Abstract
Termination by RNA polymerase III (Pol III) produces RNAs whose 3′ oligo(U) termini are bound by La protein, a chaperone that protects RNAs from 3′ exonucleases and promotes their maturation. Multiple reports indicate that yeasts use La-dependent and -independent pathways for tRNA maturation, with defective pre-tRNAs being most sensitive to decay and most dependent on La for maturation and function. The Rpc11p subunit of Pol III shows homology with the zinc ribbon of TFIIS and is known to mediate RNA 3′ cleavage and to be important for termination. We used a La-dependent opal suppressor, tRNASerUGAM, which suppresses ade6-704 and the accumulation of red pigment, to screen Schizosaccaromyces pombe for rpc11 mutants that increase tRNA-mediated suppression. Analyses of two zinc ribbon mutants indicate that they are deficient in Pol III RNA 3′ cleavage activity and produce pre-tRNASerUGAM transcripts with elongated 3′-oligo(U) tracts that are better substrates for La. A substantial fraction of pre-tRNASerUGAM contains too few 3′ Us for efficient La binding and appears to decay in wild-type cells but has elongated oligo(U) tracts and matures along the La-dependent pathway in the mutants. The data indicate that Rpc11p limits RNA 3′-U length and that this significantly restricts pre-tRNAs to a La-independent pathway of maturation in fission yeast.
The 3′ ends of tRNA and other RNA polymerase III (Pol III)-dependent genes contain dTn termination signals at which Pol III pauses and releases its RNA (6). The role of dTn extends beyond termination, since it provides a means to link Pol III transcripts to La, an abundant and ubiquitous nuclear phosphoprotein that binds these RNAs in a 3′-oligo(U) length-dependent manner and promotes their posttranscriptional processing (27, 32, 37). Although 3′-U length heterogeneity has been well documented for Pol III transcripts (27), relatively little is known about the mechanisms involved and its functional significance. A model system that can be used to alter 3′-U length and study its consequences should be helpful in understanding functional connectivity between Pol III termination and posttranscriptional processing.
La protein protects pre-tRNAs from 3′ exonucleases and imposes order on posttranscriptional processing so that 5′ processing precedes 3′ processing for many pre-tRNAs (38). Finding the reverse order suggests that different pre-tRNAs use the La-independent and -dependent pathways to various degrees (25). While La-homologous protein (Lhp1p) is nonessential in the yeast Saccharomyces cerevisiae (and in Schizosaccharomyces pombe), its deletion causes lethality or growth deficiency in combination with mutations that impair base pairing or modification of certain pre-tRNAs, indicating that defective pre-tRNAs can be salvaged by La (5, 7, 21, 38). Decay of hypomodified  occurs via 3′ adenylation and exonucleolytic degradation of
occurs via 3′ adenylation and exonucleolytic degradation of 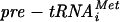 in a process called nuclear surveillance (22) but can be rescued by excess Lhp1p (1, 5).
in a process called nuclear surveillance (22) but can be rescued by excess Lhp1p (1, 5).
An opal suppressor tRNASerUGAM gene with a dT7 terminator (tRNAmSer7T), which suppresses a nonsense mutation in the ade6-704 allele and accumulation of red pigment, has been used to study tRNA biogenesis and the function of the La homolog, Sla1p, in the yeast S. pombe. tRNASerUGAM expression is dependent on accurate and efficient initiation and termination by Pol III and on La for processing (12-14, 17, 18). Mutations in pre-tRNASerUGAM that confer dependence on La also appear to cause it to be temperature sensitive, recognized poorly by RNase P, and cleaved aberrantly (13, 35). Thus, pre-tRNASerUGAM is processed inefficiently in sla1+ cells, yielding partial suppression, as reflected by pink colony color (no suppression yields red colonies, and full suppression yields white colonies), while the pre-tRNASerUGAM does not accumulate or produce mature tRNASerUGAM or any suppression in sla1 deletion cells (18). These data suggest that although pre-tRNASerUGAM is defective and cannot survive without La, it can be salvaged by La to produce mature functional tRNASerUGAM. However, any pre-tRNASerUGAM directed to the La-independent pathway would not survive to maturation, even in sla1+ cells.
Related to Pol III termination is pausing at dTn by Pol II during mRNA synthesis (10, 36). The protein TFIIS uses a zinc ribbon dipeptide, Asp-Glu, to promote 3′ cleavage of RNA stalled in the Pol II active site to resume elongation (20, 23). Rpc11p is an integral Pol III subunit, homologous to TFIIS in its Asp-Glu zinc ribbon, that mediates RNA 3′ cleavage and facilitates Pol III termination (8). Indeed, Pol III can carry out RNA 3′ cleavage and resynthesis while pausing at dTn (4), and this may help overcome kinetic barriers to termination (3, 8).
We used a partial-suppression phenotype mediated by tRNASerUGAM in S. pombe strain yYH1 to screen for dominant gain-of-suppression mutants of rpc11. Pre-tRNAs in these mutants undergo remarkably efficient processing along the La-dependent pathway of tRNA maturation. The mutant Pol III enzymes exhibit reduced 3′ cleavage activity in vitro and produce RNAs with longer 3′ Us in vitro and in vivo. We also found that Sla1p prefers RNAs with at least four Us. Since a majority of pre-tRNAs isolated from wild-type (WT) yeast have ≤3 Us, this preference helps explain the magnitude of the rpc11 mutant effects. It would appear that pre-tRNAs that would normally decay because they have too few Us for efficient La binding are instead shifted to the La-dependent pathway of tRNA maturation in the mutants. Since defective pre-tRNAs are sensitive to the loss of La, the cleavage activity of Rpc11p may also contribute to nuclear surveillance (22).
MATERIALS AND METHODS
Yeast strains are described in Table 1. yAMm4T-2, yAMm5T-1, and yYH1 carry tRNASerUGAM genes bearing dT4, dT5, and dT7, respectively, as their terminators, whose identities and integration at the leu1+ locus (13) were confirmed by colony PCR and DNA sequencing (not shown).
TABLE 1.
S. pombe strains
| Strain name | Genotype | Reference |
|---|---|---|
| yYH3282 | h+his3-D1 leu1-32 ura4-Δ18 ade6-M216 rpc53::[FH-rpc53 ura4+] | 15 |
| yYH1 | hs−leu1-32::[tRNAmSer7T-leu1+] ura4-Δ18 ade6-704 | This report |
| yAMm4T-2 | hs−leu1-32::[tRNAmSer4T-leu1+] ura4-Δ18 ade6-704 | This report |
| yAMm5T-1 | hs−leu1-32::[tRNAmSer5T-leu1+] ura4-Δ18 ade6-704 | This report |
| yAS110 | hs−leu1-32::[tRNAmSer7T-leu1+] sla1 Δ::ura4-FOA+ade6-704 | 18 |
| yAM148 | h−leu1-32::[pJK148-leu1+] ura4-Δ18 ade6-704 | This report |
| ySH9 | hs−leu1-32::[tRNApSer7T-leu1+] sla1 Δ::ura4-FOA+ade6-704 | This report |
| yRI28: | hs−leu1-32::[leu1+] ura4-Δ18 ade6-704 | This report |
Identification of mutations in rpc11 causing increased suppression.
An rpc11 mutant library was constructed by nucleotide analog-based mutagenic PCR (39). Briefly, PCR was performed in a 50-μl volume containing 10 mM Tris-Cl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 4 μM8-oxo-2′-deoxyguanosine-5′-triphosphate, 2′-deoxy-P-nucleoside-5′-triphos-phate (TriLink BioTech), 100 ng of pRep4X-spRpc11 plasmid, and 1 μM primers C11SEN and C11ANT2. A second PCR was performed using 1 μl of the first PCR product in the absence of deoxynucleoside triphosphate analogs for 15 cycles. The PCR product was digested with XhoI and BamHI, ligated into the XhoI/BamHI sites of pRep4X, and transformed into UltraMAX DH5α-FT cells (Invitrogen). About 150,000 bacterial transformants were scraped from plates and suspended in H2O, and plasmid DNA was isolated. The mutagenesis efficiency as determined by sequencing plasmids from 18 randomly chosen transformants revealed 35% with no mutation, 30% with one mutation, and 35% with more than one mutation. The rpc11 mutant library was transformed into yYH1, and cells were plated on Edinburgh minimal medium (EMM) lacking leucine and uracil and containing 10 mg of adenine/liter. Of ∼180,000 colonies screened, 0.06% exhibited increased suppression. Plasmids were recovered from these (2) and transformed into TOP10 cells (Invitrogen). Site-directed mutagenesis was done by QuikChange XL (Stratagene) using appropriate primers (Table 2), and all mutations were confirmed by sequencing.
TABLE 2.
