Abstract
To uncover factors required for transcription by RNA polymerase II on chromatin, we fractionated a mammalian cell nuclear extract. We identified the histone chaperone TAF-I (also known as INHAT [inhibitor of histone acetyltransferase]), which was previously proposed to repress transcription, as a potent activator of chromatin transcription responsive to the vitamin D3 receptor or to Gal4-VP16. TAF-I associates with chromatin in vitro and can substitute for the related protein NAP-1 in assembling chromatin onto cloned DNA templates in cooperation with the remodeling enzyme ATP-dependent chromatin assembly factor (ACF). The chromatin assembly and transcriptional activation functions are distinct, however, and can be dissociated temporally. Efficient transcription of chromatin assembled with TAF-I still requires the presence of TAF-I during the polymerization reaction. Conversely, TAF-I cannot stimulate transcript elongation when added after the other factors necessary for assembly of a preinitiation complex on naked DNA. Thus, TAF-I is required to facilitate transcription at a step after chromatin assembly but before transcript elongation.
The responsibility for transcribing protein-coding genes in eukaryotes belongs, with very rare exceptions, to RNA polymerase II (Pol II). To fulfill that role in vivo, Pol II and its accessory proteins must not only recognize transcriptional promoters accurately but must also modify the transcriptional repertoire appropriately in response to internal and external signals. Furthermore, Pol II must perform these tasks on DNA within a chromatin structure that is generally repressive. Biochemical analyses over the last 2 decades have identified a myriad of accessory factors required to initiate transcription accurately and to respond to DNA-binding transcriptional activators. It is now possible to reconstitute transcription responsive to activators in vitro with recombinant and highly purified components (21). However, the minimal system sufficient to transcribe naked DNA templates fails to support activator-dependent transcription of templates assembled into chromatin.
The basic unit of chromatin is the nucleosome, an octamer of core histones (H2A, H2B, H3, and H4) around which 147 bp of DNA wrap with a physiologic repeat length averaging 200 bp. There are several reasons why DNA assembled into chromatin requires additional factors, not needed on naked templates, to support activator-dependent transcription. First, the presence of nucleosomes on promoter DNA can inhibit recruitment of the basal transcriptional machinery to form a preinitiation complex (20). Second, chromatin structure in transcribed regions poses a physical barrier to elongation by Pol II (41). Finally, nucleosomes are substrates for covalent modifications by a variety of enzymes that can facilitate or repress transcription of nearby genes at least in part by providing docking platforms for coactivators or corepressors (17).
The earliest enzymatic systems capable of assembling correctly spaced nucleosomal arrays onto cloned DNA—a prerequisite for defining the requirements for chromatin-templated transcription—consisted of extracts derived from early embryos, purified core histones, and ATP (3, 12). A drawback of these systems, however, is that crude embryonic extracts may themselves contain the unidentified factors required for chromatin-based transcription, necessitating purification of the chromatin after assembly.
Completely recombinant chromatin assembly systems have also been described that generally contain an ATP-dependent chromatin remodeling factor of the ISWI family and a histone chaperone (23, 24). Transcription of chromatin made with recombinant assembly systems depends, however, on a crude nuclear extract. More defined systems are not competent to transcribe chromatin templates assembled with recombinant proteins, indicating a requirement for additional factors.
To reveal those requirements, we fractionated a HeLa cell nuclear extract capable of activated transcription on purified chromatin templates. We identified and purified TAF-I/SET (template-activating factor I/patient SE translocation), a member of the nucleosome assembly protein (NAP) family of histone chaperones, as a factor required to transcribe chromatin. The requirement appears to be general; TAF-I potently stimulated transcription driven by the 1,25(OH)2 vitamin D3 receptor (VDR), as well as by the GAL4-VP16 transactivator. The related NAP-1 protein, which was used to assemble the chromatin templates, can substitute for TAF-I in the transcription reaction. Conversely, TAF-I can substitute for NAP-1 in the assembly of chromatin. We show, however, that TAF-I plays distinct roles in assembling and transcribing chromatin; transcription of chromatin assembled with TAF-I instead of NAP-1, or of NAP-1-assembled chromatin preincubated with TAF-I, is still dependent on the presence of TAF-I during the polymerization reaction. Although TAF-I acts after chromatin is assembled, it is required prior to the elongation phase of the transcription cycle, distinguishing it mechanistically from FACT, a factor required for Pol II to overcome nucleosomal barriers to elongation (36). Thus, we have uncovered a novel function for the NAP family of histone chaperones at an early step in transcriptional activation on chromatin templates.
MATERIALS AND METHODS
Expression and purification of recombinant proteins.
Amino-terminally His-tagged TAF-I bacterial expression vectors were constructed by cloning the TAF-Iα, TAF-Iβ, or TAF-Iβ(1-225) open reading frame into pET-14b. A hemagglutinin (HA)-tagged version of TAF-Iβ was made by cloning the HA sequence upstream of the His tag in pET-14b. FLAG-tagged TAF-Iα was made by replacing the His tag in pET-28a with the FLAG sequence upstream of the TAF-Iα open reading frame. The His-TAF-I, HA-His-TAF-I, and Gal4-VP16 (14) constructs were expressed in Escherichia coli and purified by sequential cobalt affinity and Q anion-exchange chromatography. FLAG-TAF-Iα was purified by anti-FLAG immunoaffinity chromatography. The FLAG-TAF-Iα/His-TAF-Iβ dimer was coexpressed in E. coli and purified by sequential cobalt affinity and anti-FLAG immunoaffinity chromatography. rVDR and rRXR were overexpressed in Sf9 cells and purified as previously described (22). Recombinant ATP-dependent chromatin assembly factor (ACF) complex (rACF) and recombinant NAP-1 (rNAP-1) were expressed and purified as previously described (15). rTFIIA was purified as previously described (13).
Chromatin assembly and micrococcal nuclease digestion.
A plasmid containing four vitamin D3 response elements (VDREs) upstream of an adenovirus E1B core promoter fused to a cat reporter gene, 4 × VDRE B/INR, or a plasmid containing five Gal4-binding sites upstream of an adenovirus E4 promoter, pGEIO (14), was used as the substrate for chromatin assembly. Chromatin assembly and micrococcal nuclease digestion were performed essentially as previously described (15), except that supercoiled plasmid DNA was used instead of topoisomerase I-relaxed DNA. The final reaction conditions were as follows: 4.42 ng of plasmid DNA per μl, 16 nM rACF, 0.9 μM rNAP-1, 13 μg of purified Drosophila core histones per μl, 3.4 mM ATP, and an ATP regeneration system. In the case of chromatin assembled with rTAF-Iβ, rNAP-1 was omitted from the assembly reaction mixture. In the case of chromatin assembled with S190 extract, 2 μg of S190 extract per μl was added to the reaction mixture instead of rACF and rNAP-1. After assembly for 3 h at 27°C, the chromatin was aliquoted, frozen in liquid N2, and stored at −80°C for use in micrococcal nuclease digestion or transcription assays.
HeLa nuclear extract preparation and fractionation.
Nuclear extracts were typically prepared from 100 liters of HeLa cells harvested at a density of 5 × 105 cells per ml. Nuclear extracts were prepared essentially as previously described (8), except that KCl replaced NaCl in all buffers and a protease inhibitor cocktail (P8340; Sigma) was included during the homogenization step. The nuclear extract was dialyzed against 20 mM HEPES (pH 7.9)-0.2 mM EDTA-0.3 M KCl-20% glycerol-1 mM dithiothreitol (DTT).
