Abstract
Parafibromin, the product of the HRPT2 (hyperparathyroidism-jaw tumor syndrome 2) tumor suppressor gene, is the human homologue of yeast Cdc73, part of the yeast RNA polymerase II/Paf1 complex known to be important for histone modification and connections to posttranscriptional events. By purifying cellular parafibromin and characterizing its associated proteins, we have identified a human counterpart to the yeast Paf1 complex including homologs of Leo1, Paf1, and Ctr9. Like the yeast complex, the parafibromin complex associates with the nonphosphorylated and Ser2 and Ser5 phosphorylated forms of the RNA polymerase II large subunit. Immunofluorescence experiments show that parafibromin is a nuclear protein. In addition, cotransfection data suggest that parafibromin can interact with a histone methyltransferase complex that methylates histone H3 on lysine 4. Some mutant forms of parafibromin lack association with hPaf1 complex members and with the histone methyltransferase complex, suggesting that disruption of these complexes may correlate with the oncogenic process.
Hyperparathyroidism-jaw tumor (HPT-JT) syndrome is characterized by parathyroid tumors, as well as ossifying fibromas of the mandible and maxilla, bilateral renal cysts, hamartomas, and Wilms' tumors (2, 9, 32). The HRPT2 gene is mutated in the germ line of HPT-JT patients, causing truncation, missense, or frameshift mutations in the parafibromin open reading frame. Somatic inactivation of HRPT2 was also observed in sporadic parathyroid carcinomas (2, 7, 25).
Parafibromin, the product of the HRPT2 gene, has 32% sequence identity with yeast Cdc73 (2) in the region encompassing amino acids 419 to 523 of parafibromin, according to BLAST analysis. Cdc73 is a component of the yeast Paf1 protein complex that interacts with RNA polymerase II and is composed of five known subunits: Paf1, Cdc73, Leo1, Ctr9, and Rtf1 (14, 18, 26, 27, 30). Paf1 complex genes are nonessential (26, 27, 31); loss of PAF1 and loss of CTR9 result in similar severe phenotypes affecting many cellular processes, while deletions of CDC73, LEO1, and RTF1 lead to less prominent phenotypes (1). Deletion of Rtf1 or Cdc73 results in the loss of association of the remaining Paf1 complex members with chromatin and a significant reduction in binding of the complex to RNA polymerase II (19). In addition, loss of Paf1 complex components leads to a reduction in RNA polymerase II carboxy-terminal domain (CTD) Ser2 phosphorylation and shortened poly(A) tails on most cellular transcripts, suggesting that the Paf1 complex facilitates linkage of transcriptional and posttranscriptional events (19).
Paf1 complex components are found at promoters, as well as throughout the coding regions of genes (19, 23, 28). The complex is required for expression of several cellular genes, including genes involved in cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism (1, 3, 10, 24).
The Paf1 complex is involved in multiple aspects of histone methylation. Yeast mutants missing the Paf1, Ctr9, and Rtf1 components of the complex are defective in histone 3 lysine 4 (H3 K4) methylation (13, 22), Paf1 and Rtf1 are required for histone H3 lysine 79 methylation (13), and deletions of CDC73 and RTF1 eliminate histone H3 lysine 36 methylation on the PMA1 target gene (15). Furthermore, the Paf1 complex is required for recruitment of the yeast Set1 (COMPASS) methyltransferase to RNA polymerase II; the subunits of these three complexes have been shown to interact physically and genetically (13, 22). The Paf1 complex also mediates histone H3 methylation on lysines 36 and 79, catalyzed by the Set2 and Dot1 complexes, respectively (4, 6). In addition, it was recently shown that the Rtf1 complex component is required for histone H2B ubiquitination (21).
Methylation of specific lysine residues within histones H3 and H4 marks genes for either activation or repression of transcription (12, 29). Transcriptionally active chromatin is generally associated with methylation at H3 lysines 4, 36, and 79, whereas repression is associated with methylation of H3 lysines 9 and 27 and H4 lysine 20. Several mammalian protein complexes that are related to the yeast Set1 complex (COMPASS) have been identified and shown to methylate H3 lysine 4, including the menin (8) and MLL (17, 20, 35) complexes, the ASCOM complex (5), and the HCF-1 complex (34).
We have applied immunopurification of cellular parafibromin-containing complexes to explore the biochemical function of this tumor suppressor protein. The results suggest that parafibromin may act as part of a complex analogous to the yeast Cdc73/Paf1 assembly.
MATERIALS AND METHODS
Antibodies.
Antiparafibromin BL648(A300-170A) and BL649(A300-171A), anti-hPaf1 BL673(A300-172A) and BL674(A300-173A), anti-hLeo1 BL677(A300-175A), anti-hCtr9 BL678(A300-176A) and BL679(A300-177A), and anti-hRtf1 BL680(A300-178A) and BL681(A300-179A) are rabbit polyclonal antibodies made by Bethyl Laboratories. Anti-Rbbp5(A300-109A) and anti-Ash2L BL867 were also purchased from Bethyl. Anti-RNA polymerase II antibodies for unphosphorylated CTD (8WG16), Ser2 CTD (H5), and Ser5 CTD (H14) were from Covance. Anti-Flag M2 antibody was from Sigma. Normal rabbit immunoglobulin G (IgG) was purchased from Santa Cruz Biotechnology.
Immunopurification for mass spectrometry.
