Abstract
RNA polymerase II, and specifically the C-terminal domain (CTD) of its largest subunit, has been demonstrated to play important roles in capping, splicing, and 3′ processing of mRNA precursors. But how the CTD functions in these reactions, especially splicing, is not well understood. To address some of the basic questions concerning CTD function in splicing, we constructed and purified two fusion proteins, a protein in which the CTD is positioned at the C terminus of the splicing factor ASF/SF2 (ASF-CTD) and an RS domain deletion mutant protein (ASFΔRS-CTD). Significantly, compared to ASF/SF2, ASF-CTD increased the reaction rate during the early stages of splicing, detected as a 20- to 60-min decrease in splicing lag time depending on the pre-mRNA substrate. The increased splicing rate correlated with enhanced production of prespliceosomal complex A and the early spliceosomal complex B but, interestingly, not the very early ATP-independent complex E. Additional assays indicate that the RS domain and CTD perform distinct functions, as exemplified by our identification of an activity that cooperates only with the CTD. Dephosphorylated ASFΔRS-CTD and a glutathione S-transferase-CTD fusion protein were both inactive, suggesting that an RNA-targeting domain and CTD phosphorylation were necessary. Our results provide new insights into the mechanism by which the CTD functions in splicing.
Synthesis of mature mRNAs in the nuclei of eukaryotic cells involves a series of pre-mRNA processing events, including the addition of a cap structure to the 5′ end, removal of intronic sequences (splicing), and 3′ cleavage and polyadenylation. Recently, it has become increasingly evident that all three mRNA-processing steps are integrated with transcription in vivo, presumably to facilitate accurate and efficient processing of RNA polymerase II (RNAP II) transcripts, and that the RNAP II C-terminal domain (CTD) is important for all three processing reactions (2, 24, 36, 48). The CTD is conserved among eukaryotes and consists of multiple heptapeptide repeats with the consensus sequence YSPTSPS (9). The number of repeats varies from 26 or 27 in the yeast Saccharomyces cerevisiae to 52 in mammals. The heptapeptide consensus contains five potential phosphoacceptor amino acids, but experimental evidence suggests that the serines (or threonines) at positions 2 and 5 are the predominant sites of phosphorylation (63). Two forms of RNAP II can be distinguished based on the phosphorylation status of CTD, a hypophosphorylated IIA form, which preferentially enters the preinitiation complex at the promoter, and a hyperphosphorylated IIO form, which is associated with elongation complexes (12). More recently, different phosphorylated forms of RNAP II have been observed on genes depending on the location of the polymerase along the gene (7, 33), and specific kinases have been implicated in directing serine-specific phosphorylation of the CTD during the transcription cycle (16, 49, 56, 65).
Splicing of most mRNAs occurs in a large macromolecular complex composed of snRNPs (U1, U2, U4, U5, and U6) and non-snRNP proteins, including members of the SR protein family (23, 29, 37). In structure, SR proteins contain one or two N-terminal RNA binding domains (RBD) and a C-terminal domain enriched in arginine and serine residues (RS domain), which is extensively phosphorylated in vivo. The RBD has been demonstrated to bind RNA in a sequence-specific manner, a characteristic that allows for the interaction of SR proteins with elements such as exonic or intronic splicing enhancers. Functionally, SR proteins are believed to perform an essential role early in spliceosome assembly by stabilizing the first interactions between snRNPs and splice sites, as well as by mediating protein-protein contacts through the formation of bridges formed between 5′ and 3′ splice sites (11, 32, 52, 59). The result is the formation of a complex composed of U1 snRNP bound to the 5′ splice site and the splicing factor U2AF bound to the 3′ splice site region (E complex). In the presence of ATP, splicing proceeds as E complexes are converted to A, B, and C complexes in a well-characterized kinetic pathway that requires U2, U4, U5, and U6 snRNPs and other factors (29, 35, 42, 51)
The above model of spliceosome assembly was the result of analyses of splicing in isolation from transcription, but more-recent studies suggest an important role for the RNAP IIO CTD in spliceosome assembly. Cytological studies have suggested that splicing in vivo can occur cotranscriptionally and that essential splicing factors are localized at sites of active transcription (1, 3). RNAP IIO also colocalizes with SR proteins in large nuclear domains called “speckles” (4), and splicing of transcripts produced by RNAP II with a shortened CTD is impaired (18, 38). In biochemical studies, RNAP IIO can physically interact with snRNPs and SR-like proteins (6, 31, 41, 57, 61), and we have shown that purified RNAP IIO can enhance in vitro splicing of several pre-mRNAs at an early (E or A complex) step in spliceosome assembly, while IIA in fact has a repressive effect (26). RNAP II CTD-targeted antibodies and CTD peptides have also been shown to inhibit in vitro splicing (61). A recombinant CTD fusion protein (glutathione S-transferase [GST]-CTD) has been shown to enhance in vitro splicing of pre-mRNAs containing exons defined by splice sites (62).
