Abstract
Transcriptional coactivator p300 is required for embryonic development and cell proliferation. Valproic acid, a histone deacetylase inhibitor, is widely used in the therapy of epilepsy and bipolar disorder. However, it has intrinsic teratogenic activity through unidentified mechanisms. We report that valproic acid stimulates proteasome-dependent p300 degradation through augmentation of gene expression of the B56γ regulatory subunits of protein phosphatase 2A. The B56γ3 regulatory and catalytic subunits of protein phosphatase 2A interact with p300. Overexpression of the B56γ3 subunit leads to proteasome-mediated p300 degradation and represses p300-dependent transcriptional activation, which requires the B56γ3 interaction domain of p300. Conversely, silencing of the B56γ subunit expression by RNA interference increases the stability and transcriptional activity of the coactivator. Our study establishes the functional interaction between protein phosphatase 2A and p300 activity and provides direct evidence for signal-dependent control of p300 function.
Activation of gene expression is a dynamic and complex process in which the functional transcription machinery is recruited and assembled at target promoters (24, 45). An essential component of many transcription complexes is the transcriptional coactivator p300 (12). Structurally and functionally related to CREB binding protein (CBP), p300 has been implicated in regulating a broad array of cellular activities (10, 25, 26). Embryonic development in mice is very sensitive to gene dosage of the coactivator, and p300 knockout mice exhibit neural tube defects (51). Recent studies have demonstrated multiple molecular mechanisms of p300 function, as the coactivator has intrinsic histone acetyltransferase (HAT) and ubiquitin ligase activities (13, 37). However, the stimuli, signaling cascades, and proteins that regulate these p300 activities are poorly understood.
Degradation through the 26S proteasome pathway involves covalent modification of the target protein with multiple ubiquitin moieties prior to the destruction of the tagged protein (7, 20, 22). A growing body of evidence suggests that the 26S proteasome is not only responsible for eliminating misfolded and damaged intracellular proteins but is also responsible for selective degradation of many transcriptional activators and coactivators, including p300 (29, 34, 35, 40). During retinoic acid-induced differentiation of F9 embryonal carcinoma cells, the level of p300 was dramatically reduced as a result of the proteasome-mediated degradation (3). It has also been suggested that ubiquitination can serve as a signal for augmenting the transcriptional activation and destruction of some activator proteins via the 26S proteasome (41). Phosphorylation is known to enhance degradation of some proteins through the 26S proteasome pathway, but the regulatory signals that target each individual protein for ubiquitination remain unclear (7).
Reversible phosphorylation is critical for regulating a diverse array of cellular processes. One of the major serine-threonine phosphatases, protein phosphatase 2A (PP2A) is a heterotrimeric complex consisting of a structural subunit, a catalytic subunit, and a variable regulatory subunit. There are three distinct classes of regulatory subunits, B/B55, B′/B56, and B"/B72, which modulate substrate selectivity and intracellular localization of PP2A (49). B56 is the most diverse PP2A regulatory subunit with five isoforms (α, β, γ, δ, and ɛ) and three splice variants of the γ isoform (γ1, γ2, and γ3), which are widely expressed but are generally expressed at low levels (32, 47). Among the different isoforms of B56 regulatory subunits, B56γ1, B56γ2, B56γ3, and B56δ have nuclear localization, while B56α, B56β, and B56ɛ are cytoplasmic (31, 47). Various B56 subunits of PP2A have been shown to regulate cell growth, apoptosis, and neoplastic progression (16, 28).
Valproic acid is widely used for treatment of epilepsy, bipolar disorder, and neuropathic pain, migraine prophylaxis, and the control of a variety of seizures (17, 30). However, pregnant women treated with valproic acid early in their pregnancy can give birth to children with defects in neural tube closure (36). It has been difficult to elucidate the molecular and biochemical mechanisms underlying these therapeutic as well as teratogenic effects of valproic acid. The discovery in recent years that valproic acid acts as a histone deacetylase (HDAC) inhibitor has allowed insight into the mode of action of this short-chained fatty acid, which not only affects class 1 HDACs by inhibiting their catalytic activity but also induces proteasome-mediated HDAC2 degradation (21, 39). Therefore, alteration of gene expression through HDAC inhibition has become an interesting avenue for understanding how valproic acid affects cell differentiation and teratogenicity.
In this work, we demonstrate that the B56γ3 regulatory subunit of PP2A targets the transcriptional coactivator p300 for degradation through the 26S proteasome pathway and define the molecular mechanisms regulating this process. Valproic acid stimulates the proteasome-mediated p300 degradation by augmenting gene expression of the B56γ3 regulatory subunits of PP2A. Moreover, the selective p300 degradation is mediated by PP2A through an interaction between the B56γ3 regulatory subunit and the transcriptional coactivator p300.
MATERIALS AND METHODS
Plasmids and reagents.
The plasmids for B56γ2 and B56γ3 subunits (47), glutathione S-transferase (GST)-p300 (15), wild-type p300, p300ΔC (46), p300N, p300C (37), βRE3-Luc reporter (38) and the antibody specific to the B56γ2 and B56γ3 subunits (47) have been described previously. MG132 and valproic acid were purchased from Sigma. Okadaic acid was from Calbiochem. Antibodies against p300, SRC, p53, and retinoic acid receptor β (RARβ) were from Santa Cruz.
Cell culture and transfection.
Tissue culture cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (HyClone). Transient transfection was performed with the amount of plasmid DNA indicated in the figure legends using ExGen 500 (27). Luciferase assay was performed as previously described (5). The luciferase activities are expressed as fold induction (normalized to β-galactosidase activity) relative to untreated controls. All assays were performed in triplicate for a minimum of three times.
