Abstract
The nucleocytoplasmic transport receptor CRM1 mediates the export of macromolecules from the nucleus to the cytoplasm by forming a ternary complex with a cargo molecule and RanGTP. The in vivo mechanism of CRM1 export complex formation and its mobility throughout the nucleus have not been fully elucidated. More information is required to fully understand complex formation and the dynamics of CRM1-cargo-RanGTP complexes in space and time. We demonstrate true molecular interaction of CRM1 with its Rev cargo in living cells by using fluorescence resonance energy transfer (FRET). Interestingly, we found that the inhibitory effect of leptomycin B on this CRM1-cargo interaction is Ran dependent. Using fluorescence recovery after photobleaching (FRAP), we show that CRM1 moves at rates similar to that of free green fluorescent protein in the nucleoplasm. A slower mobility was detected on the nuclear membrane, consistent with known CRM1 interactions with nuclear pores. Based on these data, we propose an in vivo model in which CRM1 roams through the nucleus in search of high-affinity binding sites. CRM1 is able to bind Rev cargo in the nucleolus, and upon RanGTP binding a functional export complex is produced that is exported to the cytoplasm.
Molecular transport between the nucleus and cytoplasm through the nuclear pore complexes (NPCs) is mediated by soluble transport receptors, called karyopherins. Although possessing only limited sequence similarity, they have the common property of binding to proteins of the nuclear pore and to the small GTPase Ran. The first nuclear export pathway to be discovered involved the CRM1 receptor exporting proteins containing a hydrophobic nuclear export signal (NES) (16, 18, 43, 52). crm1 was originally discovered in Saccharomyces cerevisiae as a gene involved in chromosomal region maintenance (1). Evidence that CRM1 is a transport receptor came from inhibition studies with the antifungal drug leptomycin B (LMB). Because LMB was found to inhibit nuclear export of the human immunodeficiency virus (HIV) Rev protein, CRM1 was suggested to be the nuclear export receptor that exports Rev from the nucleus (57). Rev is required for transport of viral mRNA to the cytoplasm (13, 36), a process essential for virus replication (54). Rev contains an NES, a stretch of characteristically spaced hydrophobic amino acids (leucine) that is essential for its nuclear export (14, 38). Similar NESs were also found in other proteins (49, 56).
The driving force for nucleocytoplasmic transport is the RanGTP gradient over the nuclear membrane (19, 25, 35, 37, 46). The guanine nucleotide exchange factor for Ran (RanGEF) is confined within the nucleus (6), whereas the RanGTPase-activating protein (RanGAP) is located in the cytoplasm. RanGEF converts RanGDP to RanGTP, and RanGAP activates the RanGTPase to hydrolyze GTP to GDP. The compartmentalization of these factors creates a gradient of RanGTP over the nuclear membrane resulting in high concentrations of RanGTP in the nucleus and high levels of RanGDP in the cytosol. RanGTP promotes the association of cargo with nuclear export receptors, while RanGDP encourages the dissociation of the export complex. Therefore, within the nucleus the export protein, CRM1, binds export cargo proteins and RanGTP to form a functional export complex (16, 18, 26, 41, 43, 52). CRM1 has also been found to interact with the nuclear pore complex, namely nucleoporins Nup214 (17), Nup50 (22), Nup42, and Nup159 (15). However, the mechanism of how export complex finds the nuclear pores is still unknown. Subsequently, CRM1-cargo-RanGTP export complex translocates through the NPC and docks on the nucleoporin Nup214 until it is released and disassembled by RanGAP and RanBP1 or RanBP2 (2, 21, 27, 37, 42).
Previous in vitro studies have shown interaction of CRM1 with NES peptides (16, 18, 41, 43, 52). Residues Asp716 and Lys810 of CRM1 are important for NES binding (3), whereas, based on alignment studies with other Ran-binding proteins, RanGTP binding is expected to be mediated by a region near the N terminus of CRM1 (17, 20). Indeed, a region between residues 61 and 160 was shown to be essential for the interaction of RanGTP with CRM1 (44). It is clear that Ran is required for functional transport of the export complex (3, 16, 18, 43); however, the actual in vivo details of the CRM1-cargo-Ran complex formation and the kinetics behind CRM1-mediated nucleocytoplasmic transport are still largely unknown. A detailed understanding of the basic and important process of macromolecular transport from nucleus to the cytoplasm requires the understanding of timing and location of the different interactions inside living cells. Therefore, we examined the molecular mechanism of CRM1 transport complex formation and its kinetics inside living cells. We used noninvasive microscopy techniques (fluorescence resonance energy transfer [FRET] and fluorescence recovery after photobleaching [FRAP]) to demonstrate true molecular interaction between CRM1 and cargo in living cells. The HIV Rev protein was used as model cargo for the binding studies. Studying the effect of LMB on CRM1 mutants, we found that Ran binding is required for LMB to be able to disrupt the CRM1-cargo interaction, suggesting that Ran plays an essential role in LMB action. Moreover, it is not known how the functional export complex locates the nuclear pores. We found, using FRAP, that CRM1 travels in an unimpeded manner throughout the nucleoplasm. We propose a model in which CRM1 roams through the nucleus in search of high-affinity binding sites.
MATERIALS AND METHODS
Cell culture, transfections, and plasmids.
HeLa cells were plated onto glass-bottom microwell dishes (MatTek Corporation, Ashland, Mass.) and cultured in complete Dulbecco's modified Eagle's medium (DMEM) (Mediatech Inc., Herndon, Va.). Cells were transfected using SuperFect (QIAGEN, Valencia, Calif.) transfection reagent. Before imaging, cells were washed and complete medium without phenol red was added. In some cases, leptomycin B (57) was added at 50 nM for 2 to 3 h before imaging.
pBRev-green fluorescent protein (GFP) and pCRM1-GFP plasmids produce fusion proteins of HIV-1 Rev and human CRM1, respectively, fused to GFP. pBRev-blue fluorescent protein (BFP) expresses the Rev protein fused to BFP (53). pRev-yellow fluorescent protein (YFP) generates Rev protein with YFP at its N terminus. pCFP-Tat produces Tat protein with an N-terminal fusion of cyan fluorescent protein (CFP). pCFP-CRM1 produces a CFP-CRM1 fusion protein with CFP at the N terminus. pCFP-160-819CRM1 produces a fusion protein containing amino acids 160 to 819 of CRM1 fused to the C terminus of CFP. p160-819CRM1-GFP and p160-566CRM1-GFP produce fusion proteins containing amino acids 160 to 819 and 160 to 566 of CRM1, respectively, fused to the N terminus of GFP.