All fractionation was done at 4°C in HEG (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 10% glycerol, 1 mM DTT) containing the indicated concentrations of KCl. HeLa nuclear extract (600 mg) was loaded onto a 50-ml phosphocellulose P-11 (Whatman) column equilibrated at 0.3 M KCl. The PC1 fraction used as the source of general transcription factors was generated by eluting the column with HEG-1 M KCl. The PC1 fraction was then dialyzed against HEG-50 mM KCl-20% glycerol. The unbound fraction (PC.3) was the starting point for the purification of Pol II and TAF-I/SET, as described in Results. The column buffers were supplemented with 0.1% Tween 20 for all steps after DEAE chromatography.
Transcription assays.
Transcription assays were performed essentially as previously described (37), except that TFIIA was added in recombinant form and the PC1 fraction was the source of basal transcription machinery. The 25-μl reaction mixture contained 31.7 ng of template DNA assembled into chromatin, 10 nM RXR, 30 nM VDR, 3 μg of PC1, 27 ng of rTFIIA, 50 mM KCl, 20 mM HEPES, 2% polyvinyl alcohol, 10% glycerol, 2.8 μM acetyl coenzyme A, 2.7 mM MgCl2, and 1.25 mM nucleoside triphosphate. Unless otherwise indicated, 1 μM vitamin D3 was added to all reaction mixtures containing VDR. All of the transcription reaction mixtures in Fig. 5 to 8 contain 0.78 μg of immunopurified Pol II. Other factors were added as indicated. Where stated, chromatin was purified away from unbound factors by chromatography on a 500-μl S300 spin column equilibrated in R buffer (10 mM HEPES [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM β-glycerophosphate, 0.25 mM phenylmethylsulfonyl fluoride, 1 mM DTT). Typically, 80 μl of chromatin was loaded on the column and the purified chromatin was collected by centrifugation in a tabletop centrifuge. Transcription was allowed to proceed at 30°C for 1 h after nucleoside triphosphate addition. The products were analyzed by primer extension and 6% denaturing gel electrophoresis. The gel was dried and exposed to film at −80°C or quantified with a Phosphorimager.
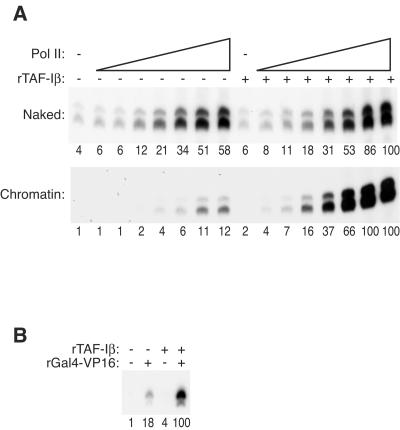
FIG. 5.
TAF-I is a chromatin-specific coactivator that stimulates transcription mediated by both VDR and Gal4-VP16. (A) VDR-mediated transcription was performed on either naked or chromatin templates in the presence or absence of 2.28 μM rTAF-Iβ, buffer alone, or 0.024, 0.049, 0.098, 0.195, 0.39, 0.78, or 1.56 μg of immunopurified Pol II, as indicated. (B) Transcription reaction mixtures were prepared with or without 20 ng of Gal4-VP16 and 3 μM rTAF-Iβ, as indicated, and Gal4-VP16-responsive promoter-containing DNA assembled into chromatin. All reaction mixtures included 0.78 μg of immunopurified Pol II. The relative level of transcription was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
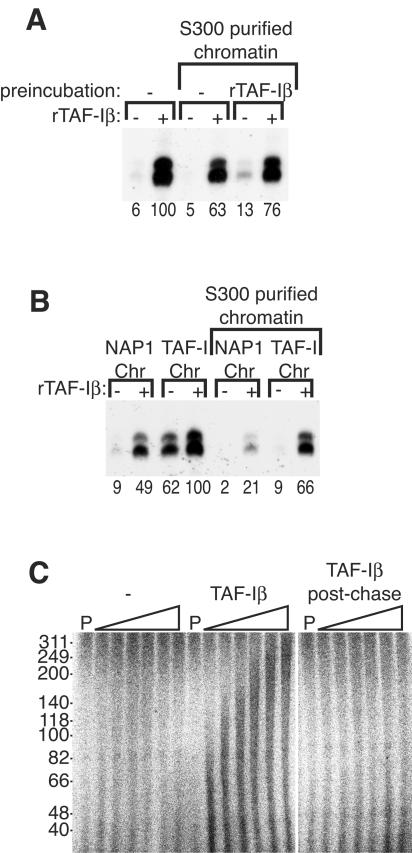
FIG. 8.
TAF-I is required at a step subsequent to chromatin assembly and prior to transcript elongation. (A) NAP-1-assembled chromatin was preincubated with or without 7.5 μM rTAF-Iβ for 30 min as indicated. The chromatin was used directly or passed over an S300 spin column. Transcription reaction mixtures were prepared with (+) or without (−) 3 μM rTAF-Iβ. (B) Chromatin (Chr) assembled with 0.9 μM NAP-1 or 8 μM rTAF-Iβ was used directly or purified over an S300 spin column, as indicated. Transcription reaction mixtures were then prepared with (+) or without (−) 3 μM rTAF-Iβ. The relative level of transcription in panels A and B was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane. (C) Transcription elongation assays were performed with buffer alone or with 4 μM TAF-Iβ added before the pulse (TAF-Iβ) or after the chase (TAF-Iβ postchase), as indicated. Samples were taken after the pulse (P) or at 0.5, 1, 1.5, 3, 6, or 18 min of chase and analyzed on a 6% denaturing gel. The smear of radioactivity extending up the gel indicates elongation of transcripts initiated during the pulse and was only seen when ΤΑF-Iβ was included prior to the chase. The values on the left are sizes in nucleotides.
Transcription elongation assays were set up essentially as described above, with the following exceptions. The transcription reactions were initiated by addition of ATP, UTP, and GTP to a 1 mM final concentration and 5 μCi of [α-32P]CTP and incubated for 2 min at 30°C. The reaction mixture was then supplemented with unlabeled CTP to a final concentration of 1 mM. Where indicated, TAF-Iβ was added at 3 μM either before the pulse-labeling or 1 min after the unlabeled CTP.
Protein identification.
Proteins excised from gels were digested with trypsin, the mixtures were fractionated on a Poros 50 R2 RP microtip, and the resulting peptide pools were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MS) with a BRUKER UltraFlex TOF/TOF instrument (Bruker Daltonics, Bremen, Germany) as previously described (9, 45). Selected experimental masses (m/z) were taken to search the human segment of the National Center for Biotechnology Information (Bethesda, Md.) nonredundant protein database (∼108,000 entries) with the PeptideSearch algorithm (Matthias Mann, Southern Denmark University, Odense, Denmark), with a mass accuracy restriction of better than 40 ppm and a maximum of one missed cleavage site allowed per peptide. Mass spectrometric sequencing of selected peptides was done by matrix-assisted laser desorption ionization-time of flight-time of flight (MS/MS) analysis on the same prepared samples with the UltraFlex instrument in LIFT mode. Fragment ion spectra were taken to search the National Center for Biotechnology Information nonredundant protein database with the MASCOT MS/MS Ion Search program (Matrix Science Ltd., London, United Kingdom). Any identification thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
Chromatin immunoprecipitation in vitro.
