Abstract
Condensins are heteropentameric complexes that were first identified as structural components of mitotic chromosomes. They are composed of two SMC (structural maintenance of chromosomes) and three non-SMC subunits. Condensins play a role in the resolution and segregation of sister chromatids during mitosis, as well as in some aspects of mitotic chromosome assembly. Two distinct condensin complexes, condensin I and condensin II, which differ only in their non-SMC subunits, exist. Here, we used an RNA interference approach to deplete hCAP-D2, a non-SMC subunit of condensin I, in HeLa cells. We found that the association of hCAP-H, another non-SMC subunit of condensin I, with mitotic chromosomes depends on the presence of hCAP-D2. Moreover, chromatid axes, as defined by topoisomerase II and hCAP-E localization, are disorganized in the absence of hCAP-D2, and the resolution and segregation of sister chromatids are impaired. In addition, hCAP-D2 depletion affects chromosome alignment in metaphase and delays entry into anaphase. This suggests that condensin I is involved in the correct attachment between chromosome kinetochores and microtubules of the mitotic spindle. These results are discussed relative to the effects of depleting both condensin complexes.
During mitosis, the two copies of replicated genomic DNA are segregated into each daughter cell. Due to the size of cells and to physical constraints linked to the segregation process, the replicated genome is converted into condensed chromosomes at the beginning of mitosis (32, 58). Among the factors involved in the chromosome assembly process, the literature has focused on a pentameric complex called condensin (28). This complex was isolated from mitotic chromosomes assembled in Xenopus egg extracts (34) and is composed of two SMC (structural maintenance of chromosomes; for a review, see reference 30) subunits (CAP-E/SMC2 and CAP-C/SMC4) and three non-SMC subunits (CAP-D2, CAP-G, and CAP-H). Homologues of each condensin subunit have been identified in all eukaryotes studied so far and were shown to be required for proper chromosome segregation (for a review, see references 28 and 42). In vitro studies showed that condensin has a DNA-stimulated ATPase activity and is able to introduce constrained positive supercoils into DNA in the presence of ATP (39, 40).
The first functional studies of condensin were made in mitotic extracts prepared from Xenopus eggs. When incubated in these extracts, sperm nuclei are converted into condensed chromosomes, and this process was found to be totally blocked upon immunodepletion of condensin (33). In living higher eukaryotes, mutating or inactivating individual condensin subunits have more discrete effects on chromosome condensation. In Caenorhabditis and Drosophila embryos lacking the condensin subunit SMC4, mitotic chromosomes are formed but adopt a dumpy morphology, indicating that lateral rather than longitudinal compaction is altered (27, 55).
It was recently revealed that two distinct complexes, condensins I and II, exist in metazoans. Condensin I is predominant in Xenopus eggs, whereas both condensins are present at similar levels in human HeLa cells (8, 48). Each of these complexes can be independently inactivated by repressing the expression of non-SMC subunits specific for either complex (48). Inactivating either condensin I or II has distinct, subtle effects on the chromosome architecture, whereas inactivating both condensin complexes severely impairs the assembly of mitotic chromosomes (48).
In mitosis, cells also have to remove topological links that exist between sister chromatids (resolution) and eventually between different chromosomes (individualization). Most of the links generated between replicated fibers are removed during replication, but sister chromatids remain largely catenated upon entry into mitosis. Resolution of these remaining links, which is necessary for proper segregation of sister chromatids in anaphase, is accounted for by topoisomerase II (TopoII) activity (13, 18). As a consequence, TopoII mutation or inactivation leads to severe segregation defects (16, 35, 59). Since TopoII activity is in equilibrium between catenation and decatenation, a mechanism must exist on chromosomes that would be able to orient this activity toward decatenation. According to a currently accepted model, this mechanism would involve step-by-step coupling between resolution and chromosome compaction activities (31, 32). Several observations suggest that such coupling could involve a functional interaction between condensins and TopoII. First, condensins and TopoII are both structural components of mitotic chromosomes (19, 22), and both participate in the assembly of condensed chromosomes (1, 33). Furthermore, condensin (9, 27, 51, 55-57) and TopoII (16, 35, 59) mutants exhibit similar segregation defects. Finally, Ycs4, the yeast homologue of CAP-D2, was shown to be indirectly involved in recruiting TopoII onto chromosomes (9). To date, the respective contributions of condensins I and II to the localization and activity of TopoII are not known.
In Caenorhabditis elegans, condensins were shown to be involved in the proper orientation of centromeres toward the spindle poles. In the absence of functional condensins, this orientation is altered, but the attachment of kinetochores to spindle microtubules remains robust (27). This is unlikely to be due to the fact that nematode chromosomes are holocentric (i.e., they have several centromeres all along their length), since early reports are in favor of a role for condensins in the bipolar attachment of chromosomes in yeast as well (41, 49). It is interesting that passenger proteins (for a review, see reference 2) are involved in both the recruitment of condensins onto chromosomes (23, 27, 44) and the control of bipolar attachment of chromosomes to spindle microtubules (3, 11, 38), possibly by transmitting microtubule tension or attachment defects to the spindle checkpoint (12, 17, 29, 50). A possibility is that such a role for passenger proteins in controlling the bipolar attachment of chromosomes could be somehow related to condensin functions. Condensins are able to introduce positive coils into DNA (7, 39, 40), and this activity could, for instance, participate in the correct assembly and orientation of centromeric structures (27).
In the present study, we used small interfering RNAs (siRNAs) specific for hCAP-D2 mRNA to specifically inactivate condensin I and to study the contribution of this complex to chromosome dynamics in mitotic HeLa cells.
MATERIALS AND METHODS
Cell culture.
HeLa cells (ECACC 93021013) were grown in Dulbecco's modified Eagle medium (Gibco-Invitrogen) supplemented with 10% fetal calf serum and penicillin-streptomycin at 37°C under a 5% CO2 atmosphere.
RNA interference.
siRNAs were provided by Dharmacon Research Inc. and used according to the provider's instructions. The human hCAP-D2 siRNA sequence (AACCAUAUGCUCAGUGCUACA ) starts at position 761 (702 from the initiation codon) in hCAP-D2/CNAP1 mRNA (accession no. NM_014865). An unrelated double-stranded RNA (dsRNA) (AACCAGCACGGACGACCUUAA ) was used for control experiments. siRNA transfections were made with Oligofectamine (Invitrogen) in OPTIMEM medium (Invitrogen) according to the manufacturer's instructions.
Antibodies.
