Abstract
Pearl millet (Pennisetum glaucum (L.) R. Br.) is a staple food and a drought-tolerant cereal well adapted to Sub-Saharan Africa agro-ecosystems. An important diversity of pearl millet landraces has been widely conserved by farmers and therefore could help copping with climate changes and contribute to future food security. Hence, characterizing its genetic diversity and population structure can contribute to better assist breeding programs for a sustainable agricultural productivity enhancement. Toward this goal, a comprehensive panel of 404 accessions were used that correspond to 12 improved varieties, 306 early flowering and 86 late-flowering cultivated landraces from Senegal. Twelve highly polymorphic SSR markers were used to study diversity and population structure. Two genes, PgMADS11 and PgPHYC, were genotyped to assess their association to flowering phenotypic difference in landraces. Results indicate a large diversity and untapped potential of Senegalese pearl millet germplasm as well as a genetic differentiation between early- and late-flowering landraces. Further, a fine-scale genetic difference of PgPHYC and PgMADS11 (SNP and indel, respectively) and co-variation of their alleles with flowering time were found among landraces. These findings highlight new genetic insights of pearl millet useful to define heterotic populations for breeding, genomic association panel, or crosses for trait-specific mapping.
Keywords: millet, Pennisetum glaucum, flowering, genetic diversity, SSR, PgMADS11, PgPHYC
Introduction
Low agricultural productivity causes food insecurity and malnutrition in Sub-Saharan Africa (SSA). This is due to climate variability, increased growth and needs of worldwide population, water demand, intensive exploitation of natural resources and environmental degradation. Therefore, better uses of natural resources could help overcoming some of these constraints and greatly contribute to improving productivity.
Pearl millet (Pennisetum glaucum (L.) R. Br.) is a major staple food for many SSA and Asian countries. Pearl millet is highly allogamous and mainly a rainfed crop, covering a wide range of different ecological zones and production systems. However, yield is low and variable, rarely reaching 1000 kg/ha. This is mainly explained by the limited exploitation of genetic resources and availability of improved varieties, but also by low soil fertility, drought, heat and highly variable rainfall (Waddington et al., 2010). Moreover, production is threatened by downy mildew disease, striga parasitic weed and predation by insects (Waddington et al., 2010). Characterization of crops genetic resources is a prerequisite to build up a breeding program for sustainable productivity enhancement.
Senegal is one of the top 10 pearl millet producers in the world (FAOSTAT, 2013), where farmers distinguish two main types of cultivars based on growth duration. Cultivars, called Souna, are sensitive to photoperiod, with a short cycle between 65 and 90 days and adapted to low (350–600 mm) rainfall regions. Cultivars, called Sanio, are a less photoperiod-sensitive type than Souna, with a long cycle between 120 and 150 days, and are adapted to high (900–1200 mm) rainfall regions. Nationwide, Souna type occupies nearly half the area sown to cereals (51%) while Sanio type, mainly cultivated in the South, represents about 15% of total millet production (ANSD, 2014).
Investigating genetic diversity and patterns of early- and late-flowering landraces is very important since flowering cycles and photoperiod sensitivity play a crucial role in the adaptation to climatic conditions. It is assumed a direct effect of selection for earliness associated with climate variations (Vigouroux et al., 2011). Genome scans and genetic association mapping have identified several genes tightly linked to adaptive traits of pearl millet in semi-arids areas. A SNP in the Phytochrome C locus (PgPHYC) and an indel variation in PgMADS11 gene were associated with flowering time variation, annual rainfall and spike length of pearl millet (Saïdou et al., 2009; Mariac et al., 2011; Vigouroux et al., 2011). From framers point of view, distinction between early- and late-flowering landraces is very clear but it is not always associated with clear genetic differentiation (Dussert et al., 2015). Flowering time in pearl millet is derived from a common domestication event and a strong gene flow between early- and late-flowering landraces is observed (Dussert et al., 2015). The level of gene flow would depend on cycle overlapping, agricultural practices and spatial distribution (Mariac et al., 2006b; Allinne et al., 2007; Lakis et al., 2012).
Because there is a critical need for adapting local agriculture to harsher future conditions, landraces and improved varieties adaptation will mostly rely on standing genetic variation available within the cultivated compartment. Recently, phenotyping (Sy et al., 2015) and genotyping by sequencing (Hu et al., 2015) studies were carried out on a set of Senegalese pearl millet landraces. Sampling was restricted to only one agro-ecological area of Senegal, the Groundnut Basin, and included only nine so-called intermediate-flowering landraces (flower between 75 to 100 days after sowing). Based on phenology, head architecture and grain color, these accessions were classified into three cultivar groups, indicating a morphological diversity between early flowering landraces (Sy et al., 2015). Using 83,875 single nucleotide polymorphisms (SNPs) on the same set of accessions in addition to 252 global accessions, a higher genetic diversity was observed in Senegal accessions compared to millet accessions in India, South and Western Africa (Hu et al., 2015). Any local structure was evidenced, therefore studies of loci that control the cycle length would be necessary to assess more accurately the evolution of cultivated millet varieties (Dussert et al., 2015).