Whole-cell extracts and immunoprecipitation.
Cells were washed with phosphate-buffered saline (PBS), harvested, and centrifuged. The pellet was then resuspended and incubated at 4°C for 30 min in whole-cell extraction (WCE) buffer containing 50 mM Tris-HCl (pH 7.6), 400 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 1% Nonidet P-40 (NP-40). Lysate was centrifuged at 14,000 × g at 4°C for 10 min. Protein concentration was determined by a Bradford assay (Bio-Rad) using bovine serum albumin as the standard. For immunoprecipitation, the NaCl concentration of whole-cell extracts was adjusted to 150 mM and the NP-40 concentration was adjusted to 0.1%. Extracts were incubated with protein A agarose and the indicated antibodies at 4°C for 2 h, washed, and subjected to Western blot analysis accordingly. All experiments were performed in duplicate at least three times.
Pulse-chase.
Cells were cultured in DMEM supplemented with 10% FBS overnight and treated with valproic acid or sodium butyrate for 6 h or left alone. The medium was then replaced with methionine-free medium, and the cells were pulsed for 3 h with [35S]methionine (100 μCi/ml) and chased for 7 h in regular medium in the presence or absence of valproic acid or sodium butyrate. The cells were then harvested for the preparation of whole-cell extracts and immunoprecipitation with a p300 antibody. The immunopurified p300 was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed with a PhosphorImager. The experiment was performed in duplicate three times.
Quantitative RT-PCR.
Total RNA from HeLa cells was isolated by using the RNeasy Mini kit (QIAGEN). Reverse transcription was performed with 1 μg of RNA and 200 U of Superscript II (Invitrogen) for 50 min at 42°C. From the resulting cDNA, 1 μl was amplified by PCR using 2.5 U of Taq DNA polymerase (Invitrogen) for 30 cycles. Primers F-1252 (GCTATTTGATGACTGTACACAAC) and R-1509 (TCCTTCTTCGGATCTTTCTGTG) were specific for B56γ3 and B56γ2 subunits, respectively, and resulted in products of different sizes. In order to quantify the PCR products, 18S rRNA (Ambion) primers and competitors were used as internal control in all PCRs to account for mRNA input and reverse transcription-PCR (RT-PCR) variations. PCR products were run on 1.5% agarose gels stained with Vistra Green (Amersham), and quantified with the Typhoon 8600 Variable Mode Imager using ImageQuant software (Molecular Dynamics).
RNA interference.
The small interfering RNA (siRNA) experiments were performed as previously described (11). A 21-bp siRNA corresponding to nucleotides 238 to 258 of the B56γ cDNA (accession no.U37352) (AAGUGACCUAAAGUGGAAGGA) was synthesized and annealed by QIAGEN, which also supplied a nonsilencing control siRNA. The siRNA (50 and 200 nM) were transfected twice at 20-h intervals using Oligofectamine according to the manufacturer's recommendation (Invitrogen). Cells were harvested for analysis 20 h after the second transfection. All experiments were performed a minimum of three times.
Immunofluorescence microscopy.
Cells were cultured on coverslips in six-well dishes. After transfections, the coverslips were washed with cold PBS supplemented with 0.05% Tween 20. The cells were fixed on ice with 4% paraformaldehyde and permeabilized at room temperature with 0.2% NP-40. The coverslips were incubated on ice for 1 h in PBS supplemented with 10% milk and 0.2% Tween 20 and then incubated at 4°C overnight with M2 antibody (1:500). The coverslips were incubated the next day first with fluorescent anti-mouse antibody (1:10,000) at room temperature for 30 min and then with PBS supplemented with Hoechst (0.5 μg/ml) for 10 min at room temperature in total darkness. The coverslips were then drained, mounted with 90% glycerol on slides, and visualized by fluorescence microscopy.
RESULTS
Valproic acid induces selective p300 degradation through the 26S proteasome.
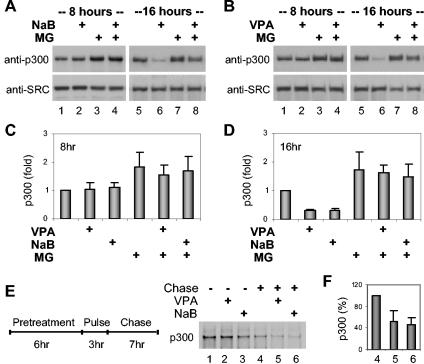
It has been reported that sodium butyrate, a HDAC inhibitor, blocks the steroid hormone action (33) and induces p300 degradation through the 26S proteasome pathway (27). To study the molecular mechanisms of selective p300 degradation, we examined the effects of sodium butyrate and other HDAC inhibitors, such as valproic acid, on the activity of the transcriptional coactivator p300 in some detail. As with the butyrate, the level of p300 protein remained constant for up to 8 h of treatment with valproic acid, whereas the level of p300 was reduced to about 30% of that in the control HeLa cells upon 16 h of exposure to valproic acid (Fig. 1A, B, C, and D). The negative effect of valproic acid on the steady-state level of p300 was similar in cells treated with valproic acid at concentrations of 1, 2, or 5 mM (Fig. 1B and data not shown). The abundance of SRC protein, however, was not affected by treatment with valproic acid or butyrate (Fig. 1A and B).
FIG. 1.
