Abstract
Ataxia-telangiectasia (A-T) mutated (ATM) kinase signals all three cell cycle checkpoints after DNA double-stranded break (DSB) damage. H2AX, NBS1, and p53 are substrates of ATM kinase and are involved in ATM-dependent DNA damage responses. We show here that H2AX is dispensable for the activation of ATM and p53 responses after DNA DSB damage. Therefore, H2AX functions primarily as a downstream mediator of ATM functions in the parallel pathway of p53. NBS1 appears to function both as an activator of ATM and as an adapter to mediate ATM activities after DNA DSB damage. Phosphorylation of ATM and H2AX induced by DNA DSB damage is normal in NBS1 mutant/mutant (NBS1m/m) mice that express an N-terminally truncated NBS1 at lower levels. Therefore, the pleiotropic A-T-related systemic and cellular defects observed in NBS1m/m mice are due to the disruption of the adapter function of NBS1 in mediating ATM activities. While H2AX is required for the irradiation-induced focus formation of NBS1, our findings indicate that NBS1 and H2AX have distinct roles in DNA damage responses. ATM-dependent phosphorylation of p53 and p53 responses are largely normal in NBS1m/m mice after DNA DSB damage, and p53 deficiency greatly facilitates tumorigenesis in NBS1m/m mice. Therefore, NBS1, H2AX, and p53 play synergistic roles in ATM-dependent DNA damage responses and tumor suppression.
Proper cellular responses to DNA double-strand breaks (DSBs) are critical for maintaining genetic stability and for tumor suppression (38). A gene consistently mutated in genetic instability syndrome ataxia-telangiectasia (A-T), called ATM (for ataxia-telangiectasia mutated), encodes a large protein kinase that is responsible for activating cellular responses to DNA damage (44). In this context, ATM is required for homologous recombination and all three cell cycle checkpoints after DNA DSB damage. In addition, ATM deficiency leads to hypersensitivity to γ-irradiation and multisystemic defects, including growth retardation, abolished germ cell development, immunodeficiency, and greatly increased cancer risk (43). As a protein kinase, ATM functions by phosphorylating and activating a number of DNA repair and checkpoint proteins, including p53, NBS1, H2AX, 53BP1, Brca1, Smc1, and Chk2 (43).
Immediately after DNA DSB damage, histone H2AX is phosphorylated at the C-terminal Ser residues (Ser136 and Ser139) (40). This phosphorylation, called γ-H2AX, can be detected within minutes after the introduction of DSBs and is involved in the recruitment of other known components of DNA repair, including Brca1, NBS1/Mre11/Rad50, and 53BP1, to the sites of the DNA damage (11, 41, 48). γ-H2AX is mainly mediated by ATM after DNA DSB damage, suggesting that H2AX is a downstream mediator of ATM function (6, 20). H2AX is also required for the recruitment of 53BP1 to sites of DNA DSB damage, and 53BP1 appears to be an upstream activator of ATM (11, 39, 48). Therefore, it remains unclear whether H2AX plays any role in activating ATM after DNA DSB damage.
NBS1, the gene product mutated in Nijmegen breakage syndrome (NBS) patients, is the p95 component of the Mre11 complex that forms foci at the sites of DNA DSBs (8, 36, 47). The importance of NBS1 in DNA repair is indicated by the multisystemic defects observed in NBS patients, including growth retardation, immunodeficiency, and increased lymphoid malignancies (46). In addition, NBS cells are hypersensitive to ionizing radiation and defective in intra-S and G2/M checkpoints. Targeted disruption of the N terminus of NBS1 in mice recapitulates most of the systemic and cellular defects observed in NBS patients (25, 50). The functional link between ATM and NBS1 is suggested by the large panel of defects shared by NBS1 and ATM mutations and is further demonstrated by the findings that ATM can phosphorylate and activate NBS1 at a consensus site, Ser343 (21, 30, 51, 55). Therefore, NBS1 appears to function as a downstream mediator of ATM function. Recent studies show that mutation of the Mre11 complex or NBS1 leads to impaired ATM activation in human cell lines, indicating a role for NBS1 in activating ATM after DNA DSB damage (9, 23, 39, 45). Since the disruption of this NBS1 function could account for the A-T-related defects observed in NBS patients, the contribution of the impaired adapter function of NBS1 in mediating ATM activities to the A-T-related defects in NBS1 mutant mice and human patients remains to be established.
The functional interaction between NBS1 and H2AX is complex. H2AX is required for the irradiation-induced focus formation of NBS1 after DNA DSB damage (11). In addition, direct interaction between γ-H2AX and NBS1 suggests a mechanism to recruit NBS1 to the sites of DNA DSB damage (27). These findings suggest that H2AX functions upstream of NBS1 in DNA damage response. However, while the defects observed in either H2AX or NBS1 mutant mice are shared by ATM mutant mice, H2AX and NBS1 mutant mice exhibit certain distinct phenotypes (4, 12, 25, 50, 52). For example, while spermatogenesis is abolished in H2AX-deficient mice, oogenesis failure is only observed in NBS1 mutant mice. Therefore, NBS1 and H2AX might have distinct roles in DNA damage responses.
It has been well established that ATM is required for the activation of p53 responses to DNA DSB damage in human and mouse cells (26, 49, 53, 54). ATM can phosphorylate p53 at Ser15 (corresponding to Ser18 of mouse p53), and this phosphorylation activates p53 responses to DNA DSB damage (13, 14, 17, 28). While it has been well established that ATM is required to activate p53-dependent cell cycle G1 arrest upon DNA DSB damage, it remains unclear whether H2AX is involved in mediating this ATM function after DNA DSB damage. We employed NBS1, H2AX, and p53 mutant mice to investigate the functional interactions among ATM, NBS1, H2AX, and p53 in DNA damage responses, maintenance of genetic stability, and tumor suppression.
