Abstract
Neutral Lipid Storage Disease with Myopathy (NLSDM) is a rare defect of triacylglycerol metabolism, characterized by the abnormal storage of neutral lipid in organelles known as lipid droplets (LDs). The main clinical features are progressive myopathy and cardiomyopathy. The onset of NLSDM is caused by autosomal recessive mutations in the PNPLA2 gene, which encodes adipose triglyceride lipase (ATGL). Despite its name, this enzyme is present in a wide variety of cell types and catalyzes the first step in triacylglycerol lipolysis and the release of fatty acids.
Here, we report the derivation of NLSDM-induced pluripotent stem cells (NLSDM-iPSCs) from fibroblasts of two patients carrying different PNPLA2 mutations. The first patient was homozygous for the c.541delAC, while the second was homozygous for the c.662G > C mutation in the PNPLA2 gene. We verified that the two types of NLSDM-iPSCs possessed properties of embryonic-like stem cells and could differentiate into the three germ layers in vitro. Immunofluorescence analysis revealed that iPSCs had an abnormal accumulation of triglycerides in LDs, the hallmark of NLSDM. Furthermore, NLSDM-iPSCs were deficient in long chain fatty acid lipolysis, when subjected to a pulse chase experiment with oleic acid. Collectively, these results demonstrate that NLSDM-iPSCs are a promising in vitro model to investigate disease mechanisms and screen drug compounds for NLSDM, a rare disease with few therapeutic options.
Keywords: NLSDM, iPSCs, PNPLA2, Lipid metabolism, Myopathy, Lipid droplets
Highlights
-
•
NLSDM is a defect of triacylglycerol metabolism, characterized by the abnormal storage of neutral lipid into lipid droplets.
-
•
The main clinical features of the disease are progressive myopathy and cardiomyopathy
-
•
No specific therapy is currently available.
-
•
We report the generation of iPSCs from patients fibroblasts with properties of embryonic- like stem cells.
-
•
These iPSCs could be differentiated into cardiomyocytes or myoblasts to screen potential therapeutic compounds.
1. Introduction
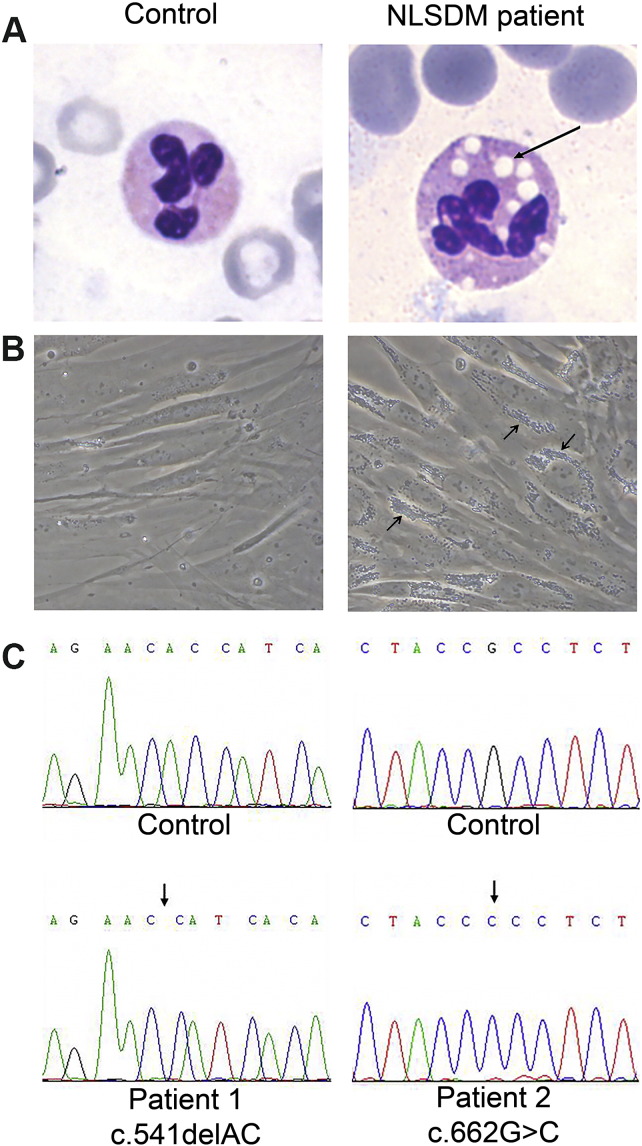
Neutral Lipid Storage Disease with Myopathy (NLSDM; MIM 610717) is a rare autosomal recessive disorder characterized by an increased accumulation of triacylglycerols (TAGs) in numerous tissues (Fig. 1A and B). TAGs are the most common neutral lipids stored in lipid droplets (LDs), the cytoplasmic organelles involved in lipid metabolism. The major clinical features of NLSDM are a progressive skeletal muscle myopathy, and elevated serum transaminases and creatine kinase in > 90% of NLSDM subjects [1], [2], [3], [4]. Approximately 40% of patients develop cardiomyopathy and in about 25% of them, the heart failure is lethal [5], [6], [7]. In addition, NLSDM subjects can develop hepatomegaly and diabetes [1], [2], [8]. The observation of lipid-containing vacuoles in leukocytes, also known as Jordans' anomaly, is an essential step for the clinical diagnosis [9] (Fig. 1A), whereas the presence of lipid inclusion in muscle biopsies confirms the muscle involvement [8], [10].
Fig. 1.
Lipid droplets abnormal storage in different tissues of NLSDM patient 1, compared to a control subject.
(A) Buffy coats stained with May-Grünwald-Giemsa (arrow indicates Jordans' anomaly in the granulocyte of patient 1). Magnification: 100 ×.