To purify parafibromin complexes, 30 10-cm-diameter semiconfluent plates of 293T cells were lysed with NP-40 lysis buffer (250 mM NaCl, 50 mM Tris [pH 8.0], 5 mM EDTA, 0.5% NP-40, complete protease inhibitor [Roche]) at 1× strength and immunoprecipitated with 60 μg of antiparafibromin antibody Ab648 or a peptide-blocked control (1:1 ratio of antibody to peptide, i.e., 60 μg of antibody and 60 μg of blocking peptide). Immunoprecipitates were separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-8% PAGE) and stained with Coomassie blue (NOVEX; Invitrogen), and specific bands were excised. Trypsin digestion and mass spectrometry were carried out at the Taplin Biological Mass Spectrometry Facility as previously described (8). Peptides from the following proteins were identified: parafibromin (NP_078805), hPaf1 or PD2 (NP_061961), hLeo1 or LOC123169 (NP_620147), and hCtr9 or SH2BP1 (NP_055448). No peptides corresponding to the human homolog of Rtf1 or KIAA0252 (NP_055953) were found. For purification of hLeo1 complexes, 10 plates of 293T cells were lysed with NP-40 lysis buffer and immunoprecipitated with either 50 μg of anti-hLeo1 Ab677 or a peptide-blocked control. Immunoprecipitates were separated by SDS-8% PAGE and silver stained (Silver Stain Plus; Bio-Rad), and the bands were excised.
Plasmids.
Full-length parafibromin and hPaf1 and putative full-length Rtf1 were cloned by performing reverse transcription-PCRs (ThermoScript; Invitrogen) on RNA extracted from K562 cells (TRIZOL; Life Technologies) with primers for parafibromin (sense, 5′ CGGAATTCGGATCCACCATGGATTACAAGGATGACGACGATAAGGTCGACATGGCGGACGTGCTTAGCGT 3′; antisense, 5′ CCGCTCGAGTCAGAATCTCAAGTGCGATTTATGCTT 3′), hPaf1 (sense, 5′ CGGGATCCATGGCGCCCACCATCCAG 3′; antisense, 5′ CCGCTCGAGTCAGTCACTGTCACTATCAGCTTCACTG 3′), and hRtf1 (sense, 5′ CCCAAGCTTATGAAGAAACAGGCCAACA 3′; antisense, 5′ CCGCTCGAGAATAAGCCCTCGTCGTTT 3′).
The primers were designed to contain restriction sites that enable cloning into expression vector pcDNA3 cut with the following enzymes: parafibromin, EcoRI and XhoI; hPaf1, BamHI and XhoI; hRtf1, HindIII and XhoI. The parafibromin sense primer also encompasses a sequence for a Flag tag. Clones representing full-length hLeo1 and hCtr9 inserted in a mammalian expression vector were purchased from Origene Technologies Inc.
Mutant versions of the HRPT2 gene, expressing Flag-tagged mutant forms of parafibromin, were generated by PCRs that resulted in insertion of a stop codon at the end of the sequence. 1238X was made with the same sense primer used to clone full-length parafibromin and the antisense primer 5′ CCGCTCGAGTCACATCTGGTCTTTTCTTCTTTG 3′ and cloned into pCDNA3. The other truncations were generated by PCR with the same sense primer (5′ CGGGATCCATGGCGGACGTGCTTAGCGT 3′) and the specific antisense primers K136X (5′ CCGCTCGAGCTATGCTTCTGCTAAAACTTC 3′) and 679x (5′ CCGCTCGAGTCAGCTGACAATATCTCGGGTCAC 3′).
The resulting fragments were cloned into pcDNA3 containing a Flag tag. The L64P mutant construct was generated with the QuikChange mutagenesis kit (Stratagene).
Transfection, coimmunoprecipitation, and immunoblotting.
For immunofluorescence assays, HeLa cells grown on coverslips in six-well plates were transfected with 1 μg of Flag-tagged wild-type (WT) parafibromin or 2 μg of Flag-tagged mutant forms of parafibromin with Lipofectamine (GIBCO) in accordance with the manufacturer's instructions. For cotransfection experiments, 293T cells in 10-cm-diameter plates were transfected with 2 μg of Flag-tagged WT parafibromin, 5 μg of Flag-L64P or Flag-413X, 10 μg of Flag-227X or Flag-136X with 2 μg of T7-hPaf1, 5 μg of hLeo1 or hCtr9 (Origene), or 3 μg of hemagglutinin-Rbbp5 or hemagglutinin-ASH2L with Lipofectamine (GIBCO) in accordance with the manufacturer's instructions.
For immunoprecipitation or reciprocal coimmunoprecipitation experiments, one 10-cm-diameter plate of cells (endogenous or transfected) was lysed with NP-40 lysis buffer. Lysates were incubated for 3 h with 3 to 5 μl of specific antibody, antibodies blocked with peptide (preincubated at a 1:1 ratio on ice for 1 h before immunoprecipitation), or rabbit IgG and 50 μl of 20% protein A Sepharose CL-4B (Amersham Bioscience). For Flag immunoprecipitation, 2 μl of antibody was used. The beads were then washed four times with NP-40 lysis buffer and resuspended in 30 μl of protein sample buffer X2. Proteins were resolved by SDS-PAGE and then subjected to immunoblot analysis. To eliminate the possibility of DNA contamination, immunoprecipitates were washed four times with NP-40 lysis buffer, washed one time with DNase buffer, and incubated with 3 μl of DNase (RQ1 Promega) in 100 μl of DNase buffer at 37°C for 30 min and washed twice with immunoprecipitation lysis buffer.
For detection by immunoblot assay, the following dilutions were used for primary antibodies: antiparafibromin antibody Ab648, 1:5,000; anti-hPaf1 antibody Ab673, 1:10,000; anti-hLeo1 antibody Ab677, 1:2,000; anti-hCtr9 antibody Ab679, 1:5,000; anti-Flag antibody, 1:2,000; anti-Rbbp5 antibody, 1:5,000; anti-Ash2L antibody, 1:2,500. RNA polymerase II antibodies were used in accordance with the manufacturer's instructions. Secondary antibodies were goat anti-rabbit antibody conjugated to horseradish peroxidase (Pierce or Bethyl) and goat anti-mouse IgG plus IgM conjugated to horseradish peroxidase (Pierce). Proteins were visualized by enhanced chemiluminescence (SuperSignal; Pierce).
Immunofluorescence assay.