Primers
| Primer name | Primer sequencea |
|---|---|
| C11sen1 | 5′-GCATCTCGAGATGCAGTTTTGTCCTACTTGTGG AA; XhoI |
| C11ant1 | 5′-CGAGGATCCCTAATTCTCACGCCATTGAAATTTGG; BamHI |
| C11ant2 | 5′-CGAGGATCCCTAATTCTCACGCCATTGAAATTTGC; BamHI |
| Seq1 | 5′-TCAATCTCATTCTCACTTTCTGAC |
| C11ant3 | 5′-CGAGGATCCCTAACCAGCGTAGTCTGGAACGTCGTATGGGTATCCACCTCCAGCATTCTCACGCCATTGAAATTTGG |
| C102f | 5′-CCTTTTATCGTTGTACCAAAtgcAAATTTCAATGGCGTG |
| C102Sr | 5′-CACGCCATTGAAATTTgcaTTTGGTACAACGATAAAAGG |
| C11DGSen | 5′-CAAATTCGTAGTGCAGGTGAACCTATGAGTACCTTTT |
| C11DGAnt | 5′-AAAAGGTACTCATAGGTTCACCTGCACTACGAATTTG |
| Hssen1 | 5′-AACAACTCGAGATGCTGCTGTTCTGCCCCGGCT; XhoI |
| Hsan1 | 5′-AACAGGATCCCTAATCCCTCCAGCGGTGTCCAC; BamHI |
| Ala102f | 5′-CCTTTTATCGTTGTACCAAAgcgAAATTTCAATGGCGTG |
| Ala102r | 5′-CACGCCATTGAAATTTcgcTTTGGTACAACGATAAAAGG |
| Asn102f | 5′-CCTTTTATCGTTGTACCAAAaatAAATTTCAATGGCGTG |
| Asn102r | 5′-CACGCCATTGAAATTTattTTTGGTACAACGATAAAAGG |
| Arg102f | 5′-CCTTTTATCGTTGTACCAAAagaAAATTTCAATGGCGTG |
| Arg102r | 5′-CACGCCATTGAAATTTtctTTTGGTACAACGATAAAAGG |
| His102f | 5′-CCTTTTATCGTTGTACCAAAcacAAATTTCAATGGCGTG |
| His102r | 5′-CACGCCATTGAAATTTgtgTTTGGTACAACGATAAAAGG |
| Ile102f | 5′-CCTTTTATCGTTGTACCAAAattAAATTTCAATGGCGTG |
| Ile102r | 5′-CACGCCATTGAAATTTaatTTTGGTACAACGATAAAAGG |
| Tyr102f | 5′-CCTTTTATCGTTGTACCAAAtatAAATTTCAATGGCGTG |
| Tyr102r | 5′-CACGCCATTGAAATTTataTTTGGTACAACGATAAAAGG |
| Asp102f | 5′-CCTTTTATCGTTGTACCAAAgacAAATTTCAATGGCGTG |
| Asp102r | 5′-CACGCCATTGAAATTTgtcTTTGGTACAACGATAAAAGG |
| Glu102f | 5′-CCTTTTATCGTTGTACCAAAgagAAATTTCAATGGCGTG |
| Glu102r | 5′-CACGCCATTGAAATTTctcTTTGGTACAACGATAAAAGG |
| Gly102f | 5′-CCTTTTATCGTTGTACCAAAggaAAATTTCAATGGCGTG |
| Gly102r | 5′-CACGCCATTGAAATTTtccTTTGGTACAACGATAAAAGG |
| Petc11s | 5′-GAACATATGCAGTTTTGTCCTACTTGTGGAAA; NdeI |
| Petc11a | 5′-GAAGGATCCCTAATTCTCACGCCATTGAAATTTGC; BamHI |
| R107Csen | 5′-CAAATGCAAATTTCAATGGTGTGAGAATTAGGGATCC |
| R107Crev | 5′-GGATCCCTAATTCTCACACCATTGAAATTTGCATTTG |
| RACE 1 | 5′-GTCCGAGTGGTTAAGGAGTTAGACTTC |
| Reverse | 5′-ACTGGAATTCGCGGTCTTTTA |
| 3′ Adapter | 5′-rArArArAdGdAdCdCdGdCdGdAdAdTdTdCdCdAdG-invdT |
Restriction sites are underlined. Lowercase letters indicate mutated codons.
Growth assays.
For liquid growth assays, logarithmically growing cells were inoculated into EMM lacking leucine and uracil and containing 200 mg of adenine/liter in the presence or absence of 500 μg of 6-azauradine (Sigma-Aldrich)/ml. The cells were grown at 32°C, and growth was monitored by optical density at 600 nm. For growth in agar, cells were plated at 10-fold dilutions; 2 μl of each was spotted onto EMM plates lacking leucine and uracil and containing 200 mg of adenine/liter, with no 6-azauradine or 500 μg of 6-azauradine/ml, and grown at 32°C for 3 to 4 days.
Plasmid construction.
spRpc11 was cloned by PCR amplification from S. pombe genomic DNA using the primers C11sen1 and C11ant2 (Table 2). The PCR product was digested with XhoI and BamHI and cloned into these sites of the ura4+ vector pRep4X (11), generating pRep4X-spRpc11. pRep4X-spRpc11(C102) was constructed by the same method except using primers C11sen1 and C11ant1. pRep4-Sla1p was as described previously (18). To create a C-terminal hemagglutinin (HA)-tagged version of spRpc11(C102S), PCR was performed using primers C11sen1 and C11ant3, and the product was digested with XhoI and BamHI and ligated into the leu2+ vector pRep3X (11), yielding pRep3X-HA-spRpc11(C102S). pRep3X-spRpc11(C102S) was made by cloning the BamHI-XhoI insert from pRep4X-spRpc11(C102S) into pRep3X (leu1+). pRep3X-spRpc11 was constructed from pRep3X-spRpc11(C102S), using primers C102f and C102f, by QuikChange XL site-directed mutagenesis (Stratagene). pRep3X-spRpc11(D90G) was made from pRep3X-spRpc11 using primers C11DGSen and C11DGAnt. pREP3X-SpRpcC11-D90GR107C was made by site-directed mutagenesis using pRep3X-SpRpc11-D90G as a template and the primers R107Csen and R107rev. For bacterial expression of spRpc11p antigen, PCR was carried out using pRep4X-spRpc11 as a template and primers Petc11s and Petc11a. The product was digested with NdeI and BamHI and inserted into the same sites of pET28a (Novagen), generating pET28a-spRpc11. All constructs were verified by sequencing.
Immunopurification and immunoblotting.
Anti-FLAG immunoprecipitation (IP) of S. pombe extract was described previously (14). Protein immunoprecipitated by anti-FLAG (Sigma-Aldrich) was resolved by 10 to 20% polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and incubated with anti-FLAG (1:5,000) to detect FLAG-His6-spRpc53, anti-spRpc11 (1:5,000), anti-Sla1p, or anti-pTR6 (1:5,000) and enhanced chemiluminescence (Amersham) (16). Anti-spRpc11p was made in rabbits from bacterially expressed affinity-purified His6-spRpc11p.
Northern blotting.
Total RNA was isolated, separated on a 6% polyacrylamide-urea gel, transferred to a nylon membrane (GeneScreen Plus; Perkin-Elmer), UV cross-linked, baked, and incubated with [32P]DNA oligonucleotides complementary to the RNA species indicated, as described previously (18).
Pol III-associated RNA 3′ cleavage assay.
RNA cleavage was performed using in vitro-reconstituted elongation complexes (ECs) (24). Briefly, affinity-purified Pol III (15) was immobilized on Ni-nitrilotriacetic acid resin (QIAGEN) (10 min; 25°C) and washed five times with TB (20 mM Tris-HCl [pH 7.9], 5 mM MgCl2, 40 mM KCl, 2 mM β-mercaptoethanol, 10 μM ZnSO4), after which the volume was adjusted to 50 μl. Complexes were assembled by incubating immobilized Pol III with 10 μM ATP and GTP plus 1 pmol [32P]RNA and a template strand of oligo-DNA for 10 min; 30 pmol of upper-strand DNA was then added and incubated for 5 min. The ECs were washed five times with TB lacking MgCl2, the volume was adjusted to 40 μl, and the cleavage reaction was initiated by adding MgCl2 to 2.5 mM. Aliquots were withdrawn at various times and added to an equal volume of 2× stop or gel-loading buffer. For extension to 20 nucleotides (nt), aliquots of the ECs were withdrawn at 0 and 5 min and incubated with 100 μM (each) ATP, CTP, and GTP and 5 mM MgCl2 for 5 min. The products were analyzed on a 20% polyacrylamide-urea gel and a Fujifilm phosphorimager and quantitated with Image Reader software.
Transcription from dT7-containing DNA.
After EC assembly, the volume of the transcription reaction was adjusted to 100 μl with TB containing 1 U of RNasin-Plus (40 U/μl; Promega)/μl, and transcription was initiated by adding nucleoside triphosphates (NTPs) to 0.8 mM and MgCl2 to 5 mM. Aliquots (10 μl) were withdrawn thereafter, added to 10 μl of 2× stop or gel-loading buffer, and analyzed by denaturing electrophoresis. Transcriptions were done at 25°C except where indicated in the figure legends.
IP of La-associated RNAs.
After a 30-min transcription reaction containing immobilized ECs and dT7-containing DNA, EDTA was increased to 5 mM, and the supernatant was separated from the Ni-nitrilotriacetic acid beads and passed through a spin column (Promega); for input, 5 μl was added to 5 μl of loading buffer. For IP, 2.5 μl of recombinant Sla1p (50 ng/μl) was added to 30 μl of the supernatant and incubated on ice for 1 h, after which 20 μl of a 50% slurry of anti-Sla1p immunoglobulin G (IgG)-protein A-agarose (PAA) beads was added, and the mixture was incubated for 1 h. The supernatant was separated from the PAA-IgG and passed through a spin column; 15 μl of loading buffer was added to 15 μl of the supernatant. The PAA-IgG was washed six times with NET-2 (150 mM NaCl, 50 mM Tris HCl [pH 7.5], 0.05% NP-40, 2 mM EDTA) and eluted with 15 μl of loading buffer. For controls, 250 ng of bovine serum albumin (BSA) and 20 μl of preimmune IgG-PAA (50% slurry) were substituted for Sla1p and anti-Sla1p PAA-IgG, respectively; 1.5 μl of the input, 4.5 μl of the supernatant, and 4.5 μl of the PAA elution were analyzed by denaturing 20% polyacrylamide electrophoresis. IgG-PAA was prepared by incubating 20 μl of 50% PAA (Sigma) with 10 μl of anti-Sla1p serum for 1 h at 4°C and then washed with 1 ml of NET-2 five times.