Because RNAP IIO is the elongating form of the polymerase and functions early to enhance spliceosome assembly, we previously proposed a model where the hyperphosphorylated CTD of RNAP IIO and its associated proteins facilitate the binding of U1 and/or U2 snRNPs to the 5′ splice site and/or branch site, respectively (26). This model is supported by in vitro transcription experiments which showed that U1 and U2 snRNPs can be recruited to elongating RNAP II by interactions involving the CTD kinase P-TEFb (20). However, despite this considerable evidence linking transcription, and specifically the CTD, to splicing, little is known about how the CTD actually functions in this process.
We have continued our examination into the relationship between the CTD and the splicing machinery by constructing chimeric proteins in which the CTD is fused to the SR protein ASF/SF2 or to an RS domain-deleted derivative of ASF/SF2. These two fusion proteins, dubbed ASF-CTD and ASFΔRS-CTD, were designed to test first whether recruitment of the CTD to the pre-mRNA, perhaps normally performed by the body of RNAP II, might be important for CTD function and second whether the CTD can functionally substitute for the RS domain. We show that, like ASF/SF2, ASF-CTD complements splicing in SR protein-deficient S100 extract. However, ASF-CTD markedly increased the reaction rate at or before the first catalytic step. Although ASFΔRS and ASFΔRS-CTD were generally much less active in S100, ASF-CTD and ASFΔRS-CTD, but not ASF/SF2 or ASFΔRS, increased the rate of splicing similarly in nuclear extracts. As with intact RNAP II, CTD phosphorylation was necessary for the observed splicing enhancement, and GST-CTD was inactive. We also provide evidence for the existence of different CTD cofactor requirements depending on the pre-mRNA substrate. These results are consistent with a model in which the phosphorylated CTD facilitates an early step(s) of spliceosome assembly but highlight the possibility that it plays a more sophisticated role in splicing regulation than previously thought.
MATERIALS AND METHODS
Oligonucleotides.
Synthetic oligonucleotides used for PCR amplification of ASF/SF2 and ASFΔRS include a common 5′ primer, NheI-ASF, CTGACTGCTAGCATGTCGGGAGGTGGTGTGATTCG, and 3′ primers BamH1-ASF, CTGACTGGATCCTGTACGAGAGCGAGATCTGCTATG, and BamH1-ASFΔRS, CTGACTGGATCCGGGCCCATCAACTTTAACCCGG.
Plasmids.
To construct pFastBac1-6HASF and pFastBac1-6HASFΔRS, EcoRI-HindIII fragments of pDS-6HASF and pDS-6HASFΔRS, which contained the coding sequence for a six-histidine tag followed by the coding sequences for ASF/SF2 and ASFΔRS, respectively (67), were cloned into the multiple cloning site of pFasBac1 (Invitrogen Life Technologies). To construct pFastBac1-6HASF-CTD and pFastBac1-6HASFΔRS-CTD, we first constructed pRSET-CTD by subcloning a BamH1-EcoRI CTD fragment from pGCTD (47), containing the murine RNAPII CTD coding sequence with a stop codon, into pRSETB (Invitrogen Life Technologies). PCR products NheI-ASF-BamHI and NheI-ASFΔRS-BamHI were generated with pDS-6HASF as the template for PCR using the NheI-ASF and BamHI-ASF primer pair or the NheI-ASF and BamHI-ASFΔRS primer pair. The NheI-ASF-BamHI and NheI-ASFΔRS-BamHI PCR products were subsequently cloned into pRSET-CTD to produce pRSET-6HASF-CTD and pRSET-6HASFΔRS-CTD, respectively. To construct pFastBac1-6HASF-CTD and pFastBac1-6HASFΔRS-CTD, ApaI-HindIII restriction fragments of pRSET-6HASF-CTD and pRSET-6HASFΔRS-CTD were cloned into pFastBac1-6HASF to produce pFastBac1-6HASF-CTD and pFastBac1-6HASFΔRS-CTD, respectively.
Recombinant proteins.