Here, a fine scale sampling strategy and genetic characterization are described that differentiate early- and late-flowering landraces of Senegalese pearl millet. Using highly polymorphic SSRs markers, genetic diversity and population structure of the landraces was assessed. Allelic diversity of PgPHYC and PgMADS11 genes, both linked with flowering time variation and rainfall, was further investigated (Saïdou et al., 2009; Mariac et al., 2011; Vigouroux et al., 2011).
Materials and Methods
Plant Materials
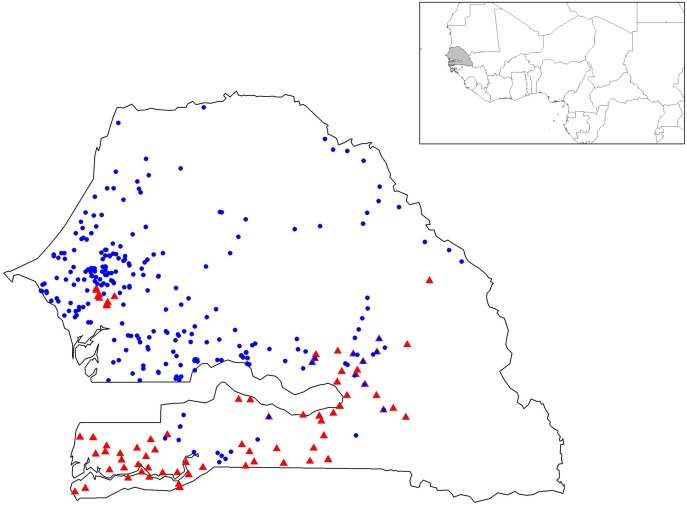
Collects were done in 1992 and 1994 in the main areas of millet production in the Groundnut Basin as previously described (Sy et al., 2015). Geographical coordinates of these accessions were partially retrieved (88%) by using village names. Additional collects were done in 2010 and 2014 to cover pearl millet production areas, except the city of Dakar and the eastern-south area where the Niokolo-Koba Wildlife Park is located. Geographical coordinates of these new accessions were recorded using a GPS. As our focus is on local landraces, villages near major roads or markets were avoided. In total, 392 accessions were collected from 316 villages, i.e., 1.24 accessions per village on average. A panel of 404 accessions was analyzed including 12 improved varieties bred locally and widely used by farmers, 306 early flowering landraces (252 villages) and 86 late-flowering landraces (74 villages). Among the 316 villages, 10 villages were sampled with both early- and late- flowering landraces (Supplementary Table S1). Cycles were recorded following farmer interviews.
DNA Extraction and SSR Genotyping
Five seeds per accession were grown in the greenhouse 3–4 weeks according to sampling date. About 200 mg of leaf sample from one individual per accession were collected and DNA extraction was carried out using the previously described protocol (Mariac et al., 2006a). Twelve highly polymorphic microsatellites distributed throughout the pearl millet genome were used (Supplementary Table S2). These markers have been previously described (Allouis et al., 2001; Qi et al., 2001; Budak et al., 2003; Mariac et al., 2006a). PCR reactions were performed using the Multiplex PCR Kit (Qiagen, Inc) following the recommended protocol. PCR were conducted using a thermal cycler TC-Plus (TECHNE): pre-denaturation of 95°C for 15 min then 35 cycles consisting of a denaturation step at 94°C for 30 s, annealing at 55°C for 90 s, elongation at 72°C for 60 s and a final extension at 60°C for 30 min). Four positive and four negative controls were repeated on each PCR plate. Samples were genotyped on an ABI 3130 Prism® (Applied Biosystems®) and read with GenemapperTM software (version 3.7; Applied Biosystems®).