Valproic acid induces p300 degradation through the 26S proteasome. (A) Western blot analysis of endogenous p300 in extracts (50 μg) from HeLa cells treated with sodium butyrate (NaB) (5 mM) and/or MG132 (MG) (5 μM). The blot was then stripped and reprobed for protein loading with an anti-SRC antibody. The treatment time (in hours) is indicated above the gel. (B) The experimental procedure was as described for panel A except that valproic acid (VPA) (2 mM) was used. (C and D) Quantitative analysis of Western blot results is expressed as fold variation compared to the values for untreated controls after the values were normalized to those of the loading controls. Values are means ± standard deviations (error bars) from a minimum of three independent experiments with each experiment performed in duplicate. (E) Cells were pretreated with valproic acid or sodium butyrate for 6 h, then pulsed for 3 h, and chased for 7 h in the different treatment conditions. The endogenous p300 protein was immunoprecipitated with a p300 antibody, separated by SDS-PAGE, and analyzed with a PhosphorImager. (F) The relative stability of p300 in cells treated with valproic acid or sodium butyrate, upon 7 h of chase (lanes 5 and 6), is presented as a percentage of the labeled p300 in untreated cells (lane 4) after being normalized to nonchased cells in the different treatment conditions (as in lanes 1 to 3 of panel E). Values are means ± standard deviations (error bars) from a minimum of three independent experiments with each experiment performed in duplicate.
Treatment of cells with a 26S proteasome inhibitor, MG132 (23), attenuated the valproic acid-induced p300 reduction, indicating that the reduction of the coactivator may be due to protein degradation in which the 26S proteasome is likely involved (Fig. 1B). A pulse-chase protocol was also employed to compare the relative stability of p300 in the presence or absence of valproic acid or sodium butyrate. The relative half-life of endogenous p300 in HeLa cells is about 11 h (5), which is similar to those in other cell types (1, 50), and the selective p300 reduction induced by valproic acid or butyrate was observed after 16 h of treatment, but not after 8 h (Fig. 1A and B). Therefore, we devised a pretreatment protocol in which the cells were treated with valproic acid or butyrate for 6 h prior to a 3-h pulse in the presence of valproic acid or butyrate to prime the cells so that the selective p300 degradation could be observed in the next 7-h chase under treatment with valproic acid or butyrate. As shown in Fig. 1E and F, the metabolic stability of endogenous p300 was reduced to about half in cells treated with valproic acid or butyrate compared to that in untreated cells. Taken together, these results suggest that valproic acid down-regulates endogenous p300 protein mainly through the 26S proteasome-mediated degradation.
Okadaic acid reverses the valproic acid-induced p300 degradation.
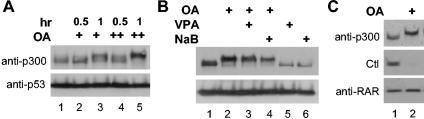
The degradation of endogenous p300 induced by valproic acid or sodium butyrate requires prolonged treatment, namely, 16 h of incubation (Fig. 1), suggesting that this might be an indirect effect, i.e., mediated through activation of a negative factor to enhance p300 turnover. The transcriptional coactivator p300 is a nuclear protein that can be massively phosphorylated at serine and threonine residues (19, 42, 50), while sodium butyrate is able to increase the cellular activity of serine-threonine phosphatase (9). These observations promoted us to investigate potential roles of serine-threonine phosphatase in the control of p300 activity. HeLa cells were treated with okadaic acid, a cell-permeable serine-threonine phosphatase inhibitor (8), for various times at different concentrations. The cellular proteins were then isolated to assess the abundance of endogenous p300 protein by Western blot analysis. As shown in Fig. 2A, the electrophoretic mobility of endogenous p300 was significantly reduced during SDS-PAGE after 1 h of okadaic acid treatment at concentrations of 0.4 and 0.8 μM, whereas the level of p300 protein was augmented notably upon 1 h of treatment at 0.8 μM, indicating that protein phosphatase may be involved in controlling p300 stability. Therefore, we wished to establish whether the effect of valproic acid or sodium butyrate on p300 degradation is mediated through activation of protein phosphatase.
FIG. 2.
Protein phosphatases play an important role in the control of the steady-state level of p300. (A) Equal amounts (50 μg) of whole-cell extracts from HeLa cells were used for Western blot analysis of endogenous p300 after treatment with okadaic acid (OA) (0.4 μM [+] or 0.8 μM [++]). The treatment time (in hours) is indicated above the lanes. The blot was then stripped and reprobed with a p53 antibody. (B) The same experimental setup as in panel A, except the cells were treated with okadaic acid (OA) (0.2 μM) in the presence or absence of valproic acid (VPA) (2 mM) or sodium butyrate (NaB) (5 mM) for 16 h. (C) The same procedure as in panel B, except the blot was probed with p300 antibody, stripped, and reprobed with a RAR antibody. A protein band cross-reacting with the RAR antibody is shown as a control (Ctl).
To this end, we examined the ability of okadaic acid to counteract the valproic acid- or butyrate-induced p300 degradation. Cells were treated with 0.2 μM okadaic acid in the absence of valproic acid or butyrate for 16 h, which reduced the electrophoretic mobility of p300 (Fig. 2B) as effectively as in shorter treatments at higher concentrations (Fig. 2A). More importantly, cotreatment with okadaic acid prevented the selective p300 degradation induced by valproic acid or sodium butyrate, which also resulted in the retardation of p300 mobility (Fig. 2B). Interestingly, the abundance of RAR was not affected by the okadaic acid treatment, whereas a protein that cross-reacts with the RAR antibody on the Western blot was destabilized (Fig. 2C), suggesting that protein degradation pathways are active under okadaic acid treatment. Taken together, these data demonstrate that serine-threonine phosphatase plays an important role in controlling the metabolic stability of transcriptional coactivator p300 and that valproic acid induces p300 degradation through the activation of phosphatase activity.