The constructs containing six-histidine-tagged ASF (pFastBac1-6HASF), ASFΔRS (pFastBac1-6HASFΔRS), ASF-CTD (pFastBac1-6HASF-CTD), and ASFΔRS-CTD (pFastBac1-6HASFΔRS-CTD) were used to construct recombinant baculoviruses with the Bac-to-Bac baculovirus expression system (Invitrogen Life Technologies). Proteins were expressed in baculovirus-infected Hi-5 cells for 48 h. Infected-cell pellets were solubilized by sonication in 6 M guanidine HCl (pH 8), and cellular debris was removed by centrifugation and filtration (0.22-μm-pore-size filter). The lysate was passed over a Ni2+-agarose column, washed in 8 M urea (pH 8) containing 10 mM imidazole, and eluted in 8 M urea (pH 8) plus 200 mM imidazole. Proteins were dialyzed at 4°C in buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT]) containing 8 M urea and were renatured during dialysis by stepwise dilution of urea with buffer D, followed by a final dialysis in pure buffer D. Precipitates were removed by centrifugation. Protein purity and concentration were assessed by Coomassie blue and silver staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Immunoblot analysis and phosphatase treatment.
ASFΔRS-CTD was dephosphorylated with calf intestinal phosphatase (CIP; New England Biolabs) at 30°C in a buffer containing 20 mM HEPES (pH 8), 50 mM KCl, 10% glycerol, 5 mM Mg2+, 0.25 mM DTT, and 0.1 mM EDTA. Approximately 10 U of CIP was required to fully dephosphorylate 1 μg of ASFΔRS-CTD after 3 h at 30°C. To prepare purified dephosphorylated ASFΔRS-CTD, 50 μg of ASFΔRS-CTD was treated with CIP and then denatured by dilution in a buffer containing 8 M guanidine HCl (pH 8). Dephosphorylated ASFΔRS-CTD was purified by Ni2+ chromatography and renatured as described for the phosphorylated protein except that final dialysis was in buffer D containing 2 M urea to increase solubility. Phosphorylated ASFΔRS-CTD was also dialyzed in buffer D containing 2 M urea as a control for functional studies involving the dephosphorylated protein. Immunoblot analysis was performed by separating phosphorylated and dephosphorylated proteins by SDS-8% polyacrylamide gel electrophoresis (PAGE), followed by immunoblotting with H14, H5, or 8WG16 antibodies (Covance). Unless specifically stated otherwise in the text, recombinant ASF-CTD and ASFΔRS-CTD proteins are referred to as ASF-CTDO and ASFΔRS-CTDO, respectively, and the recombinant ASFΔRS-CTD protein treated with CIP and repurified is referred to as ASFΔRS-CTDA.
In vitro splicing and spliceosome assembly.
32P-labeled pre-mRNA substrates were prepared as described previously (54). Nuclear extract and cytoplasmic S100 were prepared by the method of Dignam et al. (15). Fractionation of HeLa nuclear extract to obtain a 20 to 40% ammonium sulfate fraction of nuclear extract (NF20-40) was performed as described previously (53). In vitro splicing reactions were performed in either 12.5- or 25-μl reaction mixtures, which contained 12 to 16% nuclear extract or 20% S100 supplemented without or with purified proteins as described in the figure legends. The final concentrations of buffer components were 12 mM HEPES (pH 7.9), 40 to 60 mM KCl, 0.12 mM EDTA, 0.30 mM DTT, 12% glycerol, 3 mM MgCl2, 20 mM creatine phosphate (di-Tris), 1 mM ATP, 3% polyvinyl alcohol (PVA), and 0.5 U of RNasin (Promega), with the exception that, in experiments involving dephosphorylated ASFΔRS-CTD, urea was present at a final concentration of 0.16 mM. In addition, for ATP-dependent spliceosome assembly assays, PVA was omitted from the splicing reactions and heparin (0.8 mg/ml) was added prior to loading the samples on native agarose gels as described previously (13). For ATP-independent E complex assembly, PVA, ATP, creatine phosphate, and MgCl2 were omitted from splicing reactions and the samples were loaded on 1.5% native agarose gels (13). The adenovirus major late (AdML) pre-mRNA was utilized in these studies due to better resolution of E complexes. Experiments were repeated two to four times and were highly consistent with the representative data shown.
RESULTS
ASF/SF2-CTD fusion proteins expressed in baculovirus-infected insect cells are phosphorylated on Ser-2 and Ser-5 of the heptapeptide repeats.