Rabbit anti-hCAP-D2 was described previously (54). Rabbit anti-hCAP-E/SMC2 and anti hCAP-H polyclonal sera were provided by J. M. Peters (62) and U. K. Laemmli (43), respectively. Mouse monoclonal antibodies against topoisomerase IIα (clone Ki-S1) and Aurora B (clone AIM-1) were from Chemicon International and BD Transduction Laboratories, respectively. Antibodies were used at 1:2,000 for Western blotting and at 1:400 for immunofluorescence experiments.
Immunofluorescence staining.
Cells were grown on glass coverslips, rinsed in phosphate-buffered saline (PBS), fixed in methanol for 6 min at −20°C, permeabilized for 3 min in PBS containing 0.1% Triton, and blocked in PBS containing 1% bovine serum albumin (BSA). When indicated, soluble antigens were extracted by incubation in 0.1% Triton for 2 min, followed by 3 min in PBS at room temperature prior to fixation in methanol. Cells were successively incubated for 2 h at room temperature with primary antibodies (1:400 in PBS containing 1% BSA) and for 1 h at room temperature with secondary antibodies (rhodamine-coupled anti-mouse antibody and fluorescein isothiocyanate-coupled anti-rabbit antibody from goat [Sigma] at 1:400 in PBS containing 1% BSA). After each step, the cells were washed three times for 5 min each time in PBS at room temperature. The cells were counterstained in DAPI (4′,6′-diamidino-2-phenylindole) (0.5 μg/ml in PBS) and observed under a fluorescence microscope. When indicated, image stacks were acquired with a Leica DMRXA2 microscope driven by the Metavue driver. Z steps of 0.3 μm were submitted to deconvolution (nearest-neighbor method) by using Metamorph software. The pictures presented result from projections of three 0.3-μm-thick optical sections.
Flow cytometry analysis.
Cells were recovered by standard trypsin treatment, rinsed six times in PBS, and fixed in ethanol for 12 h at −20°C. The cells were rinsed in PBS and treated with RNase A (0.1 mg/ml in PBS) for 1 h at 37°C. DNA was stained with propidium iodide (0.1 mg/ml in PBS), and the cells were analyzed by fluorescence-activated cell sorting (Becton Dickinson).
Chromosome spreading.
Nonconfluent cells (∼106) were treated with nocodazole (5 μg in 10 ml of culture medium) for 6 h. The cells were recovered by mitotic shake off and sedimented by centrifugation (450 × g for 3 min). The cell pellet was resuspended in 2 ml of culture medium and mixed with 3 ml of tap water. After a 5-min incubation at room temperature, the cell suspension was mixed with 7 ml of Carnoy fixative (25% acetic acid and 75% ethanol). The cells were sedimented by centrifugation and washed in 10 ml of Carnoy fixative. This step was repeated three times before the cells were recovered in 1 ml of Carnoy fixative. Samples (80 μl) of the cell suspension were dropped from a 10-cm height onto a microscope slide and air dried. The slides were immersed for 6 min in a freshly prepared Giemsa solution (5% in PBS) and rinsed briefly in tap water. The excess liquid was removed by applying a paper towel to the slide edges. Once air dried, the slides were mounted in Entellan, and the spread chromosomes were observed under a light microscope. Alternatively, the spread chromosomes were stained in SYBR-Green (Molecular Probes), rinsed in PBS, and mounted in Vectashield (Vector Laboratories).
Chromosome preparation.
Nonconfluent cells (∼106) were treated with nocodazole (0.5 μg/ml in culture medium) for 15 h. The cells were sedimented by centrifugation (450 × g for 10 min). The cell pellet was resuspended in 10 ml of ice-cold culture medium and kept on ice for 30 min. The cells were sedimented, resuspended in 10 ml of ice-cold 75 mM KCl, and kept on ice for 20 min. After a second centrifugation, the cells were resuspended in 1 ml of ice-cold aqueous disruption buffer (10 mM Tris-HCl [pH 7.4], 120 mM KCl, 20 mM NaCl, 0.1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, 2 mM CaCl2). The cells were kept on ice for 5 min and broken by being passed 10 times through a hypodermic needle. Interphase nuclei and debris were removed by centrifugation (400 × g for 3 min at 4°C), and the supernatant (total fraction) was centrifuged again for 15 min at 3,000 × g. The chromosome pellet was resuspended in 0.2 ml of nuclear isolation buffer (10 mM HEPES [pH 7.5], 2 mM MgCl2, 250 mM sucrose, 25 mM KCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml). The chromosome suspension was centrifuged through a 0.5-ml sucrose cushion (750 mM in nuclear isolation buffer, giving a final sucrose concentration of 1 M) for 20 min at 10,000 × g. After the supernatant and sucrose cushion were removed, the chromosome pellet was solubilized in sodium dodecyl sulfate sample buffer and analyzed by polyacrylamide gel electrophoresis and Western blotting.
Western blot analysis.
Electrophoresis in sodium dodecyl sulfate-polyacrylamide gels and electrotransfer onto nitrocellulose membranes were performed following standard procedures. The membranes were blocked in TBST (25 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20 [Sigma]) containing 5% skim milk powder for 2 h at 4°C. The membranes were incubated at 4°C for 1 h with crude sera diluted 1,000-fold in TBST containing 2.5% skim milk. Immunocomplexes were revealed with alkaline phosphatase-coupled anti-rabbit antibodies (Jackson Immunoresearch Laboratories, Inc.) using a chemifluorescence assay (Amersham Biosciences) according to the manufacturer's instructions. Signals were analyzed using ImageQuant software (Amersham Biosciences).
RESULTS
Localization of hCAP-D2 during the cell cycle.
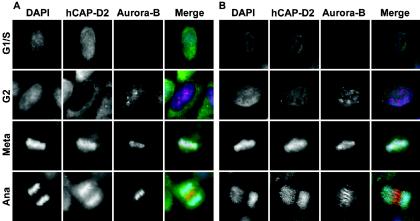
In order to test antibodies used in this study and to address hCAP-D2 behavior during the cell cycle, asynchronous HeLa cells were fixed in methanol and analyzed by immunofluorescence using a mixture of anti-Aurora B and anti-hCAP-D2 antibodies (Fig. 1A). Aurora B was reported to be undetectable in G1/S HeLa cells and to accumulate in the nucleus in G2 cells (15). In G1/S (Aurora B-negative) cells, the hCAP-D2 signal was almost uniformly distributed between the nucleus and cytoplasm. Nuclear hCAP-D2 signal strongly diminished in G2 (Aurora B-positive) cells (Fig. 1A). As a consequence, hCAP-D2 appeared to be mainly cytoplasmic, although nuclear signal remained above background. To further investigate hCAP-D2 localization, cells were treated with Triton prior to fixation in order to extract soluble antigens (Fig. 1B). When G1/S cells were examined under these conditions, hCAP-D2 signal strongly decreased in both the nucleus and cytoplasm compared with nonextracted cells, indicating that a major part of hCAP-D2 is soluble in these cells. Interestingly, nuclear hCAP-D2 signals were similar in Triton-extracted G1/S and G2 cells, indicating that most of hCAP-D2 present in G1/S nuclei is soluble. As suggested previously (53), this soluble nuclear fraction of hCAP-D2 in G1/S cells could result from condensin complexes that had dissociated from chromosomes at the end of the preceding mitosis. These observations additionally suggest that a minor part of the hCAP-D2 is associated with an insoluble nuclear component throughout interphase.