Valproic acid augments expression of the B56 regulatory subunit of PP2A.
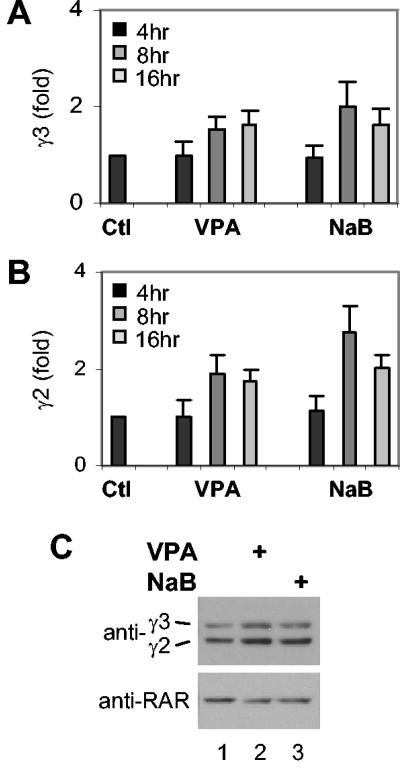
The phosphorylation status of a protein can serve as a signal for regulation through the 26S proteasome pathway. Multiple serine-threonine phosphorylation sites are present in p300, but the physiological significance of these modifications is not clear (19, 50). Therefore, we wished to identify the protein phosphatase activity that controls p300 function. Microarray analysis of gene expression was performed to screen potential candidates, and quantitative RT-PCR analysis confirmed that transcription of B56γ3 and B56γ2 regulatory subunits of PP2A was up-regulated in cells subjected to 8 and 16 h of valproic acid or sodium butyrate treatment (Fig. 3A and B). Western blot analysis showed that the protein levels of the B56γ3 and B56γ2 subunits were increased moderately upon 16 h of treatment with valproic acid or sodium butyrate (Fig. 3C). We hypothesize that the B56γ subunit may be involved in valproic acid-induced p300 degradation and wished to determine whether the B56γ regulatory subunit of PP2A is in fact responsible for the selective p300 degradation.
FIG. 3.
Valproic acid and sodium butyrate augment the expression of the B56 regulatory subunits of PP2A. (A) Cells were treated with valproic acid (VPA) (2 mM) or sodium butyrate (NaB) (5 mM) for 4, 8, and 16 h. Total RNA was isolated, and the mRNA levels of B56γ3 (γ3) were assessed by quantitative RT-PCR. Results show fold variations of the treated cells compared to those of untreated control (Ctl). Values are means ± standard deviations (error bars) from a minimum of three independent experiments with each experiment performed in duplicate. (B) The experimental setup was as in panel A, except the mRNA level of B56γ2 was examined. (C) Cells were treated as in panel A. Equal amounts of whole-cell extract (50 μg) were used for Western blotting. The blot was probed with an antibody specific to endogenous B56γ2 and B56γ3 and with an anti-RAR antibody for loading control.
B56γ regulatory subunit of PP2A enhances selective p300 degradation.
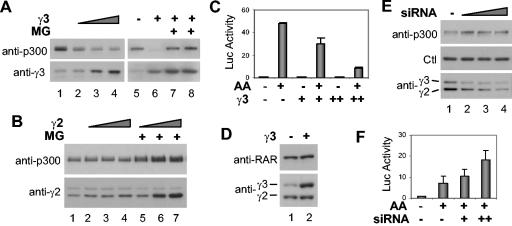
Western blotting analysis was used to assess the abundance of endogenous p300 protein in cells overexpressing the B56γ3 and B56γ2 regulatory subunits. Endogenous p300 was very sensitive to the protein dosage of the B56γ3 subunit but not to the dosage of the B56γ2 subunit (Fig. 4A and B). The level of p300 was reduced significantly even with moderate expression of the B56γ3 subunit. This negative effect of B56γ3 on the steady-state level of p300 was reversed by treating the cells with the proteasome inhibitor, MG132, indicating that the 26S proteasome is involved in the B56γ3-mediated p300 degradation (Fig. 4A). On the other hand, moderate overexpression of the B56γ2 subunit had no effect on the level of p300 protein, and high-level expression of the B56γ2 subunit was observed only in the presence of MG132 (Fig. 4B), suggesting that the B56γ2 subunit itself is tightly regulated by the 26S proteasome pathway.
FIG. 4.