Transcription reactions were allowed to proceed for 30 min in the presence of 3 μM His-TAF-Iβ or HA-His-TAF-Iβ and then diluted to 200 μl with HEG-50 mM KCl-0.1% Tween 20. Immunoprecipitations were then performed with 5 μl of HA antibody (16B12 ascites fluid; Covance) and 15 μl of protein G Sepharose (Amersham). The beads were washed four times with HEG-150 mM KCl-0.1% Tween 20 and once with HEG-50 mM NaCl. The beads were then eluted with 200 μl of 1% sodium dodecyl sulfate (SDS)-50 mM NaCl. The samples were digested with proteinase K (Sigma) and subjected to phenol-chloroform extraction and ethanol precipitation. The amount of DNA recovered was quantified by PCR with primers flanking the promoter.
HA-TAF-I protein immunoprecipitation.
The interaction of TAF-I and endogenous SNF2H was detected by incubating the indicated amounts of His-TAF-Iβ or HA-His-TAF-Iβ with PC1 for 30 min at room temperature. The reaction mixture was then rocked in the presence of 15 μl of protein G beads (Amersham) covalently coupled to HA antibody (16B12; Covance) at room temperature for 1 h. The beads were washed four times with HEG-150 mM KCl-0.1% Tween 20 and once with HEG-50 mM NaCl. The beads were eluted in SDS gel loading buffer, and bound proteins were separated by SDS-6% polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membrane, and detected with anti-hSNF2 (1B9/D12; Upstate Biotechnology).
Anti-SNF2 depletion of PC1 fraction.
Ten micrograms of anti-SNF2 (1B9/D12; Upstate Biotechnology) was prebound to 20 μl of protein G Sepharose. The anti-SNF2-coupled beads or control protein G beads were mixed with 300 μg of PC1 fraction and incubated at 4°C with agitation for 2 h. The supernatants from the control beads (mock-depleted PC1) and anti-SNF2 beads (SNF2H-depleted PC1) were collected and used in transcription reaction mixtures. The depletion was monitored by immunoblotting with the anti-SNF2 antibody.
RESULTS
Identification of a fraction required for VDR-mediated transcription on chromatin.
We set out to define the components of a HeLa cell nuclear extract required to support transcription on a chromatin template. When we incubated a reporter plasmid containing four VDREs upstream of an adenovirus E1B minimal core promoter with purified core histones and a Drosophila embryonic S190 extract, nucleosomes were deposited in a physiologically spaced array, as revealed by limited micrococcal nuclease digestion (Fig. 1A, left). In the presence of VDR, a fractionated transcription system derived from the phosphocellulose-bound fraction of a HeLa nuclear extract was capable of ligand-responsive transcription from crude chromatin assembled in the S190 extract (Fig. 1C, lanes 1 and 2). However, when the chromatin was partially purified from the S190 extract by gel filtration, ligand-dependent transcription was nearly abolished (Fig. 1C, lanes 5 and 6). Therefore, components of the S190 extract not stably incorporated into the chromatin are apparently required for efficient transcription. Transcription on the purified chromatin was restored by adding the phosphocellulose-unbound fraction of the nuclear extract (PC.3) to the reaction mixture (Fig. 1C, lanes 7 and 8), indicating that PC.3 contained an activity or activities required for VDR-activated transcription on chromatin templates. We next fractionated the PC.3 fraction by DEAE anion-exchange chromatography; transcription-stimulatory activity eluted in a single peak at ∼330 mM KCl, a fraction we designated DE330 (Fig. 2B).
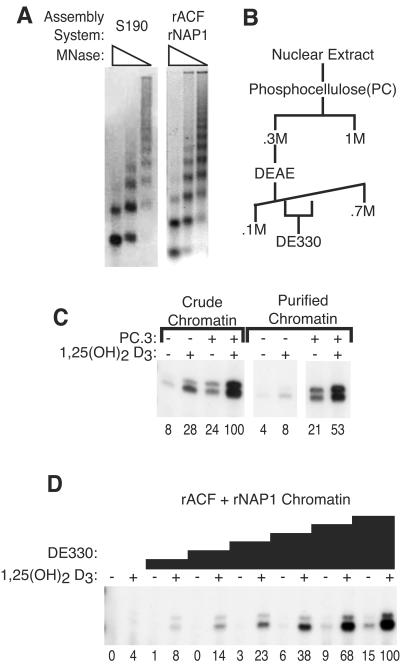
FIG. 1.
Identification of activities required for chromatin transcription. (A) Titration of micrococcal nuclease in a limited digestion of promoter-containing DNA assembled into chromatin with crude chromatin assembly extract (S190) or with a recombinant chromatin assembly system (rACF and rNAP-1). (B) Fractionation of HeLa nuclear extract. (C) VDR-mediated transcription reaction mixtures were prepared with S190-assembled chromatin left unpurified (crude chromatin) or purified over an S300 spin column (purified chromatin) in the presence or absence of the PC.3 fraction (12 μg) and vitamin D3 as indicated. (D) VDR-mediated transcription reaction mixtures were prepared on chromatin assembled in the recombinant system in the presence or absence of vitamin D3 and buffer only or 0.25, 0.5, 0.75, 1.0, 1.5, or 2 μl of the DE330 fraction, as indicated. The relative level of transcription was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
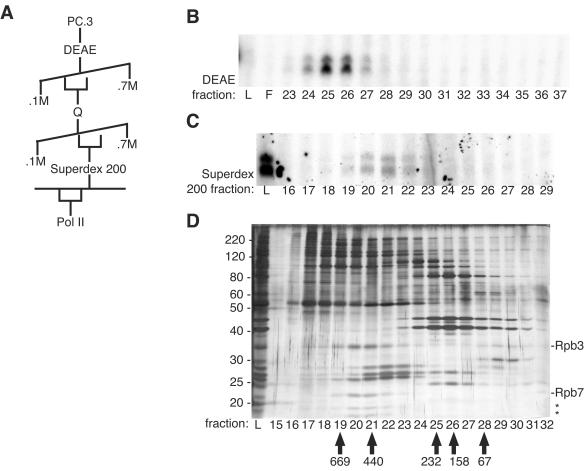
FIG. 2.
Pol II copurifies with chromatin transcription activity. (A) Scheme for the further fractionation of PC.3. (B) VDR-mediated transcription reaction mixtures were prepared with 1.5 μl of the indicated DEAE fractions. The peak of activity eluted at ∼330 mM KCl. (C) VDR-mediated transcription reaction mixtures were prepared with 2 μl of the indicated Superdex 200 fractions or 1 μl of the load. (D) The Superdex 200 fractions were resolved by SDS-10% PAGE, and proteins were visualized by silver staining. Migration of polypeptides correlating with activity is indicated by asterisks. Mass spectrometry identified two of these as Rpb3 and Rpb7, as indicated. The values on the left are molecular sizes in kilodaltons. L, load; F, flowthrough.