Live-cell imaging.
Images of live cells were acquired with a laser scanning confocal microscope (LSM 510; Carl Zeiss Inc., Thornwood, N.Y.) equipped with an Axiovert 200 microscope (Zeiss) and a 40× 1.3-numeric-aperture oil immersion Plan-Neofluar objective. For colocalization of BFP and GFP fusion proteins, BFP was excited with a multiphoton (Verdi/Mira 900; Coherent Inc., Auburn, Calif.) laser line at 780 nm and GFP was excited with an Argon laser line at 488 nm, and emissions were collected with band pass filters of 390 to 465 nm and 500 to 550 nm, respectively. Potential bleed-through was measured by acquiring dual-channel images of cells with single labels with the same setup used for cells coexpressing both labels. No bleed-through was detected.
Fluorescence resonance energy transfer.
CFP was excited with an Argon laser line at 458 nm and was detected by using a band pass filter of 480 to 520 nm. YFP was excited with an Argon laser line at 514 nm, and emission was collected with a 565- to 615-nm band pass filter. FRET was determined by the acceptor photobleaching method (4, 5, 28, 29, 58). First, prephotobleach CFP (donor) and YFP (acceptor) images were acquired. A region of interest (ROI) in the nucleolus was rendered free of YFP by repeated scanning with the 514-nm laser line until all YFP was photodestructed. A second postphotobleach CFP and YFP image was acquired. After correction for background and for the photobleaching of the donor due to imaging, the FRET efficiencies (E) in the ROI were calculated from the two CFP images by using the formula: E = 1 − D0/D1 (23, 28), where D is the mean intensity of the donor in the area where the acceptor was bleached before (D0) and after (D1) acceptor bleaching.
The image and statistical analyses were performed with Matlab (Mathworks, Inc., Natick, Mass.) and DIPimage (image processing toolbox for Matlab; Delft University of Technology, The Netherlands).
Fluorescence loss in photobleaching and cell viability.
Before and then after every 31 s of bleaching, an image was collected of a rectangle of 3 by 10 μm in the cytoplasm with the 488-nm laser line at the same power in each experiment (monitor diode). The depletion of CRM1-GFP from the nucleoli was quantified by measuring the relative fluorescence intensity. The dynamics follow an exponential decay, and the fluorescence could be reduced to an undetectable level in all nucleoli of the tested cell. The FLIP rates for all GFP fusion proteins were obtained under similar conditions, including cell size, expression level, and the area of bleaching. The loss in fluorescence in the nucleoli of the cells was quantified, and the decays were fitted by the nonlinear least-squares Gauss-Newton method. Statistical analysis was performed with Matlab (Mathworks, Inc.). For each experiment, the average half-life (t1/2) was calculated from the fitted decay curve of each cell. The standard Student's t test was used to determine the statistical significance of the results.
To assess that cells remain viable after the photobleaching period, we monitored cells by differential interference contrast (DIC) optics for changes in cellular morphology. No dramatic changes in cellular morphology were detected at up to 800 s of imaging. Therefore, our experimental conditions did not influence the collection of FRAP or FLIP data.
Fluorescence recovery after photobleaching.
Excitation of GFP was done at 488 nm, and detection was between 500 and 550 nm. The microwell dishes with coverslip bottoms were directly mounted onto an LSM 510 microscope. Live cells were imaged at 37°C (Heating Chamber; 20/20 Technology Inc., Wilmington, N.C.) in DMEM without phenol red and supplemented with 10% (vol/vol) fetal calf serum (FCS). Qualitative FRAP experiments were performed as follows: a 2-μm-wide strip throughout the cell was photobleached; bleaching was completed in 200 to 600 ms, and recovery images were acquired every 0.4 to 1 s. Quantitative FRAP experiments typically required faster acquisition rates and were therefore performed differently: a smaller field of view was scanned in the nucleus (∼20 times fewer lines), leading to a faster acquisition rate (every ∼30 ms). Three full fields of view were prescanned, followed by bleaching of a 1-μm-diameter spot with the 488-nm laser line for 100 to 200 ms at full power to create a local photobleached region. Note that the photobleaching time was optimized on a few test cells so that a minimum time was established to reach the saturated photobleached region. Quantitative FRAP was necessary to accurately sample the recoveries of the rapid movements of free GFP and CRM1-GFP in the nucleoplasm and the nuclear membrane. Because movement of CRM1-GFP in the nucleolus was much slower, recovery points were collected only every 3 s; however, the same photobleach procedure was performed. Note also that in all experiments, the laser intensity was set low enough to minimize the loss of fluorescence in the full field of view, leading to very low measurable loss during the monitoring period. The fluorescence intensity in the bleached spot of the first image collected after photobleaching was measured, and this value was used as the baseline (i.e., all intensities in the recovery curve were subtracted from this first value). The recovery curve was then normalized so that the final value converged towards 1. For this normalization, the last five time points were used to approximate the asymptotic final recovery value. The recoveries were fitted by an exponential function using the nonlinear least-squares Gauss-Newton method. Statistical analyses were performed with Matlab (Mathworks, Inc.). For each experiment, the average t1/2 and its standard deviation was calculated from the t1/2 of the individual fitted recovery curve of each cell. The standard Student's t test was used to determine the statistical significance of the results.
RESULTS
Subcellular localization and colocalization of CRM1 with Rev in living cells.
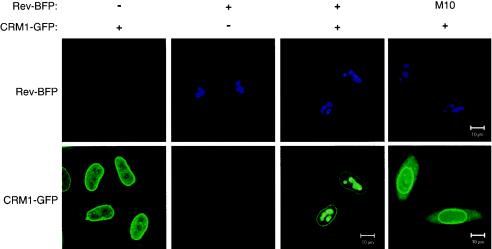
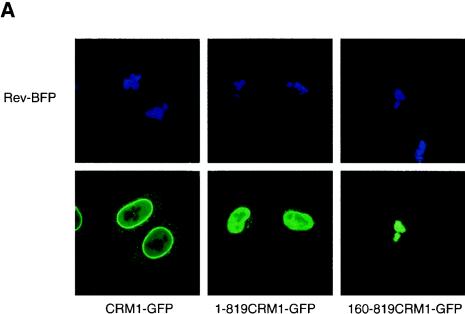
It has been shown biochemically that CRM1 interacts with leucine-rich NESs of cargo proteins (3, 16, 18, 41, 43, 45, 52, 55, 60). To demonstrate this interaction inside living mammalian cells, we studied the ability of Rev to redistribute CRM1 to the nucleoli. Therefore, we produced fusion constructs of the two proteins with either the blue fluorescent protein or the green fluorescent protein. This fluorescent pair has excitation and emission spectra that can be easily separated for detection with fluorescence microscopy. CRM1-GFP localized predominantly at the nuclear membrane as well as within the nucleus (Fig. 1, left bottom panel), in agreement with published data (17). Its predominant association with the membrane is consistent with its interaction with nucleoporins Nup214 (17), Nup50 (22), Nup42, and Nup159 of the nuclear pores (15). Rev-BFP and RevM10-BFP were found in the nucleoli (Fig. 1, top panels). RevM10 is a transdominant-negative mutant of Rev that has been mutated in its NES and is unable to interact with CRM1. When Rev-BFP and CRM1-GFP were coexpressed, a significant fraction of CRM1-GFP was found in the Rev-containing nucleoli (Fig. 1) (9, 61). Such colocalization of CRM1 with Rev in the nucleolus of living cells suggests interaction between the two proteins. In contrast, CRM1-GFP did not colocalize with RevM10-BFP, suggesting the Rev-CRM1 interaction is NES-specific.