MATERIALS AND METHODS
Generation and culture of MEFs.
Murine embryonic fibroblasts (MEFs) were derived from day 14 embryos as described previously (53). The MEFs were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 5 mM glutamine, 50 μM β-mercaptoethanol, and 50 U (each) of penicillin and streptomycin/ml at 37°C with 5% CO2. Cellular proliferation assays were performed as previously described (53). Briefly, 105 MEFs were seeded onto each 35-mm-diameter plate, and each day after being plated, MEFs from three plates of each genotype were trypsinized and counted.
Ionizing radiation (IR) treatment of MEFs and cell cycle analysis.
MEFs were synchronized at the cell cycle G0 phase by serum starvation in medium containing 0.1% fetal bovine serum for 96 h as described previously (54). The G0-synchronized MEFs were trypsinized and irradiated in suspension with a 137Cs γ-ray source. The irradiated and mock-treated MEFs were plated in 10-cm-diameter plates at a density of 1 million cells/plate in normal MEF growth medium supplemented with 10 μM bromodeoxyuridine. After 24 h of bromodeoxyuridine labeling, the cells were harvested and fixed in 70% ethanol, and the cell cycle profile was analyzed as described previously (53).
Analysis of p53-dependent apoptosis in mouse thymocytes.
Thymuses were recovered from 1- to 2-month-old mice of various genotypes, and single-cell thymocyte suspensions were treated with increasing doses of IR as described previously (15). The percentage of apoptotic cells 10 h after IR was analyzed by staining with annexin V as described previously (15).
Western blot analysis of levels of proteins and protein phosphorylation.
Protein extracts from 4 × 105 embryonic stem cells or MEFs or 5 × 106 thymocytes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 6 to 15% polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were probed with various primary antibodies and subsequently incubated with horseradish peroxidase-conjugated secondary antibody and developed with Enhanced Chemiluminescence PLUS (Amersham Pharmacia Biotech) or a Pierce SuperSignal kit. To determine the amount of protein in each lane, the filter was stripped and probed with a rabbit polyclonal antibody against actin (Santa Cruz Biotechnology, Inc.). Phosphospecific antibody against ATM was purchased from Rockland. Phosphospecific antibody against p53 was purchased from Cell Signaling Technology.
Real-time PCR analysis.
Total RNA was isolated from MEFs or thymocytes with the combination of Trizol (Invitrogen) and RNAeasy RNA Cleanup (QIAGEN). RNA was treated with RNase-free DNase (Roche Diagnostics) for 20 min at room temperature before being reverse transcribed using Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed on an ABI 7000 machine with SYBR Green PCR Master Mix (Applied Biotechnology). PCR conditions consisted of a 10-min hot start at 95°C, followed by 40 cycles of 30 s at 95°C and 1 min at 61°C. The average threshold cycle for each gene was determined from triplicate reactions, and the levels of gene expression relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were determined as previously described (5). The primers used were described previously (13).
Generation and analysis of NBS1m/m) H2AX−/− and NBS1m/m p53−/− mice.
The ATM−/−, p53−/−, NBS1 mutant/mutant (NBS1m/m), and H2AX−/− mice were described previously (4, 24, 25, 52). Because NBS1m/m females are sterile, p53+/− mice were bred with NBS1m/m males to generate p53+/− NBS1+/m mice, which were intercrossed to generate p53−/− NBS1m/m mice. The health status of various mutant mice was monitored once every 2 days. The dead or dying mice were dissected and examined for tumors. The H2AX−/− males were sterile (4), and H2AX−/− NBS1+/m female and H2AX+/− NBS1−/− males were intercrossed to screen for homozygous mutant embryos.
Analysis of chromosomes.
Pretumor thymocytes were cultured in RPM-1 medium supplemented with 12 ng of phorbol myristate acetate/ml and 0.25 μg of ionomycin/ml, and metaphases were prepared after 2 to 3 days of culture when the thymocytes reached the log phase of proliferation. Tumor cells were prepared for spectral karyotyping (SKY) analysis by culturing them in RPM-1 medium supplemented with 10% fetal calf serum, antibiotics, 100 μM β-mercaptoethanol, and 100 U of interleukin-2/ml for 2 to 3 days until they reached log-phase growth. Thymocytes and tumor cells were treating with 0.05 μg of Colcemid/ml for 2.5 h prior to standard metaphase dropping. Metaphases from pretumor thymocytes were stained with DAPI (4′,6′-diamidino-2-phenylindole) and examined. SKY analysis of chromosomes was performed as described previously (19). Chromosomes were analyzed using a Nikon Eclipse microscope equipped with an Applied Spectral Imaging charge-coupled device camera and 40 and 63× objectives.
RESULTS
H2AX is not required for ATM activation after DNA DSB damage.
