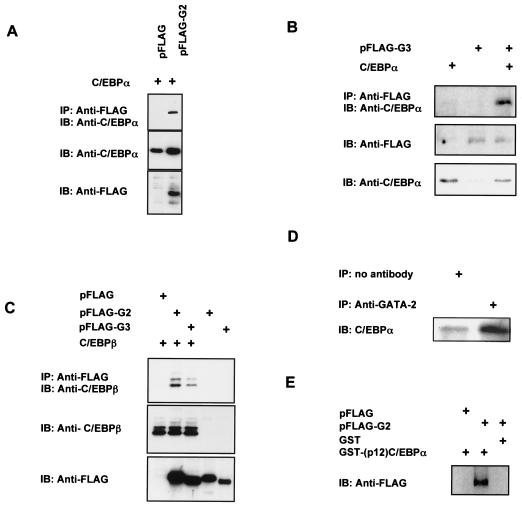
To investigate whether GATA and C/EBP proteins can form protein complexes together, we expressed FLAG-tagged GATA-2 or GATA-3 with C/EBPα or C/EBPβ in COS-7 cells by transient transfection. Complexes containing the GATA protein were immunoprecipitated with an anti-FLAG antibody immobilized on agarose beads. The presence of C/EBP factors in the immunoprecipitated protein complexes was detected by immunoblotting with isoform-specific antibodies against C/EBPα or C/EBPβ. As shown in Fig. 2, C/EBPα was readily detected in immune complexes with GATA-2 or GATA-3 (Fig. 2A and B, upper panels). Direct immunoblot analysis confirms that comparable levels of C/EBP and FLAG epitopes were present in these cells (Fig. 2A and B, middle and lower panels). The FLAG epitope was not detected when expressed as a vector control because it is much smaller than the FLAG-tagged fusion protein and ran off the gel. GATA-2 and GATA-3 can also each form protein complexes with the C/EBPβ isoform, as indicated by the presence of C/EBPβ in anti-FLAG immunoprecipitants (Fig. 2C). We also confirmed the interaction between C/EBPα and GATA-2 in intact 3T3-L1 preadipocytes by means of coimmunoprecipitation experiments (Fig. 2D). C/EBPα was observed in a complex immunoprecipitated by an antibody against GATA-2 in cell lysates isolated 24 h after cells were induced to differentiate. We next determined the domains of C/EBPα involved in this protein-protein interaction. A truncated form of C/EBPα containing only the 12-kDa carboxy-terminal region was expressed as a GST fusion protein [GST-(p12)C/EBPα]. This truncated form of C/EBPα consists of only the bZip regions that are highly conserved among all isoforms of this protein family (3) and that are critical for DNA binding and dimerization of the C/EBP proteins (9). The GST fusion proteins were immobilized on glutathione agarose beads and subsequently incubated with protein extracts prepared from COS-7 cells expressing FLAG-tagged GATA-2. The copurification of GATA-2 was detected by immunoblot analysis using an anti-FLAG antibody. As shown in Fig. 2E, GATA-2 was detected only with the GST-(p12)C/EBPα fusion protein and not with GST alone. This indicates that the bZip region of C/EBPα is sufficient for mediating physical association with GATA-2.
FIG. 2.