FIG. 1.
Colocalization of wild-type CRM1-GFP with its Rev cargo in the nucleolus of living cells. Cells expressing Rev-BFP, RevM10-BFP, and/or CRM1-GFP as indicated were analyzed by confocal fluorescence microscopy. The top panels show nucleolar localization of Rev-BFP and RevM10-BFP. The lower panels show localization of CRM1-GFP.
CRM1-GFP interacts with high-affinity sites in the nucleolus of Rev-expressing cells.
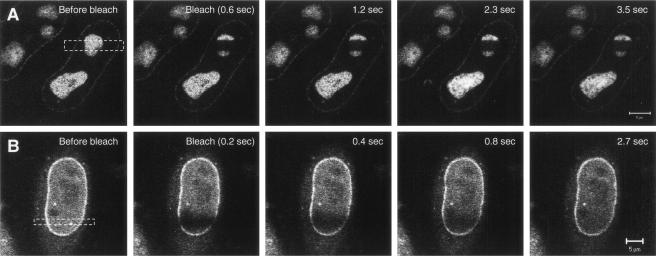
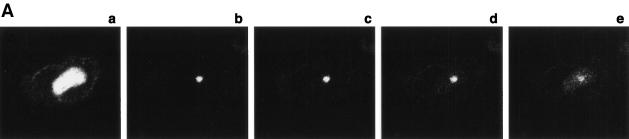
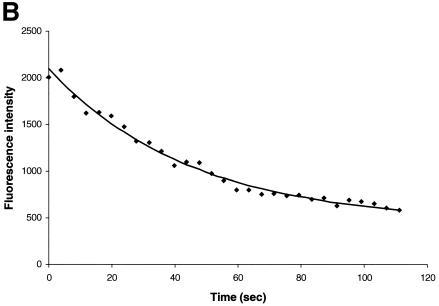
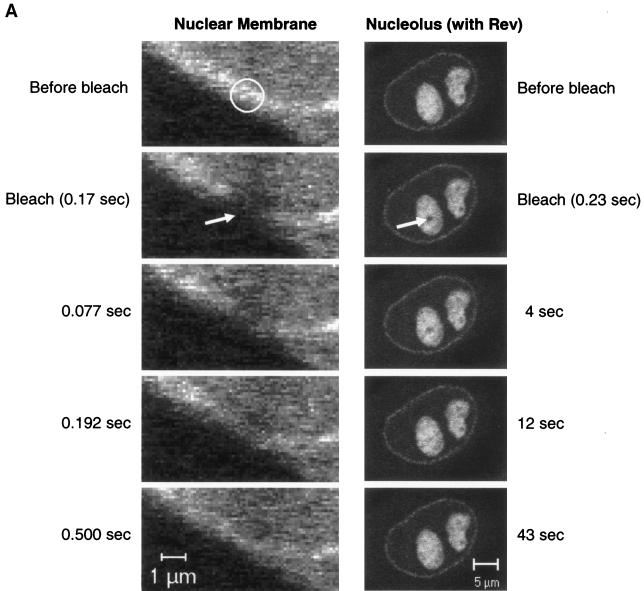
To further evaluate whether CRM1-GFP interacts with Rev-BFP in the nucleolus, we compared the mobility of CRM1-GFP in the nucleolus versus its mobility in the nucleoplasm by using FRAP-based experiments. In these experiments, a small area of a cell is rapidly photobleached by a high-intensity laser pulse. The movement of unbleached molecules from the neighboring areas into the photobleached region is then recorded by time-lapse microscopy as the recovery of fluorescence in the photobleached area (12, 34, 47, 48). The CRM1-GFP in cells expressing Rev-BFP was photobleached in a small 2-μm strip extending through the nucleoplasm and nucleolus of cells (Fig. 2A). For comparison, a similar area extending through the nucleoplasm of cells expressing only CRM1-GFP was photobleached (Fig. 2B). The CRM1 GFP fluorescence recovery in the different subcellular compartments was compared. CRM1-GFP in the nucleoplasm and nuclear membrane recovered rapidly and were uniformly followed by a slower recovery in the nucleolus. The nucleoplasm and nuclear membrane were fully recovered within 2.7 s, while the nucleolus recovered much slower, indicating that the mobility of CRM1-GFP in the nucleolus (with Rev) is very slow compared to that of nucleoplasm. The slower mobility of CRM1-GFP in the nucleolus in the presence of Rev indicates that CRM1 is in a different state than the nucleoplasm. Intracellular mobility is influenced by (i) specific and nonspecific interactions, (ii) catalytic activity, and (iii) diffusion. Therefore, the FRAP results reflect the sum of these three activities. The slower mobility of CRM1 in the presence of Rev is most likely due to high-affinity association with Rev in the nucleolus. This was also evident from the inverse experiment, termed iFRAP (10). In iFRAP, the entire cell nucleus with the exception of a small region of interest is photobleached. When iFRAP was applied on cells expressing Rev-BFP and CRM1-GFP, the GFP fluorescence signal in the unbleached region decreased very slowly (Fig. 3A), suggesting that mobility of CRM1-GFP in the nucleolus is very slow. A quantification of the fluorescence in the unbleached spot is given in Fig. 3B. When the same experiment was performed on the nucleoplasm of cells expressing CRM1-GFP, no fluorescence could be detected in the unbleached spot after photobleaching the nucleus. This is because CRM1-GFP is highly mobile in the nucleoplasm, as discussed above. During the time of photobleaching (3 s), all molecules in the unbleached spot move into the bleach area. When the same experiment was performed on the nucleus of fixed cells, a small spot of CRM1-GFP in the nucleoplasm could be observed (data not shown). Consistent with the FRAP results, these kinetics suggest that CRM1-GFP is highly mobile in the nucleoplasm and is associated with Rev in the nucleolus.