The B56γ3 regulatory subunit of PP2A inhibits p300 activity. (A) HeLa cells were transfected with increasing amounts of B56γ3 (γ3) expression plasmid (1, 1.5, and 2 μg in lanes 2, 3, and 4, respectively; 2 μg in lanes 6 to 8) and treated with MG132 (MG) (5 μM) for 16 h or left alone. The abundance of endogenous p300 protein was analyzed on a Western blot. The blot was then stripped and reprobed for the B56γ3 subunit. (B) The experimental procedure was as in panel A, except the cells were transfected with increasing amounts of expression plasmid for the B56γ2 subunit (1, 2, and 4 μg). (C) Cells were transfected with RAR luciferase reporter (0.3 μg), respiratory syncytial virus β-galactosidase (RSV-β-Gal) (0.3 μg), and increasing amounts of B56γ3 expression plasmid (0.5 and 2.0 μg). The cells were then induced with arotinoid acid (AA) (1 μM) for 24 h and assayed for luciferase (Luc) assay. Values shown were normalized to β-galactosidase activity and are expressed as the fold induction relative to the values for untreated controls. Values are means ± standard deviations (error bars) of triplicates within the representative experiments. (D) Western blot analysis of RARβ from cells transfected with 2 μg of B56γ3 expression plasmid. (E) siRNA (100, 150, and 200 nM [lanes 2, 3, and 4, respectively]) specific for the B56 subunits was transfected twice at 20-h intervals. Cells were then harvested and subjected to Western blot analysis for the abundance of p300 and B56γ3 and B56γ2 subunits. A protein band that cross-reacts with the p300 antibody is used for a loading control (Ctl). (F) Cells were first transfected with RAR luciferase reporter (0.3 μg), RSV-β-Gal (0.3 μg), and B56 siRNA (100 and 150 nM), followed by a second transfection of the siRNA alone 24 h later. The cells were then induced with arotinoid acid (AA) (1 μM) for 20 h and subjected to luciferase (Luc) assay. β-Galactosidase activity was used as an internal control. Values are means ± standard deviations (error bars) of three experiments.
Numerous studies, including those involving p300 null mouse embryos, have established that p300 is required for the transcriptional activation mediated by RAR (4, 18, 51). Therefore, we investigated whether the p300-dependent RAR signaling was also affected by the B56γ3 subunit using a retinoic acid-responsive reporter (βRE3-Luc) and a synthetic RAR-specific ligand, arotinoid acid. As shown in Fig. 4C, arotinoid acid-induced transcription was inhibited by cotransfection of B56γ3 in a concentration-dependent manner. The level of endogenous RAR protein, however, was unchanged upon overexpression of B56γ3, as assessed by the Western blot analysis (Fig. 4D). In addition, overexpression of B56γ2 subunit did not inhibit RAR signaling, and the B56γ3 subunit did not affect the transcriptional activation mediated by Gal4 (data not shown). These data suggest that the down-regulation of p300-dependent RAR signaling by the B56γ3 subunit may be attained through selective p300 degradation through the 26S proteasome pathway.
siRNA of the B56γ subunit augments p300 activity.
To further determine whether the B56γ subunit is a decisive determinant for targeting p300 degradation through the proteasome pathway, we designed an siRNA to knock down the endogenous B56γ protein. Western blot analysis demonstrated that introducing the B56γ siRNA into cells elevates the level of endogenous p300 protein while knocking down the levels of endogenous B56γ proteins (Fig. 4E). The transfection of B56γ siRNA did not alter the abundance of endogenous RAR protein or a protein that cross-reacts with the p300 antibody on the Western blot (Fig. 4E and data not shown). In addition, transfection of a nonsilencing control siRNA did not affect the level of endogenous B56γ or p300 protein (data not shown). These results indicate that the B56γ subunit of PP2A is required for down-regulation of endogenous p300 protein.
Next, we examined the effect of B56γ siRNA on p300-dependent RAR signaling. Cells were first transfected with the RAR reporter and B56γ siRNA. On the following day, the cells were again transfected with the B56γ siRNA and induced with arotinoid acid to activate the p300-dependent transcription mediated by RAR. As shown in Fig. 4F, increasing amounts of B56 siRNA enhanced the RAR-mediated transcription, which is likely accomplished through augmentation of the level of p300 protein.
The B56γ3 regulatory subunit interacts with p300.
To determine the precise role of the B56 subunit in the selective p300 turnover, we examined whether PP2A forms an interaction complex with the transcriptional coactivator p300. To this end, p300 antibody was utilized to immunoprecipitate the endogenous p300 from HeLa cell extracts, and an antibody against the catalytic subunit of PP2A was used for Western blot analysis. As shown in Fig. 5A, the endogenous catalytic subunit of PP2A was coprecipitated with p300 from nontransfected cells maintained in regular medium, indicating that p300 is able to associate with PP2A under physiological conditions.
FIG. 5.
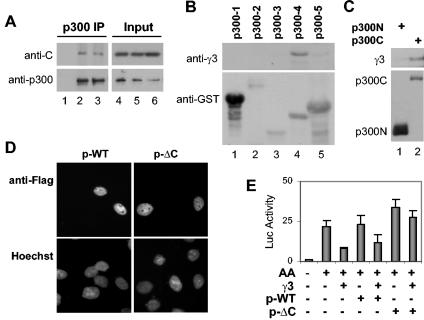
The B56γ3 regulatory subunit of PP2A associates with p300. (A) Equal amounts of whole-cell extracts were used for immunoprecipitation (IP) with a p300 antibody (lanes 2 and 3) or protein A agarose alone (lane 1) as a negative control. The precipitations were then analyzed sequentially with the antibody specific to the catalytic subunit (C) of PP2A and p300 on Western blots. Portions (10%) of the extracts used in the immunoprecipitation were subjected to Western blot analysis (lanes 4 to 6) as input control. (B) GST pull-down assays were performed with 500 μg of cell lysate from HeLa cells expressing the B56γ3 subunit and the indicated GST-p300 fusion proteins spanning the full length of p300 (p300-1, amino acids 1 to 672; p300-2, amino acids 672 to 1193; p300-3, amino acids 1069 to 1459; p300-4, amino acids 1459 to 1892; p300-5, amino acids 1893 to 2414). The blot was probed sequentially for the B56γ3 (γ3) subunit and GST tag. (C) Cells were transfected with expression plasmid for the Flag-tagged N terminus of p300 (p300N, amino acids 1 to 670; 10 μg) or the C terminus of p300 (p300C, amino acids 1135 to 2414; 10 μg). The truncated forms of p300 were purified with anti-Flag affinity gel and subjected to Western blotting with anti-Flag antibody. The blot was then stripped and reprobed for the B56γ subunits of PP2A. (D) Cells were transfected with plasmids encoding either wild-type p300 (p-WT) (0.2 μg) or p300 in which the C terminus had been deleted (p-ΔC) (2 μg) and allowed to express for 36 h. The cells were then stained with specific anti-Flag antibody and fluorophore-labeled secondary antibody for immunofluorescence microscopy. (E) Cells were transfected with RAR luciferase reporter (0.3 μg), RSV-β-Gal (0.3 μg), B56γ3 (0.3 μg), and the wild-type p300 or p300ΔC (0.2 μg) for luciferase (Luc) assay. Values shown were normalized to β-galactosidase activity and expressed as the fold induction relative to the values for untreated controls (P < 0.001 as measured by repeated analysis of variance tests). Values are means ± standard deviations (error bars) of triplicates within the representative experiments.