Interaction of GATA-2 or GATA-3 with C/EBPα or C/EBPβ. (A) COS-7 cells were transfected with expression plasmids for C/EBPα along with FLAG-tagged GATA-2 or pFLAG vector control. Cell lysates were harvested and immunoprecipitated (IP) with an anti-FLAG antibody conjugated to agarose beads, and the proper expression of C/EBPα was detected by immunoblotting (IB) with an antibody against C/EBPα. The expression of GATA-2 or C/EBPα was confirmed by direct immunoblot analysis of 10 μg of whole-cell lysate with anti-FLAG or anti-C/EBPα antibodies, respectively. (B) C/EBPα was also detected in anti-FLAG immunoprecipitants of cells expressing FLAG-tagged GATA-3 (pFLAG-G3) and C/EBPα. Coprecipitation analysis was performed as described for panel A. (C) Detection of C/EBPβ protein with an isoform-specific anti-C/EBPβ antibody in anti-FLAG immunoprecipitants of COS-7 cells expressing C/EBPβ along with either FLAG-tagged GATA-2 or GATA-3. Coprecipitation analysis was performed as described for panel A. (D) Detection of CEBP-GATA interaction in intact cells. 3T3-L1 cells were stimulated to differentiate for 24 h, at which time whole-cell lysates were collected and immunoprecipitated with anti-GATA-2 antibody, followed by immunoblot analysis with an anti-C/EBPα antibody. Immunoprecipitation of lysates with no antibody controlled for nonspecific binding of protein complexes to protein A-Sepharose beads. (E) A 12-kDa carboxy-terminal fragment of C/EBPα was expressed as a fusion protein with GST [GST-(p12)C/EBPα] and affinity purified with glutathione-Sepharose. Bound GST-(p12)C/EBPα recombinant proteins or GST alone were incubated with cell lysates of COS-7 cells expressing FLAG-tagged GATA-2. The presence of GATA-2 bound to C/EBPα was detected by immunoblot analysis with an anti-FLAG antibody.
Mapping and characterization of the interaction between GATA-2 and C/EBP.
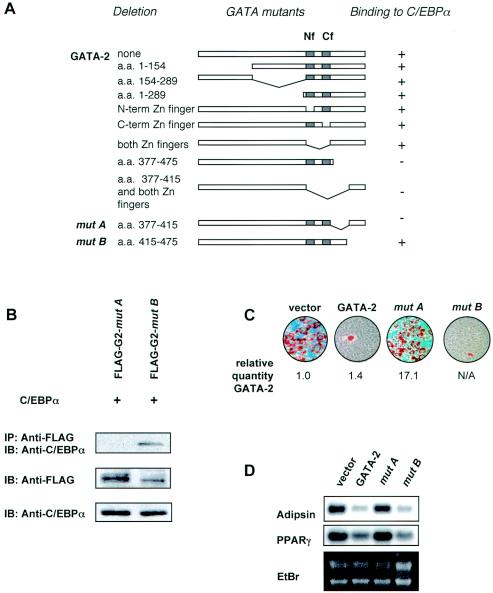
To determine the region of GATA-2 required for its interaction with C/EBPα, we tested the ability of various GATA mutants to interact with C/EBPα by means of coimmunoprecipitation experiments. For these experiments, we cotransfected into COS-7 cells plasmids expressing FLAG-tagged GATA-2 with various deletions and determined their association with C/EBPα by immunoblotting anti-FLAG immunoprecipitants with anti-C/EBPα antibodies (Fig. 3A). These experiments demonstrated that the amino-terminal region of GATA-2 was not required for interaction with C/EBP proteins, since even deletion of the entire N terminus up to the first zinc finger (deletion of aa 1 to 289 [Δ1-289]) did not hinder protein interaction with C/EBPα. Moreover, deletion of either or both zinc fingers had no effect on C/EBPα binding. In contrast, deletions of the sequence following the carboxyl zinc finger of GATA-2 (Δ377-475) resulted in a loss of interaction with C/EBPα. By making smaller deletions, i.e., deletions of either the first half (GATA-2 mutant A) or the second half (GATA-2 mutant B) of this region, we further deduced that the region most proximal to the carboxyl zinc finger (aa 377 to 415), but not the distal region (aa 415 to 475), is critical for this interaction (Fig. 3B). The proper expression of the mutant proteins was confirmed by immunoblot analysis using anti-FLAG and an isoform-specific C/EBP antibody (Fig. 3B).
FIG. 3.