Because the S190 extract contains potentially confounding activities capable of stimulating chromatin transcription, we next tested the ability of the DE330 fraction to stimulate transcription on chromatin assembled in a system consisting of purified core histones and recombinant NAP-1 and ACF (a complex of the catalytic subunit ISWI and the accessory factor Acf1). Chromatin assembled in the recombinant system also contained well-spaced nucleosomes (Fig. 1A, right). Just as PC.3 was required to transcribe partially purified chromatin assembled in the S190 extract, DE330 was required for transcription from chromatin assembled in the recombinant system (Fig. 1D), which was the source of the chromatin used in all subsequent assays. We measured ∼25-fold stimulation of activated transcription in the presence of vitamin D3 by the DE330 fraction at the maximal concentration attainable in the assay. Transcription in the absence of vitamin D3 also increased with increasing amounts of the DE330 fraction, but there was an essentially constant ratio of activated-to-basal transcription at all concentrations for which a signal could be measured above the background in the absence of ligand, indicating that the DE330 fraction facilitated but did not circumvent VDR-mediated activation of transcription on chromatin templates.
Pol II cofractionates with chromatin transcription activity.
We further purified the stimulatory activity by Q Sepharose anion-exchange and Superdex 200 gel filtration chromatography (Fig. 2A). We observed a peak of activity in Superdex 200 fractions 20 to 22, corresponding to an apparent molecular mass of ∼500 kDa (Fig. 2C), although ∼70% of the activity was lost during the gel filtration step. Four polypeptides of 38, 22, 18, and 16 kDa cofractionated with the activity (Fig. 2D). Mass spectrometric analysis identified the 38- and 22-kDa species as the Rpb3 and Rpb7 subunits of Pol II, respectively. Immunoblotting confirmed the copurification of Rpb1, the largest subunit of Pol II, with transcription-stimulatory activity (data not shown).
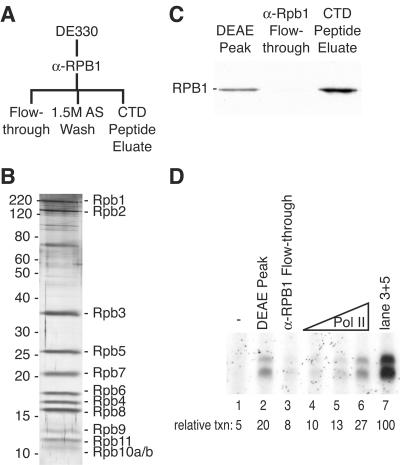
To determine whether the Pol II enzyme purified from the DE330 fraction is necessary and/or sufficient to reconstitute transcription on chromatin, we subjected the fraction to anti-Rpb1 immunoaffinity chromatography (Fig. 3A). Pol II was efficiently depleted from the DE330 fraction by passage over an anti-Rpb1 monoclonal antibody (8WG16) column and could be eluted with a peptide corresponding to the epitope: the carboxyl-terminal domain (CTD) of Rpb1 (Fig. 3B and C). The CTD peptide eluate contained highly purified Pol II, as judged by silver staining after SDS-PAGE (Fig. 3B); we detected all known subunits of Pol II in apparently equal stoichiometry, as well as several high-molecular-weight polypeptides that were apparently substoichiometric (Fig. 3B).
FIG. 3.
Pol II is required but not sufficient to reconstitute the chromatin transcription activity in the DE330 fraction. (A) Pol II immunoaffinity chromatography scheme to purify Pol II from the DE330 fraction. (B) Immunopurified Pol II was analyzed by SDS-12% PAGE and silver staining. Pol II subunits are indicated at the right. The values on the left are molecular sizes in kilodaltons. (C) Immunoblot, probed with anti-Rpb1 monoclonal antibody 8WG16, of equivalent volumes of DE330, the anti-Rpb1 flowthrough fraction, and immunopurified Pol II. (D) VDR-mediated transcription reaction mixtures prepared with buffer only (lane 1); 2 μl of DE330 (lane 2); 2 μl of the anti-Rpb1 flowthrough fraction (lane 3); 1, 2, or 4 μl of immunopurified Pol II, corresponding to roughly 1.5, 3, or 6 times the amount of Pol II present in DE330 (lane 4 to 6); or both 2 μl of the anti-Rpb1 flowthrough fraction and 2 μl of immunopurified Pol II (lane 7). The relative level of transcription (txn) was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
Depleting the DE330 fraction of Pol II nearly abolished activity, indicating that this source of Pol II was indeed required for efficient transcription from chromatin templates (Fig. 3D, compare lanes 2 and 3). Whether this represented a quantitative effect of increasing the Pol II concentration relative to those of the other PC1 components or a specific requirement for the form of Pol II that flows through phosphocellulose remains to be determined. The purified Pol II enzyme was insufficient, however, to reconstitute the full activity of the parent DE330 fraction. In fact, roughly six times more purified Pol II, relative to the Pol II in the starting material, was required to produce equivalent transcription signals (Fig. 3D, compare lanes 2 and 6).
TAF-I/SET is required for transcription from chromatin.
Because neither the anti-Rpb1 flowthrough fraction nor purified Pol II alone could fully reconstitute the activity of the starting fraction, we performed mixing experiments. When we added both the anti-Rpb1 flowthrough fraction and immunopurified Pol II, we observed full reconstitution of the input activity. The stimulation of transcription by the two fractions was more than additive (Fig. 3D, compare lanes 3 and 5 to lane 7), indicating that both Pol II and a distinct activity in the anti-Rpb1 unbound fraction are required for chromatin-templated transcription.
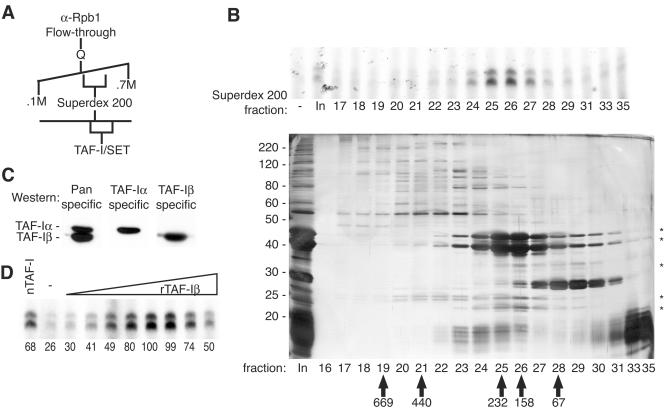
We further purified the activity in the anti-Rpb1 flowthrough fraction by Q Sepharose anion-exchange and Superdex 200 gel filtration chromatography (Fig. 4A). The activity in the anti-Rpb1 flowthrough fraction was only detected in the presence of immunopurified Pol II; we therefore assayed the column fractions in the presence of saturating amounts of Pol II. Under these conditions, we measured a >100% yield during gel filtration (Fig. 4B, upper part). Also, in contrast to the sizing column run prior to depletion of Pol II (Fig. 2C), the transcription-stimulatory activity migrated with an apparent size of ∼200 kDa (rather than ∼500 kDa). Two major polypeptides of 44 and 42 kDa cofractionated with activity during gel filtration (Fig. 4B, lower part). Mass spectrometric analysis determined that both of these polypeptides were derived from TAF-I/SET (hereafter referred to as TAF-I).
FIG. 4.