FIG. 2.
Fluorescence recovery after photobleaching of CRM1-GFP in living cells. HeLa cells coexpressing Rev-BFP and CRM1-GFP (A) or expressing only CRM1-GFP (B) were imaged by confocal fluorescence microscopy. A 2-μm line was bleached across the width of the cell nucleus as indicated, and images were gathered during the course of recovery.
FIG. 3.
iFRAP of CRM1-GFP. (A) Cells coexpressing Rev-BFP and CRM1-GFP were imaged for GFP before and after photobleaching of CRM1-GFP in the entire nucleus, with the exception of a small spot in the nucleolus. The loss of fluorescence in the unbleached spot was monitored by time-lapse confocal microscopy. Panels: a, before bleach; b, bleach; c, 3 s; d, 12 s; e, 50 s. (B) Quantitation of the iFRAP kinetics.
Direct interaction of CRM1 with Rev cargo in living cells as measured by FRET.
To demonstrate a direct molecular interaction between CRM1 and cargo in living cells, we monitored fluorescence resonance energy transfer between CFP-tagged CRM1 and YFP-tagged Rev (Fig. 4). Because of its high specificity, FRET has proven useful to investigate molecular interactions and conformational changes, even on a single-molecule scale (50). The principle of FRET is based on the ability of a higher energy fluorophore molecule (donor [CFP]) to transfer energy directly to a lower energy molecule (acceptor [YFP]) with simultaneous quenching of the donor fluorescence. Consequently, one demonstration of FRET is an increase in donor fluorescence after photobleaching the acceptor (4, 5, 28, 29, 58). In most cases, no FRET can be observed at distances greater than 10 nm. Therefore, FRET between CFP-CRM1 and Rev-YFP occurs only if true molecular interaction between the respective proteins takes place.
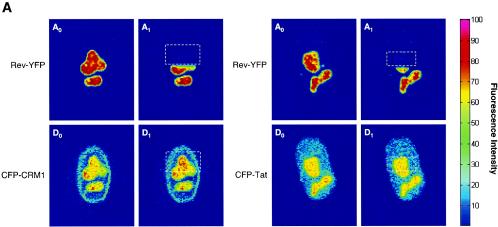
FIG. 4.
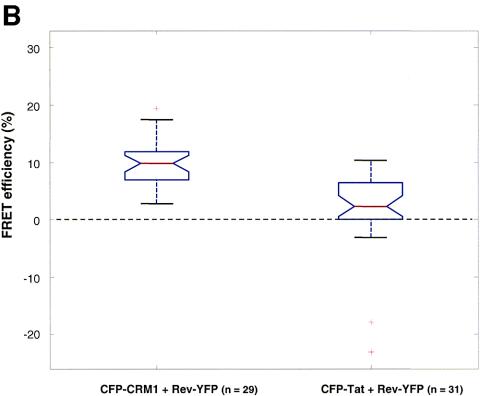
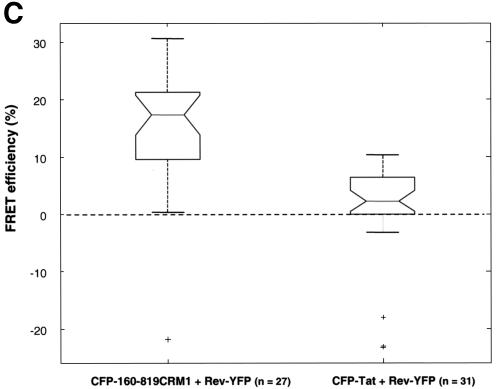
Interaction between CRM1 and cargo in living cells as measured by FRET. (A) Cells expressing CFP-CRM1 and Rev-YFP were analyzed for FRET by confocal microscopy using the photobleach method. This method is based on the principle that energy transfer is eliminated when the acceptor is bleached, thereby yielding an increase in donor fluorescence. Intensity maps of Rev-YFP (A0 and A1) and CFP-CRM1 (D0 and D1) in the nucleoli of a cell are shown. D0 indicates the fluorescence intensity distribution of CFP-CRM1 directly excited at 458 nm. A0 indicates the fluorescence intensity distribution of Rev-YFP excited at 514 nm. Rev-YFP was subsequently photobleached in a specific segment of the nucleolus (dotted box), thereby eliminating energy transfer. A second donor fluorescence image (D1) was then taken. A1 indicates the fluorescence intensity distribution of Rev-YFP after photobleaching. D1 indicates the fluorescence intensity distribution of CFP-CRM1 after photobleaching of the acceptor (Rev-YFP). As negative controls, the intensity maps of Rev-YFP (A0 and A1) and CFP-Tat (D0 and D1) in the nucleoli of a cell were also determined by the same method. (B) Quantification of FRET efficiencies between wild-type CRM1 and Rev proteins. The indicated number of cells (n) were analyzed in two different experiments, and the calculated FRET efficiency is given as a box plot distribution. The box spans the lower- to upper-quartile values. The length of the whiskers is 1.5 times the interquartile range. Outliers (indicated by plus signs) are data with values beyond the ends of the whiskers.
A first set of FRET experiments was performed by cotransfecting CFP-CRM1 and Rev-YFP in HeLa cells (Fig. 4A). FRET was monitored using the approach of acceptor photobleaching (4, 5, 28, 29, 58). The first acquired image (D0) is the fluorescence intensity distribution of the donor CFP-CRM1 directly excited at 458 nm. The second image (A0) represents the fluorescence intensity distribution of the Rev-YFP acceptor excited at 514 nm. The acceptor fluorophore (YFP) was then photobleached in part of the field (white dashed box in A1) by repeated scanning with the 514-nm laser line, thereby eliminating energy transfer. A second donor fluorescence image (D1) was then taken. If FRET is present, an increase in donor fluorescence intensity is expected in the region of acceptor photobleaching. This is apparent in the nucleolus of the cells, demonstrating a complex between Rev and CRM1 in the nucleolus (Fig. 4A, D1). As control for the FRET experiments, cells expressing CFP-Tat and Rev-YFP were used (Fig. 4A). As expected, no FRET could be observed with these proteins, as they do not interact despite their presence in the same location. The FRET efficiency derived from 29 to 31 analyzed cells was plotted as a box plot distribution showing the lower quartile, median, and upper quartile (Fig. 4B). Cells coexpressing wild-type CFP-CRM1 and Rev-YFP show an average FRET efficiency of 0.10 ± 0.04, which is significantly higher (P < 0.01 by Student's t test) than that of the CFP-Tat/Rev-YFP control (0.03 ± 0.04) (Fig. 4C). These results demonstrate a direct molecular interaction between CRM1 and Rev cargo in intact living cells.