(B) Phase contrast images of primary dermal fibroblasts obtained from a control subject and NLSDM patient (patient 1) whose fibroblasts were used to produce iPS cells. Arrows indicate abnormal TAGs accumulation in the NLSDM fibroblasts. Magnification: 40 ×.
(C) Direct sequencing of PNPLA2 gene confirmed the presence of mutations in the two disease-specific iPSC lines (c.541delAC and c.662G > C respectively).
Mutations in the PNPLA2 gene are associated with this disorder [1]. The PNPLA2 gene encodes adipose triglyceride lipase (ATGL), a member of the patatin-like phospholipase domain-containing protein family, which catalyzes the first step in the hydrolysis of TAGs that are stored within LDs. The ATGL protein consists of 504 amino acids divided into an N-terminal part, containing the patatin domain, and a C-terminal part, containing a LD-binding domain [1]. To date, 47 patients harbouring 32 different mutations have been reported [1], [2], [3], [7], [8], [10], [11], [12], [13], [14], [15].
NLSDM has been recognized only recently, and its clinical, genetic and metabolic implications have not been completely elucidated. Furthermore, for most of the human mutations in the ATGL protein, the effect on enzyme activity has not been ascertained. Fibroblasts and myocytes, obtained from patients, have been used as cellular models to investigate the pathological mechanism and/or to test compounds to treat this disorder [11]. However, the limited availability of relevant human primary cell types such as myocytes and cardiomyocytes has hampered the ability to adequately study pathways or to test potential drug therapy.
Induced pluripotent stem cells (iPSCs) are a new technology which can provide an unlimited number of human disease-affected stem cells [16], [17], [18], [19], [20], [21]. These cells can theoretically differentiate into any cell type. iPSCs have been generated from human somatic cells and have been used to investigate disease pathogenesis and to test new drugs [22], [23], [24], [25]. Although iPSCs have been generated as model for some storage diseases (Pompe disease, Gaucher disease, Niemann-Pick Type C) [22], [26], [27], NLSDM-iPSCs are not yet available.
Here, we describe the generation of NLSDM disease-specific human iPSCs from fibroblasts of previously reported patients [2], [3]. The first patient is a male who is homozygous for c.541delAC PNPLA2 gene mutation. This mutation is predicted to produce a truncated ATGL protein (I212X) lacking the LD-binding domain. The second patient is homozygous for the c.662G > C mutation. This mutation leads to the production of ATGL protein with decreased lipase activity, but able to bind to LDs [3]. In this paper we show that the iPS cells derived from these patients possess hESC-like characteristics and that the NLSDM-iPSCs recapitulate the disease phenotype of interest.
2. Material and methods
2.1. Reprogramming of human dermal fibroblasts into iPSCs
Dermal fibroblast samples were grown as previously described [3]. The enrolled subjects were 2 NLSDM patients [2], [3] carrying different mutations in the PNPLA2 gene and 2 healthy subjects. All participants gave informed consent to donate skin samples for research purpose.
iPSCs were established by infecting 2 × 105 fibroblasts with the Yamanaka reprogramming factors OCT4, SOX2, KLF4 and c-MYC contained as separated Sendai-vectors into the CytoTune™-iPS Sendai Reprogramming Kit (Life Technologies, Monza, Italy). Cells were maintained in standard growth media at 37 °C and 5% CO2. Our standard growth media consisted of DMEM-F12 (Sigma Aldrich, Milan, Italy), 2 mM l-glutamine (Euroclone, Milan, Italy), 10% Fetal Bovine Serum (FBS; GE Healthcare, Milan, Italy) and 1% penicillin/streptomycin (Euroclone, Milan, Italy). After 5 days, the cells were split and re-plated as single cells onto Matrigel coated multi-well plates (Corning, New York, USA), 35 mm diameter well, containing a MEF (mouse embryonic fibroblast) feeder layer previously inactivated for 3 h with MEF medium supplemented with 10 μg/ml mytomicin C (Mipharm SPA, Milan, Italy). On day 7, the medium was changed to hESC medium consisting of DMEM-F12, 20% Knockout serum replacement (KOSR; Life Technologies, Monza, Italy), 100 μM non-essential amino acids (Millipore, Darmstadt, Germany), 10 ng/μl basic Fibroblast Growth Factor (bFGF; Life Technologies, Monza, Italy), 1% penicillin/streptomycin, 1% l-glutamine, 1% sodium pyruvate (Sigma Aldrich, Milan, Italy) and 0.2% β-mercaptoethanol (Millipore, Darmstadt, Germany). After 24–28 days, colonies with compact human embryonic cell-like morphology were observed and clonal lines were established by manually picking colonies. Human iPSC lines were maintained with hESC medium replaced daily. Expansion and splitting of the iPSC colonies was performed as previously described [28] using exclusive mechanical action or with collagenase IV (Life Technology, Carlsbad, USA).
2.2. Karyotyping (Q-banding) of iPSC
About 5 × 104 cells (iPSCs of patient 1 at passage number 4; iPSCs of patient 2 at passage number 5) per slide flask (Thermo Scientific/NUNC 75 mm × 25 mm) were cultured for 24 h in a 37 °C incubator with 5% CO2. Colcemid solution (at a final concentration of 0.1 μg ml− 1; Sigma Aldrich, Milan, Italy) was added, and cells were placed at 37 °C for an additional 30 min to ensure cell cycle arrest at metaphase. The cells were then treated with a hypotonic solution (KCl 1.33%; CarloErba Reagents SAS, Val de Reuil, France) for 10 min and fixed using methanol:acetic acid (3:1). The chromosomes were stained with quinacrine mustard (DBA Italia, Segrate, Italy) and the QFQ banding was analyzed with a Olympus BX-41 fluorescence microscope.