TAF-I stimulates activated transcription on chromatin templates. (A) Scheme for the further fractionation of the anti-Rpb1 flowthrough fraction. (B, top) VDR-mediated transcription performed in the presence of buffer, 1 μl of load, or 4 μl of the indicated Superdex 200 fraction. All reaction mixtures included 0.78 μg of immunopurified Pol II. (B, bottom) The Superdex 200 fractions were resolved by SDS-12% PAGE, and proteins were visualized by silver staining. Migration of polypeptides copurifying with the activity—all identified by mass spectrometry as isoforms of TAF-I—is indicated by asterisks. In, input. The values on the left are molecular sizes in kilodaltons. (C) The active fractions from the Superdex 200 column were subjected to immunoblotting with a TAF-I antibody that recognizes both the α and β isoforms, an α-specific antibody, and a β-specific antibody, as indicated. (D) VDR-mediated transcription performed in the presence of 4 μl of pooled Superdex 200 fractions 25 and 26 from panel B, buffer only, or 0.14, 0.29, 0.57, 1.14, 2.28, 4.56, 9.12, or 18.24 μM rTAF-Iβ. The relative level of transcription was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
TAF-I was originally identified as the product of a gene fused to the CAN gene in an acute undifferentiated myeloid leukemia (43) and was first characterized biochemically as a cellular factor required for the replication of purified adenovirus core particles in vitro (31). TAF-I contains a central domain homologous to NAP-1 and other members of the NAP family of histone chaperones. TAF-I exists in two isoforms generated by variable splicing—TAF-Iα and TAF-Iβ—that are identical except for unique stretches of 37 and 24 amino acids, respectively, at the amino terminus. Immunoblot assays with antibodies specific for either TAF-Iα or TAF-Iβ showed that both were present in the peak fraction obtained by Superdex 200 chromatography (Fig. 4C). We purified TAF-Iβ tagged with hexahistidine (His) at the amino terminus and expressed in bacteria. The recombinant TAF-Iβ (rTAF-Iβ) obtained was sufficient to reconstitute the activity in the Superdex 200 peak fractions (Fig. 4D), indicating that TAF-I is the only required factor in those fractions and that TAF-Iβ suffices for full activity.
TAF-I is a chromatin-specific coactivator for multiple activators.
As demonstrated above (Fig. 3D), the DE330 fraction contained two components required to reconstitute chromatin-templated transcription in vitro: Pol II and TAF-I. To determine the factor(s) in which chromatin specificity resides, we tested the effect of each component, alone or in combination, on transcription performed with naked or chromatin-assembled DNA templates (Fig. 5A). The immunopurified Pol II enzyme was capable of stimulating transcription by at least 10-fold on both chromatin and naked templates, regardless of the presence of TAF-Iβ. On the other hand, rTAF-Iβ stimulated transcription on chromatin by ∼10-fold but had a less-than-2-fold effect on transcription of naked DNA over the range of Pol II concentrations tested. Thus, rTAF-Iβ preferentially stimulated transcription of chromatin templates.
We next sought to determine whether TAF-I acts as a specific coactivator for nuclear receptors or plays a more general role. We tested the effect of TAF-I on transcription activated by Gal4-VP16—a fusion of the yeast Gal4 DNA-binding domain with the activation domain of the viral transactivator VP16—of a chromatin template containing five Gal4-binding sites upstream of the E4 viral promoter. The level of Gal4-VP16-mediated transcription was enhanced about sixfold by rTAF-Iβ (Fig. 5B). Therefore, stimulation of chromatin transcription by TAF-I is not restricted to promoters activated by nuclear receptors, suggesting a more general role in facilitating transcription.
Characterization of TAF-I in transcription.
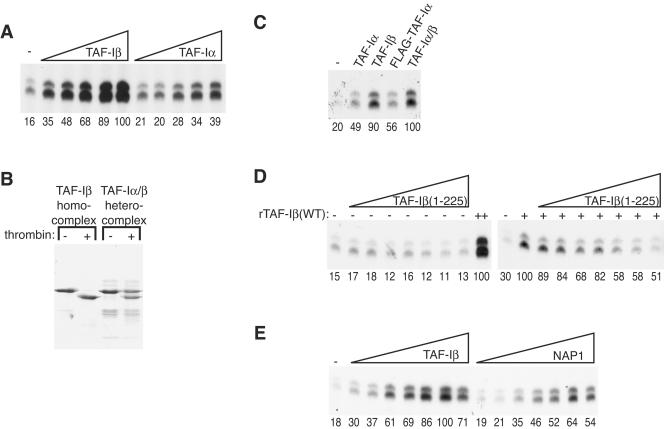
The TAF-I protein we purified from HeLa cells was a nearly stoichiometric mixture of TAF-Iα and TAF-Iβ. To examine the effect of TAF-Iα on chromatin transcription, TAF-Iα was expressed in bacteria and purified identically to rTAF-Iβ. rTAF-Iα was also able to stimulate chromatin transcription, albeit with a specific activity more than twofold lower than that of TAF-Iβ (Fig. 6A).
FIG. 6.
Characterization of the TAF-I requirement in chromatin transcription. (A) VDR-mediated transcription performed in the presence of buffer only or 0.16, 0.33, 0.67, 1.32, or 2.64 μM rTAF-Iβ or rTAF-Iα, as indicated. (B) Purification of TAF-Iα/β heterocomplexes. His-tagged TAF-Iβ and FLAG-tagged TAF-Iα migrate identically on SDS-10% PAGE, but only TAF-Iβ can be cleaved by thrombin. Thrombin cleavage of 2.5 μg of double-affinity-purified TAF-Iα/β heterocomplexes or TAF-Iβ and separation of the products by SDS-10% PAGE reveal a 1:1 ratio of TAF-Iα to TAF-Iβ in the TAF-Iα/β heterocomplex. (C) VDR-mediated transcription performed in the presence of buffer only or 500 nM His-tagged TAF-Iα, His-tagged TAF-Iβ, FLAG-tagged TAF-Iα, or TAF-Iα/β heterocomplex. (D, left) VDR-mediated transcription performed in the presence of buffer only; 0.15, 0.3, 0.6, 1.21, 2.41, 4.82, or 9.65 μM rTAF-Iβ(1-225); or 2.28 μM wild-type rTAF-Iβ (++). (D, right) VDR-mediated transcription performed in the presence of buffer only or 0.17, 0.35, 0.69, 1.39, 2.78, 5.56, or 11.11 μM rTAF-Iβ(1-255). Wild-type rTAF-Iβ (800 nM, +) was added where indicated. (E) VDR-mediated transcription performed in the presence of buffer only or 0.15, 0.29, 0.58, 1.17, 2.34, 4.67, or 9.35 μM rTAF-Iβ or rNAP-1. The relative level of transcription was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
TAF-Iα and TAF-Iβ have been shown to form dimers in extracts and when expressed in bacteria (32). To compare the activity of TAF-Iβ with that of a presumptive TAF-Iα/β dimer, we constructed rTAF-Iα in which the His tag was replaced with a FLAG tag. We expressed FLAG-rTAF-Iα together with His-TAF-Iβ in E. coli and purified, by sequential cobalt and FLAG affinity chromatography, a complex containing TAF-Iα and TAF-Iβ in equal proportions, consistent with a heterodimer (Fig. 6B). In transcription reaction mixtures, the relative activities of the three complexes were as follows: TAF-Iα/TAF-Iβ = TAF-Iβ alone > TAF-Iα alone (Fig. 6C).