Five chemically synthesized RNAs containing the sequence 5′-AUCGAGAGGGACACGN, where N varied from two to six Us, plus a control RNA, 5′AUCGAGAGGGACACGGCGAAUUU (con-U3) were labeled on the 5′ ends and combined. Aliquots of this mixture, containing ∼30 fmol of each RNA, were incubated with 100, 50, or 25 fmol of recombinant Sla1p (rSla1p) or 125, 76, or 38 fmol of recombinant human La (rhLa) in the presence of 300 fmol of Escherichia coli tRNA (Roche) for 1 h at 4°C. Then, 20 μl of anti-Sla1p or anti-hLa PAA-IgG beads (50% slurry) was added, and the mixture was incubated for 1 h at 4°C. The PAA-IgG was washed six times with NET-2, and 1.5 μl of input, 4.5 μl of the immunoprecipitates, and 2 μl of the supernatants were resolved by denaturing 15% polyacrylamide electrophoresis.
Isolation and sequencing of the 3′ ends of pre-tRNASerUGAM produced in vivo were determined using a protocol (9), modified to be gene-specific, for pre-tRNASerUGAM transcripts that bear one or more 3′ Us. Total cellular RNA (2.5 μg) in 10 μl of ligase buffer containing 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 1 U of RNasin Plus (Promega), 20% polyethylene glycol (3500), and 10% dimethyl sulfoxide was incubated with T4 RNA ligase (New England Biolabs) and 100 pmol of oligo-RNA-DNA 3′ Adapter (Dharmacon) (Table 2), which contains 5′ phosphate and an inverted dT at the 3′ end, for 1 h at 37°C. The nucleic acid was then purified and suspended in 10 μl of H2O, and 1 μl was used for one-step reverse transcription (RT)-PCR (Titan One Tube RT-PCR; Roche) with 20 pmol of primers RACE1, which is specific to pre-tRNASerUGAM, and Reverse (Table 2), which is complementary to 3′ Adapter oligo-RNA-DNA and contains an additional dA at the 3′ end to target only those ligated RNAs that bear one or more Us at the 3′ end. Annealing was done at 56°C, and 30 cycles of PCR were performed. The RT-PCR products were inserted into a TA cloning vector (Invitrogen); plasmids were isolated from multiple independent clones and sequenced.
RESULTS
Rpc11p mutations that increase tRNA-mediated suppression occur in invariant residues in its Zn ribbon domain.
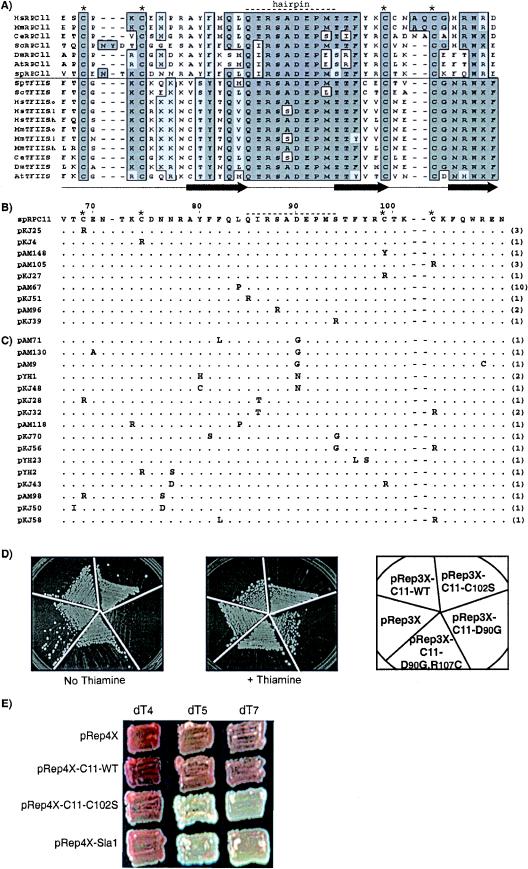
RPC11 is an essential gene that encodes a polypeptide with three domains: I, an N-terminal region containing a putative zinc-binding motif; II, a linker region; and III, a C-terminal domain that is homologous to the zinc ribbon of TFIIS (8), which consists of a three-stranded beta sheet stabilized by four cysteines that chelate zinc and an acidic hairpin between strands 1 and 2 (29) (Fig. 1A). A nonsense mutation in the ade6-704 allele of the yeast S. pombe strain yYH1 (Table 1) is partially suppressed by an integrated tRNASerUGAM gene with an efficient dT7 terminator (tRNAmSer7T) (13), as reflected by the development of pink color when grown in a limiting amount of adenine. A library of full-length rpc11 mutants in the S. pombe expression vector pRep4X, whose expression from the nmt1 promoter can be repressed by thiamine (28), was made using nucleoside analog PCR (39). Analysis of the unselected library revealed that approximately one-third of the clones contained no mutations while the remaining two-thirds contained mutations distributed in all three domains of rpc11 (data not shown; see Materials and Methods). Of ∼180,000 S. pombe colonies screened, ∼0.06% exhibited increased suppression, as reflected by white colony color. Plasmids from these were isolated and used to confirm their phenotype in yYH1. In all of several rpc11 mutants tested, suppression was repressed by thiamine (data not shown). No insertions, deletions, or silent mutations were recovered as suppressors. Of the clones bearing single substitutions, >70% were in domain III. Substitutions of the invariant hairpin residues Q85 and S88 were recovered, as was L84P, located at the junction of beta 1 and the hairpin (Fig. 1B) (29). Mutations of each of the invariant cysteines in domain III were isolated (Fig. 1B), whereas none of the four invariant cysteines in domain I were recovered. Indeed, site-directed mutation of cysteine (C24R) in domain I did not lead to suppression (data not shown). Thus, our screening assay, which monitored increased tRNA-mediated suppression, was particularly responsive to Rpc11p domain III mutations of highly conserved and invariant residues.
FIG.1.
S. pombe rpc11 mutants that increase tRNA-mediated suppression occur in invariant residues in the Zn ribbon and are predicted to decrease RNA 3′ cleavage activity. (A) Amino acid alignment of domain III of Rpc11p and the zinc ribbon domains of TFIIS of multiple species, above and below the horizontal line, respectively. The positions of invariant cysteines and a stretch of residues that corresponds to the acidic hairpin of TFIIS are indicated above by asterisks and a dashed line, respectively. Conserved residues are boxed and shaded. Filled arrows below the alignment represent beta strands 1 to 3 of TFIIS (29). (B) All Rpc11p mutants that contained single substitutions in domain III; the numbers of independent isolates are in parentheses. Wild-type S. pombe Rpc11p is shown in the first line, numbered above (8). (C) Mutants with two substitutions in domain III as their only mutations. (D) Effects of selected Rpc11p constructs on growth in nonlimiting adenine in which expression is on (No Thiamine) or repressed (+ Thiamine). (E) Suppression by C102S, C11-WT, Sla1, and pRep4X in sla1+ strains whose tRNASerUGAM gene terminators are comprised of dT4, dT5, and dT7 (yAMm4T-2, yYHm5T, and yYH1, respectively), as indicated.
Multiple mutants with two substitutions in domain III, a fraction of which were obtained as single mutants, were recovered (Fig. 1C). The majority of mutations at invariant cysteines encoded arginine (Fig. 1B and C). Additional mutations, made by site-directed mutagenesis, converted C102 to 10 other amino acids; each led to robust suppression repressible by thiamine (data not shown), and one of these, C102S, was used to characterize the system (see below).
D90 substitutions occurred only in conjunction with other substitutions (Fig. 1C). Because sequence alignment, mutagenesis, and structural analyses of TFIIS suggest that D90 may be critical for Rpc11p-mediated RNA 3′ cleavage (8, 20, 23) (Fig. 1A), we wanted to study it as a single mutation and created Rpc11-D90G (D90G) by site-directed mutagenesis. Expression of D90G caused growth inhibition, so we compared it to the double mutant Rpc11-D90GR107C recovered from our screen (D90GR107C) (Fig. 1C) and to control plasmids for growth in nonlimiting adenine (Fig. 1D). D90G impaired growth relative to D90GR107C, and repression by thiamine relieved the growth deficiency (Fig. 1D). Creation of R107C as a single mutation revealed normal growth, indicating that it was an intragenic suppressor of the growth defect of D90G (not shown). Also, R107C caused no tRNA-mediated suppression, indicating that D90G was required for suppression by D90GR107C (not shown). By comparison to TFIIS (10, 20, 36), our data suggested that the rpc11 mutants might be deficient in Pol III RNA 3′ cleavage activity, a conclusion that is supported by data described below.
Rpc11p mutants increase suppression from weak or strong tRNASerUGAM terminators.
Deletion of S. cerevisiae RPC11 was associated with impaired termination, as reflected by terminator readthrough (8). Although it seemed unlikely that rpc11 mutants that have increased nonsense suppressor function would be defective in termination, we tested this formal possibility by examining whether C102S might decrease suppression from suboptimal terminators. We previously showed that a dT≥5 terminator supports suppression while the suboptimal terminator, dT4, fails to terminate efficiently and instead produces readthrough transcripts that are not converted to tRNASerUGAM and support little if any suppression (13). We compared tRNASerUGAM genes with dT4, dT5, and dT7 terminators (Fig. 1E). Although dT4 led to less suppression than dT5 and dT7, consistent with low termination efficiency, C102S increased suppression, albeit slightly, as did Sla1p (Fig. 1E). Overexpression of Sla1p served as a positive control for this experiment (13) and also demonstrated an important feature of the system, that endogenous Sla1p is limiting for tRNASerUGAM-mediated suppression (see Discussion). The data suggest that the mutants are not deficient in termination efficiency.