2.3. Pluripotency detection of iPSC lines derived from healthy subjects and NLSDM patients
2.3.1. Stem cell markers
Morphologically intact iPSC colonies were subcloned by mechanical picking, transferred to Matrigel coated slide flasks, and maintained for 1–2 days with hESC media. Cells were fixed in 4% PFA-PBS and immunostained using standard protocols [26]. The cells (patient 1: 14th passage; patient 2: 16th passage) were characterized for the expression of the following pluripotent markers: anti-OCT4 (Mouse mAb; Sigma Aldrich, Milan, Italy), anti-SSEA-4 (Mouse mAb; Life Technology, Carlsbad, USA) and anti-TRA-1-81 (Mouse mAb; Millipore, Darmstadt, Germany); the nuclei were counter-stained with DAPI (working DAPI solution was 200 ng/ml; Sigma Aldrich, Milan, Italy) for 2 min. The secondary antibody used was goat-anti mouse Alexa 488 (Life Technologies, Monza, Italy). The dilutions adopted were 1:200 for all primary antibodies and 1:1000 for the secondary antibody.
2.3.2. Real time qRT-PCR
For Real Time qRT-PCR analysis, cell lines collected from 35 mm well were extracted with Trizol reagent (Life Technologies, Monza, Italy), according to the manufacturer's instructions. After treatment of 1 μg of RNA with DNAse I (Roche, Basel, Switzerland), the sample was reverse-transcribed into first-strand cDNA using Advantage® RT-for-PCR Kit (Diatech Lab Line Srl, Jesi, Italy). Reverse transcription was performed with a mix of random primer and poly-dT using the following protocol: incubation for 1 h at 42 °C, inactivation for 5 min at 94 °C, and cooling to 4 °C. cDNA was quantified with NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA) and diluted to a concentration of 20 ng/μl. Real Time qRT-PCR with 40 ng of cDNA in a 20 μl-reaction volume was carried out in triplicate using Light Cycler 480 SYBR Green I Master (Roche, Basel, Switzerland) in Light Cycler® 480 System (Roche, Basel, Switzerland) with the following PCR program: 95 °C for 10 min, 45 cycles at 95 °C for 10 s, 62 °C for 10 s, and 72 °C for 20 s. Primer sequences were reported in Table 1. Relative quantification was calculated using the 2− ΔΔCt method after normalization with β-actin (ACTB) expression [29] and plotted to the expression level in the fibroblast cell lines [30].
Table 1.
List of primers and their sequences used in the Real Time qRT-PCR analysis.
| Primer | Forward | Reverse |
|---|---|---|
| ACTB | GCACTCTTCCAGCCTTCC | TGTCCACGTCACACTTCATG |
| ZFP42 | GGCCTTCACTCTAGTAGTGCTCA | CTCCAGGCAGTAGTGATCTGAGT |
| SOX2 | ACCAGCTCGCAGACCTACAT | CCTGCTGCGAGTAGGACAT |
| POU5F1 | TGGGCTCGAGAAGGATGTG | TGTGCATAGTCGCTGCTTGAT |
| TERT | CGTACAGGTTTCACGCATGTG | ATGACGCGCAGGAAAAATGT |
| LIN28A | GCAGAAGCGCAGATCAAAAG | CGGACATGAGGCTACCATATG |
| DPPA2 | CATGCTTACCCTGAACAACG | GAAGCCTTGCTCTCTTGGTC |
| NANOG | GTGACGCAGAAGGCCTCAG | AGGTTCCCAGTCGGGTTCA |
| TDGF1 | GGATACCTGGCCTTCAGAGA | CAGGCAGCAGGTTCTGTTTA |
2.4. Triglyceride quantification in NLSDM and control iPSCs
NLSDM and control iPSCs were seeded onto plates 100 mm diameter at a density of 1 × 106 cells/plate. The day after the cells were homogenized in 1 ml solution containing 5% TRITON X-100 (Sigma-Aldrich, Saint Louis, MO, USA), incubated at 80 °C for 5 min and centrifuged for 2 min. Cellular triglyceride content was quantified using Triglyceride Quantification Colorimetric Kit (Biovision, Milpitas, CA, USA), according to the instructions. The absorbance was measured at 570 nm with EnVision Multilabel Reader (PerkinElmer, Waltham, MA, USA).
2.5. Embryoid bodies (EBs) formation and in vitro differentiation of NLSDM-iPSC
After treatment with collagenase at 1 mg/ml for 1 h at 37 °C, 2 wells of iPSCs (80% confluent) were harvested and transferred onto 100 mm ultra-low attachment dishes/bacteriological Petri Dishes (LP Italiana SPA, Milan, Italy) in hESC medium without bFGF to promote EB formation. After growing in suspension for 5 days, the EBs were plated on a Geltrex-coated culture (Life Technologies, Monza, Italy) slide flask (75 mm × 25 mm) for up to 20 days in DMEM containing 20% FBS [31].
After fixation in 4% PFA-PBS for 30 min, cells were characterized by immunostaining for the expression of the lineage markers of endoderm (rabbit anti-FOXA2, 1:500; ABcam, Cambridge, MA), mesoderm (mouse Anti-Actin, α-Sma; 1:100; Life Technologies, Monza, Italy) and ectoderm (rabbit anti-TUJ1, 1:500; Life Technologies, Monza, Italy). The secondary antibodies (dilution 1:250) were AlexaFluor® 488 goat anti-mouse (Life Technologies, Monza, Italy), AlexaFluor® 488 donkey anti-rabbit (Life Technologies, Monza, Italy), and AlexaFluor® 594 goat anti-mouse (Life Technologies, Monza, Italy).