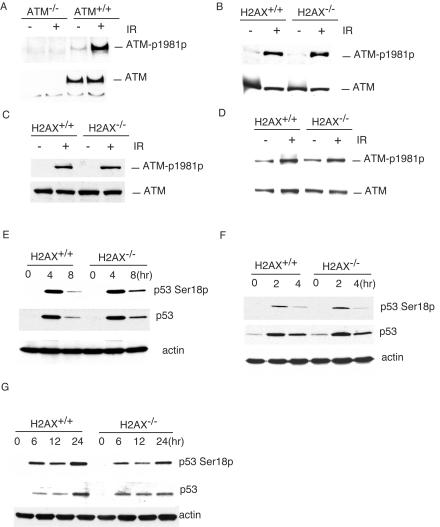
Phosphorylation of ATM at Ser1981 is required to activate ATM after DNA DSB damage (2). To control for the specificity of the commercial monoclonal antibody that recognizes the mouse ATM phosphorylated at Ser1981, we used the antibody to probe the blot of the protein extracts from ATM−/− and ATM+/+ MEFs before and 1 h after 1 Gy of IR. While similar levels of ATM protein were detected in ATM+/+ MEFs before and after IR, no ATM protein could be detected in ATM−/− samples (Fig. 1A). In addition, while no phosphorylated ATM was detected in ATM−/− MEFs before or after IR, phosphorylation of ATM was significantly increased in ATM+/+ MEFs 1 h after 1 Gy of IR (Fig. 1A). Therefore, we concluded that the monoclonal antibody specifically recognizes mouse ATM phosphorylated at Ser1981. To determine whether H2AX is required for ATM activation after DNA damage, we analyzed the Ser1981 phosphorylation of ATM in H2AX+/+ and H2AX−/− thymocytes and MEFs after IR. Normal phosphorylation of ATM at Ser1981 was observed in H2AX−/− thymocytes and MEFs 1 h after 1Gy of IR, indicating that H2AX is not required for ATM activation after DNA DSB damage (Fig. 1B and C). Consistent with this conclusion, ATM phosphorylation at Ser1981 is normal in H2AX−/− thymocytes 10 min after 1 Gy of IR (Fig. 1D). Phosphorylation of p53 at Ser18 and p53 stabilization were both normal in H2AX−/− MEFs and thymocytes after DNA DSB damage induced by IR or doxorubicin (Fig. 1E, F, and G).
FIG. 1.
Phosphorylation of ATM at Ser1981 and p53 at Ser18 after DNA DSB damage in H2AX−/− cells. (A) Phosphorylation of ATM in ATM+/+ and ATM−/− MEFs before (−) and 1 h after (+) IR (1 Gy). Phosphorylation of ATM in H2AX+/+ and H2AX−/− thymocytes (B) and MEFs (C) 1 h after 1 Gy of IR or in H2AX+/+ and H2AX−/− thymocytes 10 min after 1 Gy of IR (D). The genotypes and the treatment are indicated at the top. Phosphorylated ATM and total ATM are indicated on the right. p53 stabilization and phosphorylation at Ser18 in H2AX+/+ and H2AX−/− thymocytes after IR (E) and in MEFs after IR (F) or doxorubicin (G). The genotypes and time points after treatments are indicated at the top. Phosphorylated p53, total p53, and actin are indicated on the right.
p53-dependent responses to DNA DSB damage in H2AX−/− cells.
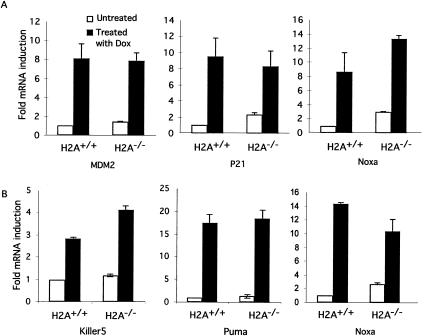
While H2AX is not required for p53 stabilization after DNA DSB damage, we analyzed p53-dependent transcription after DNA DSB damage to determine whether H2AX is required for p53 activities. Analysis of p53-dependent gene expression in H2AX−/− MEFs before and after doxorubicin treatment indicated that expression of a number of p53-dependent genes was normal or slightly increased in H2AX−/− MEFs after DNA DSB damage (Fig. 2A). In addition, the basal levels of some p53 target genes, such as p21, were increased in H2AX−/− MEFs without doxorubicin treatment (Fig. 2A). We also found normal p53-dependent apoptosis in H2AX−/− thymocytes after IR (data not shown). In addition, p53-dependent expression of apoptotic genes, including Noxa, Puma, and Killer 5 genes, was normal or higher in H2AX−/− thymocytes after IR (Fig. 2B). Similar to what was observed in H2AX−/− MEFs, the basal levels of some p53-dependent apoptotic genes were increased in H2AX−/− thymocytes in the absence of exogenous DNA damage.
FIG. 2.
p53-dependent transcription in H2AX+/+ and H2AX−/− MEFs (A) and thymocytes (B) after DNA DSB damage. The mRNA levels of each gene were determined by quantitative real-time PCR and standardized by the mRNA levels of GAPDH as described previously (13). The mRNA levels in the untreated H2AX+/+ cells are arbitrarily set to 1. The MEFs were harvested 12 h after doxorubicin (Dox) treatment (0.5 μM), and the thymocytes were harvested 4 h after treatment with 5 Gy of IR. Mean values from three independent experiments are presented, with error bars indicating standard deviations.
Various ATM-dependent phosphorylation events in NBS1m/m cells.
