Abstract
Two competing models for fibroblast growth factor (FGF) receptor (FGFR) dimerization have recently emerged based on ternary FGF-FGFR-heparin crystal structures. In the symmetric two-end model, heparin promotes dimerization of two FGF-FGFR complexes by stabilizing bivalent interactions of the ligand and receptor through primary and secondary sites and by stabilizing direct receptor-receptor contacts. In the asymmetric model, there are no protein-protein contacts between the two FGF-FGFR complexes, which are bridged solely by heparin. To identify the correct mode of FGFR dimerization, we abolished interactions at the secondary ligand-receptor interaction site, which are observed only in the symmetric two-end model, using site-directed mutagenesis. Cellular studies and real-time binding assays, as well as matrix-assisted laser desorption ionization-time of flight analysis, demonstrate that loss of secondary ligand-receptor interactions results in diminished FGFR activation due to decreased dimerization without affecting FGF-FGFR binding. Additionally, structural and biochemical analysis of an activating FGFR2 mutation resulting in Pfeiffer syndrome confirms the physiological significance of receptor-receptor contacts in the symmetric two-end model and provides a novel mechanism for FGFR gain of function in human skeletal disorders. Taken together, the data validate the symmetric two-end model of FGFR dimerization and argue against the asymmetric model of FGFR dimerization.
Fibroblast growth factors 1 to 23 (FGF1 to FGF23) regulate a wide variety of biological processes during embryonic development and adult homeostasis (17, 21). In addition, aberrant activation of the FGF signaling pathway is responsible for several craniosynostosis and dwarfing syndromes, such as Apert syndrome, Pfeiffer syndrome, and achondroplasia (11, 19, 36), and human phosphate wasting disorders, including autosomal dominant hypophosphatemic rickets and tumor-induced osteomalacia (31, 34). FGFs mediate their biological effects by binding to, dimerizing, and activating cell surface FGF receptor (FGFR) tyrosine kinases. FGFR dimerization also requires heparin or heparan sulfate proteoglycan (HSPG) (22, 28, 37). The extracellular ligand-binding portion of FGFRs consists of three immunoglobulin-like domains (D1 to D3). D2, D3, and the interconnecting linker bear the determinants for ligand binding and specificity, whereas D1 has been shown to play an autoinhibitory role in FGFR signaling (12, 14, 20, 33). Alternative splicing of exons encoding the extracellular domain of FGFR1 to -3 result in c and b isoforms with unique ligand binding specificities and tissue localizations (18, 38, 39).
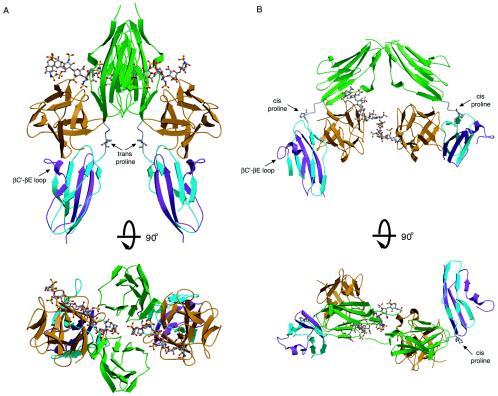
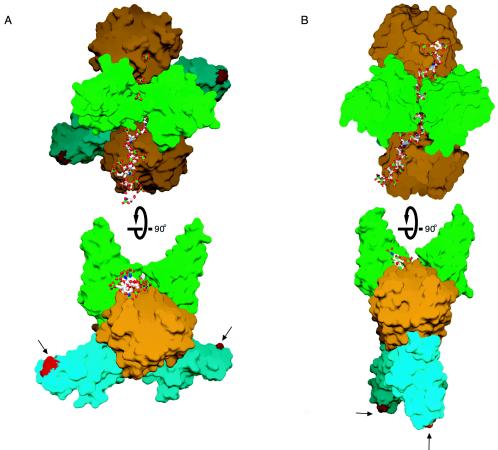
Recent crystallographic analysis of binary FGF-FGFR and ternary FGF-FGFR-heparin complexes have provided two prevailing models for FGFR dimerization (Fig. 1) (24, 27, 30, 32). The symmetric two-end model is based on the crystal structure of a symmetric 2-2 FGF2-FGFR1c dimer (Protein Data Base identifier [PDB ID], 1CVS) obtained in the absence of heparin (Fig. 1A) (27). Each ligand is bivalent, and each receptor is trivalent, with respect to protein-protein interactions. FGF and FGFR interact through an extensive primary interaction site to form a monomeric FGF-FGFR complex (Fig. 1A). In addition, ligand from one monomeric FGF-FGFR complex promotes dimerization through a distinct secondary interaction with D2 of the adjoining FGF-FGFR complex (Fig. 1A). The dimer interface is further enlarged by direct receptor-receptor contacts (Fig. 1A). A positively charged canyon is formed on the membrane distal end of the dimer and is located deep between the inward heparin binding sites of the two centrally located D2s and wanes onto the heparin binding surface of each FGF ligand (Fig. 1A). In the absence of heparin, a series of sulfate ions bind into the canyon and have been proposed to mimic the sulfate moieties of heparin. Diffusion of heparin into 2-2 FGF2-FGFR1c crystals resulted in the symmetric binding of two heparin molecules into this canyon (Fig. 1A) (PDB ID, 1FQ9) (30). Each heparin oligosaccharide interacts with one ligand and both receptors in the dimer, thus promoting FGFR dimerization by augmenting FGF-FGFR binding at primary and secondary interaction sites, as well as by stabilizing receptor-receptor contacts. Importantly, FGF1-FGFR2c (PDB ID, 1DJS) (32) and FGF10-FGFR2b (PDB ID, 1NUN) (39) complexes were also crystallized using high-ammonium sulfate buffer, which facilitated the formation of canyon dimers in both cases. The recurrence of these canyon dimers, despite different FGF and FGFR constituents, points to the physiological relevance of the mode of dimerization observed in the canyon dimers.
FIG. 1.
Two competing models for FGFR dimerization. (A) Ribbon diagram of the FGF2-FGFR1c-heparin crystal structure (PDB ID, 1FQ9) in two views related by a 90° rotation about the horizontal axis. The distance between the membrane insertion points at the end of D3 is 48 Å in the symmetric two-end model. FGF ligand and D2 are colored orange and green, respectively. The first half of D3 is colored blue, and the alternatively spliced region of D3 is colored purple. Atom coloring for heparin is as follows: oxygens red, sulfurs yellow, nitrogens blue, and carbons gray. (B) Ribbon diagram of the FGF1-FGFR2c-heparin crystal structure (PDB ID, 1E0O). Note that in the asymmetric model, heparin interacts with only the FGFR in the left half of the dimer. The distance between the membrane insertion points at the end of D3 is 74 Å in this model. Coloring is as in panel A. The location of the βC′-βE specificity loop in each model is indicated by an arrow. The location and isomerization state of the D2-D3 linker invariant proline are indicated for each structure.