2.6. Oleic acid pulse-chase experiment
Control and NLSDM-iPSCs were seeded on slide flasks (75 mm × 25 mm) and grown in a hESC medium, then incubated for 18 h in culture medium containing 200 μM oleic acid (OA/BSA). The next day, in half of the slide flasks, the medium containing OA/BSA was removed and the cells were fixed with 3% paraformaldehyde and labeled with Nile Red (NR). NR solution was prepared in PBS (1:100 v/v) from a saturated solution (1 mg/ml) dimethyl sulfoxide. After three rinses with distilled water, the cells were incubated with NR solution for 20 min in the dark and mounted with Vectashield mounting medium (Vector Laboratories, Burligame, USA). iPSC colonies in the remaining slide flasks were washed three times with PBS, and then incubated for 24 or 72 h in the hESC medium containing 5% KOSR and fatty acid-free BSA (2% w/v) to enhance cellular lipolysis (chase). At the end of each chase (24 h and 72 h), the medium was removed, and the cells were stained with NR.
2.7. Quantification of LDs number and dimension
Fluorescence images were captured with a Leica MB5000B microscope. ‘WCIF ImageJ 1.35j’ software (developed by W. Rasband, NIH, Bethesda, MD, USA) was used to isolate components having the same wavelength and to quantify some parameters like area and number of selected units (lipid droplets, in this specific case) in control and patients iPSCs lines. The statistical analysis of quantitative data of LDs identified from iPSCs by image analysis of immunofluorescent experiments was made using SPSS v.19 package (SPSS, Chigaco, IL, USA). The values were compared using Student t-test. A P-value of < 0.05 was considered to be statistically significant.
3. Results
3.1. Derivation and characterization of NLSDM-iPSCs
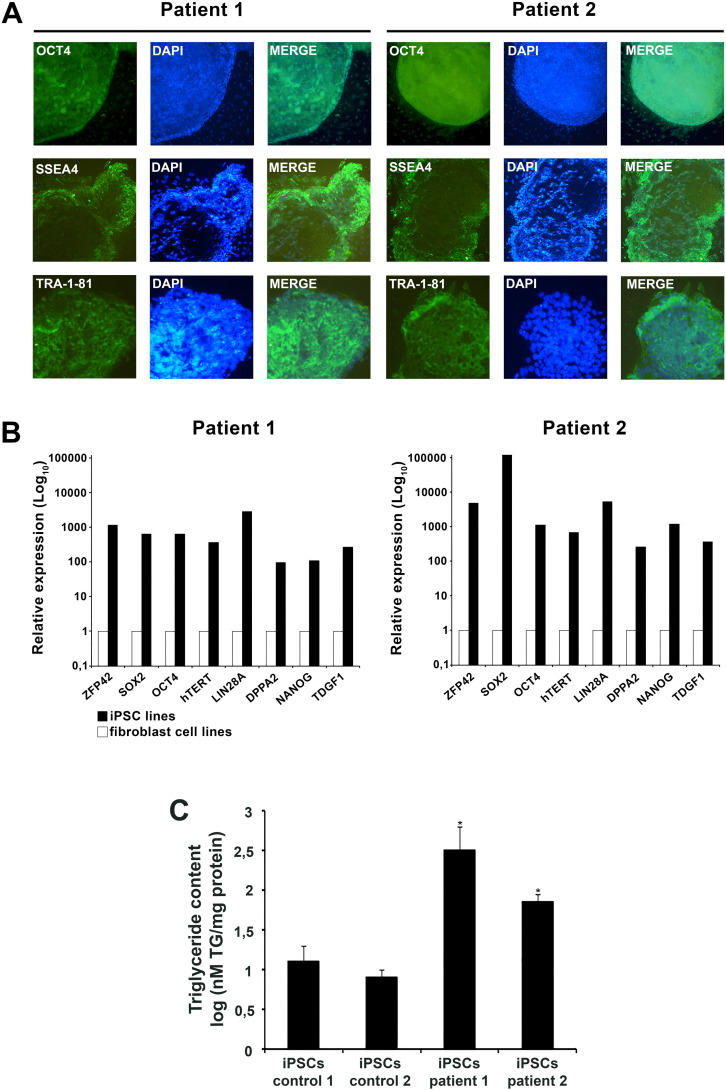
To generate iPSCs, dermal fibroblasts from two NLSDM patients and two control subjects were transduced with Sendai virus encoding for OCT4, SOX2, KLF4 and c-MYC. After about 4 weeks, the pluripotent properties of iPSCs of some NLSDM and control colonies were determined. Karyogram showed a normal 46,XY karyotype of controls and NLSDM-iPSCs (data not shown) and genomic sequencing analysis confirmed that NLSDM-iPSC lines still contained the disease-specific mutations of PNPLA2 gene (c.541delAC and c.662G > C) (Fig. 1C). To determine whether the fibroblasts were efficiently reprogrammed into iPSCs, the expression of pluripotent markers was evaluated by immunofluorescence and qRT-PCR analysis. Immunofluorescence assays versus OCT4, SSEA4 and TRA-1-81 confirmed the stemness gene expression profile (Fig. 2A). In agreement, qRT-PCR analysis indicated that NLSDM-iPSC lines, as compared to the patient fibroblast cell lines, show an increased expression of all 8 pluripotency-associated genes (ZFP42, SOX2, POU5F1 alias OCT4, hTERT, LIN28A, DPPA2, NANOG and TDGF1) (Fig. 2B). The increased expression was ranging from about 100 times (DPPA2 gene) to 100,000 times (SOX2 gene). Finally, the intracellular TG content from patients and controls iPSCs was determined in order to evaluate whether the NLSDM-iPSCs maintained the defect of neutral lipid metabolism. As expected, the TG content of NLSDM-iPSCs was significant higher than that of control iPSCs (Fig. 2C).