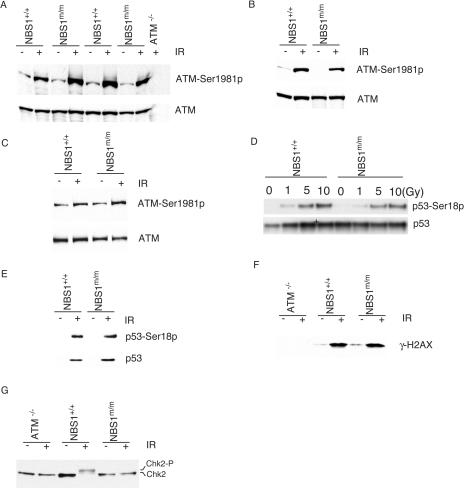
Recent studies have suggested that the Mre11/Rad50/NBS1 complex functions upstream of ATM and is required for ATM activation after DNA DSB damage (9, 23, 39, 45). To test whether the large panel of A-T-related cellular and systemic defects observed in NBS1m/m mice are due to impaired ATM activation, we analyzed the phosphorylation of ATM at Ser1981 in NBS1m/m cells after IR. Normal ATM phosphorylation was observed in NBS1m/m MEFs and thymocytes 1 h after 1 Gy of IR (Fig. 3A and B). In addition, the phosphorylation of ATM at Ser1981 is also normal in NBS1m/m thymocytes 10 min after 1 Gy of IR (Fig. 3C). ATM-dependent phosphorylation of p53 at Ser18 and γ-H2AX is normal in NBS1m/m cells after IR (Fig. 3D, E, and F). Since NBS1m/m cells expressed an N-terminally truncated NBS1 protein at low levels (25), these findings indicated that the N-terminal domain of NBS1 is not required for the activation of ATM and ATM-dependent phosphorylation of p53 and H2AX after DNA DSB damage.
FIG. 3.
Phosphorylation of ATM, p53, and Chk2 in NBS1+/+ and NBS1m/m cells after IR. Phosphorylation of ATM at Ser1981 in NBS1+/+ and NBS1m/m MEFs (A) and thymocytes (B) 1 h after treatment with 1 Gy of IR or in NBS1+/+ and NBS1m/m thymocytes 10 min after 1 Gy of IR (C). The treatment and genotypes are indicated at the top. Phosphorylated ATM and total ATM are indicated on the right. Phosphorylation of p53 in NBS1+/+ and NBS1m/m MEFs 1 h after various dosages of IR (D) and thymocytes 1 h after treatment with 1 Gy of IR (E). The treatment and genotypes are indicated at the top. Phosphorylated p53 and total p53 are indicated on the right. Phosphorylation of H2AX (F) and Chk2 (G) in ATM−/−, NBS1+/+, and NBS1m/m thymocytes 1 h after treatment with 1 Gy of IR. The genotypes and treatment are indicated at the top. Phosphorylated H2AX and Chk2 are indicated on the right.
ATM-dependent phosphorylation of Chk2 might activate Chk2 activities after IR (1, 35, 37). To test whether Chk2 is phosphorylated in NBS1m/m thymocytes after IR, we analyzed the phosphorylation status of Chk2 in NBS1+/+ and NBS1m/m thymocytes after IR. Phosphorylated Chk2 migrates more slowly than unphosphorylated Chk2 in sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and this allowed us to accurately determine the Chk2 phosphorylation status in the cells. While Chk2 was mostly phosphorylated in NBS1+/+ thymocytes after IR, little phosphorylation of Chk2 could be detected in either ATM−/− or NBS1m/m thymocytes (Fig. 3G). Therefore, a complete NBS1 is required for ATM-dependent phosphorylation of Chk2 after DNA DSB damage.
Synthetic lethality of NBS1 and H2AX mutations.
Since NBS1, but not H2AX, is required for ATM-dependent phosphorylation of Chk2 (20), these findings suggested some distinct functions of H2AX and NBS1 in mediating ATM-dependent activities. In addition, NBS1m/m and H2AX−/− mice developed some distinct defects (4, 12, 25, 50). In support of the notion that NBS1 and H2AX have distinct roles in DNA damage responses, while both NBS1m/m and H2AX−/− mice are viable, NBS1m/m H2AX−/− mice were embryonic lethal (Table 1). The embryonic lethality occurred during early embryonic development, since no homozygous mutant embryos could be detected at embryonic day 12 (Table 1). H2AX haploid deficiency has been shown to increase cancer predisposition in the p53−/− background (4, 10). Therefore, we monitored and compared tumorigenesis in 34 sets of NBS1m/m and NBS1m/m H2AX+/− mice that were >6 months of age. No increased tumorigenesis was observed in NBS1m/m H2AX+/− mice.
TABLE 1.
Genotypes of offspring derived from intercrossing of NBS1+/m H2AX−/− females and NBS1+/m H2AX+/− malesa
| Parameter | Offspring (164) from intercross of NBS1+/m H2AX−/− females and NBS1+/m H2AX+/− males | Embryos (54) from intercross of NBS1+/m H2AX−/− females and NBS1m/m H2AX+/− males | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | NBS1+/+ H2AX+/− | NBS1+/m H2AX+/− | NBS1m/m H2AX+/− | NBS1+/+ H2AX−/− | NBS1+/m H2AX−/− | NBS1m/m H2AX−/− | NBS1+/m H2AX+/− | NBS1m/m H2AX+/− | NBS1+/m H2AX−/− | NBS1m/m H2AX−/− |
| Expected frequency | 1/8 | 1/4 | 1/8 | 1/8 | 1/4 | 1/8 | 1/4 | 1/4 | 1/4 | 1/4 |
| Observed no. (frequency [%]) | 21 (12) | 46 (28) | 29 (17) | 15 (9) | 53 (32) | 0 | 24 (45) | 13 (25) | 16 (30) | 0 |
Genotypes of day 12 embryos derived from NBS1+/m H2AX−/− females and NBS1m/m H2AX+/− males are shown.
Interchromosomal rearrangements in H2AX−/− and NBS1m/m thymocytes.