Characterization of GATA-2 and C/EBP interaction in adipogenesis. (A) Schematic diagram of various GATA deletion mutants and their ability to bind C/EBPα, as evaluated by coimmunoprecipitation analyses as described in the legend to Fig. 2A. Nf and Cf, amino- and carboxy-terminal zinc fingers, respectively. N-term, N-terminal; C-term, C-terminal. (B) A DNA construct for either FLAG-tagged mutant A or B was transfected into COS-7 cells along with an expression plasmid for C/EBPα. The association of C/EBPα with these GATA mutants was detected by immunoprecipitation (IP) with anti-FLAG antibody-conjugated agarose beads, followed by immunoblotting (IB) with an anti-C/EBPα antibody. The proper expression of C/EBPα and GATA-2 mutants in this protein complex was confirmed by immunoblot analysis using anti-C/EBPα and anti-FLAG antibodies, respectively. (C) Oil red O staining of differentiated 3T3-F442A cells constitutively expressing wild-type GATA-2 or GATA-2 mutant A or B. Expression levels of GATA-2 and the mutants were confirmed by quantitative RT-PCR and numerically expressed as a difference (n-fold) of the level of the vector control. N/A, not analyzed. (D) Northern blot analysis of expression of adipogenic genes adipsin and PPARγ in differentiated 3T3-F442A cells constitutively expressing wild-type GATA-2, mutant A, or mutant B. Ethidium bromide (EtBr) staining was included to demonstrate loading in each lane and the integrity of RNA.
aa residues 377 to 415 of GATA-2 are required for suppression of adipocyte differentiation.
To analyze the functional importance of the interaction between GATA-2 and C/EBP, we performed experiments to test whether deletion of residues 377 to 415 would compromise the ability of GATA-2 to suppress adipocyte differentiation. Various GATA-2 mutants were stably expressed in 3T3-F442A cells by retroviral transduction using the pBabe expression system. Following selection with hygromycin, surviving cells were differentiated into adipocytes, and oil red O staining of lipids in differentiated adipocytes and Northern blot analysis of the adipocyte markers adipsin and PPARγ were used as criteria to assess adipocyte differentiation. Expression of GATA-2 and mutants were also quantitated by real-time RT-PCR. As seen in Fig. 3C and D, constitutive expression of wild-type GATA-2 suppressed adipocyte differentiation, whereas the GATA-2 deletion mutant A (Δ377-415), which eliminated C/EBPα binding, abolished this suppressive effect. In contrast, deletion of the adjacent region in mutant B (Δ415-475) had no effect on the ability of GATA-2 to suppress adipocyte differentiation. Notably, mutations generated in mutant A resulted in greater impairment of GATA's suppressive function than that of a mutant in which DNA binding alone was completely perturbed (GATA-2Δ2f) (Fig. 1), which suggests that interaction with C/EBPα could play a more critical role than DNA binding for proper GATA function in adipogenesis.
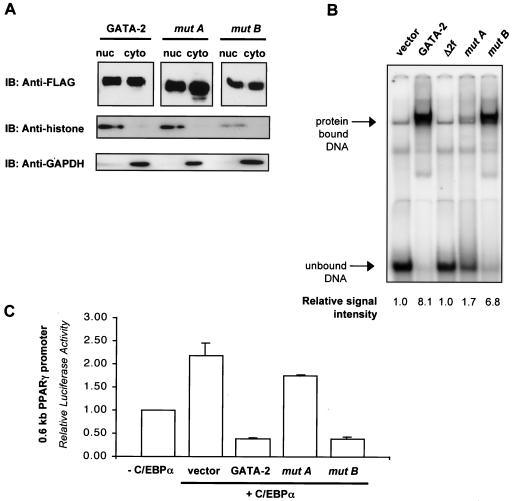
To ascertain the mechanism by which disruption of GATA interaction with C/EBPα prevents suppression of adipogenesis, we further investigated whether deletion of residues 377 to 415 would affect other intrinsic functions of GATA transcription factors. First, we examined the nuclear localization of various GATA-2 deletion mutants. FLAG-tagged GATA-2 deletion mutants were expressed in COS-7 cells, and cytosolic and nuclear fractions were prepared. Immunoblot analysis of these subcellular fractions with an anti-FLAG antibody revealed that nuclear entry was not disrupted in either mutant (Fig. 4A). The integrity of the subcellular fractionation was confirmed by detection of histone protein in the nuclear fractions and of the housekeeping protein GAPDH in the cytosolic fractions.