Fig. 2.
Characterization and differentiation of patient-derived NLSDM-iPSC.
(A) Immunostaining with primary antibodies anti-OCT4, anti-SSEA-4 and anti-TRA-1-81 revealed that NLSDM-iPSCs express markers of pluripotency, including OCT4 (red), SSEA4 (green), and TRA-1-81 (green). Nuclei were stained with DAPI. Magnification: 10 ×.
(B) Real-time quantitative RT-PCR assays for eight pluripotency-associate genes in two patient-derived iPSCs compared with fibroblast cell lines (basal level reported as 1). Data represent the mean of three independent experiments, performed in triplicate. PCR reactions were normalized against an internal control (ACTB).
(C) Quantification of TG content in iPSCs derived from 2 controls and 2 NLSDM fibroblast cell lines. The graph represents TG content obtained from three independent experiments. The analysis was performed using Student t-test. Thick bar: mean value; error bar: SD. P-values of < 0.05 and of < 0.01 were considered to be significant and indicated with “*”and “**”, respectively.
3.2. Generation of EBs from NLSDM-iPSCs
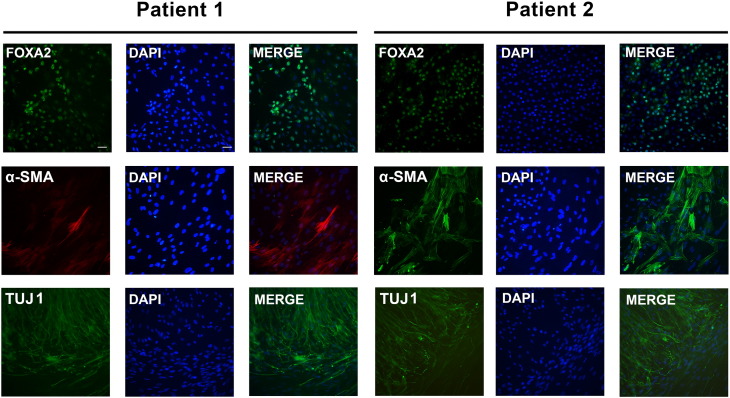
To evaluate the in vitro differentiation of NLSDM-iPSC lines, the formation of Embryoid Bodies (EBs) was promoted. After replating onto Geltrex-coated culture, the EBs spontaneously differentiated into cells with varied morphology belonging to the three germ layers. Immunofluorescence staining showed that the differentiated NLSDM-iPSCs expressed forkhead box protein A2 (FOXA2, endoderm), α-smooth muscle actin (α-SMA, mesoderm), and βIII-tubulin (TUJ1, ectoderm) (Fig. 3).
Fig. 3.
NLSDM-iPSCs differentiation into all three embryonic germ layers in vitro.
Immunostaining using primary antibodies anti-FOXA2, anti-α-SMA and anti-TUJ1 showed that markers for the three germ layers, endoderm-FOXA2, mesoderm-α-SMA and ectoderm-TUJ1, were expressed in differentiated NLSDM-iPSCs. Nuclei were stained with DAPI. Magnification: 20 ×.
3.3. Evaluation of NLSDM pathophysiological features in disease-specific iPSCs
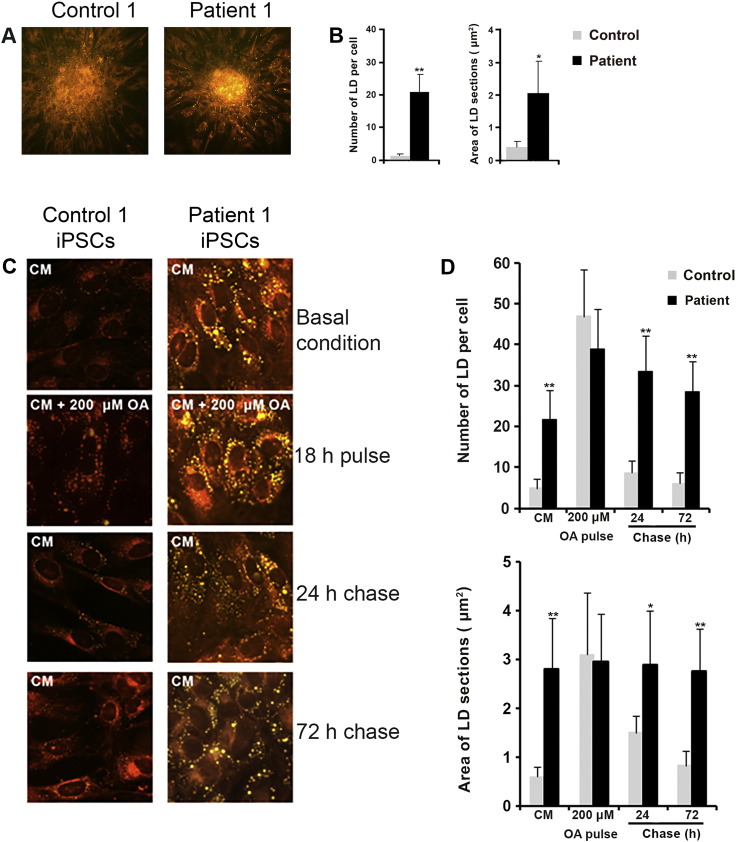
To assess whether the NLSDM-iPSCs expressed the pathological characteristics of NLSDM, we compared the retention of TAGs in control and NLSDM-iPSCs. After 3 days in culture, cells were stained with NR and the LD number and dimensions were analyzed. Compared to control cells, the NLSDM-iPSCs had 20 times more LDs and almost 5-fold larger LDs (Fig. 4A and B).