CRM1 deletion mutants interact with Rev cargo.
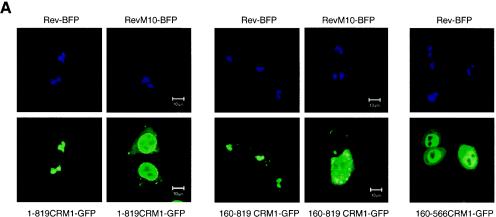
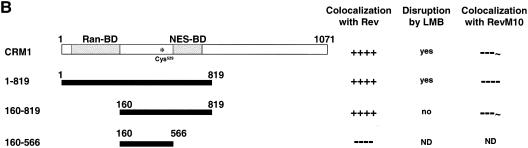
A more detailed molecular understanding of the nuclear export reaction in vivo requires a better knowledge of the regions of CRM1 accounting for NES binding. To approach this question in live cells, we monitored the colocalization of Rev-BFP with a series of N-terminal and C-terminal truncations of CRM1-GFP. We constructed GFP fusions of CRM1 that have been C-terminally deleted at amino acid 819 (1-819CRM1-GFP). Other CRM1 mutants lack both N-terminal (the first 160 amino acids) and C-terminal regions (160-819CRM1-GFP and 160-566CRM1-GFP). We found that upon expression in HeLa cells, 1-819CRM1-GFP had a localization similar to that of CRM-GFP, while 160-819CRM1-GFP and 160-566CRM1-GFP were distributed throughout the cell but not in the nucleoli; in addition, 160-819CRM1-GFP concentrated within brighter spots. When coexpressed with Rev-BFP, 1-819CRM1-GFP and 160-819CRM1-GFP translocated to the Rev-containing nucleoli (Fig. 5A). In contrast, 160-566CRM1-GFP did not localize to the nucleolus in the presence of Rev. These results provide strong evidence that amino acids 566 to 819 are essential for cargo binding in vivo. Such relocalization could not be detected by RevM10 in the majority of the cells. RevM10 is a transdominant-negative mutant of Rev that has been mutated in its NES, suggesting that inside cells the interaction between Rev and 1-819CRM1-GFP as well as Rev and 160-819CRM1-GFP is NES specific and not due to unspecific binding to Rev.
FIG. 5.
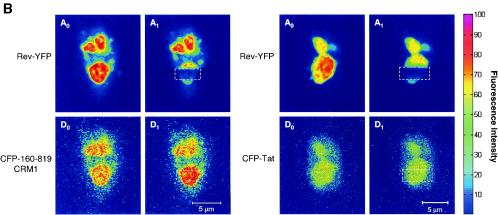
Interaction of CRM1 with Rev-cargo independent of Ran. (A) Coexpression of Rev-BFP or RevM10-BFP with 1-819CRM1-GFP, 160-819CRM1-GFP, or 160-566CRM1-GFP. The top panels show the nucleolar localization of Rev-BFP or RevM10-BFP. The lower panels display the localization of the coexpressed CRM1 mutants in the same cells. (B) Interaction between CFP-160-819CRM1, a deletion mutant unable to bind Ran (44), and Rev-YFP, as monitored by FRET. Intensity measures of CFP-160-819CRM1 and Rev-YFP as well as CFP-Tat and Rev-YFP in the nucleolus are also shown. First, a donor image, D0 (CFP-160-819CRM1), and an acceptor image, A0 (Rev-YFP), were acquired. Rev-YFP was photobleached in a specific segment of the nucleolus (A1). After photobleaching of the YFP, a second donor image was acquired (D1). An increase in donor fluorescence (CFP-160-819CRM1) after photobleaching the acceptor indicates FRET. (C) Quantification of FRET between CFP-160-819CRM1 and Rev-YFP. A number of cells (n) were analyzed in three different experiments, and the calculated FRET efficiency is given as a box plot distribution. The box spans the lower- to upper-quartile values. The length of the whiskers is 1.5 times the interquartile range. Outliers (indicated by plus signs) are data with values beyond the ends of the whiskers.
The specificity of the interaction of 160-819CRM1-GFP with Rev was confirmed by FRET measurements between CFP-160-819CRM1 and Rev-YFP (Fig. 5B). An increase in donor fluorescence intensity after acceptor photobleaching was apparent in the nucleolus of the cells expressing CFP-160-819CRM1 and Rev-YFP, demonstrating a complex between Rev-YFP and CFP-160-819CRM1. Cells expressing CFP-Tat and Rev-YFP were used as negative controls. The mean FRET efficiency between CFP-160-819CRM1 and Rev-YFP was 0.18 ± 0.075 and was significantly higher (P < 0.01, Student's t test) than that of the control (Fig. 5C). These results demonstrate direct interaction between Rev and 160-819CRM1. The FRET efficiency for the mutant CFP-160-819CRM1 and Rev-YFP was significantly higher than that of the wild-type CFP-CRM1/Rev-YFP pair. FRET efficiency depends on the distance between donor and acceptor molecules. In CFP-160-819CRM1, the CFP fluorophore may be positioned closer to Rev-YFP due to the 160-amino-acid N-terminal deletion.
CRM1 export complex disruption by LMB requires the N-terminal domain of CRM1.
To get better insight into the molecular aspects of the Rev-CRM1 interface in vivo, we investigated the effect of leptomycin B on Rev interaction with the CRM1 mutants described above. LMB disrupts the Rev-CRM1 interaction by covalently binding to Cys529 of CRM1 (16, 18, 30, 31, 43), thus disrupting Rev-CRM1 colocalization in cells (9). Cells were cotransfected with Rev-BFP and the respective CRM1-GFP mutants. Before adding LMB we verified that the CRM1 mutants colocalized with Rev in the nucleolus of the cells. After 2.5 h of incubation with 50 nM LMB, cells were analyzed for colocalization of the respective CRM1 mutants with Rev in the nucleolus (Fig. 6A). Wild-type CRM1-GFP and 1-819CRM1-GFP redistributed from the nucleolus to nucleoplasm and nuclear membrane, demonstrating that their interaction with Rev in the nucleoli was abolished by LMB. In contrast, the colocalization of 160-819CRM1-GFP with Rev in the nucleolus was unaffected. Therefore, the CRM1 molecules lacking the first 160 N-terminal amino acids (containing the Ran binding site) were not affected by LMB. A summary of these data is given in Fig. 6B. These results suggest that the first 160 N-terminal amino acids are essential for the inhibitory effect of LMB on CRM1-Rev interaction.