The mechanism by which Rpc11p mutants increase suppression requires sla1.
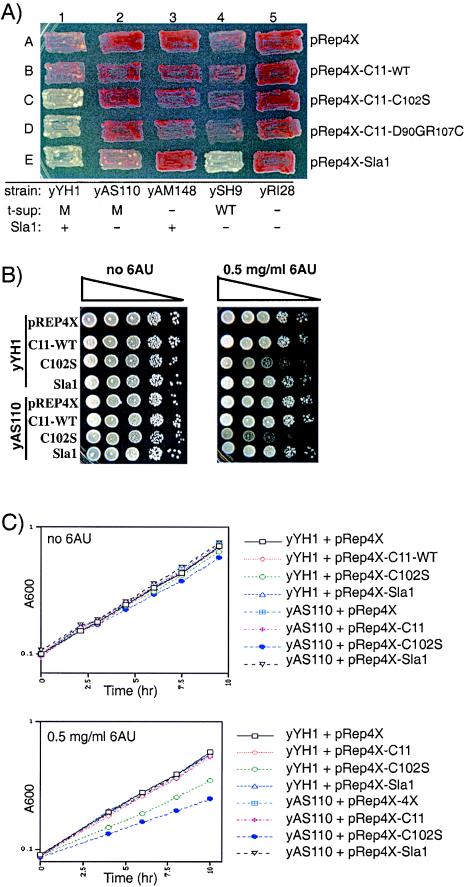
We examined C11-WT, C102S, D90GR107C, and the controls for the ability to increase suppression as mediated by two different suppressor alleles, tRNASerUGAM (M) and tRNASerUGAWT (wild type) (13), each bearing dT7 terminators, in sla1+ and sla1 deletion cells (Fig. 2A). As alluded to in the introduction, the M and WT tRNA alleles differ in their dependency on sla1+; M exhibits partial activity in the presence of sla1 and no activity in the absence of sla1 (Fig. 2A, compare patches A1 and A2), while tRNASerUGAWT exhibits partial activity in the absence of sla1 (compare patches A4 and A5) and full activity in the presence of Sla1 (Fig. 2A, patch E4).
FIG.2.
(A) The mechanism by which rpc11 mutants stimulate suppression requires La. Suppression analyses of C11-WT, C102S, D90GR107C, Sla1, and pRep4X (empty vector) in sla1+ and sla1 deletion strains that contain either tRNASerUGAM (M), tRNASerUGAWT (WT), or no tRNA (−) as their suppressor gene (t-sup), as labeled below. Patches of cells were plated on limiting adenine; red = no, pink = partial, and white = full suppression. (B and C) rpc11 mutants impair growth in a 6-azauridine (6AU)-sensitive manner that is partially rescued by La. Equal numbers of cells were plated as serial dilutions (B) or grown in liquid, and the optical density at 600 nm was monitored and plotted (C); the data points represent the average of three experiments, the standard deviations for which, for the majority of points, did not extend significantly beyond the symbol used to represent the point.
For tRNASerUGAM, C102S led to robust suppression in the sla1+ strain, yYH1, but not in the sla1 deletion strain, yAS110, while C11-WT and pRep4X were inactive (Fig. 2A, columns 1 and 2) and pRep4x-Sla1 was a positive control for both strains (Fig. 2A, patches E1 and E2) (18). Because tRNASerUGAM is dependent on La (18), it cannot be used to distinguish between the dependency of tRNASerUGAM on sla1+ and the possibility that the mechanism by which the rpc11 mutants stimulate suppression requires sla1+, whereas tRNASerUGAWT can be used (see below).
Unlike tRNASerUGAM, the tRNASerUGAWT allele is partially active in the sla1 mutant strain ySH9, as revealed by its lighter color relative to the appropriate control, yRI28, which lacks a tRNA suppressor (Fig. 2A, compare A4 with A5). The lighter color of ySH9 relative to yRI28 allows the conclusion that tRNASerUGAWT yields partial suppression in the absence of sla1 (Fig. 2A, compare A4 with A5). It is therefore significant that C102S and D90GR107C did not increase tRNASerUGAWT-mediated activity in ySH9 while Sla1 restored full activity (patch E4), similar to its activity in yYH1 (compare columns 1 and 4). Thus, failure of the rpc11 mutants to increase suppression in ySH9 indicates that they are ineffective in the La-independent pathway. These data indicate that the mechanism by which the rpc11 mutants increase suppression requires La and therefore suggest that they affect RNA 3′-U metabolism.
rpc11 mutants cause growth deficiency that is partially rescued by La.
To examine whether the mutants might manifest growth effects not revealed under suppression conditions, we monitored growth in nonlimiting adenine. C102S caused slight, if any, growth deficiency in the sla1 deletion strain yAS110 during growth in liquid (Fig. 2C). We also examined growth in 6-azauridine, which sensitizes yeast to the loss of TFIIS (10, 19, 26, 30). Growth deficiency due to C102S was apparent in the sla1+ strain yYH1 and was worse in yAS110 in 6-azauridine (Fig. 2B and C). The data indicate that sla1+ suppresses some of the growth defect caused by C102S and provide evidence of functional interaction between rpc11+ and sla1+ in vivo.
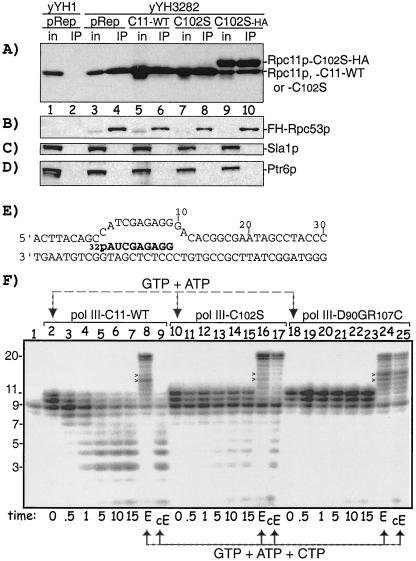
Mutated Rpc11p subunits associate with Pol III and impair its RNA 3′ cleavage activity.
To examine if mutant Rpc11p was associated with Pol III, we used an S. pombe strain, yYH3282, whose Rpc53p subunit is tagged with FLAG-His6 epitopes and from which Pol III can be isolated (15). Extracts were subjected to IP using anti-FLAG agarose. yYH3282 was transformed with pRep3X, pRep3X-C102S, and pRep3X-C102S-HA, the last of which produces HA-tagged protein and exhibits altered electrophoretic mobility. Both the input and immunoprecipitate were examined by Western blotting using anti-Rpc11p, anti-FLAG, anti-Sla1p, and anti-Ptr6p antibodies, the last two of which are controls (Fig. 3A to D). yYH1 and yYH3282 transformed with pRep3X revealed similar levels of endogenous Rpc11p (Fig. 3A, input, lanes 1 and 3), which was immunoprecipitated from yYH3282 but not yYH1, which contains untagged Rpc53p (Fig. 3A, lane 4). Expression of C11-WT and mutants led to substantial increases in Rpc11p levels (Fig. 3A, lanes 5, 7, and 9). The association of mutant or WT Rpc11p with Pol III was specific, since Ptr6p and Sla1p were not associated (Fig. 3C and D) (15, 16). Figure 3A, lane 10, shows that more C102S-HA than endogenous Rpc11p was associated with Pol III, approximately reflective of their relative expression levels (lane 9). Quantitation of endogenous Rpc11p and C102S-HA in the immunoprecipitation (Fig. 3A, lane 10) suggests that the immobilized Pol III contains ∼75% mutant and 25% wild-type Rpc11p subunits (data not shown). Since C102S-HA causes robust suppression (data not shown), the cumulative data are consistent with the increased suppression phenotype arising from a Pol III enzyme carrying the mutant Rpc11p subunit.
FIG. 3.
Mutant Rpc11p associates with and impairs the 3′ cleavage activity of Pol III. (A to D) Immunoblots of Pol III-associated Rpc11p isolated from S. pombe yYH3282 via FLAG-His6-tagged Rpc53p (lanes 3 to 10). yYH1 is a control that contains wild-type Rpc53p (lanes 1 and 2). Input (in) extracts from cells transformed with pRep3X alone, C11-WT, C102S, or C102S-HA, each in pRep3X, are in odd-numbered lanes, and anti-FLAG immunoprecipitated (IP) material is in even-numbered lanes. Antisera to Rpc11p, FLAG, Sla1p, and Ptr6p, the last of which is an mRNA-related factor in S. pombe (31) used previously (16), were used for detection as indicated; 2% of the input and 20% of the immunoprecipitate were examined (for panel A only, fivefold more input was loaded in lanes 1 and 3 than in lanes 5, 7, and 9 so that their Rpc11p levels would be comparable to the levels in the overexpressing cells on the same exposure). (E) Schematic representation of the 9-nt [32P]RNA-DNA hybrid as described previously (24), used here for Pol III-associated RNA 3′ cleavage. RNA is shown in boldface. (F) Analysis of Ni2+-agarose-immobilized Pol III containing C11-WT (lanes 2 to 9), C102S (lanes 10 to 17), and D90GR107C (lanes 18 to 25) complexes containing the [32P]RNA-DNA hybrid in panel E. Lane 1 shows [32P]RNA from the Pol III complex after incubation in buffer lacking Mg2+ and NTPs. Lanes 2, 10, and 18 show products after extension with ATP and GTP. Lanes 3 to 7, 11 to 15, and 19 to 23 show the time course of the cleavage-only reactions (Mg2+ but no NTPs), as indicated below the lanes in minutes. Lanes 8, 16, and 24 show products after extension with ATP, GTP, and CTP, indicated by E under the lanes. For lanes 9, 17, and 25, complexes were subjected to a cleavage reaction and then extended with ATP, GTP, and CTP, indicated by cE under the lanes. The [32P]RNA in lane 1 is less than in other lanes because it is less stably associated with the complex in the absence of Mg2+ and NTPs (data not shown). RNA chain lengths are indicated to the left. Arrows point to specific bands discussed in the text.