FIG. 1.
Detection of Aurora B and hCAP-D2 in interphase and mitotic cells. Asynchronous HeLa cells were treated (B) or not (A) with Triton prior to fixation in methanol (see Materials and Methods) and immunostained with a mixture of anti-Aurora B (red) and anti-hCAP-D2 (green) antibodies. The cells were counterstained with DAPI (blue) and observed by fluorescence microscopy. Cells in G1/S, G2, metaphase (Meta), and anaphase (Ana) are presented.
In mitosis, as previously described, hCAP-D2 was found to decorate chromosome arms in both metaphase and anaphase cells, and Aurora B labeling was found on centromeric regions of chromosomes in metaphase and on the spindle midzone in anaphase. These mitotic stainings were not affected by Triton treatment prior to fixation (compare Fig. 1A and B).
Depletion of hCAP-D2 by dsRNA interference in HeLa cells.
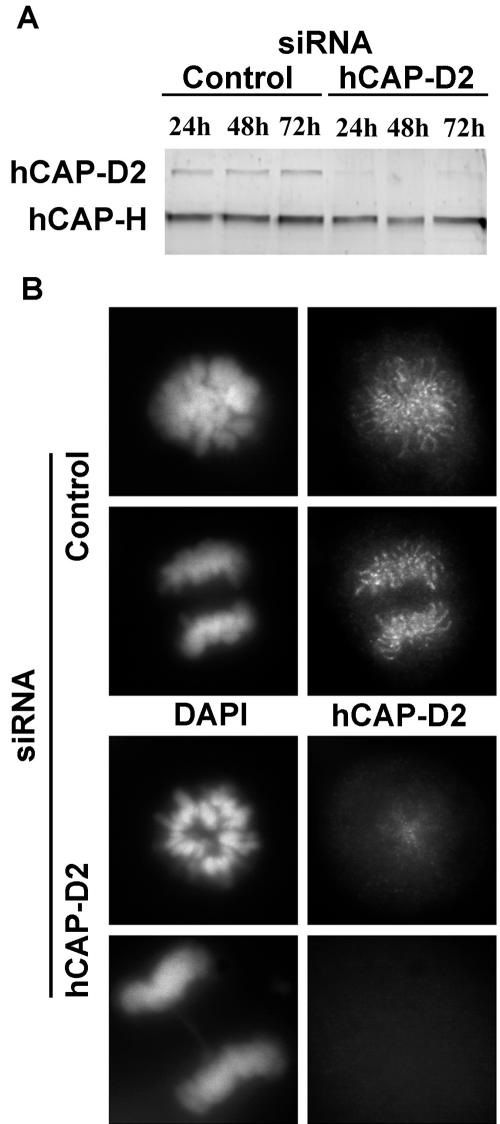
In order to deplete hCAP-D2, HeLa cells were treated with either siRNA specific for hCAP-D2 mRNA or control dsRNA (see Materials and Methods). Protein extracts were prepared at different times after treatment, and protein levels were monitored by immunoblotting with anti-hCAP-D2 antibodies (Fig. 2A). Twenty-four hours after transfection, the hCAP-D2 level had decreased up to 80% in siRNA-treated cells compared with control cells, and no further decrease was observed between 24 and 72 h. As monitored in the same experiment, the expression of hCAP-H was not affected. It should be noted that in experiments where residual hCAP-D2 was undetectable, a decrease in hCAP-H levels was sometimes observed (data not shown). This suggests that in the absence of hCAP-D2, hCAP-H no longer associates with SMC subunits and might become less stable. However, in the different experiments presented here, we used only conditions under which hCAP-D2 depletion did not affect the hCAP-H level. In some experiments, repression of hCAP-D2 was more progressive, reaching a maximal level of depletion at 72 h. The expression and localization of hCAP-D2 were further analyzed by immunofluorescence 72 h after dsRNA treatment (Fig. 2B). In control cells, consistent with previous studies, hCAP-D2 was found to decorate mitotic chromosomes. This chromosomal hCAP-D2 signal was either strongly diminished or absent in most siRNA-treated cells. However, it should be noted that this staining remained observable in ∼10% of treated cells, probably due to the fact that some of these cells escaped transfection with dsRNA (data not shown), which is consistent with the quantified depletion of hCAP-D2.
FIG. 2.
Depletion of hCAP-D2 by siRNAs. HeLa cells were treated with control or hCAP-D2 siRNAs as described in Materials and Methods. (A) Proteins were extracted at the indicated times after siRNA transfection, separated on an 8% polyacrylamide gel, and analyzed by Western blotting with anti-hCAP-D2 and anti-hCAP-H antibodies. (B) Cells were fixed 72 h after siRNA treatment, immunostained with anti-hCAP-D2 antibodies, and counterstained with DAPI. Pictures of prometaphase and anaphase cells are shown for each series.
Effects of hCAP-D2 depletion on condensin subunits and TopoII chromosome association.