HeLa cells were grown on glass coverslips in six-well dishes. For ectopic expression, cells were transfected the appropriate plasmid and grown for 48 h posttransfection. Cells were washed three times with phosphate-buffered saline (PBS) and fixed with 4% formaldehyde in PBS (Buffered Formalde-Fresh; Fisher Scientific) for 30 min. Next, cells were washed three times with PBS and permeabilized with 0.5% Triton X-100 containing 3% bovine serum albumin (BSA) for 20 min. Cells were then exposed to the relevant antibodies (anti-Flag antibody at a 1:500 dilution or anti-hLeo1 antibody Ab677 at 1:800) diluted in PBS-3% BSA for 30 min at 37°C, washed three times with PBS, and incubated with fluorescently labeled secondary antibodies diluted in PBS-3% BSA for 30 min at 37°C (fluorescein isothiocyanate [FITC]-conjugated Affinipure goat anti-mouse and Cy3-conjugated Affinipure goat anti-rabbit antibodies from Jackson Immunoresearch). Slides were washed as before and mounted on microscope slides with mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI [Vectashield]; Vector).
Slides were imaged with a Zeiss 510 confocal multiphoton fluorescence microscope equipped with laser lines for DAPI (800 nm, Ti-sapphire coherent pulsed), FITC (Argon 488), and rhodamine (HeNe 543). The images were prepared with the Zeiss multitrack mode.
HMTase assay.
Histone methyltransferase (HMTase) assays were performed as previously described (8). Anti-Flag and/or antiparafibromin immunoprecipitates were washed three times with NP-40 lysis buffer and one time with HMTase buffer (25 mM Tris [pH 8.0], 10% glycerol). Each reaction mixture included 2 μl of S-adenosyl-l-[methyl-3H]methionine (Amersham Bioscience), 2 μg of histone H3 (Upstate Biotechnology), and assay buffer (HMT Assay Reagent Kit; Upstate Biotechnology). Reaction mixtures were incubated for 1 h at room temperature, boiled with protein sample buffer, and separated by SDS-15% PAGE. The gel was then amplified for 1 h (Amplify NAMP 100V; Amersham), dried, and exposed to film. For Edman degradation, an HMTase assay was performed and proteins were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membrane with CAPS (3-cyclohexylamino-1-propanesulfonic acid) buffer. To visualize histone bands, gels were Coomassie stained (Gelcode; Pierce) and destained overnight. Histone bands were excised and subjected to 40 rounds of Edman degradation as previously described (8).
RESULTS
Identification of parafibromin-associated proteins.
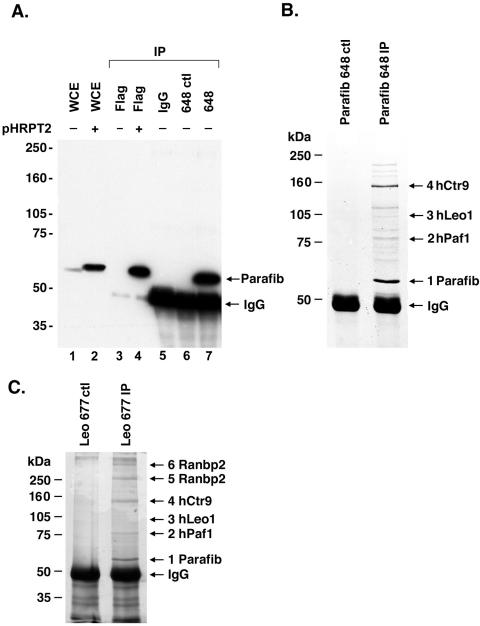
To explore the protein complexes associated with parafibromin, we generated and validated an antipeptide rabbit polyclonal antibody, Ab648, that specifically recognizes both transfected (Fig. 1A, lanes 2 and 4) and endogenous (Fig. 1A, lanes 1 and 7) parafibromin protein. To identify endogenous parafibromin-associated proteins, we immunoprecipitated protein from 293T cells with the Ab648 antiparafibromin antibody; as a control, we preincubated the antibody with a blocking peptide control (Fig. 1B). Immunoprecipitated protein bands were excised, and peptides were sequenced by tandem mass spectrometry.
FIG. 1.
Identification of parafibromin (Parafib) interacting proteins. (A) Characterization of antiparafibromin antibody Ab648 in transfected and untransfected cells. An immunoblot with Ab648 is shown. Whole-cell lysate (150 μg) of untransfected 293T cells (lane1) or 293T cells transfected with a plasmid encoding Flag-tagged parafibromin (pHRPT2; lane 2) is shown as a control. Proteins were immunoprecipitated (IP) from untransfected (lane 3) or transfected (lane 4) cell lysate with anti-Flag antibody (one-fifth of a 10-cm-diameter plate). Proteins were immunoprecipitated from lysates of untransfected cells (one 10-cm-diameter plate) with antiparafibromin antibody Ab648 (lane 7) and negative controls, normal rabbit IgG (lane 5) and antiparafibromin antibody Ab648 plus blocking peptide (lane 6). Proteins were resolved by SDS-8% PAGE and immunoblotted with antiparafibromin antibody Ab648. (B) Lysates of 293T cells were immunoprecipitated with antiparafibromin antibody Ab648 (right lane) or a peptide-blocked control (left lane), resolved by SDS-8% PAGE, and visualized by Coomassie blue staining. Bands 1 to 4 were subjected to mass spectrometric analysis. (C) Immunoprecipitation of protein from 293T cells with anti-hLeo1 antibody Ab677 (right lane) or a peptide-blocked control (left lane). Proteins were resolved by SDS-8% PAGE and visualized by silver staining. Numbered bands were identified by mass spectrometric analysis. WCE, whole-cell extract.