Fig. 4.
Evaluation of NLSDM pathophysiological characteristic in the disease-specific iPSCs.
Nile Red stained microphotographs of control and NLSDM-iPSCs (A; magnification: 20 ×) revealed more LDs (yellow) in the NLSDM cells.
(B) Analysis of LD number and dimensions observed in NLSDM-iPSCs, compared to control iPSCs (n = 3; 100 cells were analyzed for each experiment) was carried out by Student's t-test. Thick bar: mean value; error bar: SD. P-values of < 0.05 and of < 0.01 were considered to be significant and indicated with “*” and “**”, respectively.
(C) Oleic acid pulse-chase experiments on control and NLSDM-iPSCs. Culture medium (CM: DMEM-F12 containing 20% KOSR, 100 μM non-essential amino acids, 10 ng/ul bFGF, 1% penicillin/streptomycin, 1% l-glutamine, 1% sodium pyruvate and 0.2% β-mercaptoethanol) was supplemented with OA (200 μM) and, after 18 h, iPSCs were washed and chased with fresh medium containing 5% KOSR and fatty acid free BSA (2% w/v) for 24 and 72 h. Cells were stained for neutral lipids with NR. Lipid droplets are in yellow. Magnification: 40 ×.
(D) Analysis of LD number and dimension observed in NLSDM-iPSCs, compared to control iPSCs, during the oleic acid pulse-chase experiments. In two independent experiments 100 cells in each of 3 replicates were analyzed. Thick bar: mean value; error bar: SD. P-values of < 0.05 and of < 0.01 were considered to be significant and indicated with “*”and “**”, respectively.
An OA pulse-chase experiment was performed to confirm that lipase activity is impaired in NLSDM-iPSCs compared to control cells (Fig. 4C and D). When iPSCs were incubated for 18 h with 200 μM oleic acid (OA pulse) TAGs accumulated in cytoplasmic LDs of patients and control iPSCs, as demonstrated by increased LDs number and size (Pulse: control-LDnumber = 45, iPSCs-LDnumber = 38, control-LDsize = 3 μm2, iPSCs-LDsize = 2.9 μm2). In patients' cells, loss of lipid was almost completely abrogated during the chase phase (medium with 5% KOSR and fatty acid-free BSA-2% w/v), so that after 72 h, the patients' cells showed little reduction in the number of LDs (Chase-72 h: iPSCs-LDnumber = 28) and an insignificant reduction of LDs area (Chase-72 h: iPSCs-LDsize = 2.7 μm2). In contrast, control iPSCs showed an almost complete loss of LDs during the chase (Chase-72 h: control-LDnumber = 6, control-LDsize = 0,7 μm2) (Fig. 4C and D).
These results demonstrated that iPSCs recapitulate NLSDM specific-lipid metabolism defects.
4. Discussion
In this study, we report the use of Sendai-based vectors to generate iPSCs from two lines of NLSDM fibroblasts. The NLSDM-iPSC lines possess stem cells properties and are able to differentiate into three germ layers. In addition, these iPSCs show an abnormal accumulation of neutral lipids, a typical feature of neutral lipid storage disorder, caused by partial or total abrogation of the lipolytic function of ATGL. Finally, using an OA pulse-chase experiment, we demonstrate a strong impairment in TAG lipase activity of NLSDM-iPSCs compared to control iPSCs, thus confirming the validity of NLSDM-iPSCs as biochemical disease model.
Although the generation of iPSCs as cellular tool was reported for some disorders [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], this is the first time that NLSDM-iPSCs have been derived from fibroblasts of NLSDM patients. Cellular studies are needed to understand the pathophysiological mechanism of NLSDM and to develop personalized treatments that might increase fat mobilization and improve organ function. Unfortunately, it is difficult to obtain myocytes and to expand these cells in vitro. Moreover, it is even more difficult to obtain cardiomyocytes from patients. Our data shows that while maintaining NLSDM features, NLSDM-iPSCs can differentiate into germ layers, including mesoderm cells, from which skeletal and cardiac cells derive. Differentiation of NLSDM-iPS mesodermal cells into skeletal myocytes and cardiomyocytes would present a model for investigating cellular and molecular mechanisms that underlie the myopathy and cardiomyopathy of NLSDM patients. In addition, such cellular models would be useful in testing possible therapeutic agents.
Lipid-containing vacuoles in leukocytes (Jordans' anomaly) of NLSDM patients are considered to be a hallmark of the disease [9]. The diagnosis is then confirmed by the presence of abundant triacylglycerol droplets in non-adipose cells [5], [6], [7], [8], [11], [12], [14] and finally by molecular analysis of PNPLA2 gene mutations. However, clinical heterogeneity in NLSDM patients has been reported, and on occasion, abnormal storage of lipids in cytoplasmic LDs is detected before other clinical findings have occurred [32]. In contrast, in some patients the abnormal storage of lipid in cytoplasmic LDs is scarcely detected [4]. Hence, besides the static morphological evaluation of cytoplasmic LD number and dimension, it would be useful to perform a functional-dynamic assay to test cellular TAG lipase activity. As previously reported [3], [33], the ability to hydrolyze the TAGs stored in NLSDM fibroblasts can be tested using OA in a pulse-chase assay. Our previous data showed that NLSDM fibroblasts were not able to mobilize LD lipids during the chase phase [3]. In agreement, present data show that the OA pulse-chase experiment might be a useful tool to assess impaired lipase activity in iPSCs obtained from NLSDM patients.
5. Conclusion
We report the derivation of iPSCs from fibroblasts of two NLSDM patients carrying different ATGL mutations. These iPSCs exhibited defects in neutral lipid metabolism similar to those of NLSDM fibroblasts. NLSDM-iPSCs should be able to undergo directed differentiation into cardiomyocytes and could be used to investigate the cellular and molecular changes that lead to cardiomyopathy in NLSDM patients and to screen potential therapeutic compounds.