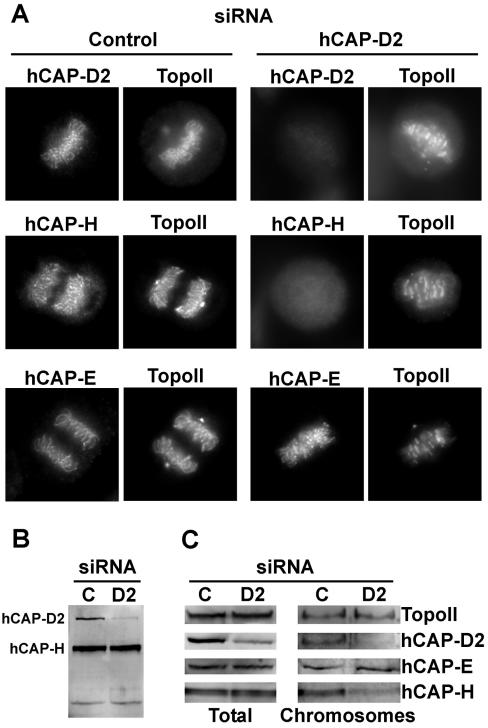
Since hCAP-D2 was proposed to play a role in the recruitment of condensin onto chromosomes (5), the localizations of two other condensin subunits, hCAP-E (one of the two SMC subunits) and hCAP-H (one of the three non-SMC subunits), were analyzed by immunofluorescence in dsRNA-treated cells (Fig. 3A). In control cells, antibodies specific for hCAP-E and hCAP-H stained chromosome arms, giving pictures very similar to that obtained with anti-hCAP-D2 antibodies. TopoII signal was also observed on chromosomes, consistent with previous studies (20, 43). It should be noted that a strong TopoII signal was present at the spindle poles, consistent with previous studies describing the presence of this protein in centrosomes (6). Whereas both TopoII and hCAP-E remained present on chromosomes in hCAP-D2-depleted cells, the chromosomal localization of hCAP-H was strongly diminished (Fig. 3A). To further analyze the effect of hCAP-D2 depletion on the chromosomal recruitment of condensin subunits, chromosomes were purified from mitotic cells, and their protein contents were analyzed by immunobloting (Fig. 3C). In siRNA-treated cells, hCAP-D2 levels were found to decrease in both the total and chromosome-associated fractions. These fractions were also probed with anti-TopoII and anti-condensin antibodies. In siRNA-treated cells, the levels of TopoII and hCAP-E remained constant in both the total and chromosomal fractions compared with fractions obtained from control cells. In contrast, although total hCAP-H levels were not affected, this condensin subunit dissociated from mitotic chromosomes in hCAP-D2-depleted cells.
FIG. 3.
Effects of hCAP-D2 depletion on condensin subunit and TopoII chromosome association. Cells were treated with control or hCAP-D2 siRNA and analyzed 72 h later. (A) Treated cells were fixed and double labeled with anti-TopoII antibodies and antibodies against either hCAP-D2, hCAP-H, or hCAP-E, as indicated. Pictures of mitotic cells are shown. (B) Total proteins were analyzed as in Fig. 2 with anti-hCAP-D2 and anti-hCAP-H antibodies. (C) Sixty hours after siRNA treatment, cells were incubated in nocodazole-containing culture medium for an additional 12 h. Mitotic cells were recovered by shake off and lysed (total extract). Chromosomes were isolated by centrifugation through a sucrose cushion. Total and chromosomal proteins were separated by polyacrylamide gel electrophoresis and analyzed by Western blotting with anti-TopoII, anti-hCAP-D2, anti-hCAP-E, and anti-hCAP-H antibodies.
TopoII and hCAP-E localization in hCAP-D2-depleted cells.
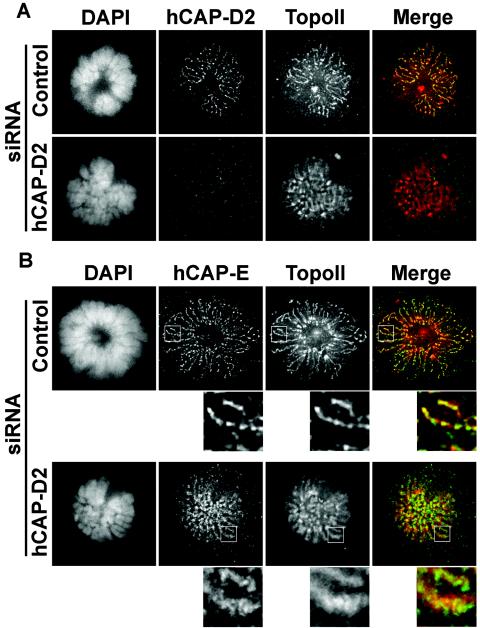
Since several reports are in favor of condensin being involved in the dynamic recruitment of TopoII onto chromosomes (9, 36), we analyzed in more detail the localization of this enzyme in hCAP-D2 siRNA-treated cells. For this purpose, immunofluorescence pictures of mitotic cells were taken at higher resolution and treated by deconvolution (Fig. 4A). In control cells, both hCAP-D2 and TopoII antibodies decorated the axial region of chromosomes, giving an interspersed staining pattern consistent with previous studies (43). In cells treated with hCAP-D2 siRNAs, immunofluorescence signals of TopoII were more diffuse, suggesting either that its localization was no longer confined to chromosome axes or that the axes themselves were disorganized. To discriminate between these hypotheses, we compared immunofluorescence signals obtained by double staining with anti-TopoII and anti-hCAP-E antibodies (Fig. 4B). In control cells, hCAP-E was found to form patches aligned along the chromosome axes. Such a patchy pattern could be reminiscent of the previously described organization of condensins as a spiral structure along chromosome axes (48). TopoII was also found along chromosome axes, although not strictly colocalized with hCAP-E. In hCAP-D2-depleted cells, hCAP-E signals, although remaining punctate, failed to form aligned patches, suggesting that condensin-defined chromosome axes were somehow perturbed. TopoII signals also failed to form aligned structures and became more diffuse than in control cells. Therefore, the spatial organization of both TopoII and condensins is altered in hCAP-D2-depleted cells.
FIG. 4.
Effects of hCAP-D2 depletion on the localization of TopoII and hCAP-E in prometaphase. Cells treated with control or hCAP-D2 siRNA were fixed and immunostained after 72 h. (A) Prometaphase cells were double labeled with anti-TopoII (red) and anti-hCAP-D2 (green) antibodies and counterstained with DAPI. (B) Prometaphase cells were double labeled with anti-hCAP-E (green) and anti-TopoII (red) antibodies. Pictures were taken with a 63× objective and treated by deconvolution. The insets are enlarged areas taken from corresponding pictures.
Effect of hCAP-D2 depletion on mitotic chromosomes.
