Abstract
The BRCT domain is a highly conserved module found in many proteins that participate in DNA damage checkpoint regulation, DNA repair, and cell cycle control. Here we describe the cloning, characterization, and targeted mutagenesis of Brctx, a novel gene with a BRCT motif. Brctx was found to be expressed ubiquitously in adult tissues and during development, with the highest levels found in testis. Brctx-deficient mice develop normally, show no pathological abnormalities, and are fertile. BRCTx binds to the C terminus of hRAD18 in yeast two-hybrid and immunoprecipitation assays and colocalizes with this protein in the nucleus. Despite this, Brctx-deficient murine embryonic fibroblasts (MEFs) do not show overt sensitivity to DNA-damaging agents. MEFs from Brctx-deficient embryos grow at a similar rate to wild-type MEF CD4/CD8 expressions, and the cell cycle parameters of thymocytes from wild-type and Brctx knockout animals are indistinguishable. Intriguingly, the BRCT domain of BRCTx is responsible for mediating its localization to the nucleus and centrosome in interphase cells. We conclude that, although highly conserved, Brctx is not essential for the above-mentioned processes and may be redundant.
The carboxyl-terminal domain (BRCT) of breast cancer gene 1 (BRCA1) is the signature domain for a number of DNA damage and checkpoint genes conserved throughout evolution (4). There are 23 genes containing BRCT domains in the human genome (www.ensembl.org). Proteins with BRCT domains function as regulators of DNA repair, the cell cycle, and transcription (4, 29). Much interest has surrounded these proteins after the cloning of the breast and ovarian cancer susceptibility genes BRCA1 and BRCA2 (18, 38). These genes have subsequently been shown to play an important role in tumor formation in other tissues (21, 35). Other proteins with BRCT motifs include NBS1 and p53BP1, which have both been shown to play a role in DNA repair. Mice carrying modified alleles of NBS1 and p53BP1 are tumor prone (12, 37). It has recently been shown that BRCT domains preferentially bind phosphopeptides in preference to unphosphorylated peptide fragments (16, 40) and in doing so can transduce signals from protein kinases operating in a variety of different signaling cascades. Thus, the physiological targets of BRCT domain proteins can be many and varied. Knockouts of BRCT domain proteins have exhibited a variety of phenotypes such as embryonic lethality, growth defects, defects in T-cell development, and infertility (8, 12, 20, 28, 36, 37). Thus, these proteins exhibit a variety of functions during development.
In the present study we set out to investigate the function of Brctx, an uncharacterized member of the BRCT domain-containing family of proteins. The human homologue of Brctx is located on chromosome 5q15, a region of the genome frequently rearranged in cancers of the breast (10), bladder (27), ovary (33), and lung (17); the tumor suppressor gene at this locus has not been identified. Here we use expression profiling, protein interaction, and targeted mutagenesis of Brctx to gain clues about its function.
MATERIALS AND METHODS
Cloning of Brctx.
BRCTx was identified in a database search for novel BRCT domain proteins that localize to regions of frequent rearrangement in cancer (www.ensembl.org) (3). We isolated Brctx from whole mouse brain cDNA by reverse transcription-PCR (RT-PCR) with the primers 5′-AAA AAA GAA TTC ATG GAA GAT AGT GCC ACA AAA C-3′ and 5′-AAA AAA GGA TCC CCC TGA AAC ATT TTT ACT AAG-3′, with PCR Supermix High Fidelity (Invitrogen, San Diego, Calif.), using 30 cycles of 20 s at 94°C, 1 min at 63°C, and 2 min at 68°C. This transcript was cloned into pBS− (Stratagene, Palo Alto, Calif.) and sequenced in full at the Australian Genome Research Facility. The full sequence of the Brctx transcript used in our study has been given the GenBank accession number AK012253.
Expression profiling of Brctx.
Northern blotting was performed on mouse tissue Northern blots containing whole RNA with the full-length Brctx cDNA as a probe. RT-PCR was performed on a mouse rapid scan panel (Origene Technologies) with the primers Akexon1-2A (5′-GGA AGA TAG TGC CAC AAA ACA TAT CAT CC-3′) and Akexon1-2B (5′-TGC TGC ACA AGC TGC TAA AAA CTT CTC-3′) spanning exons 1 and 2 of Brctx and β-actin primers (provided by the manufacturer). These reactions were performed with 50 μl of PCR Supermix (Invitrogen). For Brctx, 35 cycles of 20 s at 94°C, 1 min at 63°C, and 1 min at 72°C were performed. β-actin amplification was performed using 37 cycles of 20 s at 94°C, 1 min at 53°C, and 1 min at 72°C. Primer pairs were designed to flank introns to ensure that there was no amplification of genomic contaminants. The entire reaction was resolved in a 2% agarose gel in 1× Tris-acetate-EDTA (TAE). mRNA in situ hybridization for Brctx expression was performed with an automated Ventana Discovery instrument (Ventana Medical Instruments, Tucson, Ariz.) according to the manufacturer's instructions. Briefly, tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS). After 24 h they were washed with PBS and dehydrated through a methanol series. Tissues were then embedded in paraffin and 8-μm sections were cut. These sections were deparaffinized, fixed, acid treated, and then protease treated before being hybridized with a digoxigenin-labeled antisense probe (50 ng/slide) for 6 h. The signal was detected with an antidigoxigenin primary antibody (Jackson Immunoresearch Laboratories, West Grove, Pa.), followed by streptavidin-conjugated alkaline phosphatase secondary antibodies and finally by incubation with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate toluidine salt substrate solution for 6 h on the instrument. Sections were then counterstained with neutral red. The antisense Brctx probe (full-length cDNA) was transcribed from an EcoRI-linearized pBS plasmid by using T7 polymerase. An antibody to human BRCTx (peptide; GFKMEEKEALVKLLLK) was raised in rabbits (Alpha Diagnostics, San Antonio, Tex).