The 64-kDa band (Fig. 1B, band 1) was identified as parafibromin. Mass spectrometric analysis identified 42 peptides comprising 62.9% of the predicted parafibromin protein sequence. The proteins coimmunoprecipitated with antiparafibromin antibody include human homologs of the yeast Paf1 complex: an approximately 80-kDa band (Fig. 1B, band 2) includes peptides from hPaf1, a 105-kDa band (Fig. 1B, band 3) includes peptides from hLeo1, and an approximately 150-kDa band (Fig. 1B, band 4) includes peptides from hCtr9, with predicted amino acid sequence similarities of 40, 52, and 44%, respectively. No peptides corresponding to the human homolog of Rtf1 were identified in the immunoprecipitates. To our knowledge, the human hPaf1 and hLeo1 proteins have not been described in the literature. hCtr9 has been described as an SH2 domain binding nuclear phosphoprotein that has TPR repeats and is thought to be involved in protein-protein interaction (16). Identification of the remaining protein bands of the antiparafibromin immunoprecipitates is still in progress.
To verify interactions between parafibromin and the human homologs of the yeast Paf1 complex, we generated polyclonal antibodies against the parafibromin interacting proteins in the Paf1 complex (anti-hPaf1, anti-hLeo1, anti-hCtr9, and anti-hRtf1). These antibodies were validated for the ability to immunoprecipitate antigens and associated proteins from metabolically labeled cells (not shown). To reconfirm interactions among putative parafibromin complex members, we immunoprecipitated protein from 293T cells with anti-hLeo1 antibody Ab677, with peptide-blocked antibody as a negative control (Fig. 1C). Immunoprecipitated proteins were excised from the gel and sequenced by tandem mass spectrometry. As expected, the 105-kDa band (Fig. 1C, band 3) corresponded to hLeo1, with five peptides covering 6.8% of the predicted sequence. Multiple peptides were identified from each of the other five proteins precipitated with the anti-hLeo1 antibody. Three of these proteins are human homologues of the yeast Paf1 complex that we had found in the parafibromin-associated complex. The 64-kDa protein (Fig. 1C, band 1) is parafibromin, the ∼80-kDa protein (Fig. 1C, band 2) is hPaf1, and the 150-kDa protein (Fig. 1C, band 4) is hCtr9 (11 peptides, 7.9% coverage). Analysis of two higher-molecular-weight bands (Fig. 1C, bands 5 and 6) revealed peptides corresponding to Ranbp2, a nuclear pore complex protein (36).
Parafibromin is associated with a human Paf1 complex.
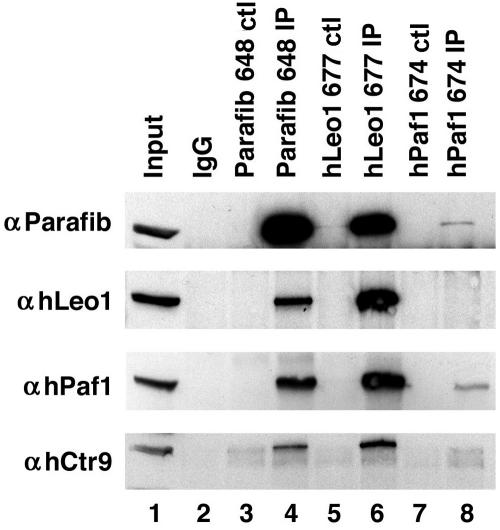
To verify interaction among the hPaf1 complex proteins more thoroughly, reciprocal coimmunoprecipitation experiments were performed with antibodies to the endogenous proteins (Fig. 2). Antiparafibromin antibody Ab648 could specifically precipitate parafibromin, hLeo1, hPaf1, and hCtr9 (Fig. 2, lane 4). These proteins were not detectable with peptide-blocked antibody, the negative control (Fig. 2, lane 3). Similarly, immunoprecipitation with anti-hLeo1 antibody but not the peptide-blocked control could isolate all four hPaf1 complex members (Fig. 2, lanes 5 and 6). The anti-hPaf1 antibodies, while effective for immunoblotting, were not as efficient for immunoprecipitation but could still precipitate hPaf1 and parafibromin (Fig. 2, lanes 7 and 8). Taken together, the above results indicate that parafibromin, hPaf1, hLeo1, and hCtr9 form a stable complex. The antiparafibromin, anti-hLeo1, and anti-hPaf1 antibodies did not immunoprecipitate a band that could be detected by anti-hRtf1 antibodies in immunoblot analysis. These interactions remained intact following treatment with DNase (not shown), indicating that the complex is likely to be assembled via protein-protein interactions rather than protein-DNA interactions.
FIG. 2.
Parafibromin (Parafib) is part of a human Paf1 complex. Antiparafibromin antibody Ab648 (lane 4), anti-hLeo1 antibody Ab677 (lane 6), and anti-hPaf1 antibody Ab674 (lane 8) immunoprecipitates from 293T cell lysates were immunoblotted with antiparafibromin antibody Ab648 (top panel), anti-hLeo1 antibody Ab677 (second panel), anti-hPaf1 antibody Ab673 (third panel), and anti-hCtr9 antibody Ab679 (bottom panel). Cell lysate was loaded in lane 1. Negative control (ctl) immunoprecipitations (IP) were performed with normal rabbit IgG (lane 2) and peptide-blocked controls (lanes 3, 5, and 7).
Parafibromin interacts with the RNA polymerase II large subunit.
Yeast Paf1 complex members have been reported to associate with RNA polymerase II (14, 18, 30). The repetitive CTD of the RNA polymerase II large subunit, Rpb1, has two important serine residues that become highly phosphorylated during transcription. Phosphorylation on Ser5 is correlated with initiation and early elongation, while phosphorylation on Ser2 is linked to established elongation (11).