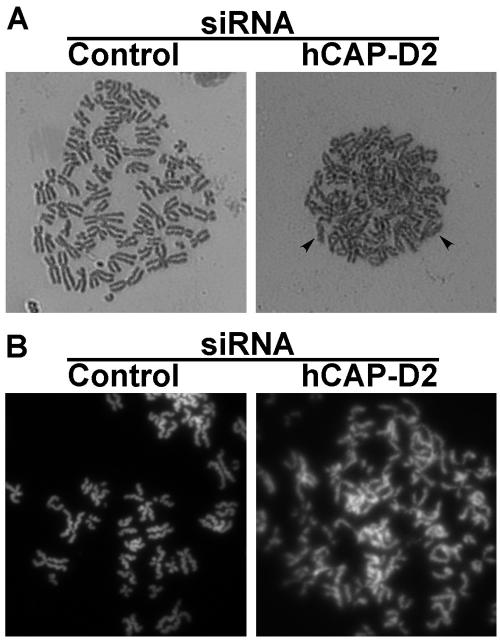
Condensins and TopoII were reported to play roles in both compaction and resolution of mitotic chromosomes. To examine chromosome structure in more detail, nocodazole-treated mitotic cells were spread and the chromosomes were stained with Giemsa (Fig. 5A). In control cells, chromosomes were individualized and had a characteristic rod-shaped structure and well-separated sister chromatids. In hCAP-D2-depleted cells, chromosomes were poorly individualized, and sister chromatids often failed to be fully separated. When stained with SYBR-Green (Fig. 5B), chromosomes from hCAP-D2-depleted cells appeared more entangled in each other, and sister chromatids were much less distinguishable than in chromosomes spread from control cells. In addition, whereas chromatids from control cells had a well-defined structure typical of supercondensed chromosomes (43), chromosomes from hCAP-D2-depleted cells adopted a diffuse and fluffy appearance, reminiscent of what was observed by depleting hCAP-G, another non-SMC subunit of condensin I (48). Therefore, hCAP-D2 depletion alters both the structure of chromosomes and the resolution of sister chromatids.
FIG. 5.
Effect of hCAP-D2 depletion on the shape, resolution, and individualization of mitotic chromosomes. Cells were treated with siRNAs. After a 66-h recovery, they were blocked in mitosis by a 6-h nocodazole treatment. Mitotic cells were recovered by shake off, and the chromosomes were spread after hypotonic treatment as described in Materials and Methods. The spread chromosomes were stained with Giemsa (A) or SYBR-Green (B).
Mitotic effects of hCAP-D2 depletion.
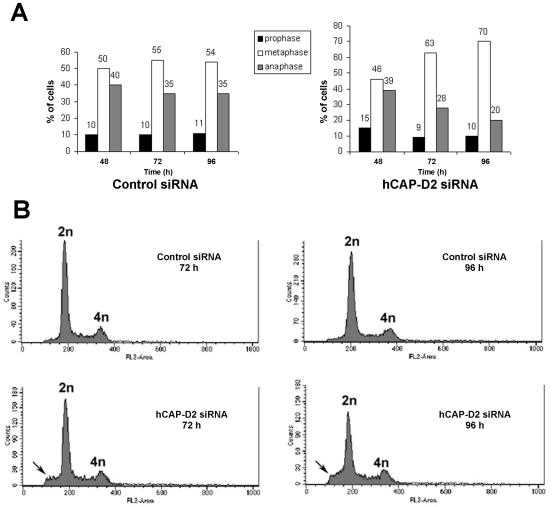
The effects of hCAP-D2 depletion on both the resolution and individualization of mitotic chromosomes were likely to have consequences for the course of mitosis. To address this issue, mitotic cells were observed at different stages in both control and hCAP-D2 siRNA series. When scored manually, the mitotic index (∼6%) was not significantly different in control and hCAP-D2-depleted cells (data not shown). The numbers of cells in different mitotic stages were scored in both series. The relative populations in each stage were constant in control cells at 48, 72, and 96 h (Fig. 6A, left). In contrast, the proportion of mitotic cells in prometaphase or metaphase increased between 48 and 96 h after treatment with hCAP-D2 siRNAs. Conversely, the proportion of cells in anaphase or telophase progressively decreased over time in these treated cells (Fig. 6A, right). This suggests that entry into anaphase was delayed in hCAP-D2-depleted cells. To estimate the possible consequences of such a delay on their DNA contents, control and depleted cells were analyzed by flow cytometry (Fig. 6B). Fluorescence-activated cell sorter profiles confirmed that the mitotic index was not significantly different between control and siRNA-treated cells. Despite the increased metaphase-to-anaphase ratio observed above, no polyploid cells were found to accumulate up to 96 h after siRNA treatment. Rather, cells with a DNA content lower than 2 N were found to accumulate in hCAP-D2-depleted cells. These cells with low DNA contents could correspond to apoptotic cells, although the possibility that aneuploid cells also exist in this population cannot be formally excluded.
FIG. 6.
Effect of hCAP-D2 depletion on the progression of mitosis. (A) Cells were treated with siRNAs, fixed at the indicated times, and stained with DAPI. Cells in prophase (black bars), prometaphase-metaphase (open bars), or anaphase-telophase (gray bars) were scored and expressed as percentages of total mitotic cells. (B) Cells were recovered by trypsin treatment 72 or 96 h after siRNA treatment. Suspended cells were fixed in ethanol and treated with RNase A. After DNA staining with propidium iodide, cells were analyzed by flow cytometry. The graphs indicate cell counts as a function of their DNA content. Note that cells with a DNA content lower than 2 N accumulate in the absence of hCAP-D2.
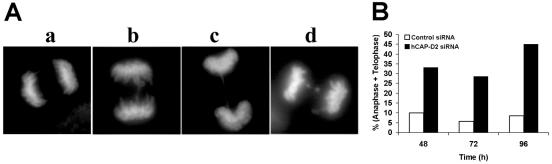
The above observations indicate that entry into anaphase is impaired in the absence of hCAP-D2. This impairment could be due to a persistence of topological links between sister chromatids that in turn would mechanically block segregation in early anaphase. On the other hand, entry into anaphase could be delayed by the persistence of metaphase defects. In order to explore these possibilities, cells in anaphase or metaphase were observed in more detail. First, we observed hCAP-D2-depleted cells that entered anaphase. Some of these cells showed segregation defects resulting in the persistence of chromatin bridges between the two sets of chromosomes (Fig. 7A). Such defects were also observed in control cells and represented 10% of all anaphases, whatever the observation time was. In hCAP-D2-depleted cells analyzed 48 and 72 h after siRNA treatment, ∼30% of all anaphases were abnormal, and this score reached 45% at 96 h (Fig. 7B). hCAP-D2 depletion therefore resulted in an increase in segregation defects.
FIG. 7.
Chromosome segregation is impaired in the absence of hCAP-D2. Cells were treated with siRNAs, fixed at different times, and stained with DAPI. (A) Examples of normal (a) and abnormal (b to d) anaphase pictures obtained in hCAP-D2-depleted cells. (B) Normal and abnormal anaphase pictures were scored at the indicated times in control (open bars) and hCAP-D2-depleted (black bars) cells. Results are expressed as a percentage of total anaphase-telophase cells.