Since use of the D90 single mutant was restricted by growth deficiency, we chose to examine the double mutant D90GR107C (Fig. 1D), along with C102S and C11-WT, for Pol III-associated RNA 3′ cleavage activity. The activity of immobilized Pol III containing WT or mutant Rpc11p was examined using reconstituted elongation complexes (24). Anti-FLAG-purified Pol III-C11-WT, -C102S, or -D90GR107C was immobilized on Ni2+-agarose and assembled with DNA and 5′-labeled 9-nt [32P]RNA as illustrated in the cartoon in Fig. 3E. In the absence of NTP and Mg2+, the Pol III-associated [32P]RNA was not extended (Fig. 3F, lane 1). Extension of the [32P]RNA to 10 and 11 nt with GTP and ATP (with Mg2+) indicates polymerization (Fig. 3F, lanes 2, 10, and 18). However, more of the fully extended 11-mer than of the 9- and 10-mers was obtained with D90GR107C and C102S than with C11-WT (compare lanes 2, 10, and 18), consistent with less 3′ cleavage by D90GR107C and C102S (24).
To initiate cleavage-only reactions, washed complexes were replenished with Mg2+ but no NTPs, and aliquots were examined thereafter. This revealed a decrease in the 9- to 11-nt [32P]RNAs with concomitant production of <8-nt [32P]RNA cleavage products by C11-WT with time (Fig. 3F, lanes 3 to 7). In contrast to this, the 9-, 10-, and 11-nt [32P]RNAs were relatively stable in the C102S and D90GR107C complexes, with less of the <8-nt [32P]RNAs produced (lanes 11 to 15 and 19 to 23), indicating that they were deficient in RNA 3′ cleavage activity relative to C11-WT. Since the immobilized enzymes contain some wild-type Rpc11p, the cleavage deficiencies attributable to the C102S and D90GR107C subunits may be more striking than is apparent in these assays.
We also compared extensions in the presence of ATP, GTP, and CTP. Although each enzyme extended the [32P]RNA to 20 nt, they differed in the relative amounts of 14- and 15-nt RNAs (Fig. 3F, lanes 8, 16, and 24), which kinetic data (not shown) indicated were due to differential pausing. Thus, although C102S may pause less while D90GR107C may pause more than C11-WT, impaired cleavage was common to both mutants.
Extension after a cleavage reaction was also examined. This revealed a large reduction of 20-mer produced by C11-WT relative to C102S and D90GR107C (Fig. 3F, lanes 9, 17, and 25), reflective of cleavage deficiency of the mutant enzymes (24). The apparent lack of extension by the WT enzyme appears to reflect the fact that most of the cleavage products were shorter than is thought to be required to maintain stable association with the polymerase (24).
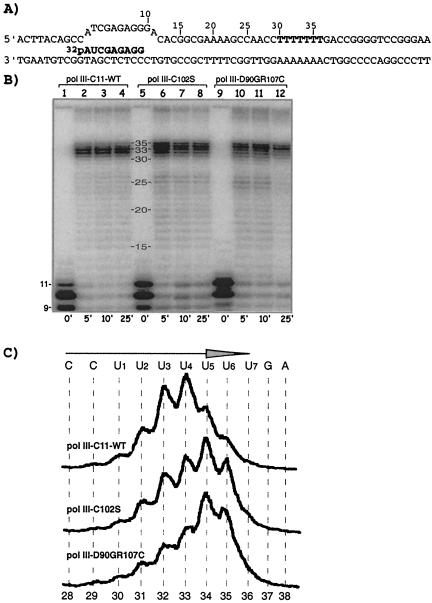
Nascent RNAs synthesized by mutant Pol III have longer 3′ oligo(U) tracts.
DNA bearing a dT7 tract (Fig. 4A) was used to examine Pol III termination in vitro. C11-WT, C102S, and D90GR107C enzymes produced prominent RNAs of 31 to 35 nt, corresponding to transcription into the dT7 (Fig. 4B). The intensities of the 31- to 35-nt RNAs relative to the 15- to 30-nt RNAs, together with the paucity of RNA above 35 nt, indicated that dT7 was efficiently recognized as a pause site by Pol III. Significant differences were observed in the distributions of bands corresponding to the dT7 tract, with C102S and D90GR107C producing RNAs slightly longer than those of C11-WT, and these differences were stable in 5- to 25-min reactions (Fig. 4B). The major RNAs from C11-WT were 32 to 33 nt long, whereas RNAs from C102S and D90GR107C included species up to 35 nt (Fig. 4B).
FIG. 4.
Nascent termination products of C102S and D90GR107C Pol III proteins have longer 3′ oligo(U) than C11-WT. (A) Schematic of the [32P]RNA-DNA hybrid containing a dT7 terminator. RNA is shown in boldface. (B) Transcription with four NTPs at 25°C using immobilized Pol III-C11-WT (lanes 1 to 4), Pol III-C102S (lanes 5 to 8), and Pol III-D90GR107C (lanes 9 to 12) for the times indicated below the lanes. Lanes 1, 5, and 9 show products using ATP and GTP only (before the addition of four NTPs), as in Fig. 3F. (C) Densitometric scans of the 28- to 38-nt regions of the 10-min time points reflect the distribution of RNAs produced as indicated. The arrow indicates the direction of transcription; the RNA sequence is indicated above, and corresponding template positions as numbered in panel A are below.
Densitometric scans of representative lanes revealed the RNA distribution patterns (Fig. 4C). The most abundant species produced by C11-WT corresponds to 3′ U4, with U3 and U5 representing the next most abundant species (Fig. 4C). The most abundant species produced by C102S was U5, with U4 and U6 as the next most abundant. The distribution was shifted further by D90GR107C, for which U5 and U6 were most abundant, with less U4 than C102S (Fig. 4C). The data indicated a correlation between the extent of cleavage deficiency and the length of the oligo(U) tract for C11-WT, C102S, and D90GR107C. Indeed, the RNA length distribution within the dT7 range roughly paralleled the patterns of the 9-, 10-, and 11-nt RNAs (Fig. 4B, lanes 1, 5, and 9), reminiscent of the differential cleavage activity seen in Fig. 3F.
To test if the RNAs produced in these reactions could associate with La, the soluble phase of a completed Pol III-C11-WT transcription reaction was separated from the immobilized enzyme, and aliquots were incubated with rSla1p or BSA and subjected to IP using anti-Sla1p IgG or nonimmune IgG or examined directly (Fig. 5A). Because a limiting amount of rSla1p was used, the different RNA species would have to compete for La binding. The specificity of rSla1p for the oligo(U)-terminated RNAs was revealed by the lack of short (∼8- to 12-nt) RNAs in the immunoprecipitate (Fig. 5A, lane 1) that remained in the supernatant (lane 2); the failure of preimmune IgG to precipitate the RNA (lane 3); and the requirement for rSla1p (lane 5) for IP. As expected, the 3′-U-terminated RNAs from the C102S and D90GR107C transcription reactions also bound specifically to rSla1p, while the short RNAs did not (Fig. 5B).
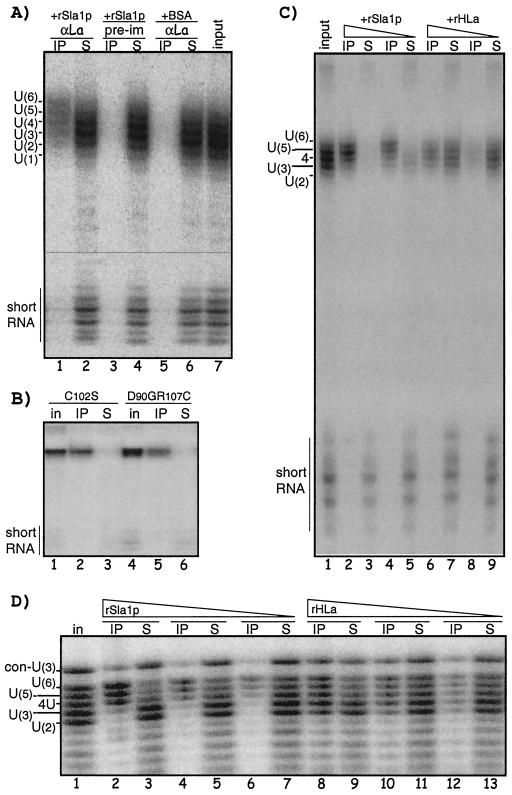
FIG. 5.