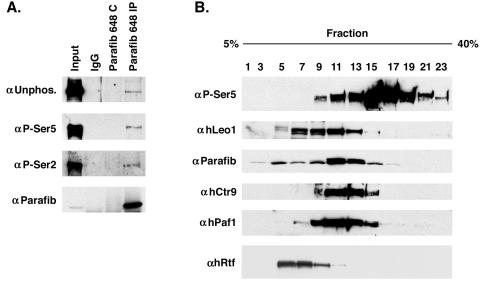
To test whether the parafibromin complex interacts with any of the phosphorylated forms of Rpb1, we immunoprecipitated parafibromin and blotted the immunoprecipitates with specific antibodies to detect the Rpb1 CTD that is unphosphorylated, phosphorylated on Ser5, or phosphorylated on Ser2. Parafibromin specifically coprecipitated all three forms of Rpb1 (Fig. 3A), suggesting that it may be involved in both initiation and elongation.
FIG. 3.
Parafibromin (Parafib) interacts with the RNA polymerase large subunit. (A) 293T cell lysates were immunoprecipitated (IP) with antiparafibromin antibody Ab648, a peptide-blocked control (C), or a normal rabbit serum control and immunoblotted with antibodies to Rpb1 with unphosphorylated (Unphos.) CTD (top part), Ser5-phosphorylated CTD (second part), Ser2-phosphorylated CTD (third part), and parafibromin (bottom part). (B) 293T cell lysate was fractioned on a glycerol gradient. Forty-microliter aliquots of the fractions depicted were used for immunoblotting with anti-Rpb1 Ser5-phosphorylated CTD (top part), anti-hLeo1 antibody Ab677 (second part), antiparafibromin antibody Ab648 (third part), anti-hCtr9 antibody Ab679 (fourth part), anti-hPaf1 antibody Ab673 (fifth part), and anti-hRtf1 antibody (bottom part).
To further explore the relationship between the hPaf1 complex and RNA polymerase II, 293T cell extract was fractionated with a glycerol gradient and fractions from the gradient were separated on a polyacrylamide gel and immunoblotted for the indicated proteins (Fig. 3B). The hPaf1 complex members mainly comigrated in fractions 11 through 13. The Rpb1 Ser5 CTD phosphorylated form overlapped with the parafibromin complex, but most of the RNA polymerase II appears to be in a higher-molecular-weight complex. In addition, we also tested the fractions for hRtf1. The hRtf1 reactivity is mainly found in fractions 5 to 9, not overlapping the parafibromin/Paf1 complex.
Parafibromin is a nuclear protein.
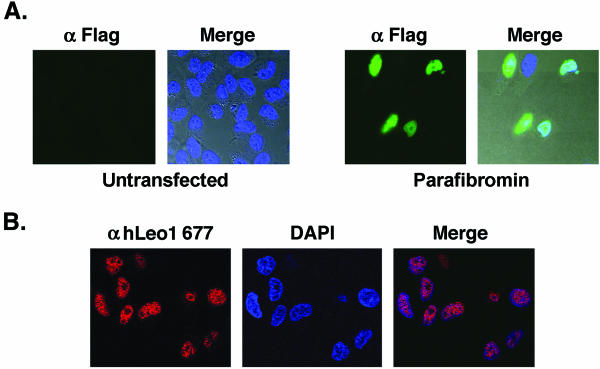
To determine the subcellular localization of parafibromin and the hPaf1 complex proteins, we performed immunofluorescence studies. As neither of the antiparafibromin antibodies could detect endogenous protein by immunofluorescence, we overexpressed WT parafibromin in HeLa cells with a Flag epitope tag on its N terminus. Confocal microscopic detection of immunofluorescence showed significant overlap between overexpressed Flag-tagged parafibromin (green fluorescence) and DAPI staining (Fig. 4A, right side), indicating that parafibromin is a nuclear protein. Control cells that were not transfected did not exhibit any staining (Fig. 4A, left side) In addition, immunofluorescence was performed with the anti-hLeo1 Ab677 antibody. Confocal microscopy shows that hLeo1 is a nuclear protein with a punctate distribution (Fig. 4B). Taken together, these results suggest that parafibromin and hPaf1 complex members localize to the nucleus.
FIG. 4.
Parafibromin and hLeo1 are nuclear proteins. (A) Immunofluorescence assay with FITC-conjugated anti-Flag antibody on untransfected HeLa cells (Untransfected) or HeLa cells transfected with Flag-tagged WT parafibromin (Parafibromin). The left side of each pair of images shows the FITC signal, while the right side, marked “Merge,” is the integration of the FITC and DAPI signals overlaid upon the Nomarski-differential interference contrast images of the cells. (B) Immunofluorescence with anti-Leo1 antibody Ab677 (Cy3 red) was used to visualize endogenous Leo1 (left image), and DAPI staining (middle image) was used to mark nuclear DNA in HeLa cells. The right image, labeled “Merge,” shows the colocalization of the two signals.
Some mutant forms of parafibromin lack association with Paf1 complex members.
Several germ line and somatic mutations have been identified in HRPT2 (2, 7, 25). Most of these are truncation and frameshift mutations that result in premature stop codons and are predicted to cause deficient or impaired parafibromin protein function.
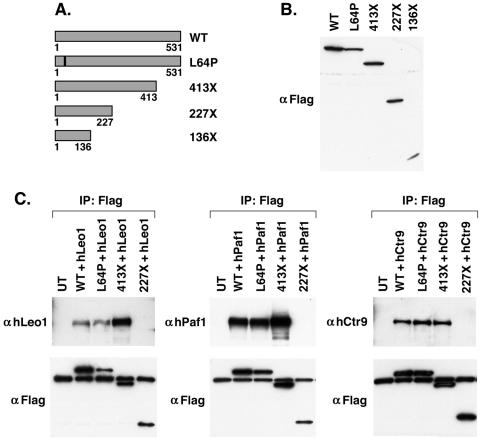
We generated expression constructs for several truncation mutant forms of parafibromin—136X, 227X, and 413X—by PCR (Fig. 5A). 136X is a truncation found in the germ line of HPT-JT patients. We chose to make the 227X and 413X truncations because some patients have germ line nucleotide insertions or deletions at codons 226 and 413 that result in a frame shift and early termination. In addition, we made an L64P point mutation that was identified in germ line HPT-JT. This leucine residue is conserved in Drosophila, Caenorhabditis elegans, mice, and humans, suggesting that it has an important function. A Flag epitope tag was introduced into the 5′ end of each parafibromin-encoding truncation mutant construct. The parafibromin-encoding truncation constructs were transfected into 293T cells. Cell lysates from these transfections were fractioned by SDS-PAGE, and expression was assessed by immunoblotting with anti-Flag antibody (Fig. 5B). All of the constructs gave rise to detectable protein corresponding to the expected size.