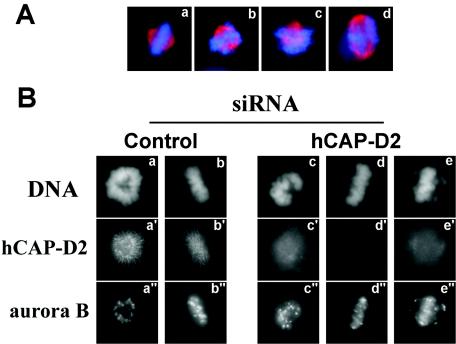
According to previous studies, a delayed entry into anaphase could result from improper attachment and/or alignment of chromosomes in metaphase, which activates the spindle checkpoint (for a review, see reference 64). This would suggest that, contrary to what is observed in SMC2- or SMC4-depleted cells (14, 36), hCAP-D2 depletion does not affect spindle checkpoint activation. Since spindle checkpoint control depends on passenger proteins, localization of these proteins should not be disrupted in hCAP-D2-depleted cells. To address this issue, control and hCAP-D2-depleted cells were stained with anti-Aurora B and anti-survivin antibodies. Aurora B and survivin are passenger proteins that localize on centromeres between prophase and metaphase. Upon entry into anaphase, they are released from centromeres to reach the midzones of mitotic spindles. In both series, Aurora B (Fig. 8B) and survivin (not shown) localized properly on mitotic chromosomes. In addition, when observed by fluorescence microscopy, hCAP-D2-depleted cells exhibited misaligned chromosomes in metaphase (Fig. 8A, b, c, and d) compared with control cells (Fig. 8A, a). This effect was further investigated by immunofluorescence experiments with anti-Aurora B antibodies; Aurora B was used as a centromeric marker that is specific for prometaphase and metaphase cells (Fig. 8B). In control cells, Aurora B staining showed discrete dots in prometaphase (Fig. 8B, a") that aligned on the metaphase plate (Fig. 8B, b"). Aurora B localization was not altered in prometaphase from hCAP-D2-depleted cells (Fig. 8B, c"). However, Aurora B-labeled centromeres appeared to be stretched in metaphase (Fig. 8B, d" and e"), and some of these seemed to escape from the metaphase plate (Fig. 8B, e") to reach spindle regions closer to the poles. This indicates that the alignment of mitotic chromosomes on the metaphase plate is impaired in hCAP-D2-depleted cells.
FIG. 8.
Effect of hCAP-D2 depletion on the metaphase plate. Cells were treated with control or hCAP-D2 siRNA and analyzed by immunofluorescence after a 72-h recovery. (A) Example of normal (a) and abnormal metaphase pictures obtained in hCAP-D2-depleted cells (b to d). Cells were immunostained with antitubulin antibodies (red) and counterstained with DAPI (blue). (B) Cells treated with control (left) or hCAP-D2 (right) siRNA were double stained with anti-Aurora B and anti-hCAP-D2 antibodies and counterstained with DAPI. Cells in prometaphase (a to a" and c to c") or metaphase (b to b", d to d", and e to e") are shown.
DISCUSSION
Condensin I (initially referred to as condensin) has been shown to play important roles in the assembly and segregation of mitotic chromosomes. Recently, a second distinct complex, called condensin II, that differs from condensin I in its non-SMC subunits, has been characterized (48). Here, we studied the effect of depleting hCAP-D2, a non-SMC subunit of condensin I, on the chromosome composition and dynamics in mitosis. First, we observed that depleting hCAP-D2 up to 80% with specific siRNAs led to a strong decrease in hCAP-D2 levels on chromosome axes. Interestingly, hCAP-D2 depletion provoked hCAP-H (the Barren homologue) to be released from chromosomes to a great extent. A two-hybrid screen performed in our laboratory identified CAP-H as a direct partner of CAP-D2 (unpublished data), and this interaction was confirmed by a direct two-hybrid assay (21). It is therefore conceivable that depleting hCAP-D2 weakens the association of hCAP-H with the condensin I core complex. hCAP-D2 was shown to contain several HEAT motifs (46), and a role for hCAP-D2 in the assembly of condensin I subunits would be consistent with a more general role for HEAT proteins in stabilizing macromolecular complexes (4, 24). hCAP-H is a member of the Kleisin protein family, which comprises CAP-H/Barren, CAP-H2/KLE2, and Scc1 in condensin I, condensin II, and cohesin, respectively (52, 62). Condensin and cohesin SMC heterodimers were shown to exhibit a V shape that would be able to embrace or trap two segments of chromatin. Accordingly, cohesin would ensure sister chromatid cohesion by embracing the two sister chromatids (25, 26), and condensins would structure mitotic chromosomes by embracing two looped adjacent segments of chromatin (63). In this model, Kleisins would act as a gate that would stably close the V-shaped SMC-containing complexes (25). Based on this model, releasing hCAP-H would directly dissociate the condensin I core complex from chromosomes. This is consistent with data suggesting that both CAP-D2 and CAP-H are involved in the recruitment of condensin I onto chromosomes (5, 21). However, under conditions where both hCAP-D2 and hCAP-H were absent from chromosomes, the levels of bound hCAP-E were not affected. One explanation would be that residual levels of hCAP-E are accounted for by the intact condensin II complex. However, HeLa cells were shown to contain equal quantities of condensin I and condensin II (48), and we did not observe any decrease in the level of chromosome-associated hCAP-E under conditions where hCAP-H was clearly diminished in purified chromosomal fractions. Furthermore, we obtained very similar results in Xenopus egg extracts (60), an experimental model in which condensin I is fivefold more abundant than condensin II (48). These observations raise the possibility that in a situation where hCAP-D2 is not completely depleted, the remaining condensin I would contribute to recruit and/or stabilize condensin core complexes (consisting of the two SMC subunits) by protein-protein interactions. This would be consistent with in vitro studies showing that 8S condensin core complexes can be processively recruited onto DNA by an aggregation process (63).
Depletion of hCAP-D2 also affected both the individualization of mitotic chromosomes and the resolution of sister chromatids. This strongly suggests that the action of TopoII is somehow perturbed in the absence of functional condensin I. We observed that chromatid axes, as defined by hCAP-E localization, are altered in the absence of hCAP-D2. Possibly as a consequence, TopoII, although remaining on the chromosomes, also fails to localize along well-defined axes in depleted cells. Such a mislocalization of TopoII has also been observed in Drosophila (14) and chicken (36) cells lacking SMC4. It is very unlikely that the observed resolution and individualization defects come from TopoII inactivation in hCAP-D2-depleted cells. Rather, we propose that its activity only fails to be oriented toward the decatenation (removal of DNA topological links between sisters) due to its failure to adopt a correct axial localization. An assay able to discriminate between catenating and decatenating activities of TopoII in living cells would be of great interest to address this issue.