Mutant Rpc11p-Pol III in vitro-synthesized RNAs terminated with 3′ oligo(U) are bound by La. (A) Aliquots of the soluble phase of a transcription reaction using immobilized Pol III-C11-WT and DNA containing dT7 as in Fig. 4B were incubated with recombinant Sla1p or BSA and subjected to IP with anti-Sla1p (αLa) or preimmune (pre-im) IgG and examined by denaturing PAGE. The lanes are numbered below; lane 7 shows input prior to IP. Supernatants (S) and immunoprecipitates are indicated above; RNA species are indicated to the left. (B) Same as in panel A except that immobilized Pol III-C102S and -D90GR107C were used for transcription, with lanes 1 and 4 showing input for each IP. (C and D) S. pombe La prefers longer 3′-U tracts than its human La counterpart. (C) As in panel A except that rSla1p and rhLa, each at two concentrations, were subjected to IP with the corresponding antisera. (D) Chemically synthesized RNAs bearing two to six Us, labeled at the 5′ ends, were combined and incubated with rSla1p or rhLa and subjected to IP. RNA (con-U3) was included as a control; it is longer but contains only three Us at its 3′ end. RNAs were resolved by denaturing 15% PAGE.
The pattern in lane 1 of Fig. 5A suggested that Sla1p preferred RNA with longer oligo(U) than described for human La (32). Therefore, the Pol III-terminated RNAs were incubated with various amounts of Sla1p or rhLa and subjected to IP with the corresponding IgG (anti-Sla1p or anti-HLa), and input, immunoprecipitate, and supernatants were examined. Comparison of input (Fig. 5C, lane 1) and Sla1p immunoprecipitate (lane 2) revealed that RNAs with five and six Us competed better for Sla1p than did RNAs with fewer Us. While hLa exhibited a preference for oligo(U) RNAs with ≥3 Us, it was not as enriched in the longest RNAs as was Sla1p (Fig. 5C).
As another approach, we used chemically synthesized RNAs that differed in 3′-U length, ranging from two to six Us (Fig. 5D), as well as a control, con-U3 RNA. Again, using various concentrations of La so that small or large fractions of the RNAs were immunoprecipitated, Sla1p selected for RNAs containing longer 3′-U tracts (Fig. 5D). This was most convincing when the immunoprecipitate and supernatants were compared as Sla1p became limiting, as in lanes 6 and 7 of Fig. 5D. When it was limiting, Sla1p appeared to prefer six Us as much or more than five Us, and both more than four Us, with relatively low preference for three and two Us. The relatively low preference for RNA with three Us did not appear to be due to shorter overall length, since the con-U3 RNA is longer but contains only three Us and was not preferentially bound by Sla1p. By contrast, hLa was clearly less selective (lanes 8 to 13), although consistent with a preference for ≥3 Us as described previously (32) (see Discussion).
Efficiency of La-dependent pre-tRNASerUGAM processing is increased in rpc11 mutants.
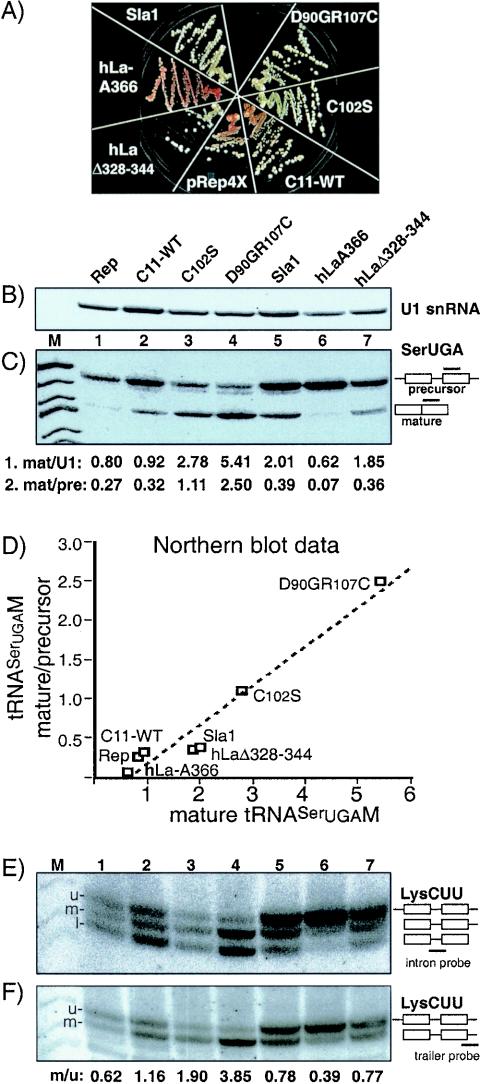
yYH1 cells expressing C11-WT, C11 mutants, and previously characterized controls that promote (Sla1p and human LaΔ328-344) or inhibit (human La-A366) pre-tRNA maturation (18) were examined for suppression (Fig. 6A) and by Northern blotting using a probe that detects precursor and mature tRNASerUGAM (Fig. 6C). We also probed the blot for U1 snRNA (Fig. 6B), which is transcribed by Pol II, to control for quantitation, as indicated in line 1 below Fig. 6C. pRep4X and C11-WT revealed basal suppression activity (Fig. 6A) and less mature tRNASerUGAM than the constructs that increased suppression, C102S, D90GR107C, Sla1p, and hLaΔ328-344 (Fig. 6C and line 1 below 6C). Consistent with inhibition of pre-tRNASerUGAM 5′ processing (18), hLa-A366 had a dominant-negative effect on suppression, as reflected by a darker red color than pRep4X (Fig. 6A), which was accompanied by low tRNASerUGAM levels (Fig. 6C, lanes 1 and 6). D90GR107C and C102S produced relatively high levels of tRNASerUGAM, albeit more so for D90GR107C (Fig. 6C). Thus, suppression by C102S and D90GR107C was accompanied by increased levels of mature tRNASerUGAM.
FIG. 6.
Efficiency of La-dependent pre-tRNA processing is increased in rpc11 mutants. (A) Suppression analysis of C11-WT, C102S, D90GR107C, Sla1p, hLa-A366, hLaΔ328-344, and pRep4X (Rep) in yYH1. (B and C) Total RNAs were analyzed on a Northern blot using probes for (B) U1 snRNA and (C) tRNASerUGAM. The bands were quantitated by phosphorimager and compiled as fractions, as mature tRNASerUGAM relative to U1 (mat/U1) and mature relative to precursor tRNASerUGAM (mat/pre), as in lines 1 and 2 under panel C. (D) The quantitation data indicated in lines 1 and 2 under panel C were plotted on the x and y axes, as indicated (see the text). (E and F) The same blot as in panels B and C was reprobed for pre-tRNALysCUU intron-containing (E) and trailer-containing (F) species. Thebands labeled u, m, and l represent upper, middle, and lower bands, respectively, and the quantification of u/m is shown below panel F. The positions of the probes are indicated as horizontal bars relative to the various tRNA species to the right of panels C, E, and F.
Direct quantitative comparison of mature and precursor tRNASerUGAM species revealed more precursor than mature species for pRep4X and C11-WT (Fig. 6C, lanes 1 and 2), as reflected by a mature/precursor ratio of ≤1.0 (line 2 below Fig. 6C). This ratio was ≥1.0 only for C102S and D90GR107C (lanes 3 and 4). Although Sla1p and hLaΔ328-344 increased tRNASerUGAM levels, they did not significantly change the precursor-to-mature-tRNA ratio relative to the pRep4X and C11-WT controls. Correlation between the mature/precursor ratio and mature tRNASerUGAM levels was observed (Fig. 6D), suggesting that the pre-tRNASerUGAM processing efficiency is a primary determinant of mature tRNASerUGAM levels in this system. According to this analysis, Rpc11p mutants exhibited a novel trait, markedly increased efficiency of functional pre-tRNASerUGAM processing.
Increased efficiency of La-dependent processing of endogenous pre-tRNALysCUU.
We wanted to know if the effects of the rpc11 mutants were specific to the suppressor tRNAs, and therefore we reprobed the above-mentioned blot for endogenous pre-tRNALysCUU. As documented previously (17, 18, 33), an intron probe detects three bands, a nascent transcript that contains a leader, trailer, and intron; a species whose leader only has been removed; and a species whose leader and trailer have been removed, as indicated on the right of Fig. 6E. The middle band results from 5′ processing of nascent pre-tRNALysCUU, and its accumulation is strictly dependent on La, as documented in previous studies (17, 18, 33). The relative amounts of upper-to-middle bands (Fig. 6E) were lower for C102S and D90GR107C than for pRep4X, C11-WT, and the La constructs, again more so for D90GR107C than C102S. Since a decrease in the upper band was accompanied by an increase in the middle band in the Rpc11p mutants, these data are consistent with their more efficient 5′ processing compared to control cells. Complete stripping (not shown) and probing for the 3′ trailer of pre-tRNALysCUU were consistent with an increase in the 5′-processed species at the apparent expense of the nascent precursor (Fig. 6F). The rpc11 mutants did not lead to the appearance of the middle band (Fig. 6E) in sla1 deletion cells (not shown). Thus, the rpc11 mutants had positive effects on the accumulation of the 5′-processed species (Fig. 6E and F, m). Since this species is dependent on La for formation (17, 18, 33), the rpc11 mutants appear to exhibit increased La-dependent processing of endogenous pre-tRNALysCUU, similar to what was found for the suppressor tRNA.
3′-cleavage-deficient rpc11 mutants exhibit altered pre-tRNA 3′-oligo(U) length in vivo.