Next, we performed GST pull-down assays to assess whether there is an interaction between p300 and the B56γ3 subunit of PP2A. Different purified truncated forms of GST-p300 fusion protein spanning the full length of p300 were used to pull down transiently expressed B56γ3 protein. As shown in Fig. 5B, the B56γ3 subunit was pulled down by GST-p300 containing amino acids 1459 to 1892, but not with any other GST-p300 fusion proteins tested. The interaction was specific to this region of p300 and was not due to the lack of GST-p300 or B56γ3 expression, since the same cell lysate was used in each pull-down assay. Purified GST protein was used as a negative control in these experiments (data not shown).
To verify the interaction domain of p300 with the B56γ3 regulatory subunit, we performed the coimmunoprecipitation experiment with Flag-tagged truncated forms of p300. The N terminus of p300 was expressed in abundance, whereas only moderate levels of the C terminus of p300 were detected in the transfected cells (Fig. 5C). Most importantly, the B56γ3 regulatory subunit of PP2A was copurified with the Flag-tagged C terminus, but not with the N terminus of p300 (Fig. 5C), whereas the B56γ2 subunit was not detected.
The B56γ3 interaction region of p300 was mapped to the C terminus (Fig. 5B and C). Therefore, we performed a transcriptional rescue assay with a p300 mutant lacking the C-terminal domain to assess the functional significance of the interaction of p300 with the B56γ3 regulatory subunit of PP2A. Both the wild-type and truncated forms of p300 were localized in the nucleus and were expressed at comparable levels judging from the immunofluorescence staining of an antibody directed against the Flag-tagged p300 protein (Fig. 5D). The RAR reporter, the B56γ3 expression plasmid, and plasmids encoding either the wild-type p300 or the truncated form of p300 were then cotransfected into cells. As shown in Fig. 5E, the B56γ3 subunit inhibited the p300-dependent RAR signaling, whereas the coexpression of the p300 mutant lacking the B56γ3 interaction region rescued the repression of the RAR-mediated transcription by the B56γ3 subunit, but not the coexpression of the wild-type p300, suggesting that the negative effect of the B56γ3 subunit on the RAR signaling requires the C terminus of p300. In conclusion, our data suggest that the control of p300 activity by PP2A is mediated through an interaction between the B56γ3 regulatory subunit of PP2A and the coactivator.
DISCUSSION
The major conclusions from this work are that the activity of the transcriptional coactivator p300 is dynamically regulated by signaling transduction and that the B56γ3 regulatory subunit of PP2A mediates valproic acid-induced p300 degradation.
Reversible phosphorylation can selectively designate proteins for degradation through the 26S proteasome pathway. Serine-threonine phosphorylation of IκBα, p27kip1, and Jak2 promotes proteasome-mediated degradation of these proteins (6, 48). Although phosphorylation often triggers protein degradation, it can also be important for maintaining protein stability. For example, phosphorylation by mitogen-activated protein kinase increases the steady-state level of c-Jun and Bcl-2 (2, 35). Interestingly, HDAC inhibitor sodium butyrate has been reported to have a stimulatory effect on the enzymatic activity of protein phosphatases (9). We found that valproic acid, also an HDAC inhibitor, induces p300 degradation through increasing gene expression of the B56γ subunit of PP2A (Fig. 1 and 3). Okadaic acid, a protein phosphatase inhibitor, reduces the electrophoretic mobility of p300 and augments the steady-state level of the coactivator (Fig. 2). In addition, okadaic acid effectively reverses the negative effect of valproic acid on the level of endogenous p300 protein (Fig. 2). Taken together, these data suggest that protein phosphatase plays an important role in the control of p300 stability and that valproic acid induces p300 degradation through activation of PP2A activity.
A role for the B56 subunit of PP2A in the 26S proteasome pathway was first revealed for β-catenin (44). All five B56 isoforms activate glycogen synthase kinase 3β (GSK3β), which phosphorylates β-catenin, targeting it for degradation. It is currently thought that various regulatory subunits of PP2A distribute the enzyme to distinct subcellular compartments (31), which argues against a substantial overlap in localization. However, other than the B56γ subunit, the spatial distributions of the other isoforms have not been thoroughly characterized. Our results indicate that the B56γ3 subunit, not the B56γ2 subunit, is an important physiological determinant for regulation of p300 activity (Fig. 4 and data not shown). Overexpression of the B56γ3 regulatory subunit of PP2A targets p300 for degradation through the 26S proteasome and inhibits the transcriptional activity of the coactivator (Fig. 4). Conversely, siRNA knockdown of the endogenous B56γ of PP2A increases p300 activity (Fig. 4).