Conflict of interest
The authors declare no potential conflict of interest.
Acknowledgments
This work was supported by Telethon [grant number GGP14066]; National Institutes of Health [grant number DK56598]; and Regione Liguria [Fondi FAS 2013]. We thank the Galliera Genetic Bank (Dr Chiara Baldo) Network of Telethon Genetic Biobanks (project no. GTB12001).
References
- 1.Fischer J., Lefèvre C., Morava E., Mussini J.M., Laforêt P., Negre-Salvayre A., Lathrop M., Salvayre R. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat. Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 2.Campagna F., Nanni L., Quagliarini F., Pennisi E., Michailidis C., Pierelli F., Bruno C., Casali C., DiMauro S., Arca M. Novel mutations in the adipose triglyceride lipase gene causing neutral lipid storage disease with myopathy. Biochem. Biophys. Res. Commun. 2008;377:843–846. doi: 10.1016/j.bbrc.2008.10.081. [DOI] [PubMed] [Google Scholar]
- 3.Tavian D., Missaglia S., Redaelli C., Pennisi E.M., Invernici G., Wessalowski R., Maiwald R., Arca M., Coleman R.A. Contribution of novel ATGL missense mutations to the clinical phenotype of NLSD-M: a strikingly low amount of lipase activity may preserve cardiac function. Hum. Mol. Genet. 2012;21:5318–5328. doi: 10.1093/hmg/dds388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennisi E.M., Missaglia S., DiMauro S., Bernardi C., Akman H.O., Tavian D. A myopathy with unusual features caused by PNPLA2 gene mutations. Muscle Nerve. 2014;51:609–613. doi: 10.1002/mus.24477. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko K., Kuroda H., Izumi R., Tateyama M., Kato M., Sugimura K., Sakata Y., Ikeda Y., Hirano K., Aoki M. A novel mutation in PNPLA2 causes neutral lipid storage disease with myopathy and triglyceride deposit cardiomyovasculopathy: a case report and literature review. Neuromuscul. Disord. 2014;24:634–641. doi: 10.1016/j.nmd.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Pasanisi M.B., Missaglia S., Cassandrini D., Salerno F., Farina S., Andreini D., Agosotoni P., Morandi L., Mora M., Tavian D. Severe cardiomyopathy in a young patient with complete deficiency of adipose triglyceride lipase due to a novel mutation in PNPLA2 gene. Int. J. Cardiol. 2016;207:165–167. doi: 10.1016/j.ijcard.2016.01.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano K., Ikeda Y., Zaima N., Sakata Y., Matsumiya G. Triglyceride deposit cardiomyovasculopathy. N. Engl. J. Med. 2008;359:2396–2398. doi: 10.1056/NEJMc0805305. [DOI] [PubMed] [Google Scholar]
- 8.Missaglia S., Tasca E., Angelini C., Moro L., Tavian D. Novel missense mutations in PNPLA2 causing late onset and clinical heterogeneity of neutral lipid storage disease with myopathy in three siblings. Mol. Genet. Metab. 2015;115:110–117. doi: 10.1016/j.ymgme.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Tavian D., Colombo R R. Improved cytochemical method for detecting Jordans' bodies in neutral-lipid storage diseases. J. Clin. Pathol. 2007;60:956–958. doi: 10.1136/jcp.2006.044917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tavian D., Missaglia S., DiMauro S., Bruno C., Pegoraro E., Cenacchi G., Coviello D., Angelini C. A late-onset case of neutral lipid storage disease with myopathy, dropped head syndrome, and peripheral nerve involvement. J. Genet. Syndr. Gene Ther. 2013;4:198. [Google Scholar]
- 11.Reilich P., Horvath R., Krause S., Schramm N., Turnbull D.M., Trenell M., Hollingsworth K.G., Gorman G.S., Hans V.H., Reimann J., MacMillan A., Turner L., Schollen A., Witte G., Czermin B., Holinski-Feder E., Walter M.C., Schoser B., Lochmüller H. The phenotypic spectrum of neutral lipid storage myopathy due to mutations in the PNPLA2 gene. J. Neurol. 2011;258:1987–1997. doi: 10.1007/s00415-011-6055-4. [DOI] [PubMed] [Google Scholar]
- 12.Hirano K., Tanaka T., Ikeda Y., Yamaguchi S., Zaima N., Kobayashi K., Suzuki A., Sakata Y., Sakata Y., Kobayashi K., Toda T., Fukushima N., Ishibashi-Ueda H., Tavian D., Nagasaka H., Hui S.P., Chiba H., Sawa Y., Hori M. Genetic mutations in the adipose triglyceride lipase and myocardial overexpression of peroxisome proliferated activated receptor- in patients with triglyceride deposit cardiomyovasculopathy. Biochem. Biophys. Res. Commun. 2014;443:574–579. doi: 10.1016/j.bbrc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Fiorillo C., Brisca G., Cassandrini D., Scapolan S., Astrea G., Valle M., Scuderi F., Trucco F., Natali A., Magnano G., Gazzerro E., Minetti C., Arca M., Santorelli S.F., Bruno C. Subclinical myopathy in a child with neutral lipid storage disease and mutations in the PNPLA2 gene. Biochem. Biophys. Res. Commun. 2013;430:241–244. doi: 10.1016/j.bbrc.2012.10.127. [DOI] [PubMed] [Google Scholar]