Genetic Diversity and Population Structure Analyses
Genetic diversity, heterozygosities (expected and observed) and F-statistics were calculated using Genalex 6.5 (Peakall and Smouse, 2012). For genetic structure, a principal component analysis (PCA) using the package ade4 (Thioulouse et al., 1997) implemented in R software (R Development Core Team, 2008) was first performed. Then, population structure was investigated using STRUCTURE software 2.3.3 (Pritchard et al., 2000). Analysis was performed with the admixture model (Falush et al., 2003) with K ancestral populations ranging from 1 to 6. We used 500,000 iterations and a burn-in period of 100,000, 10 runs for each K-value were performed. The values for the number of clusters (K) were assessed according to Evanno et al. (2005) by the (D.ΔK) criterion and the log-likelihood (Ln P (D | K)) plot. Individuals were assigned to a cluster if their ancestry was higher than 70%, q ≥ 0.7.
For spatial analysis of genetic variability, a total of 367 geo-referenced accessions including 281 early- and 86 late-flowering landraces were used. Spatial principal component analysis (sPCA) was performed using the adegenet package (Jombart, 2008) with R software. The spatial genetic structure was assessed using spatial autocorrelation analyses of kinship coefficients between individuals (Loiselle et al., 1995) following the standard procedure (Vekemans and Hardy, 2004) implemented in SPAGeDi version 1.2 (Hardy and Vekemans, 2002). Mean multilocus kinship coefficient values, Fij, i.e., genetic similarity between individuals i and j relative to the mean genetic similarity between random individuals in the sample, were regressed on both the linear (dij) and the logarithmic (ln(dij)) spatial distance between individuals. This distance was calculated as the Euclidian distances using spatial coordinates. The regression slopes bd and bLd were jointly assessed. Standard errors for the kinship coefficients were estimated using a jackknife procedure over all loci. We tested the significance of the kinship coefficients and the regression slopes bd and bLd estimates by comparing the observed values to those obtained after 10,000 random permutations.
PgPHYC and PgMADS11 Genotyping
Polymorphisms in both genes were associated with flowering time variation (Saïdou et al., 2009; Mariac et al., 2011; Vigouroux et al., 2011). The panel of accessions was genotyped with PgPHYC (Acc numbers FN376885–FN377564) (Vigouroux et al., 2011) and PgMADS11 (Acc numbers FN552468–FN552522) (Mariac et al., 2011) to test the allelic differences in genotype frequencies between early- and late-flowering landraces.
For PgPHYC, a polymorphism at the 5′ of the gene was assessed. A C/G SNP at that position is cleaved by PvuII restriction enzyme and therefore accessions scored as C/C, G/G and C/G according to their digestion pattern (Saïdou et al., 2009).
For PgMADS11, an indel polymorphism of 24 bp was assessed as previously reported (Mariac et al., 2011).
Logistic regressions between genotypes, genetic cluster, latitude and longitude for each gene were further performed.
Results
Genetic Diversity of Senegalese Germplasm
The germplasm collected through this study is the most comprehensive sampling to date of landraces from Senegal (Figure 1). Genotyping data revealed a total of 101 alleles with an average of 8.4 alleles per locus (Supplementary Table S2 and Figures S1–S3). The final data set contained only 1.8% of missing data. High levels of genetic diversity characterize both groups (Table 1). Early flowering landraces presented the highest level of genetic diversity as measured by observed HObs = 0.481 or expected heterozygosity, HExp = 0.567. In contrast, improved varieties showed the lowest, with HObs = 0.391. For early flowering landraces, only one locus (PSMP2249) was found not to be at Hardy-Weinberg equilibrium (HWE). Two loci were not at HWE (PSMP2249 and PSMP2246) in late-flowering landraces. Groups showed low levels of inbreeding with FIS values of 0.160, 0.128, and 0.258 for early and late landraces and improved varieties, respectively. Furthermore, early-, late-flowering landraces and improved varieties present 22, 8 and 1 private alleles, respectively. Low FST differentiation was found between improved varieties and early flowering landraces (0.004), between early- and late-flowering landraces (0.052) and between late-flowering landraces and improved varieties (0.063).
FIGURE 1.
Geographic positions of the 367 geo-referenced accessions of pearl millet. Early- and late-flowering landraces are represented by blue circles and red triangles, respectively.
Table 1.
Genetic diversity statistics for improved varieties, early- and late-flowering landraces.
| Nb | Na | HObs | HExp | FIS | |
|---|---|---|---|---|---|
| Improved varieties | 12 | 4.4 | 0.350 | 0.513 | 0.258 |
| Early flowering landraces | 306 | 7.7 | 0.446 | 0.542 | 0.160 |
| Late-flowering landraces | 86 | 5.8 | 0.434 | 0.494 | 0.128 |
Nb, number of accessions; Na, mean number of alleles, HObs, observed heterozygosity; HExp, expected heterozygosity; FIS, inbreeding coefficient.