FIG. 5.
Characterization of parafibromin truncation mutant constructs. (A) Schematic diagram of parafibromin truncation mutant constructs and the L64P mutant construct representing germ line alterations identified from HPT-JT patients. (B) 293T cell lysates from cells transfected with Flag-tagged WT parafibromin or Flag-tagged mutant forms of parafibromin were immunoblotted with anti-Flag antibodies. (C) Immunoprecipitations (IP) with anti-Flag antibodies from 293T cell lysates cotransfected with either hLeo1 (left), hPaf1 (middle), or hCtr9 (right) together with the indicated Flag-tagged WT parafibromin or Flag-tagged parafibromin mutant constructs. Transfectants were immunoblotted with anti-hLeo1, anti-hPaf1, or anti-hCtr9 antibodies (top) or with anti-Flag antibody (bottom) as shown. Untransfected (UT) 293T cells were used as a negative control.
To test the abilities of the mutant forms of parafibromin to associate with hPaf1 complex members, we cotransfected 293T cells with Flag-tagged WT parafibromin or parafibromin mutant constructs together with an expression plasmid encoding either hLeo1, T7-hPaf1, or hCtr9 protein and immunoprecipitated the parafibromin complex with anti-Flag antibody (Fig. 5C). A portion of this immunoprecipitation was blotted with anti-Flag antibody to verify expression and precipitation of the various parafibromin proteins. Although hPaf1, hLeo1, and hCtr9 were expressed in all of the cells (not shown), only WT parafibromin, L64P, and 413X could immunoprecipitate hPaf1 complex proteins while 227X and 136X (not shown) could not, indicating that a portion of parafibromin between amino acids 226 and 413 is required for binding to the hPaf1 complex.
Parafibromin can associate with an HMTase complex.
Because yeast Cdc73 has been reported to associate with a Set1 HMTase complex, we decided to test the association of both WT and mutant parafibromin with HMTase activity. To test the association of parafibromin and the mutant forms of parafibromin with HMTase activity, we transfected 293T cells with Flag-tagged WT parafibromin or parafibromin mutant constructs and performed an HMTase assay (Fig. 6A). WT parafibromin and the 413X mutant construct are associated with HMTase activity, while the 227X mutant construct is not. The L64P mutant form of parafibromin shows reduced activity compared to that of the WT and 413X forms (Fig. 6A); this result was obtained consistently in three experiments.
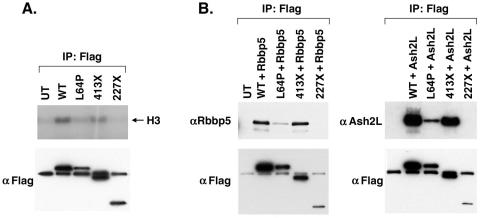
FIG. 6.
Interaction between parafibromin and HMTase activity and complex members. (A) HMTase assays were performed on immunoprecipitates (IP) with anti-Flag antibodies from 293T cell lysates transfected with the indicated Flag-tagged WT parafibromin or Flag-tagged parafibromin mutant constructs (top). Transfectants were immunoblotted with anti-Flag antibody (bottom) as shown. Untransfected (UT) 293T cells were used as a negative control. (B) Immunoprecipitations with anti-Flag antibodies from 293T cell lysates cotransfected with either Rbbp5 (left) or Ash2L (right) together with the indicated Flag-tagged WT parafibromin or Flag-tagged parafibromin mutant constructs. Transfectants were immunoblotted with anti-Rbbp5 or anti-Ash2L antibodies (top) or with anti-Flag antibody (bottom) as shown. Untransfected 293T cells were used as a negative control.
Given the HMTase activity associated with parafibromin, we tested whether WT parafibromin and the mutant forms of parafibromin can interact with components of Set1-like complexes, including Rbbp5 and Ash2L. We cotransfected 293T cells with Flag-tagged WT parafibromin or parafibromin mutant constructs together with an expression plasmid encoding either Rbbp5 or Ash2L protein and immunoprecipitated the parafibromin-containing complex with anti-Flag antibody (Fig. 6B). Similar to the association with HMTase activity, WT parafibromin and 413X could immunoprecipitate the Set1 complex proteins Rbbp5 and Ash2L while 227X could not. L64P exhibits reduced binding to these proteins, consistent with the HMTase activity of this mutant construct.
In addition, we determined the subcellular localization of the various mutant forms of parafibromin (data not shown). The L64P, 413X, and 227X mutant constructs exhibited predominantly nuclear staining, while the K136X mutant construct typically showed some even nuclear staining but also significant cytoplasmic and cell membrane localization. These results indicate that the putative classic nuclear localization signal, at amino acids 136 to 139 of parafibromin, may be functional. These results suggest that binding to hPaf1 complex components and to HMTase complex components, as well as nuclear localization, may be essential to parafibromin function and that elimination of complex assembly may contribute to tumor formation in the setting of HRPT2 mutation.