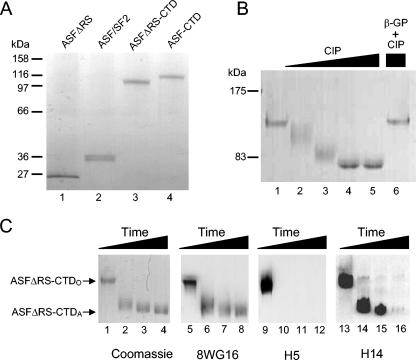
To study the functional properties of the CTD relevant to splicing, the CTD was expressed as a C-terminal fusion protein with ASF/SF2 and the RS domain deletion mutant protein, ASFΔRS (68). ASF/SF2, ASFΔRS, ASF-CTD, and ASFΔRS-CTD proteins were purified from baculovirus-infected insect cells, and aliquots were examined by SDS-PAGE (Fig. 1A). Although the expected molecular masses of ASF-CTD and ASFΔRS-CTD are 68 and 62 kDa, respectively, the apparent molecular masses observed by SDS-PAGE were approximately 116 and 110 kDa, respectively (Fig. 1A, lanes 3 and 4). Since CTD phosphorylation imparts a dramatic decrease in gel mobility (64), it was likely that the CTDs of these proteins were highly phosphorylated during expression. Treatment of ASFΔRS-CTD with CIP shifted the protein's gel mobility to approximately 80 kDa (Fig. 1B, compare lane 1 to 4 or 5) or resulted in intermediate-mobility forms at midrange concentrations of CIP (Fig. 1B, lanes 2 and 3).
FIG. 1.
Characterization of histidine-tagged ASF-CTD fusion proteins purified from baculovirus-infected insect cells. (A) Silver-stained gradient (4 to 20%) SDS-polyacrylamide gel containing 40 ng of purified ASFΔRS (lane 1), ASF (lane 2), ASFΔRS-CTD (lane 3), and ASF-CTD (lane 4). (B) Coomassie-stained SDS-8% polyacrylamide gel containing purified ASFΔRS-CTD treated with 0.0 (lane 1), 0.25 (lane 2), 1.0 (lane 3), and 5.0 U (lanes 4 to 6) of CIP for 3 h at 30°C. Phosphatase activity was inhibited by addition of 50 mM β-glycerophosphate (lane 6). (C) Time course (0, 1, 2, and 3 h) of ASFΔRS-CTD dephosphorylation analyzed by Coomassie staining and immunoblotting with 8WG16, H5, and H14 antibodies.
Since the CTD is predominantly phosphorylated at serines 2 and 5 during transcription in vivo, the CTD of ASFΔRS-CTD was tested for phosphorylation at these positions by Western blotting using H5 and H14 antibodies, which are specific for heptads phosphorylated at Ser-2 or Ser-5, respectively. A Coomassie blue stain of ASFΔRS-CTD during a CIP time course is shown in Fig. 1C. The same samples were also immunoblotted with CTD-specific antibody 8WG16, which recognizes unphosphorylated epitopes in the CTD (Fig. 1C, lanes 5 to 8), H5 (Fig. 1C, lanes 9 to 12), and H14 (Fig. 1C, lanes 13 to 16). Ser-2 phosphorylation was strongly detected at the 0-min time point of CIP incubation (Fig. 1C, lane 9) but was undetectable at later times (Fig. 1C, lanes 10 to 12). Ser-5 phosphorylation, in contrast, was detected not only at the 0-min time point (Fig. 1C, lane 13) but also at the 1- and 2-h time points and was detected in the intermediate- and even high-mobility forms of ASFΔRS-CTD (Fig. 1C, lanes 14 and 15). These results indicate that H5 reactivity can be completely lost before H14 reactivity is affected. The ability of H14, but not H5, to detect intermediate levels of CTD phosphorylation has also been observed with authentic RNAP II (44) and, together with the observation of discrete bands for the untreated protein and the dephosphorylated isoform, supports the view that these forms are analogous to the IIO and IIA isoforms of RNAP II. The CTDs of our recombinant proteins are designated below either CTDO or CTDA to differentiate the native phosphorylated and the CIP-dephosphorylated recombinant proteins, respectively.
Fusing the CTD to ASF/SF2 increases splicing rate in S100.
We showed previously that RNAP IIO can stimulate splicing of several pre-mRNAs in cell extracts but that, when GST-CTD was added to splicing reactions under similar conditions, no enhancement was observed (26). This result was somewhat unexpected since the CTD is sufficient both to activate 3′ cleavage in vitro (25) and to associate with a number of splicing factors (10, 31, 40). We speculated that GST-CTD may not enter spliceosomes formed in vitro, because it is unable to interact in the appropriate way with the pre-mRNA. Given that SR proteins both interact with the pre-mRNA and function, as appears to be the case with RNAP IIO, at a very early stage (21), it seemed reasonable to hypothesize that fusing the CTD with an SR protein may allow for the participation of the CTD in splicing. Alternatively, it could be that other regions in the multisubunit RNAP II are necessary for its function in splicing.