FIG. 4.
Analysis of nuclear localization and DNA binding properties of mutant A [GATA-2 (Δ377-415)] and mutant B [GATA-2 (Δ415-475)]. (A) Cytosolic (cyto) and nuclear (nuc) fractions were prepared from cell lysates of COS-7 cells expressing FLAG-tagged GATA-2, mutant A, or mutant B. The presence of GATA-2 in these subcellular fractions was detected by immunoblotting (IB) with an anti-FLAG antibody. The nuclear fraction was confirmed by detection of the nuclear protein histone, and the cytosolic fraction was confirmed by detection of the housekeeping protein GAPDH. (B) Nuclear extracts of COS-7 cells expressing either GATA-2, GATA-2 lacking both zinc fingers (Δ2f), or mutant A or B were analyzed by electrophoretic mobility shift assay with an oligonucleotide consisting of consensus GATA binding sites (upper panel). The signals are quantitated and expressed numerically under each lane. (C) C/EBPα-activated promoter activity of PPARγ was examined in the presence of wild-type GATA-2 or deletion mutants A or B. The values reported here represent the mean ± SEM relative luciferase activity. Renilla luciferase activity was used as an internal control.
DNA binding of GATA transcription factors has been shown to occur predominantly through the carboxy-terminal zinc finger (19). We next examined whether deletion of residues 377 to 415 from GATA would alter DNA binding. Gel shift analysis was performed using a consensus GATA binding sequence as a probe. As predicted, deletion of both zinc fingers completely disrupted DNA binding (Fig. 4B). Unexpectedly, DNA binding was also abolished in mutant A (Δ377-415), whereas mutant B (Δ415-475) was still capable of binding DNA. This indicates that residues 377 to 415 are involved in DNA binding, in addition to their contribution to protein-protein interaction with C/EBPα. Accordingly, the absence of this region also results in substantial loss of GATA-mediated suppression of PPARγ2 gene promoter activation, while deletion of residues 415 to 475 in mutant B had no effect (Fig. 4C). Since GATA mutants that are completely lacking DNA binding capacity maintain some capacity to suppress adipogenesis (Fig. 1), results with this mutant strongly support the importance of GATA-C/EBP interaction in GATA-mediated suppression of adipocyte differentiation.
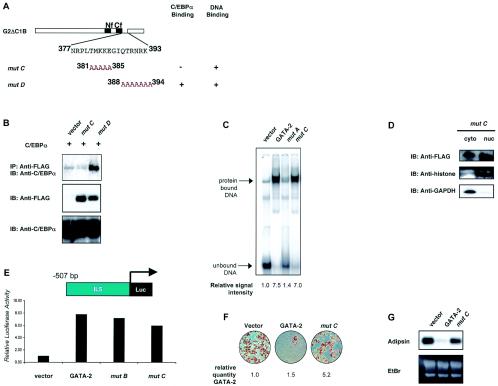
aa residues 381 to 385 of GATA-2 are required for protein interaction with C/EBPα but not for DNA binding.