Our study also revealed an interaction between p300 and the catalytic subunit of PP2A (Fig. 5). The interaction of p300 with the B56γ3 regulatory subunit is mapped to the C terminus of p300, which contains an Akt/protein kinase B phosphorylation site (Fig. 5). Akt activity is required to maintain p300 activity (5). Reversible phosphorylation might serve as a potential molecular switch to control p300 activity. Unlike β-catenin, which is indirectly targeted by PP2A for ubiquitination through GSK3β (44), direct dephosphorylation of p300 by PP2A could be the signal designating the coactivator for proteasome-mediated degradation.
The extent of serine-threonine phosphorylation of p300 can be massive (19, 42), and previously identified phosphorylation sites have been mostly tied to the transcriptional activity but not to the metabolic stability of p300 (14, 43, 52). Okadaic acid significantly reduces the electrophoretic mobility of p300 while augmenting the abundance of the coactivator (Fig. 2). However, the retardation of p300 mobility was not observed by knocking down B56γ expression with siRNA, even with increased p300 abundance (Fig. 4). This discrepancy may arise from the different specificities in terms of phosphatases targeted by okadaic acid and B56γ siRNA. It is possible that depending on the cellular environment, various phosphatases are involved in the regulation of p300 activities. Okadaic acid inhibits a broad spectrum of phosphatases and accumulates p300 in a state of massive phosphorylation, which results in significant retardation of its mobility on SDS-polyacrylamide gels. On the other hand, B56γ siRNA specifically inhibits the activity of the B56γ regulatory subunit whose target seems to be the C terminus of p300 (Fig. 5). Therefore, knocking down the B56γ subunit would not affect the global phosphorylation of p300 and therefore not its mobility in general, which also supports a high specificity of the B56γ regulatory subunit in the control of p300 activity.
Taken together, our data suggest a direct role for the B56γ3 regulatory subunit of PP2A in the control of p300 function and illustrate a molecular mechanism for valproic acid-induced p300 degradation. The transcriptional coactivator p300 is essential for embryonic development and cell proliferation (51). The level of p300 protein in the nucleus is tightly regulated to maintain normal development and cell proliferation (3, 51). Effective termination of its activity after transcriptional activation is essential for cells to ensure precisely controlled gene expression, whereas deregulation of p300 turnover would impose obstacles for embryonic development and an array of cellular processes. Selective p300 degradation may cause some of the side effects of valproic acid when used in early pregnancy. Future studies should elucidate the extra- and intracellular signals that determine the regulatory activity of the B56γ3 subunit on the transcriptional coactivator p300.
Acknowledgments
We thank V. Giguere, M. O. Hottiger, M. A. Ikeda, and T. Perlmann for plasmids. We also thank C. Kamibayashi for antibody and M. A. R. Abed for expertise.
This work was supported in part by operating grants from Canadian Institutes of Health Research (CIHR) and The Cancer Research Society, Inc. During the course of this study, Q.L. was supported by scholarships from CIHR.
REFERENCES
- 1.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 2.Breitschopf, K., J. Haendeler, P. Malchow, A. M. Zeiher, and S. Dimmeler. 2000. Posttranslational modification of Bcl-2 facilitates its proteasome-dependent degradation: molecular characterization of the involved signaling pathway. Mol. Cell. Biol. 20:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brouillard, F., and C. E. Cremisi. 2003. Concomitant increase of histone acetyltransferase activity and degradation of p300 during retinoic acid-induced differentiation of F9 cells. J. Biol. Chem. 278:39509-39516. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti, D., V. J. LaMorte, M. C. Nelson, T. Nakajima, I. G. Schulman, H. Juguilon, M. Montminy, and R. M. Evans. 1996. Role of CBP/P300 in nuclear receptor signalling. Nature 383:99-103. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., S. S. Halappanavar, J. R. St-Germain, B. K. Tsang, and Q. Li. 2004. Role of Akt/protein kinase B in the activity of transcriptional coactivator p300. Cell Mol. Life Sci. 61:1675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., J. Hagler, V. J. Palombella, F. Melandri, D. Scherer, D. Ballard, and T. Maniatis. 1995. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9:1586-1597. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover, A. 1998. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17:7151-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, P., C. F. Holmes, and Y. Tsukitani. 1990. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem. Sci. 15:98-102. [DOI] [PubMed] [Google Scholar]
- 9.Cuisset, L., L. Tichonicky, P. Jaffray, and M. Delpech. 1997. The effects of sodium butyrate on transcription are mediated through activation of a protein phosphatase. J. Biol. Chem. 272:24148-24153. [DOI] [PubMed] [Google Scholar]
- 10.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 13.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 14.Gusterson, R., B. Brar, D. Faulkes, A. Giordano, J. Chrivia, and D. Latchman. 2002. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 277:2517-2524. [DOI] [PubMed] [Google Scholar]
- 15.Hasan, S., P. O. Hassa, R. Imhof, and M. O. Hottiger. 2001. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature 410:387-391. [DOI] [PubMed] [Google Scholar]