Subcellular localization and confocal microscopy.
For BRCTx localization studies, the full-length Brctx cDNA was fused to enhanced green fluorescent protein (EGFP) in pEGFP-C2 (Clontech, Palo Alto, Calif.) to make BRCTx-EGFP. A deletion mutant containing the first 100 amino acids (100BRCTx-EGFP) of BRCTx was generated by PCR with the reverse primer 5′-AAA AAA AAA AGG ATC CCT AAT AAT GGG AGT CTT TTT CAA TTT TAT ATC CCC-3′ in combination with the forward primer 5′-AAA AAA AAA AAA GCT TGA TGG AAG ATA GTG CCA CAA AAC ATA TCA TC-3′. Reactions were performed with PCR Supermix High Fidelity as described for the cloning of the full-length Brctx. These products were cloned in frame in pEGFP-C2 and confirmed by sequencing. HeLa cells or 293HEK cells were cultured in Dulbecco modified Eagle medium with 10% fetal calf serum (Invitrogen) on poly-l-lysine-coated chamber slides (BD Biosciences, Palo Alto, Calif.). Transfections were carried out by using Superfect (Qiagen, Germantown, Md.) or Lipofectamine (Invitrogen) according to the manufacturer's instructions. After 24 h, the cells were fixed with methanol at −20°C and stained. Pericentrin and γ-tubulin staining was performed with antibodies PRB-432C from Covance (Dever, Pa.) and GTU-88 from Sigma (Dorset, United Kingdom), respectively. Imaging was performed by using a Bio-Rad scanning confocal microscope or a Zeiss Axioplan 2 imaging microscope with an Axiocam HRm camera. Images were imported into Adobe Photoshop 6.0 and adjusted to use the full range of pixel intensities. For colocalization of hRAD18 and BRCTx, a Flag-tagged hRAD18 cDNA (22) was used with the full-length BRCTx-EGFP fusion construct as described above. Flag-hRAD18 fusion proteins were detected by using M2 monoclonal antibody from Sigma.
Response of BRCTx-EGFP fusion proteins to DNA damage in V79 cells.
A hamster V79 cell line stably expressing a BRCTx-EGFP fusion protein was generated by transfection of the full-length BRCTx-EGFP construct (described previously), and selection for stable integrants in G418 (Invitrogen). Appropriate expression of the chimeric protein was confirmed by Western blotting (data not shown). Relocalization experiments were performed as described previously (6). Cell lines stably expressing RAD51-EGFP were used as a control in all experiments (6) (data not shown).
Yeast two-hybrid analysis and coimmunoprecipitation.
The budding yeast Saccharomyces cerevisiae AH109 (11) was cultured in YPAD (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 0.003% [wt/vol] adenine, 2% [wt/vol] dextrose) and transformed with the plasmid pGBKT7-Brctx containing the full-length Brctx cDNA. Activation domain plasmids, in pAct2 (Clontech), were transformed into Y187 (9). AH109 and Y187 were mated and selection performed on semisolid synthetic defined medium supplemented with kanamycin (30 μg/ml) for 1 week at 30°C (1, 2). Transformants were then transferred onto membranes for assessment of β-galactosidase expression by a filter assay (34). An hRAD18 cDNA was isolated by screening a pretransformed pAct/Y187 fetal brain cDNA library containing 2 × 106 independent cDNA clones (Clontech). Coimmunoprecipitations were performed with Flag-tagged hRAD18 cDNA or hRAD18 deletion mutants (22) and the full-length tagged Brctx-EGFP plasmid in 293T cells.
Generation of Brctx-deficient mice.