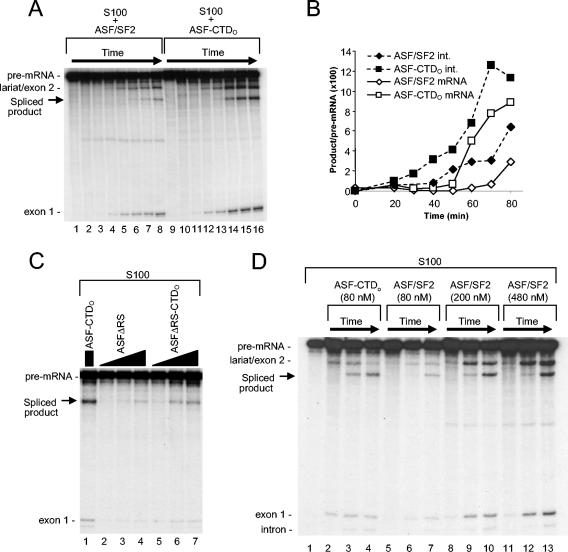
We first examined splicing of a β-globin pre-mRNA substrate in S100 extract complemented with either ASF/SF2 or ASF-CTDO. Since S100 lacks SR proteins but contains all other essential splicing factors, splicing does not occur in S100 unless supplemented with exogenous SR proteins such as ASF/SF2. To examine the effect of the CTD on ASF/SF2 activity, a splicing time course was performed with the β-globin pre-mRNA, S100, and either ASF/SF2 or ASF-CTDO (Fig. 2A). Both ASF/SF2 and ASF-CTDO at 100 nM concentrations activated splicing. Most interestingly, the reactions with ASF-CTDO showed an increase in the rate of splicing, as shown by the formation of the first-step intermediates or final products. Quantitation (Fig. 2B) revealed that reactions with ASF-CTDO displayed a reduction in splicing lag time of approximately 20 min (from 40 to 20 min) compared to reactions with ASF/SF2. Relative to ASF-CTDO, both ASFΔRS and ASFΔRS-CTDO stimulated splicing much less efficiently (Fig. 2C, compare lanes 2 to 7 with lane 1), suggesting that the RS domains in both ASF/SF2 and ASF-CTDO were functionally important in this assay. Nevertheless, like the comparison between ASF-CTDO and ASF/SF2, a comparison between ASFΔRS-CTDO and ASFΔRS showed that ASFΔRS-CTDO stimulated splicing more efficiently than did ASFΔRS (Fig. 2C, compare lanes 5 to 7 with 2 to 4). These results suggested that the CTDs of both ASF-CTDO and ASFΔRS-CTDO are stimulatory in the splicing of β-globin pre-mRNA substrate in S100 extract and that the CTDO can play a functional role in splicing that is distinct from that of the RS domain.
FIG. 2.
In vitro splicing of β-globin pre-mRNA in S100 extract complemented with ASF/SF2 or ASF-CTDO. (A) Time course in S100 complemented with 100 nM ASF/SF2 (lanes 1 to 8) or ASF-CTDO (lanes 9 to 16). Time points are at 0, 20, 30, 40, 50, 60, 70, and 80 min. (B) Quantitative analysis of β-globin spliced product (mRNA) and spliced intermediate (lariat/exon 2) shown in panel A. (C) The RS domains of both ASF/SF2 and ASF-CTD are required for efficient splicing activity in S100. Splicing with 200 nM ASF-CTDO (lane 1) was compared to that in reaction mixtures containing 20, 80, or 200 nM ASFΔRS (lanes 2 to 4) and 20, 80, or 200 nM ASFΔRS-CTDO (lanes 5 to 7). (D) High concentrations of ASF/SF2 did not compensate for the difference in early splicing product formation. Splicing kinetics for reactions with 80 nM ASF-CTDO (lanes 2 to 4) were compared to those for reactions with 80 nM ASF/SF2 (lanes 5 to 7), 200 nM ASF/SF2 (lanes 8 to 10), or 480 nM ASF/SF2 (lanes 11 to 13). Time course reactions were terminated at 30 (lanes 2, 5, 8, and 11), 60 (lanes 3, 6, 9, and 12), or 90 min (lanes 4, 7, 10, and 13).
To extend these results, we next compared the splicing kinetics of reactions with ASF-CTDO (Fig. 2D, lanes 2 to 4) with those of reactions with either the same amount (lanes 5 to 7), 2.5-fold more (lanes 8 to 10), or 6-fold more (lanes 11 to 13) ASF/SF2. We observed that, even with sixfold more ASF/SF2, the first appearance of splicing intermediates (approximately 30 min) lagged behind that seen in reactions with ASF-CTDO (Fig. 2D, compare lanes 5, 8, and 11 with 1). These data indicate that higher ASF/SF2 levels did not substantially increase the reaction rate prior to the first catalytic step. In addition, at later times in the reaction (30 to 60 min), the ratio of final spliced product to lariat/exon 2 intermediate was noticeably higher in the reactions with ASF-CTDO than in all of the reactions with ASF/SF2 and this ratio did not change significantly in the ASF/SF2 reactions regardless of the amount of ASF/SF2 included (for example, compare lanes 7, 10, and 13 in Fig. 2D). We conclude from these data that the CTD alters qualitatively the effect of ASF/SF2 on splicing.