Pellegrini et al. (24) proposed a fundamentally different model for FGFR dimerization based on the crystal structure of a 2-2-1 FGF1-FGFR2c-heparin complex (PDB ID, 1E0O) that was obtained through cocrystallization (Fig. 1B). In this structure, two FGF-FGFR complexes bind to opposing sides of a heparin oligosaccharide (24). Both FGF and FGFR are monovalent in this model, with respect to protein-protein interactions, and hence, the dimer interface lacks protein-protein contacts. Thus, the two FGF-FGFR halves are held together solely by heparin. Heparin interacts differently with each ligand and binds to one receptor only, resulting in the distinct asymmetry of this model (Fig. 1B). As a result of this asymmetry, heparin enhances ligand-receptor binding for only one of the two FGF-FGFR complexes. For the sake of comparison, we shall refer to this model as the asymmetric model. In addition to differing in the mode of dimerization, the asymmetric model also differs from the symmetric two-end model in the mode of 1-1 FGF-FGFR binding. This is because in 1E0O, the D2-D3 linker invariant proline (Pro253) is in a cis conformation, whereas in all other reported FGF-FGFR structures (PDB IDs, 1CVS, 1DJS, 1EVT, 1EV2, 1FQ9, 1NUN, and 1RY7), the analogous prolines assume a trans conformation. The cis conformation of the D2-D3 linker invariant proline does not affect the FGF-D2 interface but reorients D3 and results in completely different regions of D3 contacting FGF ligand in 1E0O than in all other FGF-FGFR structures (PDB IDs, 1CVS, 1DJS, 1EVT, 1EV2, 1FQ9, 1NUN, and 1RY7).
Pellegrini et al. (24) suggested that the trans configuration of the D2-D3 linker invariant proline in the binary FGF-FGFR complexes is inactive, and by extension, the mode of dimerization in the symmetric two-end model is not physiologically relevant. It was proposed that heparin converts these inactive complexes and dimers into active dimers by catalyzing a trans-to-cis isomerization of the D2-D3 linker invariant proline. A S252W gain-of-function mutation in FGFR2, responsible for Apert syndrome (35), was used as indirect evidence in support of this hypothesis. It was proposed that the S252W mutation results in gain of function by promoting a trans-to-cis isomerization of Pro253 (24), as aromatic amino acids preceding a proline have been shown to induce a high fraction of cis isomer in test peptides (29). However, we recently elucidated the crystal structure of S252W FGFR2c in complex with FGF2 and showed unequivocally that Pro253 remains in a trans conformation (6). Instead, we found that the S252W mutation results in receptor gain of function through the introduction of additional receptor-ligand contacts (6) that enhance 1-1 FGF-FGFR binding affinity (1, 7).
Defining the precise mechanism for FGFR dimerization is important not only for our understanding of FGFR function but also for our ability to manipulate FGFR signaling for therapeutic purposes. Hence, to resolve the uncertainty regarding the mode of FGFR dimerization, we studied the effects of abolishing the secondary ligand-receptor interaction site, observed only in the symmetric two-end model, using cellular proliferation, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF), and real-time binding experiments. The receptor and heparin binding affinities of secondary ligand-receptor interaction site FGF10 mutants are indistinguishable from those of wild-type FGF10. However, these mutations reduce the ability of FGF10 to induce receptor dimerization and signaling and provide direct biological evidence in support of a secondary ligand-receptor interaction site for FGFR signaling. We also examined the effect of a naturally occurring Pfeiffer syndrome mutation in FGFR2, Ala172 → Phe, which coincidentally maps to the receptor-receptor interaction site in the symmetric two-end model. Indeed, the crystal structure of A172F FGFR2b in complex with FGF10 demonstrates that this substitution results in receptor gain of function by increasing receptor-receptor contacts. Finally, the trans configuration of the D2-D3 linker invariant proline is validated by an FGF10 mutation that abolishes D3-FGF interactions that occur only in the symmetric two-end model. Taken together, our findings support both the mode of ligand-receptor binding and dimerization in the symmetric two-end model and are inconsistent with the asymmetric model for FGFR dimerization.
MATERIALS AND METHODS
Protein expression and purification.
The secondary ligand-receptor interface mutations (E158A, K195A, and E158A-K195A) and the Pfeiffer syndrome mutation (A172F) were introduced into full-length human FGF10 (residues 38 to 208) and the minimal ligand binding portion (D2-D3) of human FGFR2b (residues 140 to 369), respectively, using the Quick Change site-directed mutagenesis kit (Stratagene). Wild-type and mutant FGF10 proteins were expressed in Escherichia coli and purified by heparin affinity, cation exchange, and size exclusion chromatography, as previously described (39). Wild-type and A172F mutant FGFR2b were expressed in E. coli, refolded in vitro, and purified by heparin affinity and size exclusion chromatography (39).
Surface plasmon resonance analysis.
All interactions were characterized using a BIAcore (Uppsala, Sweden) 3000 instrument as previously described (8, 9, 40). To study the effects of FGF10 mutations (E158A, K195A, and E158A-K195A) or the pathogenic FGFR2 mutation (A172F) on 1-1 FGF10-FGFR2b binding, wild-type or mutant (E158A, K195A, and E158A-K195A) FGF10 was immobilized on research grade CM5 chips according to standard amine coupling protocol (Biacore AB) (8). FGF homologous factor 1b (FHF1b), formerly known as FGF12b, a structurally homologous protein that does not bind FGFR, was immobilized on control flow cells (8). Wild-type and mutant (E158A, K195A, and E158A-K195A) FGF10, as well as FHF1b, were all immobilized to similar levels (change in response units, ∼1,000). To obtain kinetic data for wild-type or mutant FGF10-FGFR2b interactions, different concentrations of analytes (wild-type and A172F FGFR2b) in HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 3 mM EDTA, 0.005% [vol/vol] polysorbate 20, pH 7.4) were injected at 50 μl/min over the sensor chip containing wild-type and mutant FGF10 ligands.
Data for FGF10-heparin and FGFR2b-heparin interactions were generated using a neoproteoglycan sensor chip, prepared by covalently immobilizing albumin-heparin conjugate (Sigma, St. Louis, Mo.) as previously described (9, 40). Albumin was immobilized on the control flow cell. To obtain kinetic data, different concentrations of analytes, wild-type and mutant (E158A, K195A, and E158A-K195A) FGF10 or wild-type and A172F FGFR2b, in HBS-EP buffer were injected over the heparin-albumin sensor chip at 50 μl/min.
Reference responses from control flow cells (FGF12 or albumin) were, respectively, subtracted from FGF10 (wild type and mutant) or albumin-heparin flow cells for each analyte injection using BiaEvaluation software (Biacore AB). The kinetic parameters were calculated by globally fitting the sensorgrams to a 1-1 interaction using BiaEvaluation software (Biacore AB). Disturbances at the beginning and end of each sensorgram were excluded. A minimum of four different analyte concentrations were used to determine the kinetic parameters for each interaction. Following curve fitting, each sensorgram was manually examined for the closeness of the model fit to the experimental data. χ2 was <10% of Rmax in all cases.
BALB/MK cellular assays.
DNA synthesis was measured by a [3H]thymidine incorporation assay using serum-starved confluent cultures of BALB/MK cells as previously described (10). To study the activation of MEK and MAP kinase, serum-starved BALB/MK cells were stimulated with different concentrations of wild-type or E158A-K195A mutant FGF10 plus or minus 0.3 μg of heparin (Sigma)/ml. Immunoblotting experiments were performed by lysing BALB/MK cells in 150 mM NaCl-50 mM Tris (pH 7.4)-1% Triton-10 μg of aprotinin/ml-2 μM phenylmethylsulfonyl fluoride-2 μM sodium orthovanadate, and the lysates were cleared of insoluble material by centrifugation at 14,000 rpm (Sorvall) for 20 min at 4°C. Proteins (100 μg) were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The membranes were blocked and probed with antibodies, including anti-MEK (Santa Cruz Biotechnology), anti-phospho-MEK, anti-MAPK, and anti-phospho-MAPK (all from Cell Signaling) and antitubulin (Sigma). The immunoreactivities of proteins were detected by chemiluminescence (Amersham).