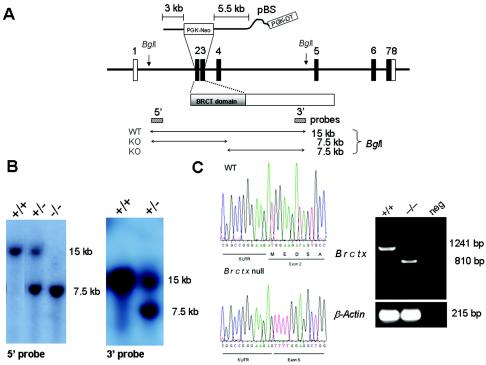
The Brctx targeting vector was constructed by using the PGK-neoF2L2DTA backbone (kindly provided by Phil Soriano, Fred Hutchinson Cancer Research Center, Seattle, Wash.). Vector arms were amplified from the PAC clone RP21-371G10, derived from 129S6/SvEvTac mouse DNA (24). Reactions were performed in 50 μl of PCR Supermix High Fidelity with 15 cycles of 20 s at 94°C, 1 min at 63°C, and 6 min at 68°C. The 5′ arm of the vector was amplified with the primers 5′-AAA AAA AAG CGG CCG CAA TAC ATT CAA ATG CTT TAA CAA TTT ATA CCT CAA AAC-3′ and 5′-AAA AAA AAG CGG CCG CCT AAC AGC TGG CCA AAA AGG CGA GCT CCA TAC AGA AGA TAG CAG ATG TTA AAC ATT ATG C-3′ and cloned into the NotI site of the vector. The 3′ arm of the targeting vector was amplified by using 5′-AAA AAA AAG CTT CAC GCT TCC TCA CCA CTG AGA CAC CCA GAG CAA AC-3′ and 5′-AAA AAA AAG CTT TCT GAT CTA ACG AAG AAG CAA CAT AAT CTA TAG GAC-3′. This fragment was cloned by using XbaI and XhoI into the NheI and SalI sites of PGK-NeoF2L2DTA. Then, 25 μg of SacII-linearized DNA was electroporated into AB2.2, 129S7 embryonic stem cells. Confirmation of targeting and genotyping was performed by Southern blotting on BglI-digested genomic DNA with a 5′ probe amplified by using the primers 5′-GTA TGT TCT GAA GTA TGG AGG ACT GAG-3′ and 5′-TCT TCA AAC ATG TGC CAG TAC TGG C-3′ and a 3′ probe amplified by using the primers 5′-CAC TTT TAG CTT CTG AAC ATC TCT CAC TCC C-3′ and 5′-TAT GTG GGT GGA CAT GCA AGC AAG CAC-3′. Blastocyst and chimeric mice were produced as described previously (26). All experiments were performed with mice on a mixed 129S7/SvEvBrd and C57BL/6J-Tyrc-Brd background. Mice were housed in accordance with Home Office regulations (United Kingdom).
Micronuclei assay, T-cell analysis, and thymocyte cell cycle studies.
For the micronucleus assay blood was collected from adult mice (6 to 8 weeks old) by cardiac puncture during terminal anesthesia and immediately fixed in −80°C methanol overnight. DNA, CD71 antibody staining, and fluorescence-activated cell sorting (FACS) analyses were performed with reagents from Litron Laboratories (Rochester, N.Y.). A total of 50,000 red cells from each animal were examined. For analysis of T cells, whole spleens and thymuses were prepared as described previously (15, 31) and stained with CD4-propidium iodide (PI) and CD8-fluorescein isothiocyanate (FITC) antibodies purchased from Pharmingen (Palo Alto, Calif.). Then, 10,000 cells were examined from each organ by FACS. For cell cycle analysis, thymuses were collected from adult (6- to 8-week-old) Brctx-null and wild-type animals, treated with RNase A, and labeled with PI (7). All FACS data analysis was performed by using WinMDI 2.8 software.
Histology and organ analysis.
Tissues were collected and fixed in 10% neutral buffered formalin, processed, and stained with hematoxylin and eosin by using standard methods (19). Organ weights were recorded from between four to six animals of each sex at 8 weeks of age.
DNA repair and cell growth assays.
Mouse embryonic fibroblasts (MEFs) were prepared as described previously (23, 28) from embryonic day 13.5 embryos. DNA repair and cell growth assays were performed on exponentially growing MEFs between passages two and five. Cells were seeded into gelatinized 96-well plates at a density of 4,000 cells/well the day before treatment. However, ionizing radiation (IR) treatment was performed on cells in suspension prior to plating. UV was delivered by using 254 nm, 40-W/m2 germicidal bulbs. Methyl methanesulfonate (MMS) was delivered to cells in warmed medium for 1 h and then washed off with warmed medium. Controls were treated with dimethyl sulfoxide, which is the vehicle for MMS. Mitomycin C (MMC) was dissolved in medium, and cells were fed daily at various doses. MMS and MMC were purchased from Sigma. MTT assays were performed on cells 5 days after treatment with CellTiter reagent (Promega, Madison, Wis.) according to the manufacturer's recommendations. A minimum of 12 wells were analyzed and averaged for each data point at each dose from three independent experiments for MMC and IR and five experiments for MMS and UV. For cell growth assays, MEFs were seeded at a density of 1.5 × 106/10-cm plate at day 0. Cells were counted daily using a Coulter counter (Beckman Coulter, Fullerton, Calif.). The data were collected from duplicate cell counts from three independent experiments.
RESULTS
BRCTx is a conserved, ubiquitously expressed, centrosomal protein.
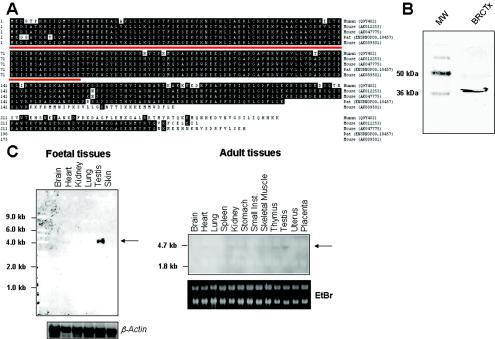
The amino acid sequence of BRCTx homologues from human, mouse, and rat sources show a high level of conservation (Fig. 1A). This conservation is particularly marked in the N terminus of the protein containing the BRCT domain. The alignment shown in Fig. 1A was compiled from protein sequences predicted from contiguous DNA fragments containing a start and stop codon and, where available, the splice variants are indicated. Several database entries for related predicted proteins, which were derived from assembled cDNAs, suggest that BRCTx is a much larger protein of between 700 and 900 amino acids. These predictions are inconsistent with our Western blot results, which show that BRCTx is a protein of ca. 36 kDa in human HCC1143 breast carcinoma cells (Fig. 1B). Northern blotting illustrates that Brctx is expressed at high levels in mouse testis (Fig. 1C). By RT-PCR we illustrate that Brctx is in fact expressed ubiquitously in all tissues and throughout the developing embryo (Fig. 1D). This correlates with expressed sequence tag data (www.ncbi.nlm.nih.gov/unigene/), which suggests a broad pattern of expression.