FIG. 6.
Effect of LMB on the colocalization of N- and C-terminal CRM1-GFP deletion mutants with Rev cargo in the nucleolus of living cells. (A) Cells coexpressing Rev-BFP and the indicated CRM1-GFP fusion proteins were analyzed by confocal fluorescence microscopy. The top panels show nucleolar localization of Rev-BFP. The lower panels show the localization of the CRM1-GFP deletion mutants 2.5 h after addition of LMB. (B) CRM1 functional domains and mutants used in this work. Ran-BD, Ran binding domain (20, 44); *, Cys529 (30); NES-BD, NES-binding domain (44). Colocalization with Rev or RevM10 and the effect of LMB on the colocalization of CRM1 mutants are summarized to the right. ND, not determined.
Nucleolar Rev-associated CRM1-GFP traffics to the cytoplasm.
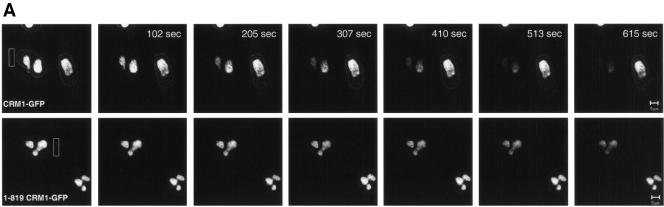
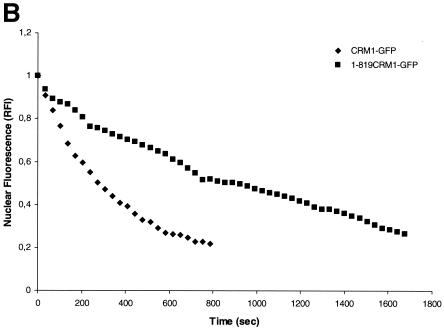
We next examined whether CRM1 interaction with Rev in the nucleolus represents a static nonfunctional aggregation or if this nucleolar CRM1 is trafficking between the nucleolus and the cytoplasm. To do so we performed fluorescence loss in photobleaching (FLIP) experiments (34, 47). In this technique, the CRM1-GFP in a defined area of the cytoplasm, distant from the nuclear membrane of the cell, is repeatedly photobleached, and the loss of fluorescence in the nucleus and nucleolus is monitored. Rapid elimination of CRM1-GFP from the nucleolus/nucleus upon repeated photobleaching of the cytoplasm would indicate a high mobility of CRM1 and trafficking to the cytoplasm. Images shown were obtained at 102-s intervals, and the depletion of CRM1-GFP and 1-819CRM1-GFP from the nucleoli was compared (Fig. 7A). The loss of fluorescence in the nucleoli is not due to the nonspecific bleaching outside of the designated area, because cells immediately adjacent to the targeted cell maintained similar fluorescence intensities throughout the period of the bleaching process. As an additional control experiment, the same measurement was performed with fixed cells. As expected, no loss in fluorescence of the nucleolus was detected in fixed cells during repeated photobleaching of the cytoplasm (data not shown). Quantitative analysis demonstrated that the majority of the wild-type CRM1-GFP has a rapid rate of depletion from the nucleus and nucleoli with a half-life of 203 ± 36 s (Fig. 7B). These results confirm that wild-type CRM1-GFP shuttles rapidly between the nucleus/nucleolus and cytoplasm and that the binding to Rev in the nucleolus is not the result of aggregation-producing complexes that are not able to be transported. We also compared the export kinetics of the mutant 1-819CRM1-GFP (Fig. 7A and B). The FLIP rate of 1-819CRM1-GFP was lower than that of the wild-type, implying a slower transport mechanism. 1-819CRM1 lacks the C-terminal domain, which contains nucleoporin-binding sites. The results suggest that the C-terminal domain of CRM1 is required for efficient transport.
FIG. 7.
Export kinetics of nucleolar Rev-associated CRM1-GFP and 1-819CRM1-GFP in cells as measured by FLIP. (A) Cells expressing Rev-BFP and CRM1-GFP or 1-819CRM1-GFP were imaged before and during sequential photobleaching of the indicated area in the cytoplasm (rectangle). Images shown were collected every ∼102 s. (B) Quantitative analyses of FLIP. The fluorescence in the nucleus was plotted versus time. The nuclear fluorescence intensity at time zero was normalized to 1.
Quantitative analysis of CRM1-GFP fluorescence recovery in the nucleolus, nucleoplasm, and nuclear membrane.
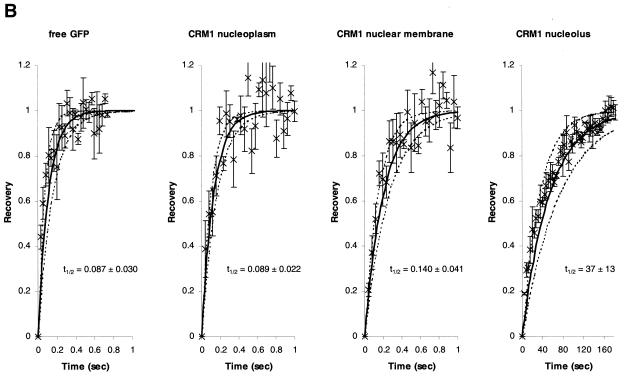
The mechanism by which CRM1 travels from the nucleolus to the nuclear membrane is not presently known. Given the highly condensed DNA and large number of proteins and RNA molecules within the nucleus, it has been assumed that the nucleus is very densely packed. On the other hand, it has been demonstrated that the nucleocytoplasmic transport system is very dynamic (51), as also confirmed by our FLIP experiments. To measure the mobility of CRM1-GFP in different cellular locations, we used quantitative FRAP analyses (12, 34, 47). As a reference we measured the fluorescence recovery rate of free GFP that diffuses freely within the nucleoplasm. Figure 8 shows the rapid recovery of CRM1-GFP after photobleaching a small circle of 1 μm in diameter. The calculated t1/2 of recovery for CRM1-GFP in the nucleoplasm was similar to that of free GFP, suggesting that CRM1 is highly mobile within the nucleus (Fig. 8). However, in this experimental setup, we measured the mobility of a mix of free CRM1-GFP and CRM1-GFP in complex with the various cellular cargo molecules. The mobility of CRM1-GFP in the nuclear membrane was significantly slower (Fig. 8A and B), in agreement with previous data on CRM1 interaction with the nuclear pore complex (15, 17, 22). Interestingly, when associated with Rev in the nucleolus, CRM1-GFP mobility was very slow (reduced by ∼400 times) (Fig. 8A and B). Our data suggest that CRM1 travels in an unimpeded manner through the nucleus in search of high-affinity binding sites, such as Rev in the nucleolus and Nup214, Nup159, Nup50, and Nup42 in the nuclear membrane (15, 17, 22).