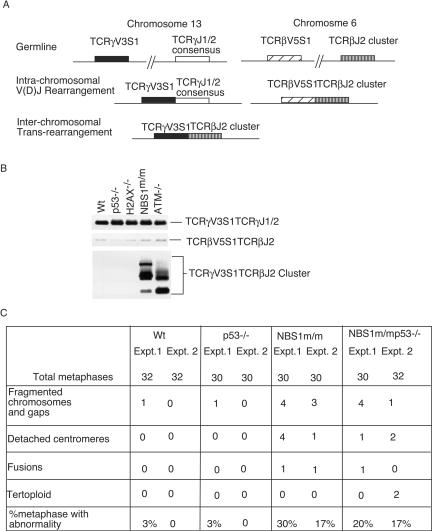
Chromosomal translocations are common in ATM−/− human and mouse cells (43). To test whether NBS1 mediates ATM functions in the maintenance of genetic stability, we analyzed the interchromosomal rearrangements in the pretumor thymocytes by detecting the interchromosomal rearrangement between T-cell receptor β (TCRβ) and TCRγ, a predictor of global chromosomal translocations in mouse thymocytes (25, 31). The thymocytes were analyzed by flow cytometry for their patterns of CD4 and CD8 expression, as well as their cell sizes, to ensure that they were not tumor cells. While little interchromosomal rearrangement can be detected in wild-type thymocytes, interchromosomal rearrangements involving these TCR loci were observed in all ATM−/− and NBS1m/m thymocytes analyzed (Fig. 4A and B). However, similar to wild-type thymocytes, little interchromosomal rearrangement was detected in thymocytes derived from 10 H2AX−/− mice and 10 p53−/− mice (Fig. 4A and B and data not shown).
FIG. 4.
Genetic instability in pretumor thymocytes derived from 1-month-old wild-type (Wt), p53−/−, H2AX−/−, NBS1m/m, and ATM−/− mice. (A) Chromosomal translocations involving TCR loci in thymocytes of various genotypes. Schematic diagram of intra- and interchromosomal rearrangements in TCRβ and -γ loci. The TCRγV3S1TCRβJ2 rearrangement represents the formation of a t (6, 13) chromosomal translocation. (B) PCR analysis of the intra- and interchromosomal rearrangements in wild-type, p53−/−, H2AX−/−, NBS1m/m, and ATM−/− thymocytes. The PCR conditions were essentially the same as those described by Kang et al. (25). One hundred nanograms of genomic DNA derived from thymocytes of various genotypes was used for PCR amplification. Representative PCR products from the three rearrangements are indicated on the right, and the genotypes are indicated at the top. (C) Chromosomal abnormalities in thymocytes of various genotypes. Data from two sets of independently prepared metaphase chromosomes of thymocytes are presented. Expt., experiment.
To further analyze the genetic instability in the pretumor thymocytes, we examined the metaphase chromosomes prepared from the proliferating thymocytes derived from mice of various genotypes. While chromosomal structural abnormalities were easily detected in both NBS1m/m and NBS1m/m p53−/− thymocytes, little chromosomal translocation was detected in the wild-type and p53−/− thymocytes (Fig. 4C).
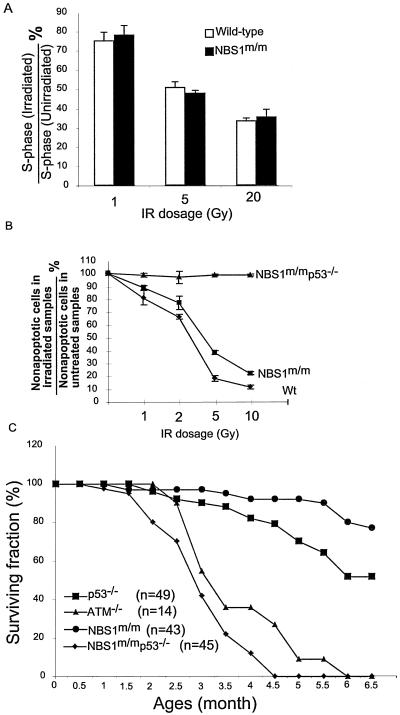
NBS1 and p53 have synergistic roles in tumor suppression.
ATM−/−, NBS1m/m, and p53−/− mice all developed thymic lymphomas (3, 16, 18, 24, 25, 52). However, ATM−/− mice developed thymic lymphomas much faster than NBS1m/m and p53−/− mice and uniformly died from thymic lymphomas by 6 months of age (3, 52). These findings suggest that ATM-dependent activation of p53 and NBS1 might play synergistic roles in tumor suppression. Consistent with this notion, p53-dependent p21 induction and cell cycle G1 arrest were both normal in NBS1m/m MEFs after various doses of IR (Fig. 5A and data not shown). In addition, p53-dependent apoptosis was modestly reduced in NBS1m/m thymocytes after IR compared with that in wild-type thymocytes (Fig. 5B). To further test the functional interactions of NBS1 and p53 in tumor suppression, NBS1m/m p53−/− mice were generated and monitored for tumorigenesis. Similar to ATM−/− mice, NBS1m/m p53−/− mice developed thymic lymphomas much faster than NBS1m/m and p53−/− mice, and all NBS1m/m p53−/− mice died from thymic lymphomas by 5 months of age (Fig. 5C). Therefore, NBS1 and p53 function synergistically in the suppression of thymic lymphomas.