Although mutant A could almost fully restore adipogenesis, the truncation introduced in GATA-2 in this mutant abolished both DNA binding ability and interaction with C/EBPα. Thus, in order to differentiate the independent contributions of each of these two functions to GATA-mediated suppression of adipogenesis, we performed further analyses of the region between residues 377 and 415 (Fig. 5A). A series of missense mutations was generated within this stretch of 17 aa residues. As shown in Fig. 5B, GATA-2 proteins with alanine substitutions at aa residues 381 to 385 (mutant C) could no longer interact with C/EBPα, while substitutions at aa residues 388 to 394 (mutant D) had no effect on the ability of GATA-2 proteins to bind C/EBPα. Gel shift analysis demonstrated that the mutations introduced in mutant C did not hinder DNA binding (Fig. 5C). Immunoblot analysis of subcellular fractions indicated that nuclear localization of the mutated transcription factor was also unimpaired (Fig. 5D). We also tested the ability of this mutant to activate a known GATA-responsive promoter (Fig. 5E). These experiments demonstrated that GATA-2 mutant C retained its transactivating capacity on a known target promoter at levels comparable to those of GATA-2 and GATA-2 mutant B. The creation of this GATA mutant, which cannot bind to C/EBPα but retains the abilities to bind DNA and transactivate a target promoter, enables us to better ascertain how C/EBPα interaction with GATA specifically contributes to the mechanism by which GATA suppresses adipogenesis.
FIG. 5.
Detailed mapping and functional analysis of GATA-C/EBP interaction. (A) Schematic diagram of specific amino acid substitutions generated in mutant C (aa 381 to 385) and mutant D (aa 388 to 394). Nf and Cf, amino- and carboxy-terminal zinc fingers, respectively. The ability of each mutant to interact with C/EBPα or to bind to target DNA has been summarized in the column next to the diagram. (B) Anti-C/EBPα antibody was used to detect C/EBPα in protein complexes immunoprecipitated (IP) with an anti-FLAG antibody from lysates of COS-7 cells expressing FLAG-tagged mutant C or mutant D along with C/EBPα. (C) An electrophoretic mobility shift assay was used to test the ability of mutant C to bind a cognate DNA sequence. Nuclear extracts from COS-7 cells expressing GATA-2, mutant A, or mutant C were used. Binding is quantitated as described in the legend to Fig. 3 and expressed numerically under each lane. (D) Cytosolic and nuclear fractions were prepared from cell lysates of COS-7 cells expressing mutant C. The presence of GATA-2 in these subcellular fractions was detected by immunoblotting (IB) with an anti-FLAG antibody. The nuclear fraction was confirmed by detection of the nuclear protein histone, and the cytosolic fraction was confirmed by detection of the housekeeping protein GAPDH. (E) GATA-mediated activation of the interleukin-5 luciferase reporter was examined in the presence of wild-type GATA-2, mutant B, or mutant C. (F) Oil red O staining of differentiated 3T3-F442A cells constitutively expressing GATA-2 or mutant C. Expression of GATA-2 and mutants were confirmed by quantitative RT-PCR and numerically expressed as described in the legend to Fig. 4. (G) Northern blot analysis of adipsin expression in differentiated 3T3-F442A cells constitutively expressing GATA-2 or mutant C. EtBr, ethidium bromide.
To investigate the functional effect of mutant C on adipogenesis, we constitutively expressed mutant C in 3T3-F442A cells by retroviral transduction and determined the differentiation capacity of these cells. Constitutive expression of GATA-2 inhibited adipocyte differentiation, while in contrast, a GATA mutant that cannot bind C/EBPα (mutant C) significantly disrupted this inhibition and permitted normal adipocyte differentiation, as determined by oil red O staining and adipsin gene expression (Fig. 5E and F). Since mutant C can still bind DNA, the failure to suppress adipogenesis observed in this experiment is attributable to the inability of this GATA-2 substitution mutant to interact with C/EBPα.
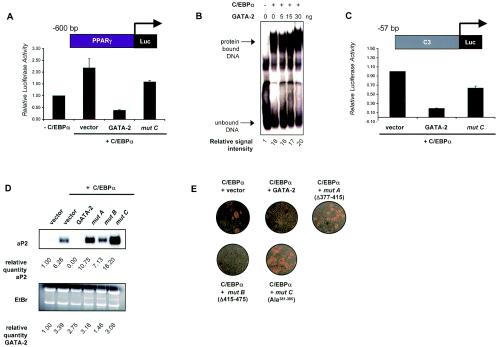
Disrupted interaction between C/EBPα and GATA-2 hinders C/EBP activation of target promoters in adipogenesis.