The acidic carboxyl terminus of TAF-I is important for several of its previously reported functions (34, 39, 40). We expressed and purified a truncation mutant form that lacked the 52 amino acids corresponding to the acidic carboxyl terminus [rTAF-Iβ(1-225)]. The truncated protein was completely inactive in transcription, indicating that the carboxyl terminus is required to stimulate transcription on chromatin (Fig. 6D, left part). The rTAF-Iβ(1-225) mutant construct was also capable of inhibiting the activity of wild-type TAF-I when added to the same reaction mixture (Fig. 6D, right part).
Because TAF-I is closely related to NAP-1, we investigated whether NAP-1 could also stimulate transcription. NAP-1 was indeed capable of substituting for TAF-I but had about twofold lower specific activity (Fig. 6E). Thus, stimulation of transcription from chromatin templates may be a general property of the NAP-1 family of histone chaperones.
TAF-I interactions with chromatin and chromatin remodeling factors.
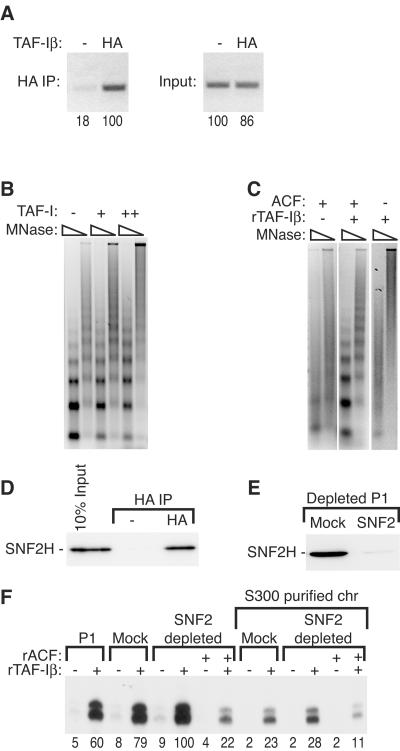
Like other histone chaperones, TAF-I is able to bind histones in solution (30). Because TAF-I is required for transcription on chromatin, we asked whether it could interact with chromatin under transcription conditions. For this experiment, we purified a version of TAF-Iβ tagged at its amino terminus with the HA epitope, as well as with His. HA-His-TAF-Iβ was as active as His-TAF-Iβ in the chromatin transcription assay (data not shown). We performed chromatin immunoprecipitations to determine if TAF-I was interacting with chromatin during transcription reactions in vitro. Transcription was allowed to proceed for 30 min in the presence of HA-tagged or untagged TAF-I. The chromatin was then digested with micrococcal nuclease to yield an average fragment size of 0.5 kb and immunoprecipitated with anti-HA antibody. We quantified the DNA recovered by PCR with primers flanking the promoter and measured about sixfold enrichment when the reaction mixture contained HA-His-TAF-Iβ, relative to control reaction mixtures containing His-TAF-Iβ (Fig. 7A). Thus, TAF-Iβ can indeed associate with chromatin in the context of transcription.
FIG. 7.
TAF-I interacts with chromatin and chromatin-remodeling factors. (A) Transcription reaction mixtures containing HA-tagged or untagged TAF-Iβ were incubated for 30 min and then subjected to chromatin immunoprecipitation (IP) in vitro with HA antibody. Coimmunoprecipitated DNA was analyzed by semiquantitative PCR and 1% agarose gel electrophoresis. (B) Preassembled chromatin was incubated with buffer alone (−) or 15.3 (+) or 53 (++) ng of HeLa TAF-I from the peak Superdex 200 fractions per μl (Fig. 4B) and then subjected to limited micrococcal nuclease (MNase) digestion. A 15.3-ng/μl concentration of this fraction is roughly equivalent to 4 μl of fractions 25 and 26 assayed in Fig. 4B. (C) Chromatin assembly reaction mixtures were prepared without rNAP-1. Where indicated, ACF and 8 μM rTAF-Iβ were present during assembly. After assembly, the chromatin was subjected to limited micrococcal nuclease digestion. (D) HA-tagged or untagged TAF-Iβ (58 pmol) was incubated with 36 μg of the PC1 fraction for 30 min and subsequently immunoprecipitated with HA antibody. The immunoprecipitates and input (10%) were analyzed by immunoblot assay with an anti-SNF2 antibody. (E) The mock-depleted or anti-SNF2-depleted PC1 fraction was analyzed by immunoblotting with the anti-SNF2 antibody. (F) Transcription reaction mixtures were prepared with 3 μg of PC1, mock-depleted PC1, or anti-SNF2-depleted PC1 on unpurified and S300 spin column-purified chromatin (chr). rACF (170 nM) and rTAF-Iβ (3 μM) were added where indicated. The relative level of transcription was quantified by Phosphorimager, with the maximal level defined as 100 U, and is indicated below each lane.
Because TAF-I is a histone chaperone, one possible explanation for its ability to enhance transcription is that it disrupts or disassembles chromatin, in effect rendering the plasmid DNA naked. To test this possibility, different amounts of TAF-I were incubated with preassembled chromatin prior to limited micrococcal nuclease digestion (Fig. 7B). At concentrations equal to or greater than those that stimulate transcription, TAF-I did not cause a gross change in chromatin structure detectable by increased accessibility to micrococcal nuclease.
TAF-I belongs to the same family of histone chaperones as NAP-1, which can participate in chromatin assembly in cooperation with the ACF complex. Indeed, TAF-I could substitute for NAP-1 in the ACF-dependent assembly of chromatin, but with a 10-fold higher optimal concentration compared to NAP-1 (Fig. 7C). We also tested whether TAF-I and ACF physically interact. In pulldown assays with purified proteins, TAF-Iβ interacted with the recombinant ISWI-containing ACF complex (data not shown). SNF2H is the human homolog of Drosophila ISWI, the catalytic subunit of the ACF complex. Incubation of HA-His-TAF-Iβ with the PC1 fraction, followed by anti-HA immunoprecipitation and immunoblotting with an SNF2H-specific antibody, confirmed an interaction of SNF2H with TAF-I (Fig. 7D). To our knowledge direct binding between a NAP family histone chaperone and an ATP-dependent chromatin-remodeling enzyme has not previously been described.
We explored a possible role of the SNF2H-TAF-I interaction in chromatin transcription by depleting SNF2H from the PC1 fraction with an anti-SNF2H antibody (Fig. 7E). Compared to mock depletion, depletion of SNF2H had a very small positive effect on TAF-I-mediated transcription (Fig. 7F, SNF2 depleted). Adding back recombinant ACF to the PC1 fraction depleted of endogenous SNF2H strongly inhibited transcription (Fig. 7F, SNF2 depleted + rACF). Therefore, SNF2H, a factor with which TAF-I cooperates to assemble chromatin, is dispensable (and possibly even inhibitory) for chromatin-templated transcription.
Defining the timing of TAF-I action.
Although chromatin incubated with TAF-I after assembly or chromatin assembled with TAF-I could not be distinguished from NAP-1-assembled chromatin by micrococcal nuclease digestion, it remained possible that TAF-I caused subtle and/or local changes in chromatin structure. To test whether TAF-I alone could stably modify the chromatin to render it transcriptionally competent, we preincubated rTAF-Iβ with preassembled chromatin for 30 min prior to S300 spin column chromatography to remove the TAF-I protein. The pretreated chromatin still required addition of TAF-I during the transcription reaction, suggesting that TAF-I was incapable of performing its function in the absence of other components of the transcription machinery (Fig. 8A).