We also showed that hCAP-D2 depletion had two major effects on the progression of mitosis: segregation defects in anaphase and failure of chromosomes to align on the metaphase plate. The effects on chromosome segregation are consistent with many studies that have highlighted a role for condensins in this process (10, 27, 41, 49, 51, 55, 56, 61). Such impaired segregation is probably a direct consequence of either altered activity (9, 14) or mislocalization (references 14 and 36 and this study) of TopoII.
We also observed a misalignment of chromosomes on the metaphase plate in the absence of hCAP-D2. Centromeric regions are stretched in metaphase cells, and some centromeres even escape from the metaphase plate in hCAP-D2-depleted cells. Similar alignment defects and kinetochore stretching were recently observed by inactivating either condensin I or II (47). Previous studies suggested that misaligned chromosomes could result from bipolar attachment defects due to an altered assembly and/or orientation of kinetochores in the absence of functional condensin (27, 41, 49). Interestingly, alignment defects were also reported in cells whose passenger proteins had been depleted or inactivated (3, 23, 38). Passenger proteins, such as Aurora B (23, 27, 38) and Bir1p/survivin (44), have been shown to be involved in the recruitment of condensins. Condensins could therefore mediate, at least partially, the role of passenger proteins in the attachment of kinetochores to spindle microtubules (11, 29, 45). Accordingly, depletion of hCAP-G, another non-SMC subunit of condensin I, was recently shown to alter the geometry of kinetochores (47). One could propose that condensins contribute to the assembly and orientation of kinetochores through their supercoiling activity. Alternatively, they could act as an intermediate platform to recruit and stabilize factors involved in the bipolar attachment of chromosomes to the spindle.
Finally, we observed that hCAP-D2 depletion led to a delayed entry into anaphase. This delay is likely to be due to the activation of the spindle checkpoint that responds to an impaired bipolar attachment of chromosomes to the spindle and/or a lack of tension at the kinetochores (64). In contrast to what we observed for CAP-D2, depleting SMC4/CAP-C, which is common to condensins I and II, also leads to alignment defects (47) but fails to delay entry into anaphase (14), implying that the spindle checkpoint is overridden under these conditions. This is reminiscent of recent observations that inactivating Aurora B impairs not only chromosome alignment but also the activation of the spindle checkpoint, due to a role of this protein in recruiting spindle checkpoint factors (17, 29, 37). Given that the recruitment of condensins onto chromosomes depends on Aurora B, it can be proposed that condensins act as intermediates for Aurora B functions in both chromosome attachment and spindle checkpoint activation. Inactivating both condensins would therefore impair chromosome attachment, as well as activation of the spindle checkpoint. Inactivating condensin I only would impair the correct attachment of chromosomes to microtubules but not the activation of the checkpoint. However, we cannot formally exclude the possibility that a complete depletion of hCAP-D2 would inactivate the spindle checkpoint as well.
Taken together with recent studies (47), these observations suggest that condensins I and II make distinct contributions to the composition, structure, or function of centromeric regions.
Acknowledgments
We thank U. K. Laemmli, T. Hirano, and J. M. Peters for providing some of the antibodies used in this study. We are also grateful to C. Kraft, S. Küng, and H. B. Osborne for critical reading of the manuscript. Microscopy analyses were performed thanks to the IFR97 microscopy service with the assistance of its manager, Stephanie Dutertre.
Our research is supported by grants from the Association pour la Recherche contre le Cancer, contract number 5711. CNRS UMR 6061 is a component of the Federative Research Institute IFR97, Functional Genomics and Health. E.W. was the recipient of a fellowship from the French Fondation pour la Recherche Médicale.
REFERENCES
- 1.Adachi, Y., M. Luke, and U. K. Laemmli. 1991. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell 64:137-148. [DOI] [PubMed] [Google Scholar]
- 2.Adams, R. R., M. Carmena, and W. C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49-54. [DOI] [PubMed] [Google Scholar]
- 3.Adams, R. R., H. Maiato, W. C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrade, M. A., C. Perez-Iratxeta, and C. P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117-131. [DOI] [PubMed] [Google Scholar]
- 5.Ball, A. R., Jr., J. A. Schmiesing, C. Zhou, H. C. Gregson, Y. Okada, T. Doi, and K. Yokomori. 2002. Identification of a chromosome-targeting domain in the human condensin subunit CNAP1/hCAP-D2/Eg7. Mol. Cell. Biol. 22:5769-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barthelmes, H. U., P. Grue, S. Feineis, T. Straub, and F. Boege. 2000. Active DNA topoisomerase IIα is a component of the salt-stable centrosome core. J. Biol. Chem. 275:38823-38830. [DOI] [PubMed] [Google Scholar]
- 7.Bazett-Jones, D. P., K. Kimura, and T. Hirano. 2002. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9:1183-1190. [DOI] [PubMed] [Google Scholar]
- 8.Bertsch, D. N., and J. E. Lindsley. 2003. Does it take two to untangle? Cell 115:4-6. [DOI] [PubMed] [Google Scholar]
- 9.Bhalla, N., S. Biggins, and A. W. Murray. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13:632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat, M. A., A. V. Philp, D. M. Glover, and H. J. Bellen. 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell 87:1103-1114. [DOI] [PubMed] [Google Scholar]
- 11.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho, A., M. Carmena, C. Sambade, W. C. Earnshaw, and S. P. Wheatley. 2003. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116:2987-2998. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, D. J., R. T. Johnson, and C. S. Downes. 1993. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J. Cell Sci. 105:563-569. [DOI] [PubMed] [Google Scholar]
- 14.Coelho, P. A., J. Queiroz-Machado, and C. E. Sunkel. 2003. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 116:4763-4776. [DOI] [PubMed] [Google Scholar]
- 15.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiNardo, S., K. Voelkel, and R. Sternglanz. 1984. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. USA 81:2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downes, C. S., A. M. Mullinger, and R. T. Johnson. 1991. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc. Natl. Acad. Sci. USA 88:8895-8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earnshaw, W. C., B. Halligan, C. A. Cooke, M. M. Heck, and L. F. Liu. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earnshaw, W. C., and M. M. Heck. 1985. Localization of topoisomerase II in mitotic chromosomes. J. Cell Biol. 100:1716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eide, T., C. Carlson, K. A. Tasken, T. Hirano, K. Tasken, and P. Collas. 2002. Distinct but overlapping domains of AKAP95 are implicated in chromosome condensation and condensin targeting. EMBO Rep. 3:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser, S. M., T. Laroche, J. Falquet, E. Boy de la Tour, and U. K. Laemmli. 1986. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188:613-629. [DOI] [PubMed] [Google Scholar]