FIG. 5.
p53 and NBS1 play synergistic roles in tumor suppression. (A) Normal p53-dependent cell cycle G1 arrest in NBS1+/+ and NBS1m/m MEFs after treatment with various doses of IR. The mean value from three independent experiments is presented with error bars indicating standard deviations. (B) Quantitative analysis of the percentages of apoptotic thymocytes in NBS1+/+ and NBS1m/m thymocytes 10 h after IR. The mean value from four independent experiments is presented with error bars indicating standard deviations. Wt, wild type. (C) Survival analysis of ATM−/−, NBS1m/m, p53−/−, and NBS1m/m p53−/− mice. Most of the data on NBS1m/m, p53−/−, and NBS1m/m p53−/− mice were derived from littermates. n represents the number of mice monitored for death caused by tumorigenesis. All NBS1m/m p53−/− mice died from thymic lymphomas within 6 months.
Chromosomal translocations in tumors derived from NBS1m/m mice.
To understand the rapid onset of tumorigenesis in NBS1m/m p53−/− mice and to determine the similarity to tumors arising in ATM−/− mice, SKY was performed to analyze the chromosomal abnormalities in tumors derived from NBS1m/m and NBS1m/m p53−/− mice. Two NBS1m/m tumors and three NBS1m/m p53−/− tumors were analyzed. Chromosomal translocations involving chromosome 12, including T(12;2), T(12;1), and T(12;11), were commonly detected in NBS1m/m tumors, with a clonal T(12;2) translocation observed in one tumor (Fig. 6A and data not shown). A subclonal T(14;12) translocation was also observed in both NBS1m/m tumors (Fig. 6A; data not shown). Similar chromosomal translocations were observed in NBS1m/m p53−/− tumors. The first tumor contained a clonal T(12;2) translocation in 12 of 13 metaphases, comprised of the centromere and most of the long arm of 12 attached to most of the distal long arm of 2 (Fig. 6B). This tumor also contained a subclonal T(3;14) translocation (3 of 13 metaphases), as well as three or four intact copies of chromosome 14 in every metaphase. The second tumor contained a T(14;2) translocation in 4 of 10 metaphases and extra chromosomes 14 and 15 in 7 metaphases (data not shown). The third tumor did not contain a detectable clonal aberration but was highly unstable, containing five separate translocations, each seen in only one metaphase. Supernumerary chromosomes were observed only in NBS1m/m p53−/− tumors (average chromosomal number, 45) but not in NBS1m/m tumors, both of which contained 40 chromosomes. These types of chromosomal translocations are also frequently observed in thymic tumors derived from ATM−/− mice, as characterized in previous publications (3, 32). These findings further support the notion that NBS1 plays an important role in mediating ATM-dependent suppression of chromosomal translocations in mouse thymocytes.
FIG. 6.
Spectral karyotype of an NBS1m/m (A) and an NBS1m/m p53−/− (B) thymic lymphoma. For each chromosome in the karyotype table, the fluorescent SKY appearance is shown on the left, a grayscale image of DAPI stain is shown in the middle, and computer-classified colon is shown on the right. The chromosomes are oriented with centromeres toward the top of the page. The translocations are shown below. Note also the presence of aneuploidy consisting of three chromosomes numbers 14 and 6, and a missing chromosome 9 in the NBS1m/m p53−/− tumor.
DISCUSSION
The kinase activity of ATM is rapidly activated after DNA DSB damage, leading to the phosphorylation and activation of a number of DNA repair proteins, including NBS1 and 53BP1 (2, 42). Recent studies have suggested that the Mre11/Rad50/NBS1 complex and 53BP1 function both upstream and downstream of ATM after DNA DSB damage (9, 20, 21, 23, 30, 39, 45, 48, 51, 55). H2AX, which is phosphorylated by ATM immediately after DNA DSB damage, is required for irradiation-induced focus formation at the site of DNA damage by a large number of DNA repair and checkpoint proteins, including 53BP1 and NBS1, suggesting that H2AX might be involved in the activation of ATM after DNA DSB damage (12, 48). However, we demonstrate here that H2AX is not required for ATM activation after DNA DSB damage. In addition, H2AX is not required for ATM-dependent phosphorylation of p53 and Chk2 after DNA DSB damage (Fig. 1) (20). Therefore, H2AX functions as a downstream mediator of ATM functions after DNA DSB damage. These findings also support the notion that ATM activation might be independent of the exact sites of DNA DSB damage (2). Therefore, while phosphorylation of substrates such as NBS1 by activated ATM might occur mostly at the sites of DNA damage (33), the MRN complex likely activates ATM at a distance from the sites of DNA damage through direct physical interaction (23, 29).
Impaired ATM activation and p53 phosphorylation have been reported in human NBS cell lines (9, 23, 39, 45). However, we found normal phosphorylation of ATM and p53 in our NBS1 mutant mice. The apparent discrepancy between our findings in NBS1 mutant mice and the human cell lines is likely due to the nature of NBS1 mutation and the expression levels of the truncated NBS1 in these two systems. Cells from NBS patients express an N-terminal 26-kDa NBS1 protein fragment and an N-terminally truncated 70-kDa NBS1 protein at low levels (34). In NBS1m/m mice, the N terminus (codons 1 to 194) of NBS1, which contains the FHA and BRCT domains, is disrupted (25). Therefore, while the 26-kDa N-terminal NBS1 protein fragment should not be produced in our NBS1 mutant mice, the N-terminally truncated NBS1 protein of ∼70 kDa is expressed at reduced levels in the cells derived from the mutant mice (25). The discrepancy in ATM activation between NBS1 mutant mice and NBS patients could be due to the different expression levels of the 70-kDa N-terminally truncated NBS1 protein. Alternatively, since the 26-kDa N-terminal NBS1 fragment is expressed only in some human NBS cells, it is also possible that the 26-kDa N-terminal NBS1 fragment plays a dominant-negative role in suppressing ATM activation after DNA DSB damage.