We also tested the possibility of a functional difference between chromatin assembled with TAF-I and chromatin assembled with NAP-1 by performing transcription of either template with or without additional TAF-I. Without purification of the templates over S300 spin columns, addition of TAF-I during the transcription reaction was only required for the chromatin assembled with NAP-1 (Fig. 8B). When the chromatin was purified to remove factors not stably incorporated during assembly, however, TAF-I was required during the transcription reaction regardless of whether NAP-1 or TAF-I mediated assembly (Fig. 8B). Therefore, TAF-I facilitates two temporally distinct processes, assembly of DNA into a chromatin structure and activation of transcription on a chromatin template.
Because neither incubation of TAF-I with preassembled chromatin nor assembly de novo with TAF-I could render chromatin competent for transcription, TAF-I must act at a subsequent step to facilitate either preinitiation complex formation, initiation of transcription, or elongation by Pol II through chromatin. Interestingly, two known Pol II elongation factors, Spt6 and FACT, are histone chaperones (6, 36). To test whether TAF-I executes its required function during transcript elongation—as does FACT—or at a prior step, we performed pulse-chase transcription on chromatin templates (Fig. 8C). When TAF-I was added prior to a pulse with [α-32P]CTP, the elongation of labeled transcripts initiated during the pulse could be observed as a smear extending up the gel after chasing with an excess of unlabeled CTP. We estimated an elongation rate of 130 nucleotides/min, comparable to rates previously measured in vitro on chromatin templates (16). However, if TAF-I was added only after the reaction mixture had been supplemented with unlabeled CTP, no elongating transcripts appeared. Therefore, the requirement for TAF-I is at a step after the assembly of chromatin but prior to the elongation phase of transcription.
DISCUSSION
TAF-I is an activator of chromatin-templated transcription.
We have purified TAF-I from HeLa cell nuclear extracts as a factor required for chromatin-templated transcription. These results establish a novel function for NAP family histone chaperones in the activation of transcription, which is at odds with previous reports suggesting a repressive role for TAF-I. There is recent evidence, however, to support a positive role in vivo. The Drosophila homolog of TAF-I has been shown to be redistributed to heat shock loci, with Pol II, upon transcriptional activation (35). Therefore, in at least one setting in vivo, TAF-I is in the right place at the right time to facilitate transcription.
Since its original genetic identification as the product of the leukemia-associated proto-oncogene SET, TAF-I has had several functions ascribed to it. TAF-I was originally purified as a factor required for the replication in vitro of adenovirus DNA core particles purified from infected cells. Adenovirus DNA is packaged into a chromatin-like nucleoprotein structure with viral basic protein; TAF-I can remodel that structure into a replication-competent form. Thus, the first and perhaps best-established role for TAF-I is to promote, rather than impede, a polymerization reaction on DNA templates assembled into higher-order chromatin-like structures.
TAF-I was also purified as a potent noncompetitive inhibitor of protein phosphatase 2A (PP2A), called I2PP2A (25, 26). Interestingly, PP2A has recently been implicated in mediating transcriptional repression in Drosophila, leading to the hypothesis that the transcriptional effect of TAF-I might be exerted, at least in part, through its inhibition of PP2A (35). TAF-I has also been identified as an inhibitor of the granzyme A-activated DNase NM23-H1 (11). Granzyme A is inserted into virus-infected cells by cytotoxic T cells and activates NM23-H1 by cleaving TAF-I, leading to NM23-H1 derepression and in turn to apoptosis.
Most recent work on TAF-I has emphasized its putative role as a transcriptional repressor (27, 33, 40, 42). This functional assignment stems from the ability of TAF-I to inhibit p300-mediated acetylation of histones in solution. Indeed, TAF-I was purified from mammalian cell extracts as INHAT (inhibitor of histone acetyltransferase) on the basis of this property. Assays of reporter gene expression after transient transfection of TAF-I seemed to support a repressive role. These findings suggest the following three possible (not mutually exclusive) scenarios: (i) TAF-I is a bona fide corepressor for at least a subset of transcription factors, (ii) overexpression of TAF-I represses transcription by promoting the assembly of transfected reporter constructs into generally repressive chromatin structures, and/or (iii) overexpression of TAF-I above the optimal level leads to squelching of transcriptional signals, a phenomenon we can recapitulate in vitro (Fig. 4D, for example). There has been no direct demonstration, however, that TAF-I represses transcription in vitro or at natural chromosomal loci in vivo. Our data, and those obtained in vivo with Drosophila (35), support a positive role for TAF-I at a subset of promoters at least. An interesting possibility is that TAF-I can be both an activator and a repressor of transcription and that which effect predominates is context dependent.
In that light, it is perhaps significant that TAF-I has been purified repeatedly as an inhibitor of one (seemingly unrelated) enzyme or another (7, 10, 11, 25, 38, 40). The abilities of TAF-I to inhibit several different enzymes and interact stably with a multitude of proteins in extracts (unpublished observations) may be functions of its highly acidic carboxyl-terminal region. Whatever the explanation, given the complexity of the protein-protein interactions possible for TAF-I, the true function(s) of this protein may be difficult to discern by considering any one interaction in isolation. We have taken a different approach to show that TAF-I plays a positive, required role in a complex biochemical process reconstituted in vitro: transcription from promoter DNA embedded in chromatin.
Requirement of a histone chaperone early in the transcription cycle.
We have shown that TAF-I not only stimulates activator-mediated transcription on chromatin but also can cooperate with ACF in the assembly of chromatin. A role for histone chaperones in the assembly of chromatin is experimentally well supported (2, 28). Histone chaperones such as CAF-I and Asf1 participate in replication-coupled pathways, whereas others, such as NAP-1, participate with ISWI family members in replication-independent chromatin assembly.
Histone chaperones have also recently been implicated directly in transcriptional activation as components of a transcription-coupled chromatin assembly or maintenance pathway. In vivo, regions of DNA actively transcribed by Pol II retain a chromatin structure. At a physiologic salt concentration, however, even a single nucleosome is a strong barrier to Pol II transcription (19). In the presence of either a high salt concentration or the chromatin-specific transcription elongation factor and histone chaperone FACT, Pol II transcription disrupts chromatin structure by displacement of an H2A-H2B dimer from the nucleosome (4, 19). Mutation of SPT16 (which encodes a component of FACT) in yeast leads to disruption of chromatin structure specifically in actively transcribed regions, leading to aberrant initiation from cryptic start sites and misregulation of certain transcripts (18). We attempted to discern a similar effect of TAF-I on elongation, but because of its required role at an earlier step, any effect of TAF-I analogous to that of FACT could not be measured.
We have identified another step in the transcription cycle at which a histone chaperone is required to relieve chromatin-mediated repression: after chromatin assembly and prior to the elongation phase. TAF-I may be needed, for example, to remodel chromatin into a state competent for preinitiation complex assembly. Indeed, recent evidence from two groups supports a role for histone chaperones in remodeling the chromatin of promoters to activate transcription in vivo. At the yeast PHO5 and PHO8 promoters, the chromatin conformational changes that occur upon activation of transcription are due to nucleosome disassembly rather than the sliding of nucleosomes mediated by remodeling enzymes (5). Furthermore, the histone chaperone Asf1 is required both for the creation of the nuclease-hypersensitive chromatin structure at these promoters and for their transcriptional activation (1).