Splicing activity of ASF-CTDO is not specific to β-globin pre-mRNA and S100 extracts.
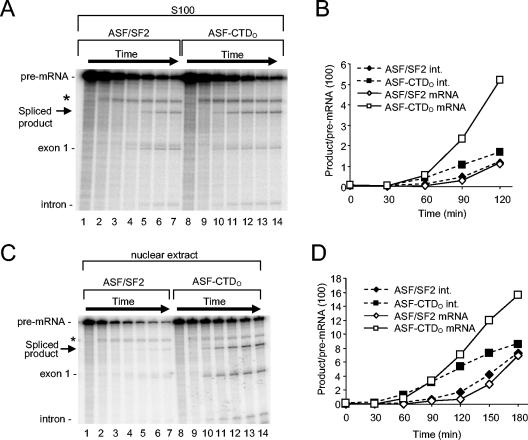
To test both the generality of the above findings and the response of a pre-mRNA with a more specific requirement for ASF/SF2, we examined the splicing of the human immunodeficiency virus type 1 (HIV-1) tat pre-mRNA (34). In contrast to that of β-globin, the tat pre-mRNA can be specifically committed to splicing by ASF/SF2, but not by other SR proteins (21). A time course of splicing in S100 extract (Fig. 3A) shows that ASF-CTDO (lanes 8 to 14) again increased the splicing rate compared to ASF/SF2 at an identical concentration (lanes 1 to 7). Quantitation revealed that the splicing lag time was decreased by approximately 30 min when either splicing intermediates or final products were quantitated (Fig. 3B). The splicing of tat pre-mRNA in nuclear extracts, which unlike S100 extracts contain all of the essential splicing factors, is unusual because it is inhibited by endogenous hnRNP A1 (5, 66). However, this inhibition can be overcome, at least in part, by addition of exogenous ASF/SF2 (34) or RNAP IIO (26). Addition of ASF-CTDO to nuclear extract not only enabled tat pre-mRNA splicing but again increased the splicing rate compared to that for ASF/SF2 (Fig. 3C, compare lanes 8 to 14 with 1 to 7). Remarkably, ASF-CTDO decreased the long lag time of tat pre-mRNA splicing in nuclear extract by nearly 60 min (from 120 to 60 min) compared to ASF/SF2 (Fig. 3D).
FIG. 3.
In vitro splicing of the HIV tat pre-mRNA substrate is stimulated more efficiently by ASF-CTDO than ASF/SF2 in both S100 and nuclear extracts. (A) Time course of tat pre-mRNA splicing in S100 containing 280 nM ASF/SF2 (lanes 1 to 7) or ASF-CTDO (lanes 8 to 14). Time points are at 0, 30, 60, 90, 120, 150, and 180 min. (B) Quantitative analysis of tat pre-mRNA spliced product (mRNA) and splicing intermediate (exon 1) shown in panel A. (C) Time course of tat pre-mRNA splicing in nuclear extract supplemented with 240 nM ASF/SF2 (lanes 1 to 7) or ASF-CTDO (lanes 8 to 14). Time points are at 0, 30, 45, 60, 120, and 150, and 180 min. A band unrelated to splicing (*) is an artifactual cleavage product (34). (D) Quantitative analysis of tat pre-mRNA splicing products shown in panel C.
CTDO-dependent splicing enhancement correlates with ATP-dependent spliceosome assembly, but not ATP-independent prespliceosome assembly.
We next wished to investigate at which step the CTD functioned to accelerate splicing. To this end, we utilized an AdML pre-mRNA, which has frequently been used to study spliceosomal complex formation in reaction mixtures containing nuclear extracts (13). As with other pre-mRNAs tested, the AdML pre-mRNA was also spliced with increased kinetics when nuclear extracts were supplemented with ASF-CTDO (Fig. 4A and B). When spliceosome assembly was examined by native gel electrophoresis, the formation of prespliceosomal A complexes as well as early spliceosomal B complexes was more efficient in reactions with ASF-CTDO than in reactions with ASF/SF2 or nuclear extract alone (Fig. 4C and D). This was most clearly seen with the A complexes at the earliest (2- and 5-min) time points (Fig. 4C [compare lanes 14 and 15 to 8 and 9] and D), and with B complexes at later (15- and 30-min) time points (Fig. 4C [compare lanes 16 and 17 to 10 and 11] and D).