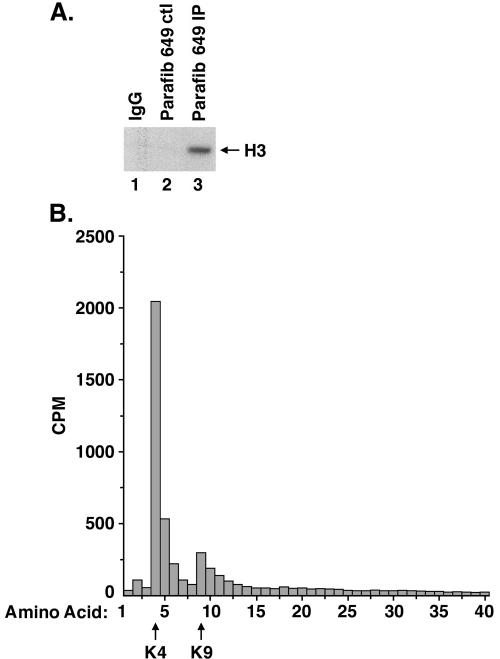
Recently we have generated a second antiparafibromin polyclonal antibody, Ab649. After checking the specificity of this antibody, we found that in addition to parafibromin it precipitates two other major bands, possibly corresponding to the angiomotin protein (Amot). Angiomotin has been reported to bind to angiostatin and to regulate endothelial cell migration and tube formation and has not been shown to be involved in chromatin remodeling (33). Since the overexpressed parafibromin protein is associated with HMTase complex components and activity, we assayed parafibromin-containing complexes precipitated by Ab649 for HMTase activity. To that end, we immunoprecipitated protein from 293T cell lysates with antiparafibromin Ab649 or peptide-blocked or IgG controls and performed an HMTase assay. The antiparafibromin immunoprecipitate specifically methylated histone H3 (Fig. 7A, lane 3), while the controls did not (Fig. 7A, lanes 1 and 2).
FIG. 7.
Parafibromin (Parafib) interacts with an HMTase complex that methylates histone H3 on lysine 4. (A) Antiparafibromin antibody Ab649 (lane 3), normal rabbit IgG (lane 1), and peptide-blocked antibody Ab649 control (ctl; lane 2) immunoprecipitates were incubated with histone H3 and the methyl donor S-adenosyl-l-[methyl-3H]methionine, resolved by SDS-15% PAGE, amplified, dried, and fluorographed. (B) Scintillation counting of sequential Edman degradation products of histone H3, labeled by incubation with S-adenosyl-l-[methyl-3H]methionine and with immunoprecipitates of antiparafibromin antibody Ab649. The arrows at the bottom indicate lysines 4 and 9.
To determine which lysine residue in histone H3 is methylated by the parafibromin-associated HMTase complex, we performed Edman degradation on labeled histone H3 with antiparafibromin Ab649, peptide-blocked, or IgG immunoprecipitates prepared from lysates of 293T cells. The radioactivity released by the degradation products was counted (Fig. 7B). Histone H3 incubated with the antiparafibromin immunoprecipitate was specifically labeled on lysine 4 (Fig. 7B), while the control was not specifically methylated (not shown). Some methylation was also detected on H3 lysine 9, but this appeared to be nonspecific. Taken together, theses results suggest that endogenous parafibromin is associated with a Set1-like HMTase complex that methylates histone H3 on Lys4.
DISCUSSION
The biological role of the parafibromin tumor suppressor protein encoded by the HRPT2 gene (2) has not been defined previously. A hint as to parafibromin's function comes from its homology to yeast Cdc73, part of the Paf1 complex that is involved in transcription and histone methylation (4, 6, 13, 22).
We show that parafibromin is part of a complex that contains the human homologues of the yeast Paf1 complex, the hPaf1, hLeo1, and hCtr9 proteins. These interactions were confirmed by mass spectrometry, endogenous coimmunoprecipitation, comigration on a glycerol gradient, and immunofluorescence assay. In yeast, the Paf1 complex was isolated via its interaction with RNA polymerase II and shown to be composed of five subunits (Paf1, Rtf1, Cdc73, Leo1, and Ctr9) (14, 18, 30). To our surprise, the human complex did not contain the human homolog of Rtf1. We do not know whether the human Rtf1 homolog is in fact associated with the hPaf1 complex, but multiple assays including glycerol gradients do not support such an association.
The function of the human parafibromin/hPaf1 complex remains to be elucidated. While various possible functions for the yeast complex have been identified, including regulation of transcriptional elongation, histone methylation, and RNA processing, the critical biochemical activities of the complex are somewhat unclear. Further experiments are required to determine the cellular functions of the hPaf1 complex and their relationship to parafibromin tumor suppression.
Our data also suggest that, like Cdc73 (13, 22), parafibromin is associated with a Set1-like HMTase complex that methylates histone H3 on lysine 4, an activity that is associated with transcriptional activation. Transfected and endogenous parafibromin are associated with a histone methylation activity that is absent or reduced in several mutant forms of parafibromin. We also have some preliminary evidence that endogenous cellular parafibromin is associated with a cellular Set1 family HMTase protein; further investigation is needed to confirm these observations.
Interestingly, the menin tumor suppressor protein, which is also mutated in multiple endocrine tumors, has been shown to associate with a Set1-like HMTase complex and RNA polymerase II (8, 35). The menin complex includes the MLL2 or MLL1 protein as the SET domain protein responsible for histone methylation. Mutations in both parafibromin and menin give rise to parathyroid tumors. This may indicate that the two proteins might regulate similar genes involved in cellular differentiation or proliferation or that these complexes might interact. Despite the similarity between the two complexes, we have not been able to show a direct interaction between parafibromin and menin. Since mutations in HRPT2 do not account for all cases of parathyroid carcinomas (2, 7, 25), it would be interesting to sequence genes for other hPaf1 complex members in these tumors since their loss might also promote tumorigenesis.
Like Cdc73, parafibromin is associated with RNA polymerase II that is phosphorylated on Ser5 and Ser2. This suggests that parafibromin is associated with the entire active mRNA coding region and might also be traveling with the elongated polymerase II enzyme throughout the entire gene. Data on yeast suggest that Set1 recruitment to the promoter occurs after transcriptional initiation and is dependent on Ser5 phosphorylation of the RNA polymerase II CTD and on the presence of the Paf1 complex (4, 6, 13, 22).