Suppression of C/EBPα-mediated activation of a 0.6-kb 5′-end flanking region of the PPARγ gene by GATA was almost completely restored by a loss of interaction between GATA-2 and C/EBPα (Fig. 6A). This indicates that the ability of GATA-2 to regulate a target promoter in the presence of C/EBP, such as that of PPARγ, was not completely dependent on GATA binding to cognate DNA sequences. In addition, the ability of C/EBPα to bind to DNA was not altered by GATA. Mobility shift assays demonstrated that C/EBP binding to its cognate sequence was not altered by increasing amounts of recombinant GATA-2 (Fig. 6B). We also tested the ability of GATA to modulate C/EBPα activation of a target promoter devoid of GATA binding sites. For this assay, we used the C3Luc reporter, which is transactivated by C/EBPα but does not contain GATA binding sites (8) (Fig. 6C). Coexpression of mutant C with C/EBPα does not lead to or suppression of C3Luc (Fig. 6C), suggesting that the interaction of GATA-2 with C/EBPα partially limits the transactivation potential of C/EBPα. While mutant C lacks the potency of wild-type GATA-2 to suppress a C/EBP-responsive reporter, it also does not permit full activation.
FIG. 6.
Effect of various GATA-2 mutants on C/EBPα-activated adipogenesis in NIH 3T3 cells. (A) C/EBPα-activated promoter activity of PPARγ was examined in the presence of wild-type GATA-2 or mutant C. (B) Electrophoretic mobility shift assay was used to test the ability of C/EBPα to bind a cognate DNA sequence in the presence of increasing amounts of GATA-2. Nuclear extracts from COS-7 cells expressing C/EBPα were used along with various amounts of recombinant GATA-2. (C) C/EBPα-activated promoter activity of C3Luc in the presence of wild-type GATA-2 or mutant C. For panels A and C, the values reported represent the mean luciferase activity values ± SEM. Renilla luciferase activity was used as an internal control. (D) Northern blot analysis of total RNA from the same cells was used to detect the expression of the late adipogenic marker aP2. Ethidium bromide (EtBr) staining was included to demonstrate equal loading in each lane as well as the integrity of RNA. Signal intensities of aP2 were calculated using Personal FX phosphorimaging software (Bio-Rad). Exogenous expression of GATA-2 and mutants were confirmed by quantitative RT-PCR. Both values are numerically expressed as a difference (n-fold) of the value for the vector control. (E) Phase-contrast micrographs of differentiated NIH 3T3 cells constitutively expressing C/EBPα along with various GATA-2 mutants.
To underscore the functional significance of GATA interaction with C/EBPα, we studied adipogenesis in NIH 3T3 cells, a fibroblast cell line that expresses very low endogenous levels of C/EBPα and PPARγ. In order to properly differentiate into adipocytes, these cells are dependent on exogenously supplied C/EBPα or PPARγ (6, 17, 23). Various GATA mutants and C/EBPα were ectopically expressed in NIH 3T3 cells by retroviral transduction and subsequently differentiated and assayed for their adipogenic capacities. GATA expression in these cells was quantitated by real-time RT-PCR (Fig. 6D). Exogenous C/EBPα expression alone can induce differentiation in the presence of appropriate reagents, while coexpression with either wild-type GATA-2 or mutant B, a functionally intact mutant, suppresses differentiation-dependent gene expression (Fig. 6D) as well as lipid accumulation (Fig. 6E). However, mutant A, which cannot bind DNA nor interact with C/EBPα, and mutant C (G2Ala381-385), which cannot interact with C/EBP but can still bind DNA, both lose the ability to suppress adipogenesis.