Population Genetic Structure
Bayesian clustering analyses showed a clear structure between early- and late-flowering landraces (Figure 2). The value of the Evanno criterion (ΔK) was the highest for K = 2 (Supplementary Figure S1), supporting the evidence of two major clusters. A total of 89% of the early flowering landraces were assigned to a single cluster, and 90% of the late-flowering landraces were assigned to the other cluster. Thirty-four accessions of early- and nine of late-flowering landraces were misassigned (q < 0.7), but showed intermediate ancestries. For the 10 villages where both early- and late-flowering landraces were sampled, all accessions of similar phenotype were assigned to their respective cluster.
FIGURE 2.
STRUCTURE results for K = 2 based on 404 accessions analyzed with 12 SSRs. Each bar represents an individual. It shows the proportion of genome belonging to each of the two genetic groups (blue for early flowering accessions and red for late-flowering accessions) identified with STRUCTURE.
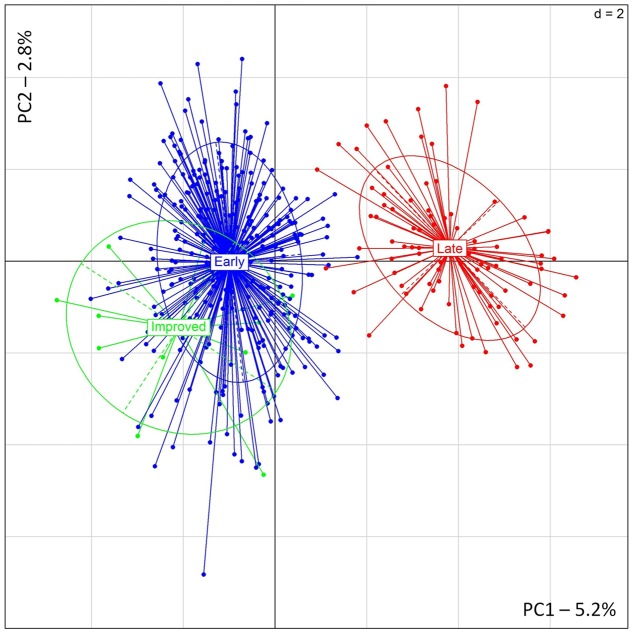
Similar results were found with PCA showing a clear distinction between early- and late-flowering landraces (Figure 3). The two first principal components explained, respectively, 5.2 and 2.8% of the inertia. PSMP2247 and PSMP2202 showed the highest contribution to the PC1. Two alleles, PSMP2247-199 and PSMP2202-146, showed high frequencies in late-flowering landraces (0.92 and 0.83, respectively). However, removing these two loci did not affect PCA results. Finally, any genetic differentiation was found between improved and early accessions.
FIGURE 3.
PCA results obtained for 12 improved, 306 early- and 86 late-flowering landraces accessions.
Spatial Analysis of Genetic Variability
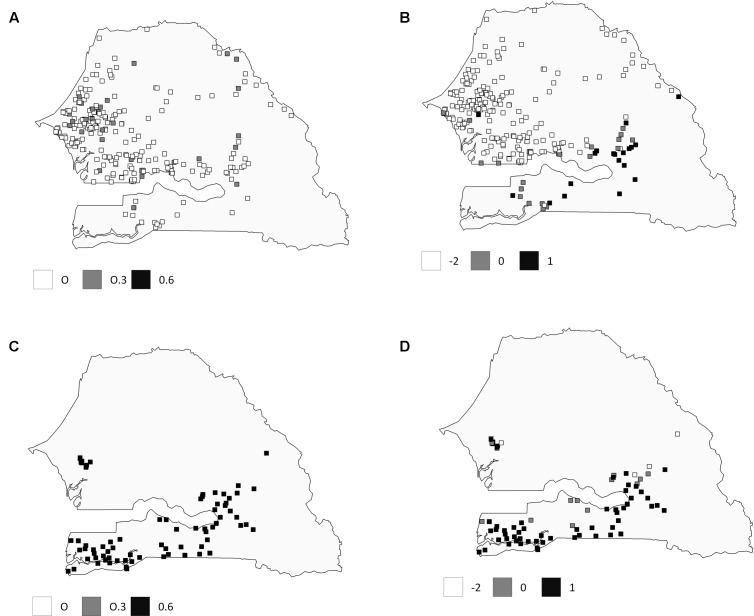
Spatial principal component analysis revealed a more cryptic genetic structure (Figure 4). Global structures, i.e., large spatial scale, were significant (p-value = 0.0001) with the first principal component, showing a high autocorrelation (Morran’s I = 0.50). In contrast, local structures were not significant (p-value = 0.77). The first axis of the sPCA identified two clusters. Considering spatial information, early flowering landraces from Southern Senegal showed a more admixed pattern than observed with STRUCTURE, while no differences were observed considering late-flowering landraces. Bayesian and multivariate approaches confirm genetic assignments for late-flowering landraces from Central Senegal.