FIG. 1.
BRCTx is highly conserved and ubiquitously expressed. (A) Alignment of BRCTx homologues and splice variants from human mouse and rat reveals a high level of conservation. This alignment was generated by using contiguous cDNA sequences rather than assembled cDNAs. The BRCT domain is underlined in red. (B) Human BRCTx is a protein of ca. 36 kDa. (C) Northern blotting of mouse RNA illustrates strong expression of Brctx in the testis. (D) RT-PCR expression profiling of Brctx shows ubiquitous expression. β-actin is the internal control.
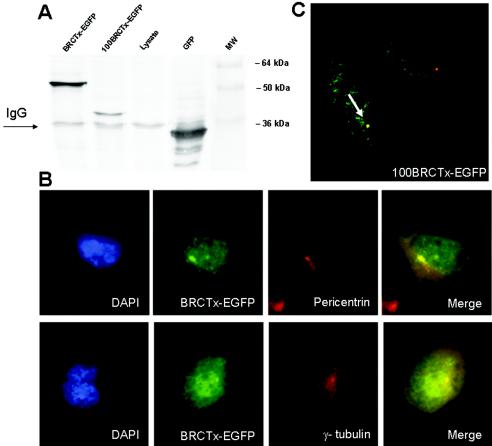
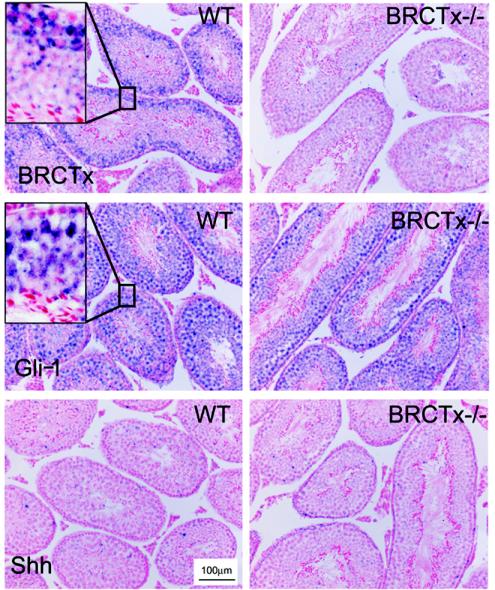
To reveal the subcellular localization of BRCTx, we tagged the N terminus of the full-length Brctx cDNA with EGFP. This revealed that BRCTx is localized mainly to the nucleus with scant expression in the cytoplasm, Fig. 2. Double labeling of cells with either γ-tubulin or pericentrin, which are markers of the centrosome, and BRCTx shows that these proteins colocalize in ca. 10 to 15% of cells (Fig. 2). To determine the domain of the protein controlling localization, we generated a truncation mutant (100BRCTx-EGFP) of BRCTx containing the first 100 amino acids (BRCT domain) of the protein. This truncation mutant colocalized with γ-tubulin (Fig. 2), in some cells, suggesting that the BRCT domain of BRCTx is responsible for its centrosomal localization. In situ hybridization for Brctx in mouse testis revealed that it is mainly expressed in spermatocytes (Fig. 3). Sections were also hybridized with probes against Gli-1, which mainly stains germ cells and a subset of pachytene spermatocytes, and SSh, which does not stain the testis.
FIG. 2.
BRCTx localizes mainly to the nucleus and in some cells to the centrosome. (A) BRCTx fusions are appropriately expressed as illustrated by this Western blot. (B) BRCTx-EGFP fusion proteins localize to the nucleus and in some cells (10 to 15%) colocalizes with γ-tubulin and pericentrin, which are centrosomal proteins. Some scant staining of BRCTx-EGFP was observed in the cytoplasm. (C) The BRCT domain of BRCTx (100BRCTx-EGFP) mediates the localization of this protein to the centrosome (arrow).
FIG. 3.
In situ hybridization for Brctx in adult testis. In situ hybridization revealed expression of Brctx in spermatocytes. (Inset) High-power view of testis section showing blue cytoplasmic RNA staining. Expression of Gli-1 which stains spermatocytes and germ cells, and Shh which does not stain the testis, was also assessed. BRCTx−/− refers to tissue from a Brctx knockout animal.
BRCTx interacts and colocalizes with the post-replication-repair protein hRAD18.