FIG. 8.
Comparison of recovery rates of free GFP and CRM1-GFP in different cellular compartments during FRAP. (A) Cells expressing CRM1-GFP or coexpressing Rev-BFP and CRM1-GFP were imaged for GFP fluorescence before and after photobleaching a small spot of 1 μm in diameter either on the nuclear membrane (left) or in the nucleolus (right) (indicated by arrows). Fluorescence recovery in the bleached spot was monitored by time-lapse confocal microscopy. (B) Quantitative analyses of FRAP demonstrated that the mobility of CRM1-GFP is similar to that of free GFP in the nucleoplasm but is slower in the nuclear membrane and nucleolus (in the presence of Rev). Cells expressing free GFP or CRM1-GFP or coexpressing Rev-BFP and CRM1-GFP were examined by FRAP. The fluorescence intensity in the bleached spot of the first image collected after photobleaching was measured, and this value was normalized to zero; the intensity in the bleached spot at the end of the recovery (last five time points) was normalized to 1. The average normalized fluorescent values for each time point was plotted (×). Bars represent standard errors. Lines represent fits of determinations from more than six different cells using the least-squares method. The solid black line represents the mean fit. Dashed lines represent standard deviations.
DISCUSSION
CRM1 nucleocytoplasmic transport has been studied with a variety of different techniques. However, more information is required to fully understand complex formation and the dynamics in space and time of CRM1-cargo-RanGTP complexes in living cells. In this study, we have applied different approaches to study CRM1 interaction with its cargo inside living cells. Specific relocalization of CRM1 by Rev and not by RevM10 suggests an NES-specific interaction in the nucleolus. FRET measurements between CRM1 and Rev in live cells resulted in the detection of true molecular interactions existing in vivo in specific cellular locations with great resolution. Under similar conditions, the FRET efficiency between CRM1 and Rev was significantly higher than that of the negative controls Tat and Rev. FLIP experiments demonstrated that nucleolar CRM1 rapidly traffics to the cytoplasm, excluding the possibility that CRM1 found in the nucleolus is simply a nonfunctional aggregate. The fact that CRM1 associates with Rev in the nucleoli of cells suggests that the nucleolus may play a role in Rev function. Rev is required for export of viral mRNA to the cytoplasm (13, 36), a process essential for virus replication (54). It has been suggested previously that HIV-1 RNA is trafficking through the nucleoli of cells, giving the nucleolus a critical role in HIV-1 RNA export (39). In addition, the nucleolus has also been implicated in the route of CRM1 based on its nucleolar localization upon treatment with actinomycin D (17). These results suggest that the nucleolus may be part of the normal CRM1 routing.
In vitro RanGTP cooperative cargo binding to CRM1 has been shown for different proteins, like snurportin (45), Nmd3 (55), and p27 (8). The N-terminal motif of CRM1 has been proposed to account for interaction with RanGTP (7, 32, 59). Previous in vitro showed that deletion of the first 160 residues of CRM1 prevented the interaction of CRM1 with RanGTP in the presence of NES (44). Here we have demonstrated interaction of Rev with N-terminal and C-terminal deletion mutants of CRM1 by colocalization and by FRET. This interaction is specific and NES dependent, because the mutants did not interact with RevM10 that has been mutated in its NES. Although it has been established that Ran-binding is essential for cargo transport, the N-terminal deletion mutant 160-819CRM1-GFP specifically interacts with Rev in the nucleolus, suggesting that in cells CRM1 could interact with Rev independently of Ran. This property might be specific only for Rev. Moreover, the affinity of the Ran-independent complex might be very low. Therefore, it would be interesting to compare inside cells the CRM1-Rev affinity in the presence and absence of Ran. Variation in the amino acid sequence of different NESs might result in different binding affinities to CRM1. Rev has low affinity for CRM1 in vitro (2), but it has the ability to multimerize, which might increase the number of NESs per complex. The Ran-independent interaction of Rev was also proposed by Askjaer et al. on the basis of in vitro gel-shift assays and protein footprinting data (3). However, in vitro this Ran-independent interaction was NES independent, as M10 and M32 mutants obtained an identical Ran-independent footprint (3).
Another important issue addressed here is the molecular basis for LMB inhibition. Interestingly, LMB did not disrupt complexes of CRM1 from which the first 160 N-terminal amino acids were deleted, even though this mutant contains the LMB interaction site (Cys529), suggesting that the N-terminal region of CRM1 plays a role in LMB action. One hypothesis is that transport to the cytoplasm and disruption of the complex in the cytoplasm is required for LMB action. In fact, N-terminal deletion mutants of CRM1 are not trafficking to the cytoplasm. However, this hypothesis would suggest a high-affinity binding of the 160-819 CRM1 to Rev. Another possibility is that binding of RanGTP to CRM1 induces a conformational change in the complex that makes it accessible to LMB or that enables LMB to discharge the NES. A conformational change upon binding of RanGTP to CRM1 was suggested earlier on the basis of site-specific cross-linking data, which demonstrated that CRM1 was only cross-linked to Cys89 of Rev in the presence of RanGTP (3).
The exact mechanism by which CRM1 moves from the nucleolus to the nuclear membrane is not presently known. Therefore, we studied the mobility of CRM1-GFP in different nuclear locations by FRAP. We demonstrate that CRM1-GFP is highly mobile within the nucleus. It has similar mobility as free GFP, which diffuses freely in the cell. However, in this experimental setup we anticipate a mix of free and cargo-bound CRM1. These data imply that CRM1-GFP travels unimpeded through the nucleoplasm. The high mobility of CRM1 in the nucleoplasm is not unprecedented. It has been shown in earlier studies that proteins can move by a passive, diffusion-based mechanism throughout the nucleus (11, 40). Diffusion provides an efficient, rapid mode of transport. The data support a model in which CRM1 roams through the nucleus in search of high-affinity binding sites, e.g., Rev in the nucleolus and Nup214, Nup159, Nup50, and Nup42 in the NPCs. This roaming behavior has been clearly demonstrated for DNA repair factors and DNA replication factors (24, 33). Our data were obtained upon CRM1-GFP overexpression, suggesting that most of the molecules are indeed not in complexes in the nucleoplasm. It would be of interest to further examine the specific movement and mobility of the CRM1-cargo-RanGTP transport complex in the nucleoplasm.