DISCUSSION
Previously, we demonstrated that GATA transcription factors suppress adipocyte differentiation, partially through inhibition of PPARγ expression by direct suppression of the basal PPARγ2 promoter activity. In the present study, we examined additional pathways that GATA utilizes to suppress adipogenesis and demonstrated that both GATA-2 and GATA-3 are capable of forming protein complexes with either C/EBPα or C/EBPβ. Through deletion analysis, we mapped the interaction domain to the bZip region of C/EBPα and to aa residues 381 to 385, which follow the carboxyl zinc finger of GATA-2. This domain is critical for C/EBP interaction but not for nuclear localization or DNA binding. Finally, we demonstrated that the interaction between GATA and C/EBP is critical for GATA suppression of adipocyte differentiation, since isolated disruption of the C/EBP-GATA interaction impairs the ability of GATA to suppress adipocyte differentiation.
Both GATA-2 and GATA-3 can form protein complexes with either C/EBPα or C/EBPβ. Our previous data indicated that the expression of both GATA-2 and GATA-3 are significantly down-regulated at the onset of adipogenesis, while expression of C/EBPα increases during differentiation. However, expression of C/EBPβ and C/EBPδ remains relatively low in preadipocytes and surges during the early stages of differentiation upon hormonal stimulation (3, 25). The temporal coexpression of GATA with these members of the C/EBP protein family in adipocytes therefore suggests that suppression of the C/EBPβ or C/EBPδ expression function may also be the target of the antagonism between GATA and the C/EBP family of transcription factors. The ability of GATA to interact with C/EBPα may result from interactions at homologous regions of the C/EBP family or indicate that constitutive expression of GATA during adipogenesis may impair differentiation by inappropriately interacting with C/EBPα. Since GATA down-regulation partially overlaps with C/EBPα induction, it remains possible that under normal conditions GATA factors can interact with C/EBPα, perhaps in order to sequester its ability to activate the adipogenic program until derepression is appropriate. It is also possible that interaction of GATA with C/EBPα could be triggered at other times upon exposure to specific antiadipogenic stimuli. The ability of GATA to interact with multiple C/EBP isoforms with various temporal expression patterns suggests that their proadipogenic contribution may be counterregulated by GATA in a stage-specific fashion.
Our work collectively demonstrates that GATA suppresses adipocyte differentiation through at least two pathways: inhibition of PPARγ expression by binding to its promoter, and interference with C/EBP function through protein-protein interaction. When various deletion mutants were investigated for their ability to suppress the PPARγ gene promoter, we found that loss of DNA binding (in mutant A) rendered GATA incapable of suppressing PPARγ promoter activation, whereas a functionally intact mutant (mutant B) exerted the same effect as wild-type GATA-2. More detailed analysis indicated that a GATA protein which retained its ability to bind DNA but could not interact with C/EBP (mutant C) likewise failed to suppress adipocyte differentiation as well as C/EBPα-activated PPARγ promoter activity, indicating that protein interaction with C/EBP was independent of DNA binding and critical for GATA suppression of differentiation and PPARγ gene transcription in the presence of C/EBP, which can both interact with C/EBP and bind its cognate DNA sequence, has little or no effect on PPARγ promoter activity, although when constitutively expressed in NIH 3T3 cells, it was able to hamper lipid accumulation and adipogenic gene expression (data not shown). Taken together, these findings highlight the significance of GATA interaction with C/EBP, which allows GATA to exert its full effect on adipogenesis. The dual mechanism by which GATA acts underscores the necessity for tight control of the initiation of adipogenesis in preadipocytes, so that perturbation of either pathway may result in only partial loss of GATA suppression of adipocyte differentiation. It is not yet known whether temporal release of these suppressive effects is critical for proper adipogenesis to proceed.
GATA transcription factors interact with many proteins, such as FOG (18), PU.1 (11, 26), Rb (21), Trap220 (5), and CBP (1, 2). Interestingly, many of these GATA-interacting factors have been implicated in the process of adipogenesis. For example, Rb and Trap220 were shown to be necessary for adipocyte differentiation (4, 7), and CBP heterozygous mice are lipodystrophic (24). These proteins may serve as integral partners for the action of GATA during adipogenesis to impart the specificity of its action in various cellular, temporal, or spatial contexts. The involvement of other cofactors interacting with GATA in adipogenesis remains to be investigated and may elucidate additional pathways by which GATA factors regulate adipogenesis. This study introduces a new mechanism by which GATA interferes with adipocyte differentiation. In addition to its classical role as a DNA-binding transcription factor, we have shown here that GATA interaction with C/EBP proteins is also a critical component of this regulation.