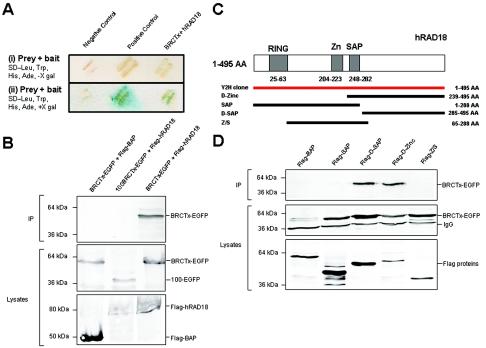
We performed a yeast two-hybrid screen to isolate proteins that interact with BRCTx. We identified a full-length cDNA encoding the post-replication-repair protein hRAD18 (32) (Fig. 4A). This cDNA was found to be in frame with the Gal4 activation domain. We confirmed this interaction by growth on selective medium, by LacZ assay, and by coimmunoprecipitation in 293T cells (Fig. 4A and B). We reveal that the C terminus of BRCTx is critical for the interaction (Fig. 4B). This domain does not contain any known protein interaction motifs. Further analysis revealed that residues in the C terminus of hRAD18, between amino acids 285 and 495, mediate its interaction with BRCTx (Fig. 4D). Thus, the well-characterized ring, zinc (Zn), and SAP domains do not appear to participate in the interaction between these proteins. The C-terminal end of hRAD18 was recently shown to be important for its interaction with polη and was implicated in recruiting polη to sites of DNA damage by protein-protein interaction and by PCNA monoubiquitination (39).
FIG. 4.
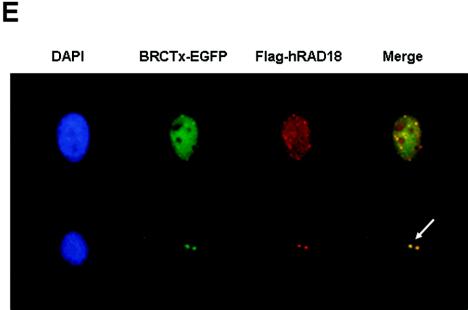
BRCTx binds to hRAD18 in yeast two-hybrid and immunoprecipitation assays, and these proteins colocalize in the nucleus. (A) Screening of a fetal brain cDNA library isolated a cDNA clone of hRAD18. This interaction was confirmed by growth on selective medium (i) and by the LacZ assay (ii). (B) The interaction between BRCTx and hRAD18 was confirmed by coimmunoprecipitation. Complexes were precipitated with Flag antibodies on agarose beads. A specific product of the correct size was observed in three independent experiments. hRAD18 does not interact with the N-terminal BRCT domain (100BRCTx-EGFP). Flag-BAP (bacterial alkaline phosphatase) was used as a negative control. (C) Structure of hRAD18 deletion mutants. (D) BRCTx interacts with the hRAD18 Flag d-SAP and Flag D-Zinc fusion proteins. (E) BRCTx and hRAD18 colocalize in the nucleus. Both proteins appeared to localize to the centrosome in some cells but in the majority of cases are distributed evenly throughout the nucleus. Arrow indicates centrosomes.
We also show that BRCTx colocalizes with hRAD18 in the nucleus (Fig. 4E). Strong interactions between BRCTx and HDAC2, RAN-BPM, and TACC2 were also observed in our yeast two-hybrid screen (data not shown), but these interactions could not be confirmed by coimmunoprecipitation from 293T cells.
Targeted disruption of Brctx.
We knocked out Brctx in embryonic stem cells by using a vector designed to delete the first two coding exons (Fig. 5A). Homozygous null animals were obtained at expected Mendelian ratios from interbreeding Brctx heterozygotes (Fig. 5B). Test matings revealed that both Brctx-null males and females were fertile. Disruption of the gene was confirmed by RT-PCR performed on testis cDNA (Fig. 5C) and by in situ hybridization on testis sections (Fig. 3). RT-PCR revealed that Brctx transcripts were produced from the mutant allele but these were fusions between the 5′ untranslated region (UTR; exon 1) and exon 5. This mutant mRNA transcript does not produce a truncated form of BRCTx because it lacks an in-frame start codons.
FIG. 5.
Targeted disruption of Brctx in mice. (A) Brctx was disrupted by recombination of a PGK-neo cassette with the first two coding exons resulting in disruption of the BRCT domain. (B) Diagnostic Southern blotting performed on mouse tail DNA by using external probes 5′ and 3′ of the targeting vector. (C) RT-PCR performed on testis cDNA from wild-type WT (+/+) and Brctx-null (−/−) animals revealed that −/− animals had a truncated Brctx transcript formed by the fusion of the 5′UTR (exon 1) with exon 5. This transcript lacks in-frame start codons for BRCTx. β-actin and ethidium bromide were used as internal controls for RNA quality.
Assessment of T-cell compartment and body and organ weights.
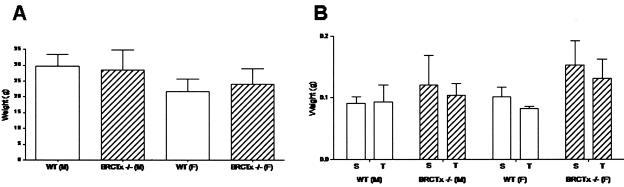
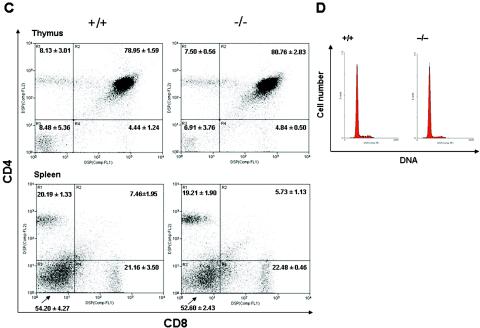
Given that knockouts of other proteins with BRCT domains show severe defects in T-cell maturation, growth impairment, and abnormal spleen and thymus weights (20), we investigated these parameters in Brctx-null mice. Body weight, and the weights of spleens and thymuses from 6- to 8-week-old Brctx-null and wild-type animals were similar (Fig. 6A and B). Histopathologic analysis of spleens revealed a slight increase in extramedullary hematopoiesis in Brctx-null animals, which may account for their slightly larger size compared to wild-type spleens. Additional histopathologic analysis on Brctx-null tissues from all other organ systems revealed no major defects compared to tissues from littermate controls. No significant differences in the CD4/CD8 profiles of spleens and thymuses from wild-type and knockout animals (Fig. 6C) was observed. The cell cycle profiles of thymuses from Brctx-null and wild-type animals were comparable (Fig. 6D).