MALDI-TOF mass spectrometry.
Protein samples (0.5 μl), containing 25 pmol of wild-type or mutant FGF10-FGFR2b complex in the presence or absence of 25 pmol of heparin dodecasaccharide, were mixed with 0.5 μl of matrix solution (10 mg of α-cyano-4-hydroxycinnamic acid/ml in 0.1% trifluoroacetic acid and 50% acetonitrile). The mixtures were allowed to dry and crystallize at room temperature on top of a preformed thin crystal layer of matrix alone (4). Positive ion mass spectra were acquired in linear mode using a Micromass TofSpec-2E MALDI-TOF mass spectrometer with time lag focusing. Ions were formed by irradiation with a 337-nm-wavelength N2 laser for a pulse time of 4 ns and then accelerated at a potential of 22.5 kV. A matrix suppression lens was set to suppress ions below m/z 5,000, and ions were detected using a high-mass detector. Two hundred laser shots were summed per spectrum. Data were acquired and processed using manufacturer-supplied MassLynx software.
Crystallization, data collection, structure determination, and refinement.
Crystals of the A172F FGFR2b-FGF10 complex were grown using the previously reported crystallization conditions (2 M ammonium sulfate) for the wild-type FGFR2b-FGF10 (39). A172F FGFR2b-FGF10 crystals belong to space group P6422 and are isomorphous to wild-type FGFR2-FGF10 crystals (39). The unit cell dimensions of the A172F FGFR2b-FGF10 crystals are as follows: a = b = 115.109 Å and c = 161.957 Å. The asymmetric unit contains a single A172F FGFR2b-FGF10 complex. A 2.8-Å data set was collected from a flash-frozen crystal (in a dry nitrogen stream) on a charge-coupled device detector at beamline X-4A at the National Synchrotron Light Source, Brookhaven National Laboratory. The data were processed using DENZO and SCALEPACK software (23).
Rigid body, positional and B-factor refinement, and simulated annealing were performed using CNS software (3). Tight noncrystallographic symmetry restraints were imposed throughout the refinement for the backbone atoms of FGF10, D2, and D3. Model building into 2Fo-Fc and Fo-Fc electron density maps was performed with program O (15). The final model consists of one FGF10 (residues 72 to 207), one FGFR2b (residues 151 to 359), two sulfate ions, and one PEG-400 molecule. The average B factor for all of the protein atoms is 64 Å2.
RESULTS AND DISCUSSION
Interactions at the crystallographic FGF10-FGFR2b dimer interface.
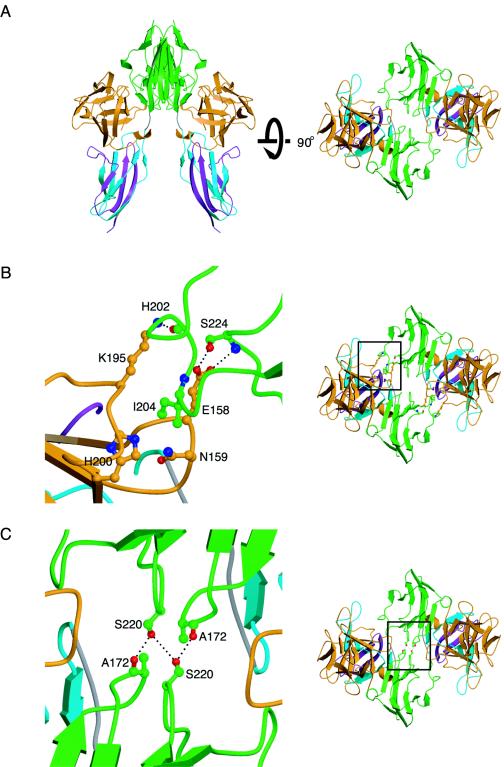
We recently reported the crystal structure of the FGF10-FGFR2b complex (PDB ID, 1NUN) (39). This crystal was grown in high ammonium sulfate and contains a twofold crystallographic canyon dimer (Fig. 2A) similar to the noncrystallographic FGF2-FGFR1c (PDB ID, 1CVS) (27) and the crystallographic FGF1-FGFR2c (PDB ID, 1DJS) (32) canyon dimers. The dimer interface consists of secondary ligand-receptor interactions and direct receptor-receptor contacts.
FIG. 2.
Crystallographic FGF10-FGFR2b dimer interface. (A) Ribbon diagram of the twofold crystallographic FGF10-FGFR2b canyon dimer in two views related by a 90° rotation about the horizontal axis. (B) Secondary interaction site at the dimeric interface. (C) Interactions at the receptor-receptor interface. D2, D3, and FGF are colored as in Fig. 1. The side chains of selected interacting residues are displayed. Oxygen atoms are colored red, nitrogen is blue, and carbon atoms are the same color as the molecules to which they belong. On the right are views of the whole structure in the exact orientation that the detailed views show, with the regions of interest boxed.
The secondary ligand-receptor interaction site, with a buried surface area of 500 Å2, consists of the β8-β9 and β11-β12 loop regions of FGF10 and the βE-βF and βC′-βD loops of D2 of FGFR2b (Fig. 2B). As in the case of the FGF2-FGFR1c and FGF1-FGFR2c dimeric structures, the majority of secondary ligand-receptor interactions are van der Waals contacts. A major van der Waals contact involves FGFR-invariant Ile204 (from the βC′-βD loop) of FGFR2b, which is surrounded by Glu158 and Asn159 (from the β8-β9 loop) and Lys195 and His200 (from the β11-β12 loop) of FGF10. Additionally, hydrogen bonds fortify this interface. In the FGF2-FGFR1c and FGF1-FGFR2c structures, backbone atoms of FGF ligand mediate the hydrogen bonds, whereas in the FGF10-FGFR2b structure, the side chains of FGF10 engage in hydrogen bonds with the receptor (Fig. 2B). Specifically, the side chain of Glu158 (from the β8-β9 loop of FGF10) engages in two hydrogen bonds with Ser224 (from the βE-βF loop) and the side chain of Lys195 (from the β11-β12 loop of FGF10) engages in one hydrogen bond with the backbone of His202 (Fig. 2B). The involvement of side chain atoms of FGF10 in the secondary ligand-receptor interaction site makes the FGF10-FGFR2b dimer ideal for studying the biological relevance of secondary ligand-receptor interactions observed in the symmetric two-end model.
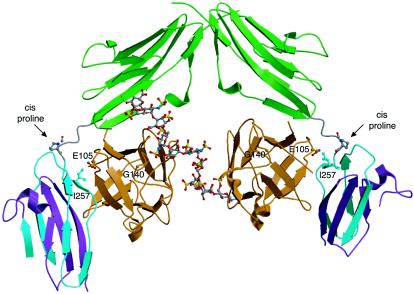
As mentioned above, there are no protein-protein interfaces that facilitate dimerization in the asymmetric model (PDB ID, 1E0O). We have highlighted the locations of Glu105 and Gly140 of FGF1, which correspond to Glu158 and Lys195 of FGF10, respectively, in the asymmetric model (Fig. 3). Due to the cis-configured D2-D3 linker invariant proline, Glu105 of FGF1 contributes to 1-1 FGF1-FGFR2c binding by making a van der Waals contact with Ile257 of D3 (Fig. 3). Gly140 of FGF1 does not interact with receptor or heparin (Fig. 3). The distinct roles of these two residues in each model provide an attractive opportunity to identify the correct model for FGFR dimerization. The symmetric two-end model predicts that the E158A and K195A mutations should diminish the ability of FGF10 to promote FGFR2b signaling due to a decrease in dimerization efficiency. However, the E158A and K195A mutations should not affect the 1-1 binding affinity of FGF10 for FGFR2b in the symmetric two-end model. The asymmetric model also predicts that the E158A mutation in FGF10 will diminish the ability of FGF10 to activate FGFR2b but, in contrast, will be due to diminished 1-1 FGF10-FGFR2b binding affinity. The asymmetric model predicts that the K195A mutation should have no effect on 1-1 FGF10-FGFR2b binding affinity or FGFR2b signaling.