Acknowledgments
This work was supported by National Research Service Association (NRSA) grant F32 DK09940 and a National Institutes of Health (NIH) postdoctoral training fellowship (Q.T.), National Institute of Environmental Health Sciences (NIEHS) training grant EK56894 (J.T.), and NIH grant DK56894 (G.S.H.).
REFERENCES
- 1.Blobel, G. A., T. Nakajima, R. Eckner, M. Montminy, and S. H. Orkin. 1998. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl. Acad. Sci. USA 95:2061-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 3.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P. L., D. J. Riley, Y. Chen, and W. H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, S. E., C. Qi, P. Misra, V. Stellmach, M. S. Rao, J. D. Engel, Y. Zhu, and J. K. Reddy. 2002. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 277:3585-3592. [DOI] [PubMed] [Google Scholar]
- 6.Freytag, S. O., D. L. Paielli, and J. D. Gilbert. 1994. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 8:1654-1663. [DOI] [PubMed] [Google Scholar]
- 7.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 8.Juan, T. S., D. R. Wilson, M. D. Wilde, and G. J. Darlington. 1993. Participation of the transcription factor C/EBPδ in the acute-phase regulation of the human gene for complement component C3. Proc. Natl. Acad. Sci. USA 90:2584-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1989. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 243:1681-1688. [DOI] [PubMed] [Google Scholar]
- 10.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU. 1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 13.Tanaka, T., N. Yoshida, T. Kishimoto, and S. Akira. 1997. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 16:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong, Q., G. Dalgin, H. Xu, C. N. Ting, J. M. Leiden, and G. S. Hotamisligil. 2000. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science 290:134-138. [DOI] [PubMed] [Google Scholar]
- 15.Tong, Q., and G. S. Hotamisligil. 2001. Molecular mechanisms of adipocyte differentiation. Rev. Endocr. Metab. Disord. 2:349-355. [DOI] [PubMed] [Google Scholar]
- 16.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224-1234. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 18.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 19.Visvader, J. E., M. Crossley, J. Hill, S. H. Orkin, and J. M. Adams. 1995. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Mol. Cell Biol. 15:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, N. D., M. J. Finegold, A. Bradley, C. N. Ou, S. V. Abdelsayed, M. D. Wilde, L. R. Taylor, D. R. Wilson, and G. J. Darlington. 1995. Impaired energy homeostasis in C/EBPα knockout mice. Science 269:1108-1112. [DOI] [PubMed] [Google Scholar]
- 21.Whyatt, D. J., A. Karis, I. C. Harkes, A. Verkerk, N. Gillemans, A. G. Elefanty, G. Vairo, R. Ploemacher, F. Grosveld, and S. Philipsen. 1997. The level of the tissue-specific factor GATA-1 affects the cell-cycle machinery. Genes Funct. 1:11-24. [DOI] [PubMed] [Google Scholar]
- 22.Wu, Z., E. D. Rosen, R. Brun, S. Hauser, G. Adelmant, A. E. Troy, C. McKeon, G. J. Darlington, and B. M. Spiegelman. 1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 3:151-158. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBPβ in NIH-3T3 cells induces PPARγ and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi, T., Y. Oike, J. Kamon, H. Waki, K. Komeda, A. Tsuchida, Y. Date, M. X. Li, H. Miki, Y. Akanuma, R. Nagai, S. Kimura, T. Saheki, M. Nakazato, T. Naitoh, K. Yamamura, and T. Kadowaki. 2002. Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat. Genet. 30:221-226. [DOI] [PubMed] [Google Scholar]
- 25.Yeh, W. C., Z. Cao, M. Classon, and S. L. McKnight. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU. 1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]