FIG. 6.
Analysis of thymic and splenic T-lymphocytes and of organ and body weights in Brctx-null and wild-type animals. Body (A) and organ (B) weights from Brctx-null and wild-type animals are indicated. Adult animals, 6 to 8 weeks of age, were weighed at necropsy when organs were collected. T, thymus; S, spleen. The data are presented as means ± standard deviations. (C) Spleens and thymuses were taken from adult animals and stained with CD4-PI and CD8-FITC. A minimum of 10,000 events were analyzed from each organ. The data are presented as means ± the standard deviations. (D) PI cell cycle of thymocytes.
Assessment of the role of Brctx in DNA repair.
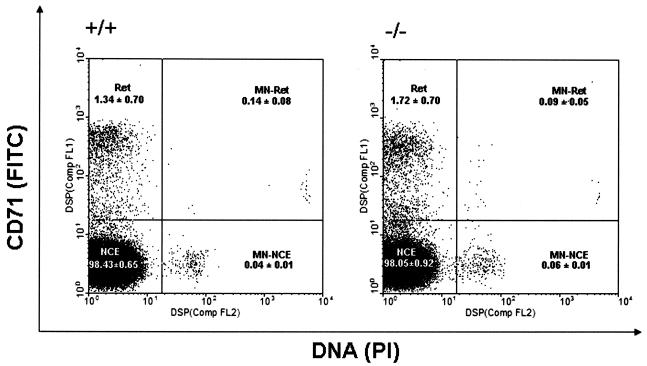
We used the flow cytometric micronucleus assay to determine whether Brctx-null mice exhibit an overt DNA repair defect or have unstable genomes (Fig. 7). These data showed that Brctx-null animals were indistinguishable from their wild-type littermates.
FIG. 7.
Assessment of genomic instability. Micronucleus assays on blood from Brctx-null (−/−) and wild-type (+/+) animals were performed. Peripheral blood was stained with CD71-FITC antibodies and with PI, which stains DNA. The data represent analyses of blood from five animals, 8 to 10 weeks of age, of each genotype (mean ± the standard deviation). A minimum of 50,000 events was analyzed for each sample. Red cell subcompartments are indicated. Results for reticulocytes (Ret), normochromatic erythrocytes (NCE), nucleated reticulocytes (MN-Ret), and nucleated normochromatic erythrocytes (MN-NCE) are shown. Bloom (Blm)-deficient mice (14), were used as a positive control (data not shown).
Assays for DNA repair, growth, and cell cycle analysis of Brctx-null cells.
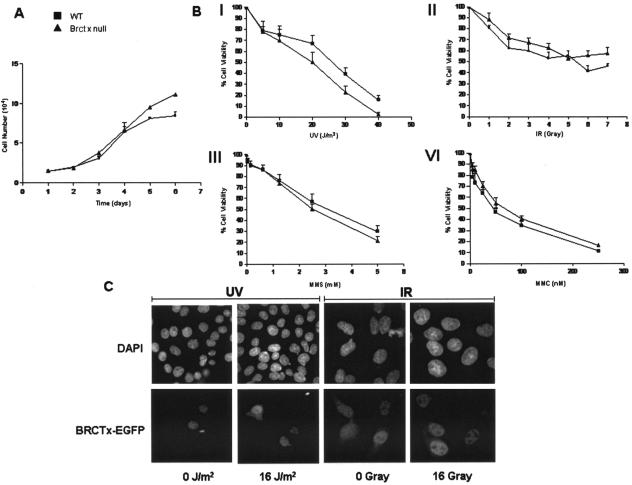
We observed no significant differences in the growth of Brctx-null cells compared to wild-type cells (Fig. 8A).
FIG. 8.
Growth and DNA repair characteristics of Brctx-null and wild-type cells. (A) Growth curve of P2 MEFs derived from Brctx-null and wild-type embryonic day 13.5 embryos. Symbols: ▪, wild-type (WT) cells; ▴, Brctx-null cells. Cell counts were performed in duplicate at intervals of 24 h. The data represent means ± the standard deviations of three independent experiments. (B) Role of Brctx in DNA repair. The data for MEF experiments represent the mean ± the standard errors of the mean for MTT assays in 96-well plate format. Treatments: I, UV irradiation; II, IR; III, MMS; IV, MMC. The data were normalized to untreated cells of the same genotype. (C) BRCTx-EGFP fusion proteins do not relocalize in response to DNA damage. Stable hamster V79 cells expressing BRCTx-EGFP fusion proteins were treated with DNA-damaging agents as indicated. At 24 h after exposure, cells were stained with DAPI and examined by immunofluorescence. The data are representative of two independent experiments.