In conclusion, the data suggest a model for CRM1-cargo-RanGTP complex formation. We propose that CRM1 roams through the nucleus in search of high-affinity binding sites, such as Rev cargo in the nucleolus and nucleoporins in the NPCs. In the case of Rev cargo binding in the nucleolus, CRM1 binds to Rev in anticipation of Ran. However, it would be of importance to measure the binding affinities of the different complexes inside cells. In addition, data suggest a new role for Ran in the dissociation of the export complex from the nucleoli. It will be of interest to apply this methodology to investigate this phenomenon in more detail and to determine the general applicability of these conclusions.
Acknowledgments
We thank Edward Hui Cho and Rebecca Erwin-Cohen for technical assistance and the CCR/NCI Fellows Editorial Board for critical comments.
This work has been funded in part by the National Cancer Institute, National Institutes of Health, under contract number N01-CO-12400.
REFERENCES
- 1.Adachi, Y., and M. Yanagida. 1989. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J. Cell Biol. 108:1195-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273:33414-33422. [DOI] [PubMed] [Google Scholar]
- 4.Bastiaens, P. I., and T. M. Jovin. 1996. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proc. Natl. Acad. Sci. USA 93:8407-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastiaens, P. I., I. V. Majoul, P. J. Verveer, H. D. Soling, and T. M. Jovin. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15:4246-4253. [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, F. R., and H. Ponstingl. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354:80-82. [DOI] [PubMed] [Google Scholar]
- 7.Chi, N. C., E. J. Adam, and S. A. Adam. 1997. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J. Biol. Chem. 272:6818-6822. [DOI] [PubMed] [Google Scholar]
- 8.Connor, M. K., R. Kotchetkov, S. Cariou, A. Resch, R. Lupetti, R. G. Beniston, F. Melchior, L. Hengst, and J. M. Slingerland. 2003. CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell 14:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daelemans, D., E. Afonina, J. Nilsson, G. Werner, J. Kjems, E. De Clercq, G. N. Pavlakis, and A. M. Vandamme. 2002. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc. Natl. Acad. Sci. USA 99:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 11.Dundr, M., and T. Misteli. 2001. Functional architecture in the cell nucleus. Biochem. J. 356:297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edidin, M., Y. Zagyansky, and T. J. Lardner. 1976. Measurement of membrane protein lateral diffusion in single cells. Science 191:466-468. [DOI] [PubMed] [Google Scholar]
- 13.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 15.Floer, M., and G. Blobel. 1999. Putative reaction intermediates in Crm1-mediated nuclear protein export. J. Biol. Chem. 274:16279-16286. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod, M., J. van Deursen, S. van Baal, A. Reynolds, D. Davis, K. G. Murti, J. Fransen, and G. Grosveld. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich, D. 1998. Transport into and out of the cell nucleus. EMBO J. 17:2721-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 22.Guan, T., R. H. Kehlenbach, E. C. Schirmer, A. Kehlenbach, F. Fan, B. E. Clurman, N. Arnheim, and L. Gerace. 2000. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell. Biol. 20:5619-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha, T., T. Enderle, D. F. Ogletree, D. S. Chemla, P. R. Selvin, and S. Weiss. 1996. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. USA 93:6264-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houtsmuller, A. B., S. Rademakers, A. L. Nigg, D. Hoogstraten, J. H. Hoeijmakers, and W. Vermeulen. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284:958-961. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde, E., U. Kutay, C. von Kobbe, I. W. Mattaj, and D. Gorlich. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16:6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kehlenbach, R. H., A. Dickmanns, and L. Gerace. 1998. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J. Cell Biol. 141:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenworthy, A. K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289-296. [DOI] [PubMed] [Google Scholar]
- 29.Kenworthy, A. K., and M. Edidin. 1998. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J. Cell Biol. 142:69-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 32.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Gorlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 35.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 37.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 38.Meyer, B. E., J. L. Meinkoth, and M. H. Malim. 1996. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J. Virol. 70:2350-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michienzi, A., L. Cagnon, I. Bahner, and J. J. Rossi. 2000. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 97:8955-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 41.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 42.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 43.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 44.Ossareh-Nazari, B., and C. Dargemont. 1999. Domains of Crm1 involved in the formation of the Crm1, RanGTP, and leucine-rich nuclear export sequences trimeric complex. Exp. Cell Res. 252:236-241. [DOI] [PubMed] [Google Scholar]
- 45.Paraskeva, E., E. Izaurralde, F. R. Bischoff, J. Huber, U. Kutay, E. Hartmann, R. Luhrmann, and D. Gorlich. 1999. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pemberton, L. F., G. Blobel, and J. S. Rosenblum. 1998. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10:392-399. [DOI] [PubMed] [Google Scholar]
- 47.Phair, R. D., and T. Misteli. 2001. Kinetic modelling approaches to in vivo imaging. Nat. Rev. Mol. Cell Biol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 48.Reits, E. A., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145—E147. [DOI] [PubMed] [Google Scholar]
- 49.Richards, S. A., K. M. Lounsbury, K. L. Carey, and I. G. Macara. 1996. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J. Cell Biol. 134:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selvin, P. R. 2000. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7:730-734. [DOI] [PubMed] [Google Scholar]
- 51.Smith, A. E., B. M. Slepchenko, J. C. Schaff, L. M. Loew, and I. G. Macara. 2002. Systems analysis of Ran transport. Science 295:488-491. [DOI] [PubMed] [Google Scholar]
- 52.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 53.Stauber, R. H., E. Afonina, S. Gulnik, J. Erickson, and G. N. Pavlakis. 1998. Analysis of intracellular trafficking and interactions of cytoplasmic HIV-1 Rev mutants in living cells. Virology 251:38-48. [DOI] [PubMed] [Google Scholar]
- 54.Terwilliger, E., R. Burghoff, R. Sia, J. Sodroski, W. Haseltine, and C. Rosen. 1988. The art gene product of human immunodeficiency virus is required for replication. J. Virol. 62:655-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, F., and U. Kutay. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116:2409-2419. [DOI] [PubMed] [Google Scholar]
- 56.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 57.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 58.Wouters, F. S., P. I. Bastiaens, K. W. Wirtz, and T. M. Jovin. 1998. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. EMBO J. 17:7179-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wozniak, R. W., M. P. Rout, and J. D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8:184-188. [DOI] [PubMed] [Google Scholar]
- 60.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zolotukhin, A. S., and B. K. Felber. 1999. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120-127. [DOI] [PMC free article] [PubMed] [Google Scholar]