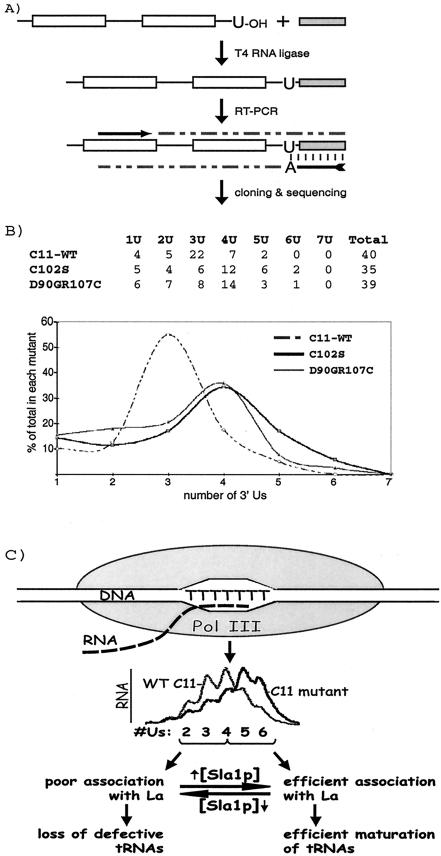
The data indicated a correlation between oligo(U) length and tRNA maturation. Accordingly, nascent pre-tRNAs with the longest 3′-U tracts may be converted to mature tRNA faster in rpc11 mutants than in wild-type cells, suggesting that these pre-tRNAs might be underrepresented in total RNA in vivo. We determined the 3′-U lengths of pre-tRNASerUGAM transcripts isolated from C11-WT, C102S, and D90GR107C cells, using a pre-tRNASerUGAM-specific RT-PCR directed at RNAs bearing at least one U at the 3′ end, followed by cloning and sequencing (Fig, 7A). Sequences of ∼40 independent clones, each containing an intron, a 3′ trailer, and an internal pre-tRNASerUGAM-specific sequence, were obtained from each strain. The number of sequences bearing a specific number of 3′ Us for each strain was compiled in table and graph formats (Fig. 7B). C11-WT was clearly distinguished from the C102S and D90GR107C mutants in their 3′-U length profiles (Fig. 7B). Although these distributions did not precisely match that obtained after transcription in vitro, the data indicate altered pre-tRNASerUGAM 3′-oligo(U) length in vivo.
FIG.7.
Pre-tRNASerUGAM isolated from Rpc11p mutants exhibit lengthened 3′ oligo(U). (A) Outline of experimental strategy for isolation of the 3′ sequences of nascent pre-tRNASerUGAM transcripts synthesized in vivo. Note that only RNAs that bear one or more 3′ Us will be preferentially amplified because the antisense oligonucleotide used for RT-PCR contains a dA at its 3′ end. The sense oligonucleotide for RT-PCR (horizontal arrow) is specific for the tRNASerUGAM sequence. Open boxes represent exons, shaded boxes represent the 3′ adapter, the A with the arrow tail represents the DNA primer used for RT-PCR, the arrow represents the sense primer for PCR, and dashed lines represent the PCR product. (B) Compilation of the 3′-end sequences determined from 40, 35, and 39 pre-tRNASerUGAM transcripts from C11-WT, C102S, and D90GR107C cells, respectively, shown in table format and also as percentages of the total (y axis), as indicated, in graphic format. The compiled sequences contain introns and 3′ trailers and were 100% identical to the pre-tRNASerUGAM sequence (not shown). (C) Model of proposed links among Pol III termination, RNA 3′-oligo(U) length, binding of the nascent RNA products to S. pombe La protein, Sla1p, and consequent effects on pre-tRNA processing in fission yeast as described in the text (see Discussion).
DISCUSSION
A major conclusion that can be drawn from these data is that mutations that impair the RNA 3′ cleavage activity of Rpc11p lead to pre-tRNAs with longer 3′-U tracts. In addition, the data provide the first in vivo evidence that RNA 3′-U length heterogeneity can have functional effects on posttranscriptional processing of a pre-tRNA and that this reflects association with La. This study reveals novel functional connectivity between the role of Rpc11p in RNA 3′-end formation by Pol III and posttranscriptional processing. The biological system described here should be useful for future studies of RNA 3′-end formation by Pol III.
Our data are consistent with published characteristics of Pol III-associated RNA 3′ cleavage (3, 4, 8, 34). In S. cerevisiae, an rpc11 deletion mutant Pol III exhibited lack of RNA 3′ cleavage and severe terminator readthrough (8). However, these defects could be uncoupled, since reconstitution with recombinant Rpc11p restored cleavage activity but did not prevent readthrough (8), reminiscent of some TFIIS mutants that uncouple cleavage and readthrough (10).
The rpc11 mutants illustrate a link between Pol III termination and posttranscriptional RNA accumulation.
It is important for this discussion to refer to termination efficiency as conventionally defined, as the fraction of polymerases that terminate within a dTn site rather than read through it to a downstream site. Multiple observations support the idea that extension beyond the dT7 terminator of the tRNASerUGAM gene, as occurs in conventional readthrough transcription (13), does not appear to be a significant feature of, or to contribute to, the phenotype of the rpc11 mutants characterized here. It has been documented that tRNASerUGAM genes produce functional suppressor tRNA only if termination occurs within the dTn terminator, since readthrough beyond the dTn produces long transcripts that are not processed to mature suppressor tRNASerUGAM (13). Thus, in this system, production of functional suppressor decreases as termination efficiency decreases. Accordingly, since rpc11 mutants increase suppression from tRNASerUGAM genes bearing dT4 and dT5 terminators (Fig. 1E), their positive effects on suppression would not appear to be due to decreased termination efficiency.
Considering the possibility that the rpc11 mutants increased termination efficiency was also inappropriate, because the increase in tRNASerUGAM levels was higher than could be accounted for by simply increasing the efficiency of a dT7 terminator (13). For example, prior data suggested that the termination efficiency of S. pombe Pol III was ∼90% at the dT7 terminator of the tRNASerUGAM gene that is present in the yYH1 strain used here (13). Increasing the termination efficiency to 100% should therefore lead to only a 10% increase in transcript levels, whereas the rpc11 mutants expressed 300 to 600% of the tRNASerUGAM present in the C11-WT control cells (Fig. 6C). Thus, we were left with explaining the conundrum of how the rpc11 mutants can dramatically increase tRNASerUGAM levels from an efficient terminator in a La-dependent manner. It is important to emphasize here that little if any tRNASerUGAM accumulates in sla1 mutant cells. This is presumably because pre-tRNASerUGAM is defective (see the introduction) and succumbs to nuclear surveillance, a contention supported by increased suppression in S. pombe strains with Rrp6p, an exonuclease involved in nuclear surveillance, deleted (22; Y. Huang and R. J. Maraia, unpublished observation). The finding that the majority of pre-tRNASerUGAM transcripts in wild-type cells contained too few 3′ Us for efficient Sla1p binding in vitro was significant, because it helped to explain the large elevation of tRNASerUGAM levels in the mutants (see below).
A proposal that satisfies the quantitative and qualitative issues raised above would be that 65 to 80% of pre-tRNASerUGAM succumbs to nuclear surveillance in yYH1 but can be salvaged by the La-dependent pathway in the rpc11 mutants through their increased 3′-U lengths. A question that arises is why these transcripts are not functionally engaged by Sla1p in yYH1. The answer, we believe, is supported by two sets of data. First, Sla1p is limiting for tRNASerUGAM-mediated suppression, as suggested by the fact that overexpression of Sla1p increases suppression (Fig. 1E and 6A) (reference 18 and data not shown). This suggests that pre-tRNASerUGAMs have to compete, presumably with all other cellular Pol III transcripts, for Sla1p. Second, a substantial fraction of pre-tRNASerUGAM transcripts bear only three or fewer 3′ U residues, which is insufficient for efficient binding by Sla1p (Fig. 5 and 7). The data suggest that by conferring longer 3′ oligo(U) tracts on these transcripts, the rpc11 mutants produce pre-tRNASerUGAMs that can compete more productively for Sla1p. Thus, it would appear that by shunting otherwise nonproductive pre-tRNASerUGAMs to La, the mutants produce higher tRNASerUGAM levels. The results suggest that the efficiency of La-dependent pre-tRNA processing may be increased either by increasing 3′-U length, as in the rpc11 mutants, or by increasing Sla1p levels by overexpression (Fig. 7C).
Cleavage-deficient rpc11 mutants alter RNA 3′-end formation with relatively less if any defect in readthrough.
The above considerations suggest a model in which the mutant phenotype results not from changes in the quantity of nascent pre-tRNASerUGAM produced by Pol III termination in the dT7 terminator but from the quality of the 3′ ends of the pre-tRNASerUGAM produced, which in turn affects the posttranscriptional fate of the pre-tRNA.
Consistent with this model, direct examination revealed that the rpc11 mutant Pol III proteins terminated within the dT7 tract with little evidence of readthrough, although this remains to be examined in more extensive studies (Fig. 4). No readthrough transcripts were detected in the rpc11 mutants using our standard probes, as well as readthrough-specific probes that do detect readthrough transcripts in cells carrying inefficient terminators (reference 13and data not shown). These observations suggest that readthrough beyond the dTn terminator does not appear to be a significant feature of the rpc11 mutants examined here.
The cumulative data argue that the major effect of the rpc11 mutants is not readthrough as conventionally defined (see above). Rather, the rpc11 mutant Pol III proteins terminate more distally within the dT7 tract than does wild-type Pol III, producing nascent RNAs with longer oligo(U) tracts. Considering that the immobilized rpc11 mutant Pol III proteins contained some wild-type Rpc11p subunit (Fig. 3), the differences in oligo(U) length observed between wild-type and mutant Pol III may have been even more striking in a homogeneous mutant rpc11 background. In any case, the data indicate that the rpc11 mutants alter the Pol III termination process by a novel mechanism, extension farther within, but not beyond, the dT7 tract.