FIG. 4.
ASF-CTDO facilitates spliceosome assembly and splicing product formation in reaction mixtures containing AdML pre-mRNA. (A) Time course of AdML pre-mRNA splicing in reaction mixtures containing nuclear extract (N.E.; lanes 1 to 5) and in reaction mixtures supplemented with 100 nM ASF/SF2 (lanes 6 to 10) or ASF-CTDO (lanes 11 to 15). Reaction mixtures were incubated for 0, 20, 45, 60, or 90 min. (B) Quantitative analysis of AdML spliced product (mRNA) shown in panel A. (C) Time course of AdML pre-mRNA spliceosome assembly in reaction mixtures containing nuclear extract (lanes 1 to 6) and in reaction mixtures supplemented with 100 nM ASF/SF2 (lanes 7 to 12) or ASF-CTDO (lanes 13 to 18). Incubation times were 0, 2, 5, 15, 30, and 60 min, after which heparin (0.8 μg/μl) was added, and reaction mixtures were electrophoresed on a native 2% agarose gel (13). (D) Quantitative comparison of spliceosomal A and B complex formation shown in panel C for ASF/SF2 reactions (lanes 7 to 11) and ASF-CTDO reactions (lanes 13 to 17). (E) ASF-CTDO has no effect on ATP-independent spliceosomal-complex formation. AdML pre-mRNA was incubated in ATP-depleted nuclear extract in the absence (lanes 1 to 5) or presence of ASF/SF2 (lanes 6 to 10) or ASF-CTDO (lanes 7 to 15). Reaction mixtures were incubated for 0, 10, 20, 40, or 60 min, after which they were chilled on ice and loaded directly onto a 1.5% native agarose gel.
The data with ASF-CTDO were very similar to results obtained with purified RNAP IIO (26) and are consistent with the possibility that the CTD may facilitate U1 and/or U2 binding to the pre-mRNA. However, the first discrete splicing complex detectable in HeLa extracts is the E complex (39, 50), which forms in the absence of ATP and which is believed to be a functional precursor of the A complex. Therefore, we next examined whether ASF-CTDO could specifically enhance E complex assembly by examining prespliceosome formation in nuclear extract in the absence of ATP and creatine phosphate. Unlike nonspecific H complexes, E complexes require incubation at 30°C and can be separated from H complexes by native agarose gel electrophoresis as long as heparin is not added to the reactions (13). Surprisingly, we observed no differences in the rate of E complex assembly between ASF-CTDO and ASF/SF2 (Fig. 4E, compare lanes 11 to 15 to lanes 6 to 10). This evidence suggests that the CTDO may not be involved in ATP-independent U1 binding to the 5′ splice site, as has been suggested for ASF/SF2 (28, 32), and rather may facilitate the stable binding of U2 to the branch site (see Discussion).
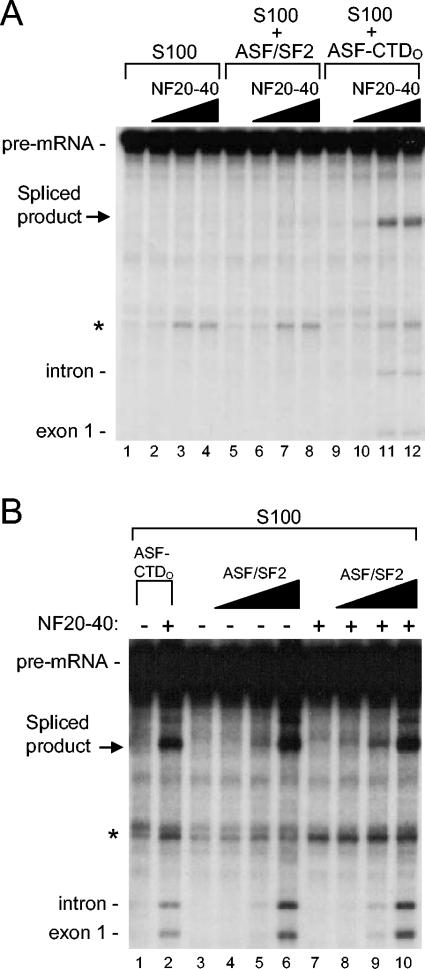
Addition of a nuclear fraction to S100 stimulates IgM-A3 splicing in a CTDO-dependent manner
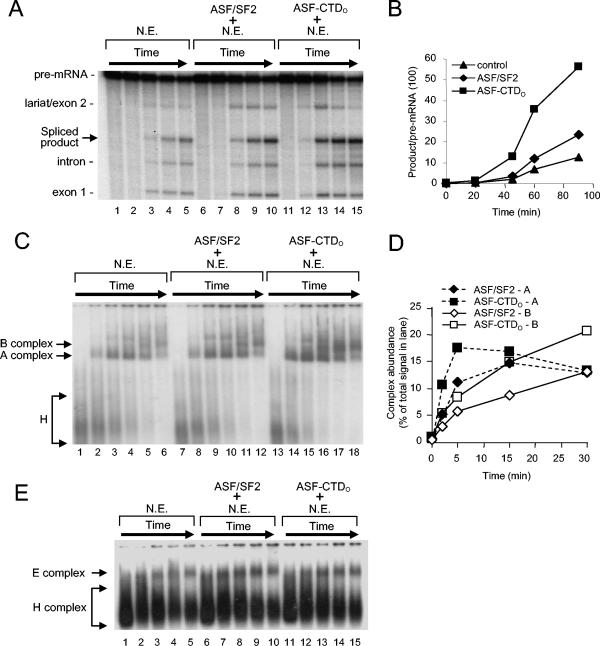