FIGURE 4.
Comparison of genetic assignment between Structure and SPCA results for early- and late-flowering landraces. The top row represents values for early flowering landraces: (A) with q-values obtained with Structure analysis, and (B) lagged scores from the sPCA. The bottom row represents values for late-flowering landraces (C) with q-values obtained with Structure analysis and (D) lagged scores from sPCA. Values are depicted in grayscale.
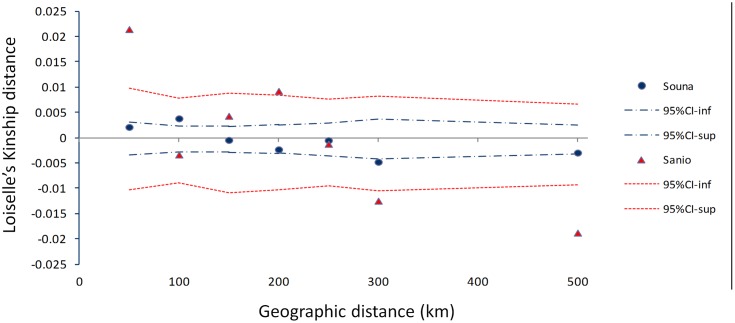
The pattern of isolation by distance (IBD) for early- and late-flowering landraces was investigated. Low IBD slopes were obtained for early flowering (bE = -2.41E-05) and late-flowering (bL = -9.89E-05) landraces (Figure 5). Similar results were found for logarithmic distances. This suggests no significant pattern of isolation-by-distance.
FIGURE 5.
Isolation by distance pattern for early- (blue dots) and late-flowering (red triangles) landraces as measured by the mean multilocus Loiselle’s kinship genetic distance, plotted against geographical distance (km). Upper and lower 95% confidence limits are indicated by colored dotted lines. Each point represents averaged Loiselle’s kinship for each class distance.
PgPHYC and PgMADS11 Allele Diversity
Landraces were genotyped for both PgPHYC and PgMADS11 alleles to assess genetic diversity in relation to flowering time (Table 2 and Supplementary Figures S4, S5). Only early landraces carried early flowering allele (G) at the PgPHYC locus leading to a significant difference in genotypes frequencies (F-test, p = 0.006). For PgMADS11, a significant difference in genotypes frequencies (F-test, p = 0.004) was observed with early flowering landraces having higher frequency of the allele (363 bp fragment). A significant correlation between allele G from PgPHYC with latitude (p-value = 0.0348) was observed. However, significance disappeared when taking into consideration genetic clustering (q ≤ 0.7). For PgMADS11, data reveal a significant correlation with longitude (p-value < 0.001) even when considering genetic clustering in the mode.
Table 2.
Genotypes counts for improved varieties, early- and late-flowering landraces with q-values ≤ 0.7.
|
PgPHYC alleles |
PgMADS11 alleles |
|||||
|---|---|---|---|---|---|---|
| C/C | C/G | G/G | 363/363 | 363/387 | 387/387 | |
| Improved varieties | 8 | 3 | 1 | 1 | 2 | 9 |
| Early flowering landraces | 239 | 21 | 0 | 18 | 65 | 185 |
| Late-flowering landraces | 79 | 0 | 0 | 2 | 16 | 62 |
Discussion
Large and Untapped Diversity in Senegalese Germplasm
In this study, a relatively high genetic diversity (HExp: 0.516) of pearl millet germplasm of Senegal is reported. Previous studies have reported a higher (HExp: 0.69) (Stich et al., 2010) and a lower (HExp = 0.49) genetic diversity in Niger pearl millet (Mariac et al., 2006a). These differences are due to the number of SSRs used. In the meanwhile, a similar number of alleles per locus (6 alleles per locus for Senegal vs. 6.2 alleles per locus for Niger) was observed. Comparing heterozygosity levels, observation showed a higher coefficient (FIS = 0.30) in Niger germplasm than in Senegal germplasm (FIS = 0.18). However, these results are consistent with data from others studies carried out on accessions across SSA regions and India (Oumar et al., 2008; Dussert et al., 2015; Hu et al., 2015). Pearl millet shows a high genetic diversity that can be explained by its strong outcrossing rate (75%) and the still on-going gene flow with its wild relative (Mariac et al., 2006a; Lewis, 2010). This high genetic diversity is in line with its high phenotyping diversity observed in Senegal (Sy et al., 2015) as in Western Africa (Pucher et al., 2015). Together, these findings highlight the untapped potential of Senegalese pearl millet germplasm for breeding.