RNA 3′-oligo(U) length is inversely related to cleavage activity by Rpc11p.
With regard to the relationship between RNA 3′ cleavage and 3′-oligo(U) length, a simple explanation would be that lengthening results from the deficiency of the rpc11 mutants in cleaving the 3′ end of the nascent RNA, although our data do not directly demonstrate this. In support of this idea is the finding that the mutant with the more severe RNA 3′ cleavage deficiency, D90GR107C, produced the longest 3′-oligo(U) in vitro. These data provide evidence to suggest links among RNA 3′ cleavage, 3′-oligo(U) length, La-dependent pre-tRNA processing, and tRNA accumulation. For each of these parameters, the relative activities of C11-WT, C102S, and D90GR107C were maintained, so that the stronger the defect for RNA 3′ cleavage, the longer the 3′ oligo(U), and the more efficient was pre-tRNA processing and accumulation of mature tRNA. Specifically, D90GR107C was most defective in 3′ cleavage, produced the longest 3′ oligo(U), showed the most efficient pre-tRNA processing, and produced the most mature tRNA, whereas C102S was intermediate between it and C11-WT. This correlation among the three samples for these parameters suggests that these aspects of RNA biogenesis are linked.
The 3′-oligo(U) length is a determinant of La binding that can partition nascent Pol III transcripts to alternative pathways of maturation.
An unexpected notion that emerged from this study is that nascent pre-tRNAs appear to be partitioned in fission yeast into substantial fractions that do and do not engage Sla1p (Fig. 7C). The question arises as to what role would be served by the partitioning of a large fraction of nascent pre-tRNAs to the La-independent pathway. It is noteworthy here that, as noted in the introduction, La is nonessential in yeast and that pre-tRNASerUGAM is defective and functionally null in sla1 deletion cells. Together, these observations suggest that while pre-tRNASerUGAM is a specific probe for the La-dependent pathway, it does not represent functional cellular pre-tRNAs in their lesser dependence on Sla1p for maturation. Thus, in wild-type cells that contain tRNAs that survive in the absence of engaging La, the cleavage activity of Rpc11p is not detrimental. What might happen to tRNAs with compromising mutations? By limiting oligo(U) length and consequent access to Sla1p, wild-type Rpc11p might limit the abilities of defective pre-tRNAs to survive to maturity even in sla1+ cells. A positive aspect of this is that most defective pre-tRNAs would be eliminated and only mostly nondefective tRNAs would accumulate. In this regard, Rpc11p may be considered a positive factor that can effect quality control over tRNA accumulation.
La function is linked to Pol III termination.
The notion that overexpression of mutant Rpc11ps is detrimental to growth is consistent with the idea that Rpc11p is vital in S. pombe. We note that while the present study does not indicate a role for La in transcription per se, the demonstration that La can partially suppress growth deficiency due to mutant C11, as well as the species-specific parallel between La preference for RNA 3′-Un length described here (Fig. 5) and the species-specific Pol III requirement for minimum dTn length for termination documented previously (13), strengthens the link between Pol III termination and La action in eukaryotes.
We note that while the rpc11 mutants are RNA 3′ cleavage deficient and produce longer oligo(U) tracts, this does not exclude the possibility that they may do so, at least in part, by causing structural alterations in other Pol III subunits that affect termination. Likewise, although the mutants lead to longer 3′ oligo(U), we cannot exclude the possibility that they may also affect other aspects of Pol III function that lead to increased tRNASerUGAM levels, and we leave this possibility open.
Acknowledgments
We thank M. Blum for medium preparation, M. Kawano for advice on RNA ligation, M. Cashel for advice on mutagenesis and yeast growth in liquid, H. Levin for the plasmid rescue protocol, K. Joshipura for plasmid rescue, and T. Kokubo for anti-Ptr6p. We are deeply grateful to M. Kashlev and M. Kireeva for teaching us the cleavage assay and for advice and discussion. We thank C. Kane for critical reading; A. Hinnebusch, The Friday Seminar, and SMCB members for discussion; and reviewers for comments.
R.J.M. receives support from the Commissioned Corps, USPHS. A.M. and S.H. were supported by the NICHD postbaccalaureate program.
REFERENCES
- 1.Anderson, J., L. Phan, R. Cuesta, B. A. Carlson, M. Pak, K. Asano, G. R. Bjork, M. Tamame, and A. G. Hinnebusch. 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12:3650-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwood, A., J. Choi, and H. L. Levin. 1998. The application of a homologous recombination assay revealed amino acid residues in an LTR-retrotransposon that were critical for integration. J. Virol. 72:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobkova, E. V., N. Habib, G. Alexander, and B. D. Hall. 1999. Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. J. Biol. Chem. 274:21342-21348. [DOI] [PubMed] [Google Scholar]
- 4.Bobkova, E. V., and B. D. Hall. 1997. Substrate specificity of the RNase activity of yeast RNA polymerase III. J. Biol. Chem. 272:22832-22839. [DOI] [PubMed] [Google Scholar]
- 5.Calvo, O., R. Cuesta, J. Anderson, N. Gutierrez, M. T. Garcia-Barrio, A. G. Hinnebusch, and M. Tamame. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4167-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, F. E., and D. R. Setzer. 1992. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12:2260-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakshusmathi, G., S. D. Kim, D. A. Rubinson, and S. L. Wolin. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 22:6562-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chedin, S., M. Riva, P. Schultz, A. Sentenac, and C. Carles. 1998. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 12:3857-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fish, R. N., and C. M. Kane. 2002. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta 1577:287-307. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamada, M., Y. Huang, T. M. Lowe, and R. J. Maraia. 2001. Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol. Cell. Biol. 21:6870-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamada, M., A. L. Sakulich, S. B. Koduru, and R. Maraia. 2000. Transcription termination by RNA polymerase III in fission yeast: a genetic and biochemically-tractable model system. J. Biol. Chem. 275:29076-29081. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., M. Hamada, and R. J. Maraia. 2000. Isolation and cloning of four subunits of a fission yeast TFIIIC complex that includes an ortholog of the human regulatory protein TFIIICβ. J. Biol. Chem. 275:31480-31487. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Y., M. Hamada, and R. J. Maraia. 2003. RNA polymerase III from the fission yeast, Schizosaccharomyces pombe. Methods Enzymol. 370:165-173. [DOI] [PubMed] [Google Scholar]
- 16.Huang, Y., E. McGillicuddy, M. Weindel, S. Dong, and R. Maraia. 2003. The fission yeast TFIIB-related factor limits RNA polymerase III to a TATA-dependent pathway of TBP recruitment. Nucleic Acids Res. 31:2108-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Intine, R. V., M. Dundr, T. Misteli, and R. J. Maraia. 2002. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell 9:1113-1123. [DOI] [PubMed] [Google Scholar]
- 18.Intine, R. V. A., A. L. Sakulich, S. B. Koduru, Y. Huang, E. Pierstorrf, J. L. Goodier, L. Phan, and R. J. Maraia. 2000. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol. Cell 6:339-348. [DOI] [PubMed] [Google Scholar]
- 19.Ishiguro, A., Y. Nogi, K. Hisatake, M. Muramatsu, and A. Ishihama. 2000. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol. Cell. Biol. 20:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon, C., H. Yoon, and K. Agarwal. 1994. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl. Acad. Sci. USA 91:9106-9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson, M. J., and A. S. Bystrom. 2002. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA 8:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kettenberger, H., K. J. Armache, and P. Cramer. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114:347-357. [DOI] [PubMed] [Google Scholar]
- 24.Komissarova, N., M. L. Kireeva, J. Becker, I. Sidorenkov, and M. Kashlev. 2003. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 370:233-251. [DOI] [PubMed] [Google Scholar]
- 25.Kufel, J., and D. Tollervey. 2003. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA 9:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 27.Maraia, R. J., and R. V. Intine. 2002. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Expr. 10:41-57. [PMC free article] [PubMed] [Google Scholar]
- 28.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 29.Qian, X., C. Jeon, H. Yoon, K. Agarwal, and M. A. Weiss. 1993. Structure of a new nucleic-acid-binding motif in eukaryotic transcriptional elongation factor TFIIS. Nature 365:277-279. [DOI] [PubMed] [Google Scholar]
- 30.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibuya, T., S. Tsuneyoshi, A. K. Azad, S. Urushiyama, Y. Ohshima, and T. Tani. 1999. Characterization of the ptr6+ gene in fission yeast: a possible involvement of a transcriptional coactivator TAF in nucleocytoplasmic transport of mRNA. Genetics 152:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefano, J. E. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36:145-154. [DOI] [PubMed] [Google Scholar]
- 33.Van Horn, D. J., C. J. Yoo, D. Xue, H. Shi, and S. L. Wolin. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3:1434-1443. [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehall, S. K., C. Bardeleben, and G. A. Kassavetis. 1994. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J. Biol. Chem. 269:2299-2306. [PubMed] [Google Scholar]
- 35.Willis, I., D. Frendewey, M. Nichols, A. Hottinger-Werlen, J. Schaack, and D. Soll. 1986. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J. Biol. Chem. 261:5878-5885. [PubMed] [Google Scholar]
- 36.Wind, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 38.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393-402. [DOI] [PubMed] [Google Scholar]
- 39.Zaccolo, M., D. M. Williams, D. M. Brown, and E. Gherardi. 1996. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 255:589-603. [DOI] [PubMed] [Google Scholar]