FIG. 3.
Analysis of the FGF1-FGFR2c-heparin structure (PDB ID, 1E0O). Locations of FGF1 residues corresponding to Glu158 and Lys195 of FGF10 in 1E0O are shown. D2, D3, and FGF are colored as in Fig. 1. Oxygen atoms are colored red, nitrogen is blue, and carbon atoms are the same color as the molecules to which they belong. The location and isomerization state of the D2-D3 linker invariant proline are indicated.
E158A and K195A mutants and the E158A-K195A double mutant do not affect FGF10-FGFR2b or FGF10-heparin interactions.
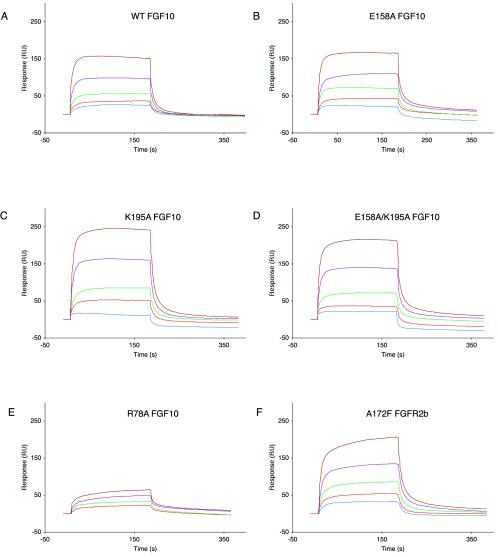
We mutated Glu158 and Lys195 of FGF10 to alanine individually and in combination. Wild-type and mutant (E158A, K195A, and E158A-K195A) FGF10 proteins were expressed to similar levels in E. coli and purified over heparin affinity, ion-exchange, and size exclusion chromatography. To measure 1-1 FGF10-FGFR2b binding, we performed our surface plasmon resonance (SPR) studies in the absence of heparin. All three FGF10 mutants bind FGFR2b as well as wild-type FGF10 does (Fig. 4A to D and Table 1). These data are consistent with the involvement of Glu158 and Lys195 in the secondary ligand-receptor interaction site in the symmetric two-end model. Importantly, the lack of effect of the E158A mutation on 1-1 FGF10-FGFR2b binding affinity is inconsistent with the observed mode of 1-1 FGF-FGFR binding in the asymmetric model. We also examined the binding of E158A, K195A, and E158A-K195A FGF10 mutants to heparin using SPR. The SPR data show that E158A, K195A, and E158A-K195A FGF10 mutants bind heparin with affinities that are similar to that of wild-type FGF10 to heparin (Fig. 5A to D and Table 2).
FIG. 4.
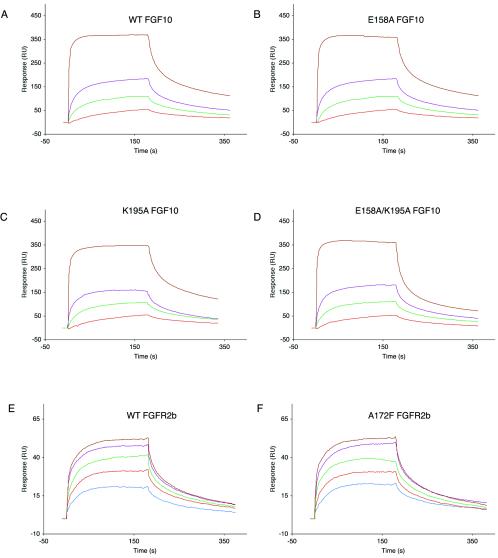
Surface plasmon resonance analysis of wild-type (WT) and mutant FGF10-FGFR2b interactions. Sensorgrams of FGFR2b binding to (A) wild-type FGF10, (B) E158A FGF10, (C) K195A FGF10, (D) E158A-K195A FGF10, and (E) R78A FGF10 are shown. In panel F, a sensorgram of A172F FGFR2b binding to wild-type FGF10 is shown. Analyte concentrations are colored as follows: 50 nM, blue; 100 nM, red; 200 nM, green; 400 nM, violet; and 800 nM, brown. Kinetic data are summarized in Table 1. RU, response units.
TABLE 1.
Summary of kinetic data for wild-type and mutant FGF10-FGFR2b interactions
| FGF10-FGFR2b | Parameter | Value |
|---|---|---|
| FGF10-FGFR2b | Kon (/M/s)a | 1.38 × 105 |
| Koff (/s)a | 8.58 × 10−2 | |
| KD (M)b | 6.23 × 10−7 | |
| E158A FGF10-FGFR2b | Kon (/M/s) | 1.25 × 105 |
| Koff (/s) | 7.57 × 10−2 | |
| KD (M) | 6.04 × 10−7 | |
| K195A FGF10-FGFR2b | Kon (/M/s) | 1.22 × 105 |
| Koff (/s) | 7.46 × 10−2 | |
| KD (M) | 6.13 × 10−7 | |
| E158A-K195A FGF10-FGFR2b | Kon (/M/s) | 1.43 × 105 |
| Koff (/s) | 8.69 × 10−2 | |
| KD (M) | 6.08 × 10−7 | |
| R78A FGF10-FGFR2b | Kon (/M/s) | 2.24 × 104 |
| Koff (/s) | 4.99 × 10−2 | |
| KD (M) | 2.23 × 10−6 | |
| A172F FGFR2b-FGF10 | Kon (/M/s) | 1.27 × 105 |
| Koff (/s) | 7.88 × 10−2 | |
| KD (M) | 6.22 × 10−7 |
Kon and Koff were derived as described in Materials and Methods. χ2 was <10% of Rmax in all cases.
The apparent affinity, KD, is equal to Koff/Kon.
FIG. 5.
Surface plasmon resonance analysis of wild-type (WT) and mutant FGF10 or FGFR2b interactions with heparin. Sensorgrams of heparin binding to (A) wild-type FGF10, (B) E158A FGF10, (C) K195A FGF10, and (D) E158A-K195A FGF10 are shown. In panels E and F, sensorgrams of heparin binding to wild-type FGFR2b and A172F FGFR2b, respectively, are shown. Analyte concentrations are colored as follows: 50 nM, blue; 100 nM, red; 200 nMm, green; 400 nM, violet; and 800 nM, brown. Kinetic data are summarized in Table 2. RU, response units.
TABLE 2.
Summary of kinetic data of wild-type and mutant FGF10-heparin and wild-type and mutant FGFR2b-heparin interactions
| Protein-heparin | Parameter | Value |
|---|---|---|
| FGF10 | Kon (/M/s)a | 6.33 × 104 |
| Koff (/s)a | 2.27 × 10−2 | |
| KD (M)b | 4.38 × 10−7 | |
| E158A FGF10 | Kon (/M/s) | 6.36 × 104 |
| Koff (/s) | 2.89 × 10−2 | |
| KD (M) | 4.55 × 10−7 | |
| K195A FGF10 | Kon (/M/s) | 5.13 × 104 |
| Koff (/s) | 2.52 × 10−2 | |
| KD (M) | 4.90 × 10−7 | |
| E158A/K195A FGF10 | Kon (/M/s) | 6.71 × 104 |
| Koff (/s) | 2.86 × 10−2 | |
| KD (M) | 4.22 × 10−7 | |
| FGFR2b | Kon (/M/s) | 6.85 × 104 |
| Koff (/s) | 1.58 × 10−2 | |
| KD (M) | 2.31 × 10−7 | |
| A172F FGFR2b | Kon (/M/s) | 6.67 × 104 |
| Koff (/s) | 1.65 × 10−2 | |
| KD (M) | 2.48 × 10−7 |
Kon and Koff were derived as described in Materials and Methods. χ2 was <10% of Rmax in all cases.