- 23.Giet, R., and D. M. Glover. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groves, M. R., and D. Barford. 1999. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9:383-389. [DOI] [PubMed] [Google Scholar]
- 25.Gruber, S., C. H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell 112:765-777. [DOI] [PubMed] [Google Scholar]
- 26.Haering, C. H., J. Lowe, A. Hochwagen, and K. Nasmyth. 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9:773-788. [DOI] [PubMed] [Google Scholar]
- 27.Hagstrom, K. A., V. F. Holmes, N. R. Cozzarelli, and B. J. Meyer. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagstrom, K. A., and B. J. Meyer. 2003. Condensin and cohesin: more than chromosome compactor and glue. Nat. Rev. Genet. 4:520-534. [DOI] [PubMed] [Google Scholar]
- 29.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 31.Hirano, T. 1995. Biochemical and genetic dissection of mitotic chromosome condensation. Trends Biochem. Sci. 20:357-361. [DOI] [PubMed] [Google Scholar]
- 32.Hirano, T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69:115-144. [DOI] [PubMed] [Google Scholar]
- 33.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 34.Hirano, T., and T. J. Mitchison. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449-458. [DOI] [PubMed] [Google Scholar]
- 35.Holm, C., T. Stearns, and D. Botstein. 1989. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 9:159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudson, D. F., P. Vagnarelli, R. Gassmann, and W. C. Earnshaw. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 5:323-336. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, V. L., M. I. Scott, S. V. Holt, D. Hussein, and S. S. Taylor. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577-1589. [DOI] [PubMed] [Google Scholar]
- 38.Kaitna, S., P. Pasierbek, M. Jantsch, J. Loidl, and M. Glotzer. 2002. The Aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798-812. [DOI] [PubMed] [Google Scholar]
- 39.Kimura, K., and T. Hirano. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90:625-634. [DOI] [PubMed] [Google Scholar]
- 40.Kimura, K., V. V. Rybenkov, N. J. Crisona, T. Hirano, and N. R. Cozzarelli. 1999. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98:239-248. [DOI] [PubMed] [Google Scholar]
- 41.Lavoie, B. D., K. M. Tuffo, S. Oh, D. Koshland, and C. Holm. 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell 11:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legagneux, V., F. Cubizolles, and E. Watrin. 2004. Multiple roles of condensins: a complex story. Biol. Cell 96:201-213. [DOI] [PubMed] [Google Scholar]
- 43.Maeshima, K., and U. K. Laemmli. 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev. Cell 4:467-480. [DOI] [PubMed] [Google Scholar]
- 44.Morishita, J., T. Matsusaka, G. Goshima, T. Nakamura, H. Tatebe, and M. Yanagida. 2001. Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells 6:743-763. [DOI] [PubMed] [Google Scholar]
- 45.Murata-Hori, M., and Y. L. Wang. 2002. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12:894-899. [DOI] [PubMed] [Google Scholar]
- 46.Neuwald, A. F., and T. Hirano. 2000. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 10:1445-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ono, T., Y. Fang, D. L. Spector, and T. Hirano. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15:3296-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ono, T., A. Losada, M. Hirano, M. P. Myers, A. F. Neuwald, and T. Hirano. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109-121. [DOI] [PubMed] [Google Scholar]
- 49.Ouspenski, I. I., O. A. Cabello, and B. R. Brinkley. 2000. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell 11:1305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen, J., and I. M. Hagan. 2003. S. pombe Aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13:590-597. [DOI] [PubMed] [Google Scholar]
- 51.Saka, Y., T. Sutani, Y. Yamashita, S. Saitoh, M. Takeuchi, Y. Nakaseko, and M. Yanagida. 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13:4938-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schleiffer, A., S. Kaitna, S. Maurer-Stroh, M. Glotzer, K. Nasmyth, and F. Eisenhaber. 2003. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell 11:571-575. [DOI] [PubMed] [Google Scholar]
- 53.Schmiesing, J. A., H. C. Gregson, S. Zhou, and K. Yokomori. 2000. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol. 20:6996-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steen, R. L., F. Cubizolles, K. Le Guellec, and P. Collas. 2000. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J. Cell Biol. 149:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steffensen, S., P. A. Coelho, N. Cobbe, S. Vass, M. Costa, B. Hassan, S. N. Prokopenko, H. Bellen, M. M. Heck, and C. E. Sunkel. 2001. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 11:295-307. [DOI] [PubMed] [Google Scholar]
- 56.Strunnikov, A. V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587-599. [DOI] [PubMed] [Google Scholar]
- 57.Sutani, T., T. Yuasa, T. Tomonaga, N. Dohmae, K. Takio, and M. Yanagida. 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13:2271-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swedlow, J. R., and T. Hirano. 2003. The making of the mitotic chromosome. Modern insights into classical questions. Mol. Cell 11:557-569. [DOI] [PubMed] [Google Scholar]
- 59.Uemura, T., H. Ohkura, Y. Adachi, K. Morino, K. Shiozaki, and M. Yanagida. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50:917-925. [DOI] [PubMed] [Google Scholar]
- 60.Watrin, E., F. Cubizolles, H. B. Osborne, K. Le Guellec, and V. Legagneux. 2003. Expression and functional dynamics of the XCAP-D2 condensin subunit in Xenopus laevis oocytes. J. Biol. Chem. 278:25708-25715. [DOI] [PubMed] [Google Scholar]
- 61.Wignall, S. M., R. Deehan, T. J. Maresca, and R. Heald. 2003. The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J. Cell Biol. 161:1041-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeong, F. M., H. Hombauer, K. S. Wendt, T. Hirota, I. Mudrak, K. Mechtler, T. Loregger, A. Marchler-Bauer, K. Tanaka, J. M. Peters, and E. Ogris. 2003. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr. Biol. 13:2058-2064. [DOI] [PubMed] [Google Scholar]
- 63.Yoshimura, S. H., K. Hizume, A. Murakami, T. Sutani, K. Takeyasu, and M. Yanagida. 2002. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 12:508-513. [DOI] [PubMed] [Google Scholar]
- 64.Zhou, J., J. Yao, and H. C. Joshi. 2002. Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115:3547-3555. [DOI] [PubMed] [Google Scholar]