- 14.Higashi M., Hirano K., Kobayashi K., Ikeda Y., Issiki A., Otsuka T., Suzuki A., Yamaguchi S., Zaima N., Hamada S., Hanada H., Suzuki C., Nakamura H., Nagasaka H., Miyata T., Miyamoto Y., Kobayashi K., Naito H., Toda T. Distinct cardiac phenotype between two homozygotes born in a village with accumulation of a genetic deficiency of adipose triglyceride lipase. Int. J. Cardiol. 2015;192:30–32. doi: 10.1016/j.ijcard.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Missaglia S., Maggi L., Mora M., Gibertini S., Blasevich F., Agostoni P., Moro L., Cassandrini D., Santorelli F.M., Gerevini S., Tavian D. Late onset of neutral lipid storage disease due to novel PNPLA2 mutations totally abrogating lipase activity in a patient with myopathy and slight cardiac involvement. Neuromuscul. Disord. 2017 doi: 10.1016/j.nmd.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Saha K., Jaenisch R. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell. 2009;5:584–595. doi: 10.1016/j.stem.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L., Geva L., Roberts A.E., Ma Q., Ding J., Chen J., Wang D.Z., Li K., Wang J., Wanders R.J., Kulik W., Vaz F.M., Laflamme M.A., Murry C.E., Chien K.R., Kelley R.I., Church G.M., Parker K.K., Pu W.T. Modeling the mitochondrial cardiomyopathy of Barth syndrome with iPSC and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin Y., Gao W.Q. Concise review: patient-derived stem cell research for monogenic disorders. Stem Cells. 2015;34:44–45. doi: 10.1002/stem.2112. [DOI] [PubMed] [Google Scholar]
- 20.Haile Y., Nakhaei-Nejad M., Boakye P.A., Baker G., Smith P.A., Murray A.G., Giuliani F., Jahroudi N. Reprogramming of HUVECs into induced pluripotent stem cells (HiPSCs), generation and characterization of HiPSC-derived neurons and astrocytes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spitalieri P., Talarico V.R., Murdocca M., Novelli G., Sangiuolo F. Human induced pluripotent stem cells for monogenic disease modelling and therapy. World J. Stem Cells. 2016;8:118–135. doi: 10.4252/wjsc.v8.i4.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H.P., Chuang C.Y., Kuo H.C. Induced pluripotent stem cell technology for disease modeling and drug screening with emphasis on lysosomal storage diseases. Stem Cell Res. Ther. 2012;3:34. doi: 10.1186/scrt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper O., Seo H., Andrabi S., Guardia-Laguarta C., Graziotto J., Sundberg M., McLean J.R., Carrillo-Reid L., Xie Z., Osborn T., Hargus G., Deleidi M., Lawson T., Bogetofte H., Perez-Torres E., Clark L., Moskowitz C., Mazzulli J., Chen L., Volpicelli-Daley L., Romero N., Jiang H., Uitti R.J., Huang Z., Opala G., Scarffe L.A., Dawson V.L., Klein C., Feng J., Ross O.A., Trojanowski J.Q., Lee V.M., Marder K., Surmeier D.J., Wszolek Z.K., Przedborski S., Krainc D., Dawson T.M., Isacson O. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura K., Hirano K., Wu S.M. iPS cell modeling of cardiometabolic diseases. J. Cardiovasc. Transl. Res. 2013;6:46–53. doi: 10.1007/s12265-012-9413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiscornia G., Vivas E.L., Matalonga L. Neuronopathic Gaucher's disease: induced pluripotent stem cells for disease modelling and testing chaperone activity of small compounds. Hum. Mol. Genet. 2013;22:633–645. doi: 10.1093/hmg/dds471. [DOI] [PubMed] [Google Scholar]
- 27.Maetzel D., Sarkar S., Wang H., Abi-Mosleh L., Xu P., Cheng A.W., Gao Q., Mitalipova M., Jaenisch R. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick type C patient-specific iPS cells. Stem Cell Reports. 2014;2:866–880. doi: 10.1016/j.stemcr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerou P.H., Yabuuchi A., Huo H., Takeuchi A., Shea J., Cimini T., Ince T.A., Ginsburg E., Racowsky C., Daley G.Q. Human embryonic stem cell derivation from poor-quality embryos. Nat. Biotechnol. 2008;26:212–214. doi: 10.1038/nbt1378. [DOI] [PubMed] [Google Scholar]
- 29.Degiorgio D., Corsetto P.A., Rizzo A.M., Colombo C., Seia M., Costantino L., Montorfano G., Tomaiuolo R., Bordo D., Sansanelli S., Li M., Tavian D., Rastaldi M.P., Coviello D.A. Two ABCB4 point mutations of strategic NBD-motifs do not prevent protein targeting to the plasma membrane but promote MDR3 dysfunction. Eur. J. Hum. Genet. 2014;22:633–639. doi: 10.1038/ejhg.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 31.Ricciardi S., Ungaro F., Hambrock M., Rademacher N., Stefanelli G., Brambilla D., Sessa A., Magagnotti C., Bachi A., Giarda E., Verpelli C., Kilstrup-Nielsen C., Sala C., Kalscheuer V.M., Broccoli V. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat. Cell Biol. 2012;14:911–923. doi: 10.1038/ncb2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akman H.O., Davidzon G., Tanji K., Macdermott E.J., Larsen L., Davidson M.M., Haller R.G., Szczepaniak L.S., Lehman T.J., Hirano M., DiMauro S. Neutral lipid storage disease with subclinical myopathy due to a retrotransposal insertion in the PNPLA2 gene. Neuromuscul. Disord. 2010;10:397–402. doi: 10.1016/j.nmd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Igal R.A., Coleman R.A. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. J. Biol. Chem. 1996;271:16644–16651. doi: 10.1074/jbc.271.28.16644. [DOI] [PubMed] [Google Scholar]