The apparent affinity, KD, is equal to Koff/Kon.
Mutagenesis of FGF10 confirms the importance of Glu158 and Lys195 for FGFR2b signaling.
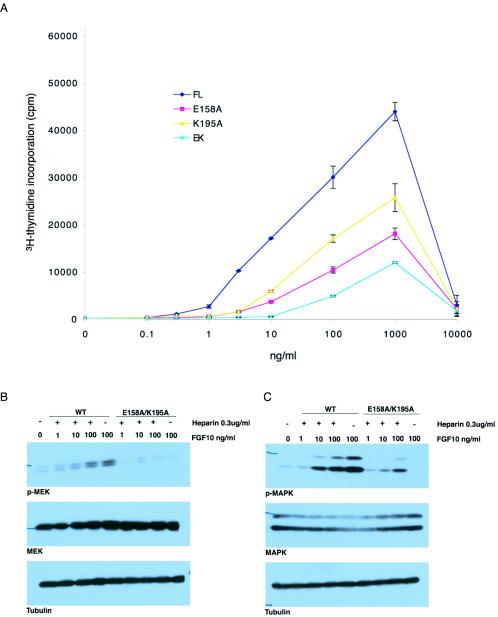
It is well known that FGFR dimerization is a prerequisite for the promotion of all FGF biological activity, including mitogenesis. Hence, to assess the effects of these mutations on receptor dimerization, we examined the mitogenic activities of wild-type and mutant FGF10 ligands on BALB/MK cells that naturally express FGFR2b. Consistent with the role of Glu158 and Lys195 residues in receptor dimerization in the symmetric two-end model, all three FGF10 mutants have reduced capacities to induce DNA synthesis relative to wild-type FGF10 (Fig. 6A). The greater effect of the E158A mutation than the K195A mutation is consistent with the number of hydrogen bonds Glu158 and Lys195 (two hydrogen bonds versus one) make in the secondary ligand-receptor interaction site (Fig. 2B). Accordingly, the E158A-K195A double mutant exhibits the lowest mitogenic activity and reflects the additive effects of the two mutations on FGFR activation. Next, we compared the ability of wild-type and E158A-K195A FGF10 to stimulate activation of both MEK and MAP kinases (Fig. 6B and C). Consistent with the mitogenic data, the E158A-K195A double mutation impairs the ability of FGF10 to activate MEK and MAP kinases (Fig. 6B and C) by 1 to 2 orders of magnitude. It is important to mention that these data rule out the possibility that the E158A and K195A mutations lead to diminished mitogenic activity by destabilizing the tertiary structure of FGF10. Mitogenic assays require extended (overnight) exposure of ligand at 37°C, while the phosphorylation studies were done with short (5-min) exposure times. Taken together with the SPR data, these data provide physiological evidence that these residues participate in secondary receptor-ligand interactions and are important for FGFR2b dimerization and signal transduction. Importantly, the diminished biological activities of E158A FGF10, in light of the SPR data, and K195A FGF10 are not consistent with the asymmetric model.
FIG. 6.
Structure-based mutagenesis of FGF10 confirms the importance of Glu158 and Lys195 for FGF10 biological activity. (A) Serum-starved BALB/MK cells were stimulated with increasing concentrations of wild-type FGF10 or E158A, K195A, or E158A-K195A FGF10 mutants plus 0.3 μg of heparin (Sigma)/ml. Sixteen hours later, [3H]thymidine was added for 6 h, and incorporation was determined as described previously (39). The error bars indicate standard deviations. FL, full length; EK, E158A-K195A. (B and C) Confluent BALB/MK cultures were incubated overnight in serum-free medium, followed by exposure to purified wild-type (WT) FGF10 or E158A-K195A FGF10 mutant at the indicated concentrations for 5 min at 37°C either in the presence (+) or absence (−) of 0.3 μg of heparin/ml. Around 100 μg of cell lysates was prepared as described in Materials and Methods, and immunoblot analysis was performed for (B) MEK and P-MEK and (C) MAPK and P-MAPK to assess the activation of these downstream components of the FGF10 pathway. Immunoblotting for tubulin was performed as a loading control.
MALDI-TOF analysis demonstrates that E158A and K195A mutations directly impair dimerization efficiency.
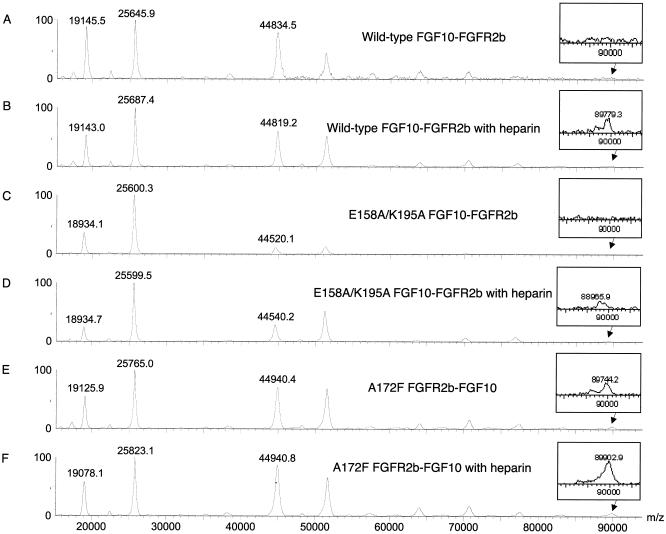
MALDI-TOF has recently emerged as a useful tool to study noncovalent protein-protein interactions (2, 13). Depending on the affinity of interaction, different amounts of protein complexes can survive the ionization process and fly together in the electromagnetic field of the mass spectrometer, thus allowing for semiquantitative assessment of complexes. We developed a semiquantitative MALDI-TOF method to study the dimerization of FGF10-FGFR2b complexes. MALDI-TOF analysis of wild-type FGF10-FGFR2b complex in the absence of heparin (Fig. 7A) shows major peaks for FGF10 (∼20 kDa) and FGFR2b (∼25 kDa) and also reveals a peak at a mass of ∼45 kDa, corresponding to a 1-1 monomeric FGF10-FGFR2b complex peak. The addition of one equivalent of heparin dodecasaccharide to wild-type FGF10-FGFR2b complex results in the appearance of a consistently reproducible peak at ∼90 kDa (Fig. 7B), which corresponds to a dimeric 2-2 FGF10-FGFR2b complex. Having established that MALDI-TOF is capable of detecting heparin-induced dimerization of FGF-FGFR complexes, we next studied the effects of E158A and K195A mutations on FGFR dimerization in the presence and absence of heparin (Fig. 7C and D). Analysis of the E158A-K195A FGF10-FGFR2b complex in the presence of one equivalent of heparin dodecasaccharide (Fig. 7D) reveals a significantly smaller dimer peak than observed in the wild-type FGF10-FGFR2b complex in the presence of heparin (Fig. 7B). Hence, these data provide direct evidence that Glu158 and Lys195 of FGF10 participate in FGF10-FGFR2b dimerization.