Genetic proximity between early flowering varieties and improved varieties highlight a history of breeding programs and agricultural practices. In Senegal, few breeding programs have been undertaken on pearl millet but all focused on reducing the flowering cycle and were “population” varieties whose parental seeds were collected from local landraces. In addition, farmers still grow improved varieties jointly with landraces in their field, increasing gene flow and thus genetic proximity.
Genetic Structure of Early- and Late-Flowering Landraces
Bayesian results clearly help identifying genetic structure associated with early- and late-flowering landraces. This genetic structure was partly explained by the geographic distribution of landraces as shown through the sPCA analysis, making it difficult to entirely disentangle spatial and genetic structure. More contrast in admixture patterns might have been obtained with a higher number of SSR markers. However, this was compensated by our sampling effort and we were able to reveal a clear structure in Senegal landraces where other studies failed with a higher number of markers (Hu et al., 2015).
At a local scale, more contrasted admixture patterns were observed. In Central Senegal, clear differentiation between early- and late-flowering landraces was found. In contrast, more admixed patterns were observed in the Southern Senegal. The results might be related to the agricultural practices of the farmers, such as spatial cropping and seed circulation (Mariac et al., 2006b; Lakis et al., 2012; Kouakou et al., 2013).
Our data indicated two distinct genetic clusters in Senegalese pearl millet germplasm. In addition, comparing early- and late-flowering landraces, we found a slight genetic differentiation between the two groups of 0.052, which is of the same magnitude (0.053) found in Niger (Lakis et al., 2012). A single domestication event led to early- and late-flowering landraces, partly explaining the limited genetic differentiation observed at a regional scale (Dussert et al., 2015).
Flowering Traits Diversity
Photoperiod-sensitivity of pearl millet landraces and thus variation of flowering cycle constitute a key response for adaptation to future climate conditions (Sultan et al., 2013). Indeed, pearl millet landraces from Niger show reduced flowering cycle associated with drought episodes from 1976 to 2003 (Vigouroux et al., 2011). This reduction was correlated with changes in allele frequencies for PgPHYC. Correlation between allele frequencies and rainfall were also found with PgMADS11 (Saïdou et al., 2009; Mariac et al., 2011). Our data showed significant genetic differences for both genes with early flowering landraces enriched in precocity alleles.
Flowering time has been correlated with latitude, early flowering landraces being grown in northern latitude where environmental conditions are more arid (Haussmann et al., 2006; Pucher et al., 2015). In our study, the latitude effect was confounded with spatial genetic structure for PgPHYC. On the other hand, a correlation with longitude was found for PgMADS11. Further investigation would be needed to fully address this correlation. In any case, the use of flowering genes PgPHYC and PgMADS11 in marker assisted-selection programs presents some interest.
Challenges for Adaptation of Pearl Millet and Breeding Strategies
Sub-Saharan Africa recorded long dry spells in the 1970s and 1980s that led to breeding for short cycles improved varieties. Indeed, pearl millet breeding programs were predominantly built on restricted genetic resources of early flowering landraces. Strong differentiation between early- and late-flowering landraces from Senegal suggests the existence of an important gene pool that has not been exploited yet. The high genetic diversity could explain the wider range of pearl millet adaptation to dry areas and this potential may further contribute to breeding programs in response to the specific needs or target areas (Kouressy et al., 2004). For instance, a key strategy to cope against climate changes within SSA agrosystems is to tap into diversity of flowering time (Haussmann et al., 2012; Sultan et al., 2013) and resilience (Prieto et al., 2015).
Analyses of allelic variation of PgPHYC and PgMADS11 indicate fine-scale genetic difference (SNP and indel, respectively) among individuals and/or genotypes. Knowing that responses to photoperiod and rainfall were genetically associated with both genes, implication could be their use to detect/track climate adaptive changes to environment variations. For example, earliness of flowering and latitude correlation observed in early landraces support assumption that a direct effect of selection for that trait which is associated with climate variations such as photoperiod and rainfall. Both traits are key targets in selection for millet genotypes to be cultivated in rainfed areas.
Conclusion
The genetic diversity and population structure of Senegalese pearl millet landraces were assessed using a large panel of accessions and a limited number of SSRs markers. Results highlight a high genetic diversity and an untapped potential of the germplasm. However, two clusters were clearly distinguished as revealed by differentiation between early- and late-flowering landraces. Further, genetic difference and allelic co-variation in flowering genes PgPHYC and PgMADS11 were found among individuals. These findings give new insights into Senegalese pearl millet germplasm and are promising for developing new cultivars and heterotic groups that can be used to breed synthetic and hybrid varieties with higher degrees of heterozygosity in order to intensify yield production under harsh semiarids environments.