The sensitivity of Brctx-null MEFs to DNA-damaging agents was examined (Fig. 8B). These assays revealed that Brctx-null MEFs are not overtly sensitive to any of the tested mutagens, although they did appear to show marginal sensitivity to UV irradiation. We also investigated the response of BRCTx-EGFP fusion proteins to UV irradiation and IR (Fig. 8C). BRCTx-EGFP fusions did not relocalize in response to DNA damage, unlike RAD51-EGFP fusion proteins, which were used as a control (data not shown).
DISCUSSION
We show here that BRCTx is a highly conserved, hRAD18 interacting protein that is dispensable for fertility and viability. Although other hRAD18 interacting proteins have been reported to participate in DNA repair (13), assays performed on Brctx-null murine embryonic fibroblasts suggest that these cells are not more sensitive to DNA-damaging agents than are wild-type cells. In addition, we have been unable to detect any major defects in embryonic development, in T-cell development, or in the genomic stability of these mice. MEFs from Brctx-null embryos appear to grow at a normal rate, and Brctx-null thymocytes do not show any major cell cycle defects (Fig. 6). We have had Brctx-null mice in our facility for at least 18 months. These mice have failed to exhibit any of the characteristic signs of premature aging, which have been attributed to other DNA repair knockouts (5). Collectively, these observations and the observation that Brctx is highly conserved in humans, mice, and rats appears to be in conflict. One explanation may be that Brctx is redundant. Although there are no Ensembl proteins that show >40% amino acid similarity to BRCTx, it is possible that there are genes in the genome that encode proteins that may compensate for the loss of Brctx. In an attempt to reconcile this discrepancy, we performed microarray analysis to look for differences in the expression profile of Brctx-null MEFs compared to wild-type MEFs (data not shown). These experiments revealed only two genes, H19 (25) and CXCL12 (30), to be >2-fold overexpressed in Brctx-null cells (P < 0.05). Neither of these genes have been implicated in DNA repair or the cell cycle. Thus, it seems unlikely that these expression differences are of functional significance.
At this stage it is not possible to determine whether Brctx plays a role in tumorigenesis. It is interesting that chromosome 5q15, where BRCTX resides, has recently been shown to be a hotspot for rearrangements in p53-negative tumors (10). It is compelling to speculate that there is some interaction between BRCTX and p53, which may explain the rearrangement of 5q15 observed in tumor samples.
Acknowledgments
D.J.A. and L.V.D.W. were supported by C. J. Martin and C. J. Martin/RG Menzies fellowships, respectively, from the NHMRC (Australia). F.V.G. is a Dorothy Hodgkin fellow of the Royal Society (United Kingdom). This study was supported by the Wellcome Trust (United Kingdom).
We thank Lubbertus Mulder, Mandy van der Rakt, Lucy Martin, LuAnn McKinney, Hideo Hishitani, John Schimenti, Jos Jonkers, K. J. Patel, William Wang, Graham Bignell, Fergus Lippi, Laura Harris, and Alan Lehmann for reagents and helpful discussions; Evelyn Grau for microinjection; and Kate Abbey for animal care.
REFERENCES
- 1.Adams, D. J., L. van der Weyden, A. Mayeda, S. Stamm, B. J. Morris, and J. E. Rasko. 2001. ZNF265: a novel spliceosomal protein able to induce alternative splicing. J. Cell Biol. 154:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, D. J., D. J. Beveridge, L. van der Weyden, H. Mangs, P. J. Leedman, and B. J. Morris. 2003. HADHB, HuR, and CP1 bind to the distal 3′-untranslated region of human renin mRNA and differentially modulate renin expression. J. Biol. Chem. 278:44894-44903. [DOI] [PubMed] [Google Scholar]
- 3.Birney, E., D. Andrews, P. Bevan, M. Caccamo, G. Cameron, Y. Chen, L. Clarke, G. Coates, T. Cox, J. Cuff, V. Curwen, T. Cutts, T. Down, R. Durbin, E. Eyras, X. M. Fernandez-Suarez, P. Gane, B. Gibbins, J. Gilbert, M. Hammond, H. Hotz, V. Iyer, A. Kahari, K. Jekosch, A. Kasprzyk, D. Keefe, S. Keenan, H. Lehvaslaiho, G. McVicker, C. Melsopp, P. Meidl, E. Mongin, R. Pettett, S. Potter, G. Proctor, M. Rae, S. Searle, G. Slater, D. Smedley, J. Smith, W. Spooner, A. Stabenau, J. Stalker, R. Storey, A. Ureta-Vidal, C. Woodwark, M. Clamp, and T. Hubbard. 2004. Ensembl 2004. Nucleic Acids Res. 32(Database Issue):D468-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul, and E. V. Koonin. 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11:68-76. [PubMed] [Google Scholar]
- 5.de Boer, J., J. O. Andressoo, J. de Wit, J. Huijmans, R. B. Beems, H. van Steeg, G. Weeda, G. T. van der Horst, W. van Leeuwen, A. P. Themmen, M. Meradji, and J. H. Hoeijmakers. 2002. Premature aging in mice deficient in DNA repair and transcription. Science 296:1276-1279. [DOI] [PubMed] [Google Scholar]
- 6.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray, J. W., and Z. Darzynkiewicz. 1987. Techniques in cell cycle analysis. Humana Press, London, England.