FIG. 7.
MALDI-TOF analysis of FGF10-FGFR2b dimerization. Twenty-five picomoles of 1-1 FGF10-FGFR2b complexes (wild type and mutant) with and without 25 pmol of heparin dodecasaccharide were analyzed by MALDI-TOF mass spectrometry in linear ion mode as described in Materials and Methods. (A) Wild-type FGF10-FGFR2b complex without heparin. (B) Wild-type FGF10-FGFR2b complex with one equivalent of heparin. (C) E158A-K195A FGF10-FGFR2b complex without heparin. (D) E158A-K195A FGF10-FGFR2b complex with one equivalent of heparin. (E) A172F FGFR2b-FGF10 complex without heparin. (F) A172F FGFR2b-FGF10 complex with one equivalent of heparin.
Structural and biophysical analysis of a pathogenic FGFR2 mutation supports the physiological significance of the symmetric two-end model.
In addition to the secondary ligand-receptor interaction site, dimerization in the symmetric two-end model is also facilitated by direct receptor-receptor contacts. In the FGF10-FGFR2b dimer, direct FGFR2b-FGFR2b contacts (buried surface area, 240 Å2) occur at the bottom of D2 and involve the βA′-βB and βE-βF loops of both monomers (Fig. 2C). Ala172 from one receptor engages the corresponding Ala from the other receptor in a hydrophobic interaction. Additionally, the side chain of Ser220 from one receptor hydrogen bonds with the corresponding Ser side chain and the backbone of Ala172 in the adjacent receptor (Fig. 2C).
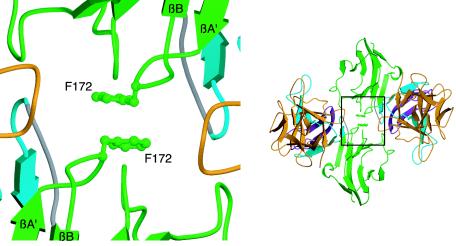
Gain-of-function mutations in FGFR2 are responsible for a variety of craniosynostosis syndromes (11, 19). Coincidentally, a novel pathogenic FGFR2 mutation, A172F, was recently detected in a kindred originally described by Pfeiffer in 1964 (16, 25) and maps to the receptor-receptor interface in the symmetric two-end model. Based on this model, the introduction of a Phe at residue 172 in FGFR2 should increase the magnitude of the hydrophobic interaction and further stabilize the dimer. To test this hypothesis, we crystallized the A172F FGFR2b-FGF10 complex. A172F FGFR2b-FGF10 complex crystals were grown under conditions similar to those for the wild-type FGFR2b-FGF10 complex and are isomorphous to wild-type FGFR2b-FGF10 crystals (PDB ID, 1NUN) (39). The overall conformation of the A172F FGFR2b-FGF10 complex is identical to that of the wild-type FGFR2b-FGF10 complex, and two A172F FGFR2b-FGF10 complexes form a symmetric canyon dimer. Data collection and refinement statistics are given in Table 3. Examination of the receptor-receptor interface reveals that Phe172 residues from both receptors engage in a hydrophobic interaction, with their aromatic benzyl groups optimally stacking against each other (Fig. 8). The closest approach between the two Phe172 molecules is 3.3 Å, with a total of 187 Å2 of surface area buried between the two residues, whereas in the wild-type FGFR2b-FGF10 structure, the closest approach between the two Ala172 molecules is 3.8 Å and only 76 Å2 of surface area is buried between the two alanine residues. Thus, the crystal structure provides evidence that the A172F mutation results in receptor gain of function by promoting receptor-receptor contacts, as observed in the symmetric two-end model. Consistent with the observed mode of gain of function, SPR analysis shows that the A172F mutation does not affect 1-1 FGF10-FGFR2b or FGFR2b-heparin binding (Fig. 4F and 5F and Tables 1 and 2).
TABLE 3.
Summary of crystallographic analysis
| Data collection statistics
|
Refinement statisticsc
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Resolution (Å) | Reflections (total/unique) | Completeness (%) | Rsyma (%) | Signal (〈I/σI〉) | Resolution (Å) | Reflections | Rcryst/Rfreed (%) | Root-mean-square deviations
|
||
| Bonds (Å) | Angles (degrees) | B factorse (Å2) | ||||||||
| 30-2.8 | 179,609/15,732 | 96.7 (100)b | 6.3 (26.8)b | 26.9 | 25-2.8 | 15,252 | 25.3/28.8 | 0.014 | 1.64 | 64 |
Rsym = 100 × ΣhklΣI|II(hkl) − 〈I(hkl)〉| ΣhklΣIII(hkl).
Value in parentheses is for the highest resolution shell: 2.9 to 2.8 Å.
Atomic model: 2,678 protein atoms, 1 PEG-400 molecule, and 2 sulfate ions.
Rcryst/free = 100 × hkl Fo(hkl)| |Fc(hkl)/hkl |Fo(hkl)|, where Fo (>0) and Fc are the observed and calculated structure factors, respectively. Ten percent of the reflections were used for calculation of Rfree.
For bonded protein atoms.
FIG. 8.
Gain-of-function interactions in the A172F FGFR2b-FGF10 dimer. Detailed view of the receptor-receptor interface. The side chains of interacting residues leading to increased receptor-receptor contacts are displayed. On the right is a view of the whole structure in the exact orientation that the detailed view shows, with the region of interest boxed. D2, D3, and FGF10 are colored as in Fig. 1.
To further confirm these structural findings, we performed a MALDI-TOF analysis of the A172F FGFR2b-FGF10 complex in the presence and absence of heparin (Fig. 7E and F). A significantly larger dimer complex peak is observed in this mutant sample with heparin (Fig. 7F) than in the wild-type FGFR2b-FGF10 complex in the presence of heparin (Fig. 7B), thus providing further evidence that the A172F mutation directly enhances receptor dimerization. Interestingly, a peak corresponding to a dimeric complex is present in the A172F FGFR2b-FGF10 sample in the absence of heparin (Fig. 7E), which suggests that the A172F mutation diminishes the requirement for heparin in dimerization of the FGFR2b-FGF10 complex. This is consistent with our structural findings, as the A172F mutation strengthens the dimerization interface, thereby lessening the need for heparin in dimerization. Hence, the mechanism by which the A172F mutation results in FGFR gain of function is entirely novel and consistent with the symmetric two-end model. Importantly, the asymmetric model cannot provide a mechanism for receptor gain of function by the A172F mutation, as Ala172 does not participate in any functional interactions in 1E0O.
SPR analysis of an FGF10 mutant reaffirms a trans configuration for the D2-D3 linker invariant proline.
In addition to differing in the mode of receptor dimerization, the asymmetric model differs from the symmetric two-end model in the mode of 1-1 FGF-FGFR binding. In the FGF10-FGFR2b crystal structure (PDB ID, 1NUN), as well as in all other FGF-FGFR crystal structures with the exception of 1E0O, the D2-D3 linker invariant proline is in a trans conformation. We have previously shown that Asp76, Arg78, and Arg155 of FGF10 make specific contacts with D3 within the primary ligand-receptor interface and that the D76A, R78A, and R155A mutations diminish the mitogenic activity of FGF10 (39). Using a model of FGF10-FGFR2b with the D2-D3 linker invariant proline in a cis configuration, we showed that the reorientation of D3 prevents an interaction between these residues and D3 (39). These data argue against a cis configuration for the D2-D3 linker invariant proline. To provide further evidence that the D2-D3 linker invariant proline adopts a trans configuration, we analyzed the binding of the R78A FGF10 mutant to FGFR2b using SPR (Fig. 4E and Table 1). The R78A mutation results in an ∼4-fold reduction in 1-1 FGF10-FGFR2b binding affinity due to the loss of several hydrogen bonds that occur within the primary ligand-receptor interface in the FGF10-FGFR2b complex (39). Taken together with the lack of effect of the E158A mutation on FGF10-FGFR2b binding affinity (Fig. 4B), these findings confirm that the D2-D3 linker invariant proline must adopt a trans configuration.