- 16.Ito, A., T. R. Kataoka, M. Watanabe, K. Nishiyama, Y. Mazaki, H. Sabe, Y. Kitamura, and H. Nojima. 2000. A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J. 19:562-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johannessen, C. U., and S. I. Johannessen. 2003. Valproate: past, present, and future. CNS Drug Rev. 9:199-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki, H., R. Eckner, T. P. Yao, K. Taira, R. Chiu, D. M. Livingston, and K. K. Yokoyama. 1998. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393:284-289. [DOI] [PubMed] [Google Scholar]
- 19.Kitabayashi, I., R. Eckner, Z. Arany, R. Chiu, G. Gachelin, D. M. Livingston, and K. K. Yokoyama. 1995. Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells. EMBO J. 14:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleijnen, M. F., A. H. Shih, P. Zhou, S. Kumar, R. E. Soccio, N. L. Kedersha, G. Gill, and P. M. Howley. 2000. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6:409-419. [DOI] [PubMed] [Google Scholar]
- 21.Kramer, O. H., P. Zhu, H. P. Ostendorff, M. Golebiewski, J. Tiefenbach, M. A. Peters, B. Brill, B. Groner, I. Bach, T. Heinzel, and M. Gottlicher. 2003. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22:3411-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam, Y. A., T. G. Lawson, M. Velayutham, J. L. Zweier, and C. M. Pickart. 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416:763-767. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 24.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 25.Li, Q., M. Herrler, N. Landsberger, N. Kaludov, V. V. Ogryzko, Y. Nakatani, and A. P. Wolffe. 1998. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 17:6300-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q., A. Su, J. Chen, Y. A. Lefebvre, and R. J. Hache. 2002. Attenuation of glucocorticoid signaling through targeted degradation of p300 via the 26S proteasome pathway. Mol. Endocrinol. 16:2819-2827. [DOI] [PubMed] [Google Scholar]
- 28.Li, X., A. Scuderi, A. Letsou, and D. M. Virshup. 2002. B56-associated protein phosphatase 2A is required for survival and protects from apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 22:3674-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 30.Loscher, W. 1999. Valproate: a reappraisal of its pharmacodynamic properties and mechanisms of action. Prog. Neurobiol. 58:31-59. [DOI] [PubMed] [Google Scholar]
- 31.McCright, B., A. M. Rivers, S. Audlin, and D. M. Virshup. 1996. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271:22081-22089. [DOI] [PubMed] [Google Scholar]
- 32.McCright, B., and D. M. Virshup. 1995. Identification of a new family of protein phosphatase 2A regulatory subunits. J. Biol. Chem. 270:26123-26128. [DOI] [PubMed] [Google Scholar]
- 33.McKnight, G. S., L. Hager, and R. D. Palmiter. 1980. Butyrate and related inhibitors of histone deacetylation block the induction of egg white genes by steroid hormones. Cell 22:469-477. [DOI] [PubMed] [Google Scholar]
- 34.Molinari, E., M. Gilman, and S. Natesan. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musti, A. M., M. Treier, and D. Bohmann. 1997. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275:400-402. [DOI] [PubMed] [Google Scholar]
- 36.Nau, H., R. S. Hauck, and K. Ehlers. 1991. Valproic acid-induced neural tube defects in mouse and human: aspects of chirality, alternative drug development, pharmacokinetics and possible mechanisms. Pharmacol. Toxicol. 69:310-321. [DOI] [PubMed] [Google Scholar]
- 37.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 38.Perlmann, T., and L. Jansson. 1995. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 9:769-782. [DOI] [PubMed] [Google Scholar]
- 39.Phiel, C. J., F. Zhang, E. Y. Huang, M. G. Guenther, M. A. Lazar, and P. S. Klein. 2001. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276:36734-36741. [DOI] [PubMed] [Google Scholar]
- 40.Poizat, C., V. Sartorelli, G. Chung, R. A. Kloner, and L. Kedes. 2000. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol. Cell. Biol. 20:8643-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz, C., K. Beck, S. Mink, M. Schmolke, B. Budde, D. Wenning, and K. H. Klempnauer. 2003. Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 22:882-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.See, R. H., D. Calvo, Y. Shi, H. Kawa, M. P. Luke, and Z. Yuan. 2001. Stimulation of p300-mediated transcription by the kinase MEKK1. J. Biol. Chem. 276:16310-16317. [DOI] [PubMed] [Google Scholar]
- 44.Seeling, J. M., J. R. Miller, R. Gil, R. T. Moon, R. White, and D. M. Virshup. 1999. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283:2089-2091. [DOI] [PubMed] [Google Scholar]
- 45.Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98:1-4. [DOI] [PubMed] [Google Scholar]
- 46.Suganuma, T., M. Kawabata, T. Ohshima, and M. A. Ikeda. 2002. Growth suppression of human carcinoma cells by reintroduction of the p300 coactivator. Proc. Natl. Acad. Sci. USA 99:13073-13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tehrani, M. A., M. C. Mumby, and C. Kamibayashi. 1996. Identification of a novel protein phosphatase 2A regulatory subunit highly expressed in muscle. J. Biol. Chem. 271:5164-5170. [DOI] [PubMed] [Google Scholar]
- 48.Ungureanu, D., P. Saharinen, I. Junttila, D. J. Hilton, and O. Silvennoinen. 2002. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol. Cell. Biol. 22:3316-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Virshup, D. M. 2000. Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12:180-185. [DOI] [PubMed] [Google Scholar]
- 50.Yaciuk, P., and E. Moran. 1991. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kDa product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol. Cell. Biol. 11:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, L. W., and J. E. Gambee. 2000. Phosphorylation of p300 at serine 89 by protein kinase C. J. Biol. Chem. 275:40946-40951. [DOI] [PubMed] [Google Scholar]