- 8.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 9.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 10.Jain, A. N., K. Chin, A. L. Borresen-Dale, B. K. Erikstein, P. Eynstein Lonning, R. Kaaresen, and J. W. Gray. 2001. Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival. Proc. Natl. Acad. Sci. USA 98:7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang, J., R. T. Bronson, and Y. Xu. 2002. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 21:1447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunapuli, P., R. Somerville, I. H. Still, and J. K. Cowell. 2003. ZNF198 protein, involved in rearrangement in myeloproliferative disease, forms complexes with the DNA repair-associated HHR6A/6B and RAD18 proteins. Oncogene 22:3417-3423. [DOI] [PubMed] [Google Scholar]
- 14.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 15.Maclean, K. H., U. B. Keller, C. Rodriguez-Galindo, J. A. Nilsson, and J. L. Cleveland. 2003. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol. Cell. Biol. 23:7256-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manke, I. A., D. M. Lowery, A. Nguyen, and M. B. Yaffe. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302:636-639. [DOI] [PubMed] [Google Scholar]
- 17.Mendes-da-Silva, P., A. Moreira, J. Duro-da-Costa, D. Matias, and C. Monteiro. 2000. Frequent loss of heterozygosity on chromosome 5 in non-small cell lung carcinoma. Mol. Pathol. 53:184-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 19.Mills, A. A., B. Zheng, X. J. Wang, H. Vogel, D. R. Roop, and A. Bradley. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398:708-713. [DOI] [PubMed] [Google Scholar]
- 20.Morales, J. C., Z. Xia, T. Lu, M. B. Aldrich, B. Wang, C. Rosales, R. E. Kellems, W. N. Hittelman, S. J. Elledge, and P. B. Carpenter. 2003. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 278:14971-14977. [DOI] [PubMed] [Google Scholar]
- 21.Moynahan, M. E. 2002. The cancer connection: BRCA1 and BRCA2 tumor suppression in mice and humans. Oncogene 21:8994-9007. [DOI] [PubMed] [Google Scholar]
- 22.Mulder, L. C., L. A. Chakrabarti, and M. A. Muesing. 2002. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277:27489-27493. [DOI] [PubMed] [Google Scholar]
- 23.Nagy, A., M. Gertenstein, K. Vintersten, and R. Behringer 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Osoegawa, K., M. Tateno, P. Y. Woon, E. Frengen, A. G. Mammoser, J. J. Catanese, Y. Hayashizaki, and P. J. de Jong. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10:116-128. [PMC free article] [PubMed] [Google Scholar]
- 25.Pachnis, V., C. I. Brannan, and S. M. Tilghman. 1988. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 7:673-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez-Solis, R., P. Liu, and A. Bradley. 1995. Chromosome engineering in mice. Nature 378:720-724. [DOI] [PubMed] [Google Scholar]
- 27.Richter, J., U. Wagner, P. Schraml, R. Maurer, G. Alund, H. Knonagel, H. Moch, M. J. Mihatsch, T. C. Gasser, and G. Sauter. 1999. Chromosomal imbalances are associated with a high risk of progression in early invasive (pT1) urinary bladder cancer. Cancer Res. 59:5687-5691. [PubMed] [Google Scholar]
- 28.Sharan, S. K., M. Morimatsu, U. Albrecht, D. S. Lim, E. Regel, C. Dinh, A. Sands, G. Eichele, P. Hasty, and A. Bradley. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386:804-810. [DOI] [PubMed] [Google Scholar]
- 29.Shin, S., and I. M. Verma. 2003. BRCA2 cooperates with histone acetyltransferases in androgen receptor-mediated transcription. Proc. Natl. Acad. Sci. USA 100:7201-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shirozu, M., T. Nakano, J. Inazawa, K. Tashiro, H. Tada, T. Shinohara, and T. Honjo. 1995. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics 28:495-500. [DOI] [PubMed] [Google Scholar]
- 31.Smith, K. G., U. Weiss, K. Rajewsky, G. J. Nossal, and D. M. Tarlinton. 1994. Bcl-2 increases memory B-cell recruitment but does not perturb selection in germinal centers. Immunity 1:803-813. [DOI] [PubMed] [Google Scholar]
- 32.Tateishi, S., H. Niwa, J. Miyazaki, S. Fujimoto, H. Inoue, and M. Yamaizumi. 2003. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23:474-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsao, S. W., N. Wong, X. Wang, Y. Liu, T. S. Wan, L. F. Fung, W. D. Lancaster, L. Gregoire, and Y. C. Wong. 2001. Nonrandom chromosomal imbalances in human ovarian surface epithelial cells immortalized by HPV16-E6E7 viral oncogenes. Cancer Genet. Cytogenet. 130:141-149. [DOI] [PubMed] [Google Scholar]
- 34.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkitaraman, A. R. 2003. A growing network of cancer-susceptibility genes. N. Engl. J. Med. 348:1917-1919. [DOI] [PubMed] [Google Scholar]
- 36.Ward, I. M., K. Minn, J. van Deursen, and J. Chen. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12:648-653. [DOI] [PubMed] [Google Scholar]
- 38.Wooster, R., G. Bignell, J. Lancaster, S. Swift, S. Seal, J. Mangion, N. Collins, S. Gregory, C. Gumbs, and G. Micklem. 1995. Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789-792. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, K., S. Tateishi, M. Kawasuji, T. Tsurimoto, H. Inoue, and M. Yamaizumi. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, X., C. C. Chini, M. He, G. Mer, and J. Chen. 2003. The BRCT domain is a phospho-protein binding domain. Science 302:639-642. [DOI] [PubMed] [Google Scholar]