Ligand binding specificity can only be explained by a trans conformation of the D2-D3 linker invariant proline.
By comparing the crystal structures of FGF2-FGFR2c and FGF10-FGFR2b (26, 39), we have shown that contacts between FGF and the splice isoform-specific βC′-βE loop are the basis for each ligand binding preferentially to one splice isoform of FGFR2 (26, 39). These structural data argue against the asymmetric model, as the cis conformation of the D2-D3 linker invariant proline in 1E0O orients the βC′-βE loop away from FGF (Fig. 1B), and therefore, this loop does not play any role whatsoever in ligand binding in the asymmetric model. Furthermore, analysis of the effects of pathogenic FGFR mutations mapping to the βC′-βE loop of FGFR2c provides physiological evidence against the cis conformation of the D2-D3 linker invariant proline (7). We have shown that these βC′-βE loop pathogenic mutations differentially affect the binding of various FGFs and are even capable of altering the FGF binding specificity of FGFR2c (7). Interestingly, Harmer and coworkers recently realized that the D2-D3 linker invariant proline must assume a trans configuration, as they were unable to explain the unique specificity of the FGF19-FGFR4 complex using a FGFR4 model containing a cis-configured D2-D3 linker invariant proline (5). Instead, by using an FGFR4 model containing a trans-configured D2-D3 linker invariant proline, they also concluded that specific interactions between FGF19 and the βC′-βE loop of FGFR4 play a central role in determining the exquisite specificity of the FGF19-FGFR4 complex (5).
A canyon dimer also forms in the FGF1-FGFR2c-heparin crystal.
The FGF1-FGFR2c-heparin complex (PDB ID, 1E0O) reported by Pellegrini et al. (24) was also crystallized under high-sulfate-ion conditions (1 M lithium sulfate). Interestingly, examination of the crystal packing in 1E0O reveals that, in addition to the asymmetric dimer reported by Pellegrini et al. (24), the crystal lattice also contains a symmetric canyon-like dimer, which is stabilized by secondary ligand-receptor interactions, as well as direct receptor-receptor contact (Fig. 9). We note that in this symmetric canyon-like dimer (PDB ID, 1E0O), the D3 domains are coplanar rather than being perpendicular (as in other canyon dimers) to the plasma membrane due to the cis conformation of the D2-D3 linker invariant proline (Fig. 9). Moreover, a single heparin molecule binds into one half of the canyon, and ordered sulfate ions decorate the other half of the canyon. The presence of a single heparin molecule in this canyon is not unexpected and reflects the 2-2-1 FGF-FGFR-heparin stoichiometry used in the crystallization of the FGF1-FGFR2c-heparin complex. The formation of this symmetric canyon-like dimer, despite the incorrect conformation of the D2-D3 linker invariant proline, is remarkable and reiterates the physiological significance of the mode of FGFR dimerization in the symmetric two-end model. Paradoxically, the incorrect cis configuration of the D2-D3 linker invariant proline must have dissuaded Harmer and coworkers from considering the symmetric canyon-like dimer observed in their crystal.
FIG. 9.
A symmetric canyon-like dimer is observed in 1E0O. (A) GRASP (graphical representation and analysis or structural properties) representation of the canyon dimer in 1E0O. The two views are related by a 90° rotation about the horizontal axis. D2, D3, FGF1, and heparin are colored as in Fig. 1. Note that the membrane insertion points (indicated by arrows) at the end of D3 are coplanar to the plasma membrane, due to the incorrect conformation of the D2-D3 linker invariant proline. The distance between the membrane insertion points at the end of D3 is 84 Å in this representation. Only sulfate ions bound to the canyon are shown, and they are colored as follows: oxygens red, sulfurs yellow, and nitrogens blue. (B) For comparison, GRASP representation of the canyon dimer observed in the FGF2-FGFR1c-heparin structure (PDB ID, 1FQ9). The two views are related by a 90° rotation about the horizontal axis. The distance between the membrane insertion points (indicated by arrows) at the end of D3 is 48 Å in this representation. Coloring is as in panel A.
Concluding remarks.
Receptor dimerization is a universal mechanism for receptor tyrosine kinase activation. Recent crystallographic studies show that the mode of receptor dimerization varies greatly among receptor tyrosine kinase subfamilies and is tailored to meet their individual biological functions. The mode of receptor dimerization observed in the symmetric two-end model is highly cooperative and well suited to execute the essential roles of FGF signaling in development. This model is based on synergistic binding events, including secondary ligand-receptor and receptor-receptor interactions, which are facilitated by HSPGs. Thus, this mode of dimerization is capable of sensing and responding to the dynamic changes in HSPGs that occur during development. Future challenges include structural and biochemical studies of how these changes modulate dimer assembly and will provide greater insight into the role of FGF signaling in physiological and pathophysiological settings.
Acknowledgments
We thank C. Ogata and R. Abramowitz for synchrotron beamline assistance and T. Neubert, Y. Lu, and the NYU Protein Analysis Facility for assistance with the MALDI-TOF analysis. We are grateful to S. Hubbard and C. Basilico for comments and helpful discussions.
Beamline X4A at the National Synchrotron Light Source, a DOE facility, is supported by the Howard Hughes Medical Institute. The NYU Protein Analysis Facility is supported by NIH shared instrumentation grant RR14662. This work was funded by NIH grants DE13686 (to M.M.), HL52622 (to R.J.L.), and CA71672 (to S.A.A.).
REFERENCES
- 1.Anderson, J., H. D. Burns, P. Enriquez-Harris, A. O. Wilkie, and J. K. Heath. 1998. Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Hum. Mol. Genet. 7:1475-1483. [DOI] [PubMed] [Google Scholar]
- 2.Borchers, C., and K. B. Tomer. 1999. Characterization of the noncovalent complex of human immunodeficiency virus glycoprotein 120 with its cellular receptor CD4 by matrix-assisted laser desorption/ionization mass spectrometry. Biochemistry 38:11734-11740. [DOI] [PubMed] [Google Scholar]
- 3.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Cadene, M., and B. T. Chait. 2000. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal. Chem. 72:5655-5658. [DOI] [PubMed] [Google Scholar]
- 5.Harmer, N. J., L. Pellegrini, D. Chirgadze, J. Fernandez-Recio, and T. L. Blundell. 2004. The crystal structure of fibroblast growth factor (FGF) 19 reveals novel features of the FGF family and offers a structural basis for its unusual receptor affinity. Biochemistry 43:629-640. [DOI] [PubMed] [Google Scholar]
- 6.Ibrahimi, O. A., A. V. Eliseenkova, A. N. Plotnikov, K. Yu, D. M. Ornitz, and M. Mohammadi. 2001. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc. Natl. Acad. Sci. USA 98:7182-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahimi, O. A., F. Zhang, A. V. Eliseenkova, N. Itoh, R. J. Linhardt, and M. Mohammadi. 2004. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum. Mol. Genet. 13:2313-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahimi, O. A., F. Zhang, A. V. Eliseenkova, R. J. Linhardt, and M. Mohammadi. 2004. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum. Mol. Genet. 13:69-78. [DOI] [PubMed] [Google Scholar]
- 9.Ibrahimi, O. A., F. Zhang, S. C. Lang Hrstka, M. Mohammadi, and R. J. Linhardt. 2004. Kinetic model for FGF, FGFR, and proteoglycan signal transduction complex assembly. Biochemistry 43:4724-4730. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi, M., P. W. Finch, and S. A. Aaronson. 1998. Characterization of recombinant human fibroblast growth factor (FGF)-10 reveals functional similarities with keratinocyte growth factor (FGF-7). J. Biol. Chem. 273:13230-13235. [DOI] [PubMed] [Google Scholar]
- 11.Jabs, E. W. 2002. Genetic etiologies of craniosynostosis, p. 125-146. In M. K. Mooney and M. I. Siegel (ed.), Understanding craniofacial anomalies: the etiopathogenesis of craniosynostoses and facial clefting. Wiley-Liss, New York, N.Y.