Author Contributions
NK, AF, DD, MG, YV, and AB designed the study. OD, AF, MG, MP, HT, BD, and OS collected samples. OD, MC, MP, and LZ performed DNA extraction, PCR and sequencing. AB, CB-S, and OD performed the genetic analyses. OD, AB, CB-S, DD, and NK drafted the manuscript. All authors contributed to the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We also thank farmers and local authorities for their consent and collaboration for seeds sharing. We acknowledge Claire Billot and Monique Deu (CIRAD) for sampling collection and Cédric Mariac (IRD) for scientific and technical assistance.
Footnotes
Funding. We are thankfully to the West Africa Agricultural Productivity Program (WAAPP/PPAAO 2A) for funding research activities (Grant No 51350 SE to NK) and OD thesis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00818/full#supplementary-material
References
- Allinne C., Mariac C., Vigouroux Y., Bezançon G., Couturon E., Moussa D., et al. (2007). Role of seed flow on the pattern and dynamics of pearl millet (Pennisetum glaucum [L.] R. Br.) genetic diversity assessed by AFLP markers: a study in south-western Niger. Genetica 133 167–178. 10.1007/s10709-007-9197-7 [DOI] [PubMed] [Google Scholar]
- Allouis S., Qi X., Lindup S., Gale M. D., Devos K. M. (2001). Construction of a BAC library of pearl millet, Pennisetum glaucum. Theor. Appl. Genet. 102 1200–1205. 10.1007/s001220100559 [DOI] [Google Scholar]
- ANSD (2014). Bulletin Mensuel des Statistiques Économiques de Mars 2014. Dakar: Agence Nationale de la Statistique et de la Démographie. [Google Scholar]
- Budak H., Pedraza F., Cregan P. B., Baenziger P. S., Dweikat I. (2003). Development and utilization of SSRs to estimate the degree of genetic relationships in a collection of pearl millet germplasm. Crop Sci. 43 2284–2290. 10.2135/cropsci2003.2284 [DOI] [Google Scholar]
- Dussert Y., Snirc A., Robert T. (2015). Inference of domestication history and differentiation between early- and late-flowering varieties in pearl millet. Mol. Ecol. 24 1387–1402. 10.1111/mec.13119 [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT (2013). Millets Production in de World. Rome: FAOSTAT. [Google Scholar]
- Hardy O. J., Vekemans X. (2002). spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2 618–620. 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- Haussmann B. I. G., Boubacar A., Boureima S. S., Vigouroux Y. (2006). Multiplication and preliminary characterization of west and central African pearl millet landraces. Int. Sorghum Millets Newsl. 47 110–112. [Google Scholar]
- Haussmann B. I. G., Fred Rattunde H., Weltzien-Rattunde E., Traoré P. S. C., vom Brocke K., Parzies H. K. (2012). Breeding strategies for adaptation of pearl millet and sorghum to climate variability and change in West Africa. J. Agron. Crop Sci. 198 327–339. 10.1111/j.1439-037X.2012.00526.x [DOI] [Google Scholar]
- Hu Z., Mbacké B., Perumal R., Guèye M. C., Sy O., Bouchet S., et al. (2015). Population genomics of pearl millet (Pennisetum glaucum (L.) R. Br.): Comparative analysis of global accessions and Senegalese landraces. BMC Genomics 16:1048 10.1186/s12864-015-2255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24 1403–1405. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Kouakou P. K., Muller B., Guissé A., Yao R. N., Fofana A., Cissé N. (2013). Étude et prise en compte en modélisation de l’effet de la latitude sur la réponse à la photopériode chez divers génotypes de mil (Pennisetum glaucum (L.) R. Br.) du Sénégal. J. Appl. Biosci. 67 5289–5301. 10.4314/jab.v67i0.95051 [DOI] [Google Scholar]
- Kouressy M., Vaksmann M., Niangado O., Sanogo M. D. (2004). Valorisation et Préservation de la Diversité Génétique du Mil au Mali. Paris: IRD Éditions; 10.4000/books.irdeditions.539 [DOI] [Google Scholar]
- Lakis G., Ousmane A. M., Sanoussi D., Habibou A., Badamassi M., Lamy F., et al. (2012). Evolutionary dynamics of cycle length in pearl millet: the role of farmer’s practices and gene flow. Genetica 139 1367–1380. 10.1007/s10709-012-9633-1 [DOI] [PubMed] [Google Scholar]
- Lewis L. R. (2010). Biogeography and genetic diversity of pearl millet (Pennisetum glaucum) from Sahelian Africa. Prof. Geogr. 62 377–394. 10.1080/00330124.2010.483640 [DOI] [Google Scholar]