Model for TAF-I function in transcriptional activation.
Might TAF-I act in a pathway analogous to that of Asf1 to disassemble chromatin at a promoter destined for transcriptional activation? Support for a role in chromatin disassembly comes from TAF-I's ability to bind and trigger the decondensation of sperm chromatin, with simultaneous release of sperm-specific, histone-like basic proteins from the chromatin (29, 30). Furthermore, two independent reports have demonstrated that the related (and in our assays, functionally equivalent) NAP-1 protein can mediate activator-dependent disruption of nucleosomes (14, 44). Our experiments indicate, however, that incubation of TAF-I by itself with chromatin is insufficient to render the chromatin transcriptionally competent (Fig. 8). Therefore, TAF-I is likely to act in concert with other transcription factors or chromatin-remodeling activities to activate transcription. In summary, we have uncovered a positive role for TAF-I and its relatives in transcribing chromatin and have defined a window for that action early in the transcription cycle. We may now ask whether this is a general requirement in vivo and how these conserved histone chaperones activate transcription on chromatin.
Acknowledgments
We thank Stewart Shuman and Stéphane Larochelle for critical review of the manuscript. We are grateful to James Kadonaga, who provided the NAP-1, ISWI, and Acf1 baculoviruses; Kyosuke Nagata, who provided the anti-TAF-I antibodies; Jeffrey Parvin, who provided the TFIIA expression vector; and W. Lee Kraus, who provided the Gal4-VP16 expression vector and the pGEIO plasmid. HeLa cells were cultured by the National Cell Culture Center. We thank Brian Lemon for helpful discussion and Chao-Pei Chang for experimental support.
This work was supported by NIH grant DK45460 to L.P.F. and R.P.F.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Akey, C. W., and K. Luger. 2003. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 13:6-14. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P. B., and C. Wu. 1992. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol. Cell. Biol. 12:2241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. [DOI] [PubMed] [Google Scholar]
- 5.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 7.Canela, N., A. Rodriguez-Vilarrupla, J. M. Estanyol, C. Diaz, M. J. Pujol, N. Agell, and O. Bachs. 2003. The SET protein regulates G2/M transition by modulating cyclin B-cyclin-dependent kinase 1 activity. J. Biol. Chem. 278:1158-1164. [DOI] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdjument-Bromage, H., M. Lui, L. Lacomis, A. Grewal, R. S. Annan, D. E. McNulty, S. A. Carr, and P. Tempst. 1998. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826:167-181. [DOI] [PubMed] [Google Scholar]
- 10.Estanyol, J. M., M. Jaumot, O. Casanovas, A. Rodriguez-Vilarrupla, N. Agell, and O. Bachs. 1999. The protein SET regulates the inhibitory effect of p21(Cip1) on cyclin E-cyclin-dependent kinase 2 activity. J. Biol. Chem. 274:33161-33165. [DOI] [PubMed] [Google Scholar]
- 11.Fan, Z., P. J. Beresford, D. Y. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659-672. [DOI] [PubMed] [Google Scholar]
- 12.Glikin, G. C., I. Ruberti, and A. Worcel. 1984. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell 37:33-41. [DOI] [PubMed] [Google Scholar]
- 13.Haile, D. T., and J. D. Parvin. 1999. Activation of transcription in vitro by the BRCA1 carboxyl-terminal domain. J. Biol. Chem. 274:2113-2117. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izban, M. G., and D. S. Luse. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267:13647-13655. [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 19.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 20.Kornberg, R. D., and Y. Lorch. 2002. Chromatin and transcription: where do we go from here. Curr. Opin. Genet. Dev. 12:249-251. [DOI] [PubMed] [Google Scholar]
- 21.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 22.Lemon, B. D., J. D. Fondell, and L. P. Freedman. 1997. Retinoid X receptor:vitamin D3 receptor heterodimers promote stable preinitiation complex formation and direct 1,25-dihydroxyvitamin D3-dependent cell-free transcription. Mol. Cell. Biol. 17:1923-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 24.Levenstein, M. E., and J. T. Kadonaga. 2002. Biochemical analysis of chromatin containing recombinant Drosophila core histones. J. Biol. Chem. 277:8749-8754. [DOI] [PubMed] [Google Scholar]
- 25.Li, M., H. Guo, and Z. Damuni. 1995. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry 34:1988-1996. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., A. Makkinje, and Z. Damuni. 1996. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 271:11059-11062. [DOI] [PubMed] [Google Scholar]
- 27.Loven, M. A., N. Muster, J. R. Yates, and A. M. Nardulli. 2003. A novel estrogen receptor alpha-associated protein, template-activating factor Iβ, inhibits acetylation and transactivation. Mol. Endocrinol. 17:67-78. [DOI] [PubMed] [Google Scholar]
- 28.Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677:3-11. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto, K., K. Nagata, M. Miyaji-Yamaguchi, A. Kikuchi, and M. Tsujimoto. 1999. Sperm chromatin decondensation by template activating factor I through direct interaction with basic proteins. Mol. Cell. Biol. 19:6940-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, K., K. Nagata, M. Okuwaki, and M. Tsujimoto. 1999. Histone- and chromatin-binding activity of template activating factor-I. FEBS Lett. 463:285-288. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, K., K. Nagata, M. Ui, and F. Hanaoka. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582-10587. [PubMed] [Google Scholar]
- 32.Miyaji-Yamaguchi, M., M. Okuwaki, and K. Nagata. 1999. Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity. J. Mol. Biol. 290:547-557. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto, S., T. Suzuki, S. Muto, K. Aizawa, A. Kimura, Y. Mizuno, T. Nagino, Y. Imai, N. Adachi, M. Horikoshi, and R. Nagai. 2003. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 23:8528-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak, S. J., C. Y. Pai, and V. G. Corces. 2003. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23:6129-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 37.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Naar, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 38.Ruggiero-Lopez, D., C. Manioc, C. Geourjon, P. Louisot, and A. Martin. 1994. Purification and partial amino acid sequence of fuctinin, an endogenous inhibitor of fucosyltransferase activities. Eur. J. Biochem. 224:47-55. [DOI] [PubMed] [Google Scholar]
- 39.Saito, S., M. Miyaji-Yamaguchi, T. Shimoyama, and K. Nagata. 1999. Functional domains of template-activating factor-I as a protein phosphatase 2A inhibitor. Biochem. Biophys. Res. Commun. 259:471-475. [DOI] [PubMed] [Google Scholar]
- 40.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 41.Studitsky, V. M., W. Walter, M. Kireeva, M. Kashlev, and G. Felsenfeld. 2004. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29:127-135. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, T., S. Muto, S. Miyamoto, K. Aizawa, M. Horikoshi, and R. Nagai. 2003. Functional interaction of the DNA-binding transcription factor Sp1 through its DNA-binding domain with the histone chaperone TAF-I. J. Biol. Chem. 278:28758-28764. [DOI] [PubMed] [Google Scholar]
- 43.von Lindern, M., S. van Baal, J. Wiegant, A. Raap, A. Hagemeijer, and G. Grosveld. 1992. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 12:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, P. P., T. A. Owen-Hughes, J. Cote, and J. L. Workman. 1995. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler, G. S., A. Kristjuhan, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99:3517-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]