- 12.Jaye, M., J. Schlessinger, and C. A. Dionne. 1992. Fibroblast growth factor receptor tyrosine kinases: molecular analysis and signal transduction. Biochim. Biophys. Acta 1135:185-199. [DOI] [PubMed] [Google Scholar]
- 13.Jespersen, S., W. M. Niessen, U. R. Tjaden, and J. van der Greef. 1998. Basic matrices in the analysis of non-covalent complexes by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 33:1088-1093. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. E., L. T. Williams, A. Gritli-Linde, P. Lewis, A. P. McMahon, and A. Linde. 1993. Structural and functional diversity in the FGF receptor multigene family. Adv. Cancer Res. 60:1-41. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. A., J. Y. Zou, S. W. Cowan, and G. Kjeldgaard. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 16.Kan, S. H., N. Elanko, D. Johnson, L. Cornejo-Roldan, J. Cook, E. W. Reich, S. Tomkins, A. Verloes, S. R. Twigg, S. Rannan-Eliya, D. M. McDonald-McGinn, E. H. Zackai, S. A. Wall, M. Muenke, and A. O. Wilkie. 2002. Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am. J. Hum. Genet. 70:472-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, G. R. 1998. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 12:1571-1586. [DOI] [PubMed] [Google Scholar]
- 18.Miki, T., D. P. Bottaro, T. P. Fleming, C. L. Smith, W. H. Burgess, A. M. Chan, and S. A. Aaronson. 1992. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA 89:246-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muenke, M., and A. O. Wilkie. 2001. Craniosynostosis syndromes, p. 6117-6146. In C. R. Scriver, A. L. Beaudet, D. Valle, W. S. Sly, B. Childs, K. Kinzler, and B. Vogelstein (ed.), The metabolic and molecular bases of inherited disease, vol. IV. McGraw-Hill, New York, N.Y. [Google Scholar]
- 20.Olsen, S. K., O. A. Ibrahimi, A. Raucci, F. Zhang, A. V. Eliseenkova, A. Yayon, C. Basilico, R. J. Linhardt, J. Schlessinger, and M. Mohammadi. 2004. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. USA 101:935-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ornitz, D. M., and N. Itoh. 2001. Fibroblast growth factors. Genome Biol. 2:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ornitz, D. M., A. Yayon, J. G. Flanagan, C. M. Svahn, E. Levi, and P. Leder. 1992. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell. Biol. 12:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 24.Pellegrini, L., D. F. Burke, F. von Delft, B. Mulloy, and T. L. Blundell. 2000. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407:1029-1034. [DOI] [PubMed] [Google Scholar]
- 25.Pfeiffer, R. A. 1964. Dominant erbliche Akrocephalosyndaktylie. Z. Kinderheilk. 90:301. [PubMed] [Google Scholar]
- 26.Plotnikov, A. N., S. R. Hubbard, J. Schlessinger, and M. Mohammadi. 2000. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101:413-424. [DOI] [PubMed] [Google Scholar]
- 27.Plotnikov, A. N., J. Schlessinger, S. R. Hubbard, and M. Mohammadi. 1999. Structural basis for FGF receptor dimerization and activation. Cell 98:641-650. [DOI] [PubMed] [Google Scholar]
- 28.Rapraeger, A. C., A. Krufka, and B. B. Olwin. 1991. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252:1705-1708. [DOI] [PubMed] [Google Scholar]
- 29.Reimer, U., G. Scherer, M. Drewello, S. Kruber, M. Schutkowski, and G. Fischer. 1998. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J. Mol. Biol. 279:449-460. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger, J., A. N. Plotnikov, O. A. Ibrahimi, A. V. Eliseenkova, B. K. Yeh, A. Yayon, R. J. Linhardt, and M. Mohammadi. 2000. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6:743-750. [DOI] [PubMed] [Google Scholar]
- 31.Shimada, T., S. Mizutani, T. Muto, T. Yoneya, R. Hino, S. Takeda, Y. Takeuchi, T. Fujita, S. Fukumoto, and T. Yamashita. 2001. Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl. Acad. Sci. USA 98:6500-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauber, D. J., A. D. DiGabriele, and W. A. Hendrickson. 2000. Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl. Acad. Sci. USA 97:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, F., M. Kan, G. Yan, J. Xu, and W. L. McKeehan. 1995. Alternately spliced NH2-terminal immunoglobulin-like loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 270:10231-10235. [DOI] [PubMed] [Google Scholar]
- 34.White, K. E., K. B. Jonsson, G. Carn, G. Hampson, T. D. Spector, M. Mannstadt, B. Lorenz-Depiereux, A. Miyauchi, I. M. Yang, O. Ljunggren, T. Meitinger, T. M. Strom, H. Juppner, and M. J. Econs. 2001. The autosomal dominant hypophosphatemic rickets (ADHR) gene is a secreted polypeptide overexpressed by tumors that cause phosphate wasting. J. Clin. Endocrinol. Metab. 86:497-500. [DOI] [PubMed] [Google Scholar]
- 35.Wilkie, A. O., S. F. Slaney, M. Oldridge, M. D. Poole, G. J. Ashworth, A. D. Hockley, R. D. Hayward, D. J. David, L. J. Pulleyn, P. Rutland, et al. 1995. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet. 9:165-172. [DOI] [PubMed] [Google Scholar]
- 36.Wilkin, D. J., J. T. Hecht, and C. A. Francomano. 2001. Achondroplasia and pseudoachondroplasia, p. 5379-5395. In C. R. Scriver, A. L. Beaudet, D. Valle, W. S. Sly, B. Childs, K. Kinzler, and B. Vogelstein (ed.), The metabolic and molecular bases of inherited disease, vol. IV. McGraw-Hill, New York, N.Y. [Google Scholar]
- 37.Yayon, A., M. Klagsbrun, J. D. Esko, P. Leder, and D. M. Ornitz. 1991. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64:841-848. [DOI] [PubMed] [Google Scholar]
- 38.Yayon, A., Y. Zimmer, G. H. Shen, A. Avivi, Y. Yarden, and D. Givol. 1992. A confined variable region confers ligand specificity on fibroblast growth factor receptors: implications for the origin of the immunoglobulin fold. EMBO J. 11:1885-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh, B. K., M. Igarashi, A. V. Eliseenkova, A. N. Plotnikov, I. Sher, D. Ron, S. A. Aaronson, and M. Mohammadi. 2003. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc. Natl. Acad. Sci. USA 100:2266-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, F., M. Fath, R. Marks, and R. J. Linhardt. 2002. A highly stable covalent conjugated heparin biochip for heparin-protein interaction studies. Anal. Biochem. 304:271-273. [DOI] [PubMed] [Google Scholar]