The finding of normal ATM activation in NBS1m/m mice after DNA DSB damage extends the observation by Horejsi et al. that the N-terminal FHA domain of NBS1 is not required for ATM activation and indicates that neither the FHA nor the BRCT domain of NBS1 is required for ATM activation (23). In addition, consistent with the finding that NBS1 plays important roles in mediating ATM activities (22, 45), this finding indicates that the A-T-related defects observed in NBS1 mutant mice are mainly due to the disruption of the adapter function of NBS1 in mediating ATM activities, such as the phosphorylation of Chk2.
Our studies also provide an improved understanding of the functional interactions among H2AX, NBS1, and p53. An earlier report showed that H2AX is required for the irradiation-induced focus formation of NBS1 after DNA DSB damage, suggesting that H2AX and NBS1 function in overlapping pathways in the cellular responses to DNA DSB damage (11). However, several lines of evidence presented here and elsewhere indicate that NBS1 and H2AX also function in distinct pathways in DNA damage responses. In this context, NBS1, but not H2AX, is required for ATM-dependent Chk2 phosphorylation after DNA DSB damage (Fig. 3) (7, 20). In addition, NBS1, but not H2AX, is required for the ATM-dependent suppression of interchromosomal rearrangement, indicating that NBS1 and H2AX have distinct roles in DNA damage responses. In support of this notion, the chromosomal abnormalities observed in the thymic tumors derived from NBS1m/m and NBS1m/m p53−/− mice are different from those derived from H2AX−/− p53−/− mice (4). The distinct roles of H2AX and NBS1 in DNA repair could also account for the synthetic lethality of NBS1 mutation and H2AX mutation.
ATM is required to activate p53 responses to DNA DSB damage (42). Our finding that H2AX and NBS1 are not important for p53 responses to DNA DSB damage indicates that H2AX and NBS1 function largely in distinct pathways of p53 in DNA DSB damage responses. In the context of H2AX−/− cells, increased basal levels of p53 activities, possibly due to the presence of endogenous genetic instability, might account for the very modest cancer risk in these mutant mice. Consistent with these notions, inactivation of p53 greatly facilitates tumorigenesis in H2AX−/− mice and our NBS1 mutant mice, as well as independently generated NBS1 mutant mice (4, 10, 50). Considering the greatly increased genetic instability in H2AX and NBS1 mutant mice, these findings indicate that p53 can efficiently suppress tumorigenesis in the presence of significant endogenous chromosomal abnormalities. Therefore, cancer predisposition in ATM−/− mice, in which both p53 inactivation and genetic instability are present, is greatly increased compared with that in NBS1 mutant mice.
Acknowledgments
This work was supported by an NIH grant to Y.X. (CA77563).
REFERENCES
- 1.Ahn, J. Y., J. K. Schwarz, H. Piwnica-Worms, and C. E. Canman. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60:5934-5936. [PubMed] [Google Scholar]
- 2.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, C., S. Hirotsune, R. Paylor, M. Liyanage, M. Eckhaus, F. Collins, Y. Shiloh, J. N. Crawley, T. Ried, D. Tagle, and A. Wynshaw-Boris. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86:159-171. [DOI] [PubMed] [Google Scholar]
- 4.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359-370. [DOI] [PubMed] [Google Scholar]
- 5.Boley, S. E., V. A. Wong, J. E. French, and L. Recio. 2002. p53 heterozygosity alters the mRNA expression of p53 target genes in the bone marrow in response to inhaled benzene. Toxicol. Sci. 66:209-215. [DOI] [PubMed] [Google Scholar]
- 6.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 7.Buscemi, G., C. Savio, L. Zannini, F. Micciche, D. Masnada, M. Nakanishi, H. Tauchi, K. Komatsu, S. Mizutani, K. Khanna, P. Chen, P. Concannon, L. Chessa, and D. Delia. 2001. Chk2 activation dependence on Nbs1 after DNA damage. Mol. Cell. Biol. 21:5214-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 9.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 12.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao, C., M. Hergenhahn, M. D. Kaeser, Z. Wu, S. Saito, R. Iggo, M. Hollstein, E. Appella, and Y. Xu. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 8:8. [DOI] [PubMed] [Google Scholar]
- 14.Chao, C., S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2000. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 97:11936-11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao, C., S. Saito, J. Kang, C. W. Anderson, E. Appella, and Y. Xu. 2000. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 19:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 17.Dumaz, N., D. M. Milne, and D. W. Meek. 1999. Protein kinase CK1 is a p53-threonine 18 kinase which requires prior phosphorylation of serine 15. FEBS Lett. 463:312-316. [DOI] [PubMed] [Google Scholar]
- 18.Elson, A., Y. Wang, C. J. Daugherty, C. C. Morton, F. Zhou, J. Campos-Torres, and P. Leder. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA 93:13084-13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson, D. O., J. M. Sekiguchi, S. Chang, K. M. Frank, Y. Gao, R. A. DePinho, and F. W. Alt. 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 97:6630-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 21.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 22.Girard, P. M., E. Riballo, A. C. Begg, A. Waugh, and P. A. Jeggo. 2002. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene 21:4191-4199. [DOI] [PubMed] [Google Scholar]