- Loiselle B. A., Sork V. L., Nason J., Graham C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82 1420–1425. 10.2307/2445869 [DOI] [Google Scholar]
- Mariac C., Jehin L., Saïdou A.-A., Thuillet A.-C., Couderc M., Sire P., et al. (2011). Genetic basis of pearl millet adaptation along an environmental gradient investigated by a combination of genome scan and association mapping: adaptation to climatic variation. Mol. Ecol. 20 80–91. 10.1111/j.1365-294X.2010.04893.x [DOI] [PubMed] [Google Scholar]
- Mariac C., Luong V., Kapran I., Mamadou A., Sagnard F., Deu M., et al. (2006a). Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor. Appl. Genet. 114 49–58. [DOI] [PubMed] [Google Scholar]
- Mariac C., Robert T., Allinne C., Remigereau M. S., Luxereau A., Tidjani M., et al. (2006b). Genetic diversity and gene flow among pearl millet crop/weed complex: a case study. Theor. Appl. Genet. 113 1003–1014. [DOI] [PubMed] [Google Scholar]
- Oumar I., Mariac C., Pham J.-L., Vigouroux Y. (2008). Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsatellite loci. Theor. Appl. Genet. 117 489–497. 10.1007/s00122-008-0793-4 [DOI] [PubMed] [Google Scholar]
- Peakall R., Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I., Violle C., Barre P., Durand J.-L., Ghesquiere M., Litrico I. (2015). Complementary effects of species and genetic diversity on productivity and stability of sown grasslands. Nat. Plants 1:15033 10.1038/nplants.2015.33 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucher A., Sy O., Angarawai I. I., Gondah J., Zangre R., Ouedraogo M., et al. (2015). Agro-morphological characterization of west and central african pearl millet accessions. Crop Sci. 55 737–748. 10.2135/cropsci2014.06.0450 [DOI] [Google Scholar]
- Qi X., Lindup S., Pittaway T. S., Allouis S., Gale M. D., Devos K. M. (2001). Development of simple sequence repeat markers from bacterial artificial chromosomes without subcloning. Biotechniques 31 355–362. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Saïdou A.-A., Mariac C., Luong V., Pham J.-L., Bezançon G., Vigouroux Y. (2009). Association studies identify natural variation at PHYC linked to flowering time and morphological variation in pearl millet. Genetics 182 899–910. 10.1534/genetics.109.102756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stich B., Haussmann B. I., Pasam R., Bhosale S., Hash C. T., Melchinger A. E., et al. (2010). Patterns of molecular and phenotypic diversity in pearl millet [Pennisetum glaucum (L.) R. Br.] from West and Central Africa and their relation to geographical and environmental parameters. BMC Plant Biol. 10:216 10.1186/1471-2229-10-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan B., Roudier P., Quirion P., Alhassane A., Muller B., Dingkuhn M., et al. (2013). Assessing climate change impacts on sorghum and millet yields in the Sudanian and Sahelian savannas of West Africa. Environ. Res. Lett. 8 014040 10.1088/1748-9326/8/1/014040 [DOI] [Google Scholar]
- Sy O., Fofana A., Cisse N., Noba K., Diouf D., Ndoye I., et al. (2015). Étude de la variabilité agromorphologique de la collection nationale de mils locaux du Sénégal. J. Appl. Biosci. 87 8030–8046. 10.4314/jab.v87i1.1 [DOI] [Google Scholar]
- Thioulouse J., Chessel D., Dolédec S., Olivier J.-M. (1997). ADE-4: a multivariate analysis and graphical display software. Stat. Comput 7 75–83. 10.1023/A:1018513530268 [DOI] [Google Scholar]
- Vekemans X., Hardy O. J. (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 13 921–935. 10.1046/j.1365-294X.2004.02076.x [DOI] [PubMed] [Google Scholar]
- Vigouroux Y., Mariac C., De Mita S., Pham J.-L., Gérard B., Kapran I., et al. (2011). Selection for earlier flowering crop associated with climatic variations in the sahel. PLoS ONE 6:e19563 10.1371/journal.pone.0019563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington S. R., Li X., Dixon J., Hyman G., de Vicente M. C. (2010). Getting the focus right: production constraints for six major food crops in Asian and African farming systems. Food Secur. 2 27–48. 10.1007/s12571-010-0053-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.