However, the Paf1 complex associates with the entire mRNA coding region, indicating that it probably has an additional role(s) (19, 23, 28). No parafibromin target genes have been published to date. It would be interesting to test whether parafibromin and Paf1 complexes are associated with the transcription of all, most, or only several genes.
The observation that some of the patient-derived parafibromin truncation mutant constructs fail to coimmunoprecipitate Paf1 complex members and HMTase activity suggests that complex formation is related to parafibromin tumor suppressor function. Further detailed study of these complexes should lead to a better understanding of HPT-JT and to the role of histone modifications in cancer, most particularly, endocrine cancer. Recent data (19) suggest that the Paf1 complex has an additional role(s) in the cell. Loss of Paf1 factors results in a reduction of RNA polymerase II Ser2 phosphorylation and shortened poly(A) tails, suggesting that the complex might facilitate linkage of transcriptional and posttranscriptional events. Identification of the other proteins that were immunoprecipitated with the antiparafibromin antibody could reveal other hPaf1 complex functions.
Acknowledgments
We thank E. McIntush at Bethyl Laboratories for antibody design, E. Kort and E. Hudson for immunofluorescence assay assistance, and J. W. Lee for the ASC-2 antibody. We are grateful to the Taplin Biological Institute for mass spectrometry analysis and Bethyl Laboratories for preparation of polyclonal antibodies. We also thank J. Jaehning and G. David for critical comments on the manuscript and helpful discussions.
This work was supported by a gift from Raymond and Beverly Sackler.
REFERENCES
- 1.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. [DOI] [PubMed] [Google Scholar]
- 2.Carpten, J. D., C. M. Robbins, A. Villablanca, L. Forsberg, S. Presciuttini, J. Bailey-Wilson, W. F. Simonds, E. M. Gillanders, A. M. Kennedy, J. D. Chen, S. K. Agarwal, R. Sood, M. P. Jones, T. Y. Moses, C. Haven, D. Petillo, P. D. Leotlela, B. Harding, D. Cameron, A. A. Pannett, A. Hoog, H. Heath III, L. A. James-Newton, B. Robinson, R. J. Zarbo, B. M. Cavaco, W. Wassif, N. D. Perrier, I. B. Rosen, U. Kristoffersson, P. D. Turnpenny, L. O. Farnebo, G. M. Besser, C. E. Jackson, H. Morreau, J. M. Trent, R. V. Thakker, S. J. Marx, B. T. Teh, C. Larsson, and M. R. Hobbs. 2002. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 32:676-680. [DOI] [PubMed] [Google Scholar]
- 3.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 5.Goo, Y. H., Y. C. Sohn, D. H. Kim, S. W. Kim, M. J. Kang, D. J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, V. T. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P. G. Suh, E. J. Song, K. J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 7.Howell, V. M., C. J. Haven, K. Kahnoski, S. K. Khoo, D. Petillo, J. Chen, G. J. Fleuren, B. G. Robinson, L. W. Delbridge, J. Philips, A. E. Nelson, U. Krause, K. Hammje, H. Dralle, C. Hoang-Vu, O. Gimm, D. J. Marsh, H. Morreau, and B. T. Teh. 2003. HRPT2 mutations are associated with malignancy in sporadic parathyroid tumours. J. Med. Genet. 40:657-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes, C. M., O. Rozenblatt-Rosen, T. A. Milne, T. D. Copeland, S. S. Levine, J. C. Lee, D. N. Hayes, K. S. Shanmugam, A. Bhattacharjee, C. A. Biondi, G. F. Kay, N. K. Hayward, J. L. Hess, and M. Meyerson. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13:587-597. [DOI] [PubMed] [Google Scholar]
- 9.Jackson, C. E., R. A. Norum, S. B. Boyd, G. B. Talpos, S. D. Wilson, R. T. Taggart, and L. E. Mallette. 1990. Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: a clinically and genetically distinct syndrome. Surgery 108:1006-1013. [PubMed] [Google Scholar]
- 10.Koch, C., P. Wollmann, M. Dahl, and F. Lottspeich. 1999. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 27:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 13.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 14.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malek, S. N., C. H. Yang, W. C. Earnshaw, C. A. Kozak, and S. Desiderio. 1996. p150TSP, a conserved nuclear phosphoprotein that contains multiple tetratricopeptide repeats and binds specifically to SH2 domains. J. Biol. Chem. 271:6952-6962. [DOI] [PubMed] [Google Scholar]
- 17.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 18.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447-456. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 21.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 22.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 23.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 24.Porter, S. E., T. M. Washburn, M. Chang, and J. A. Jaehning. 2002. The yeast Pafl-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot. Cell 1:830-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shattuck, T. M., S. Valimaki, T. Obara, R. D. Gaz, O. H. Clark, D. Shoback, M. E. Wierman, K. Tojo, C. M. Robbins, J. D. Carpten, L. O. Farnebo, C. Larsson, and A. Arnold. 2003. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N. Engl. J. Med. 349:1722-1729. [DOI] [PubMed] [Google Scholar]
- 26.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, X., A. Finkelstein, A. J. Wolf, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sims, R. J., III, K. Nishioka, and D. Reinberg. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet. 19:629-639. [DOI] [PubMed] [Google Scholar]
- 30.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szabo, J., B. Heath, V. M. Hill, C. E. Jackson, R. J. Zarbo, L. E. Mallette, S. L. Chew, G. M. Besser, R. V. Thakker, V. Huff, M. F. Leppert, and H. Heath. 1995. Hereditary hyperparathyroidism-jaw tumor syndrome: the endocrine tumor gene HRPT2 maps to chromosome 1q21-q31. Am. J. Hum. Genet. 56:944-950. [PMC free article] [PubMed] [Google Scholar]
- 33.Troyanovsky, B., T. Levchenko, G. Mansson, O. Matvijenko, and L. Holmgren. 2001. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J. Cell Biol. 152:1247-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, M. Fukui, and T. Nishimoto. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]