- 23.Horejsi, Z., J. Falck, C. J. Bakkenist, M. B. Kastan, J. Lukas, and J. Bartek. 2004. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23:3122-3127. [DOI] [PubMed] [Google Scholar]
- 24.Jacks, T., L. Remington, B. O. Williams, E. M. Schmitt, S. Halachmi, R. T. Bronson, and R. A. Weinberg. 1994. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 25.Kang, J., R. T. Bronson, and Y. Xu. 2002. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 21:1447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 28.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J. H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93-96. [DOI] [PubMed] [Google Scholar]
- 30.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Lin, J. H. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404:613-617. [DOI] [PubMed] [Google Scholar]
- 31.Lista, F., V. Bertness, C. J. Guidos, J. S. Danska, and I. R. Kirsch. 1997. The absolute number of trans-rearrangements between the TCRG and TCRB loci is predictive of lymphoma risk: a severe combined immune deficiency (SCID) murine model. Cancer Res. 57:4408-4413. [PubMed] [Google Scholar]
- 32.Liyanage, M., A. Coleman, S. du Manoir, T. Veldman, S. McCormack, R. B. Dickson, C. Barlow, A. Wynshaw-Boris, S. Janz, J. Wienberg, M. A. Ferguson-Smith, E. Schrock, and T. Ried. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat. Genet. 14:312-315. [DOI] [PubMed] [Google Scholar]
- 33.Lukas, C., J. Falck, J. Bartkova, J. Bartek, and J. Lukas. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5:255-260. [DOI] [PubMed] [Google Scholar]
- 34.Maser, R. S., R. Zinkel, and J. H. Petrini. 2001. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 27:417-421. [DOI] [PubMed] [Google Scholar]
- 35.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuura, S., H. Tauchi, A. Nakamura, N. Kondo, S. Sakamoto, S. Endo, D. Smeets, B. Solder, B. H. Belohradsky, V. M. Der Kaloustian, M. Oshimura, M. Isomura, Y. Nakamura, and K. Komatsu. 1998. Positional cloning of the gene for Nijmegen breakage syndrome. Nat. Genet. 19:179-181. [DOI] [PubMed] [Google Scholar]
- 37.Melchionna, R., X. B. Chen, A. Blasina, and C. H. McGowan. 2000. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2:762-765. [DOI] [PubMed] [Google Scholar]
- 38.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77-95. [DOI] [PubMed] [Google Scholar]
- 39.Mochan, T. A., M. Venere, R. A. DiTullio, Jr., and T. D. Halazonetis. 2003. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 63:8586-8591. [PubMed] [Google Scholar]
- 40.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 41.Sedelnikova, O. A., D. R. Pilch, C. Redon, and W. M. Bonner. 2003. Histone H2AX in DNA damage and repair. Cancer Biol. Ther. 2:233-235. [DOI] [PubMed] [Google Scholar]
- 42.Shiloh, Y. 2003. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer 3:155-168. [DOI] [PubMed] [Google Scholar]
- 43.Shiloh, Y., and M. B. Kastan. 2001. ATM: genome stability, neuronal development, and cancer cross paths. Adv. Cancer Res. 83:209-254. [DOI] [PubMed] [Google Scholar]
- 44.Shiloh, Y., and G. Rotman. 1996. Ataxia-telangiectasia and the ATM gene: linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J. Clin. Immunol. 16:254-260. [DOI] [PubMed] [Google Scholar]
- 45.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Burgt, I., K. H. Chrzanowska, D. Smeets, and C. Weemaes. 1996. Nijmegen breakage syndrome. J. Med. Genet. 33:153-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska, K. Saar, G. Beckmann, E. Seemanova, P. R. Cooper, N. J. Nowak, M. Stumm, C. M. Weemaes, R. A. Gatti, R. K. Wilson, M. Digweed, A. Rosenthal, K. Sperling, P. Concannon, and A. Reis. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93:467-476. [DOI] [PubMed] [Google Scholar]
- 48.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 49.Westphal, C. H., S. Rowan, C. Schmaltz, A. Elson, D. E. Fisher, and P. Leder. 1997. atm and p53 cooperate in apoptosis and suppression of tumorigenesis, but not in resistance to acute radiation toxicity. Nat. Genet. 16:397-401. [DOI] [PubMed] [Google Scholar]
- 50.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12:648-653. [DOI] [PubMed] [Google Scholar]
- 51.Wu, X., V. Ranganathan, D. S. Weisman, W. F. Heine, D. N. Ciccone, T. B. O'Neill, K. E. Crick, K. A. Pierce, W. S. Lane, G. Rathbun, D. M. Livingston, and D. T. Weaver. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature 405:477-482. [DOI] [PubMed] [Google Scholar]
- 52.Xu, Y., T. Ashley, E. E. Brainerd, R. T. Bronson, M. S. Meyn, and D. Baltimore. 1996. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10:2411-2422. [DOI] [PubMed] [Google Scholar]
- 53.Xu, Y., and D. Baltimore. 1996. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 10:2401-2410. [DOI] [PubMed] [Google Scholar]
- 54.Xu, Y., E. M. Yang, J. Brugarolas, T. Jacks, and D. Baltimore. 1998. Involvement of p53 and p21 in cellular defects and tumorigenesis in atm−/− mice. Mol. Cell. Biol. 18:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, S., Y. C. Weng, S. S. Yuan, Y. T. Lin, H. C. Hsu, S. C. Lin, E. Gerbino, M. H. Song, M. Z. Zdzienicka, R. A. Gatti, J. W. Shay, Y. Ziv, Y. Shiloh, and E. Y. Lee. 2000. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature 405:473-477. [DOI] [PubMed] [Google Scholar]