An important question is whether CTD function involves cofactors distinct from components of the general splicing machinery. To begin to address this, we took advantage of the fact that, while high concentrations of ASF/SF2 can activate splicing of the IgM-A3 substrate (an immunoglobulin M pre-mRNA-based substrate containing consensus ASF/SF2 binding sites) in S100 (54, 58), at lower concentrations, neither ASF/SF2 nor ASF-CTDO had any effect on splicing (Fig. 5A, compare lanes 5 and 9 to 1). However, when we added increasing amounts of NF20-40 (53), ASF-CTDO, but not ASF/SF2, activated IgM-A3 splicing (Fig. 5A, compare lanes 10 to 12 to 6 to 8). The effect of NF20-40 on the splicing of other pre-mRNAs was also examined, but the stimulation was significantly less than that observed with the IgM-A3 pre-mRNA (data not shown). These findings suggest that a splicing factor or cofactor that cooperates specifically with the CTD and not with ASF/SF2 is present in NF20-40. To confirm this, we performed an ASF/SF2 titration in the absence or presence of NF20-40. Increasing amounts of ASF/SF2 resulted in a dose-dependent increase in splicing of IgM-A3 pre-mRNA (Fig. 5B, lanes 4 to 6), but addition of a constant amount of NF20-40 had no significant effect at any ASF/SF2 concentration (Fig. 5B, compare lanes 8 to 10 with 4 to 6). This is in sharp contrast to the significant effect that NF20-40 had on ASF-CTDO-activated splicing (Fig. 5B, compare lanes 1 and 2). These data provide strong evidence that a splicing cofactor(s) can specifically cooperate with the CTDO to activate splicing.
FIG. 5.
Addition of NF20-40 to S100 strongly stimulates in vitro splicing of IgM-A3 in the presence of ASF-CTDO but not ASF/SF2. (A) Titration of NF20-40 into splicing reactions does not complement S100 alone (lanes 1 to 4) or S100 supplemented with 200 nM ASF/SF2 (lanes 5 to 8), but NF20-40 stimulates splicing of IgM-A3 in the presence of 200 nM ASF-CTDO (lanes 9 to 12). The amounts of NF20-40 used in the titrations were 0, 1, 3, and 6 μl. (B) Three microliters of NF20-40 stimulates splicing in S100 in the presence of 100 nM ASF-CTDO (lane 2). Larger amounts of ASF/SF2 stimulate splicing of IgM-A3 equally well in either the absence (lanes 3 to 6) or presence (lanes 7 to 10) of 3 μl of NF20-40. Concentrations of ASF/SF2 used in titrations were 0, 200, 400, and 600 nM.
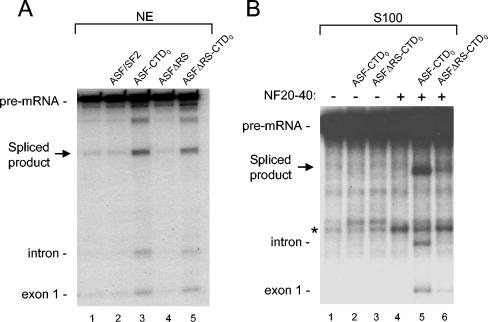
The RS domain and CTD have distinct functions in splicing.
The above data suggest that the CTD and RS domain perform qualitatively distinct functions in splicing. However, because both the ASF/SF2 RS domain and the RNAP IIO CTD consist of highly phosphorylated repetitive motifs, we wished to further examine differences and similarities between them. In nuclear extract, both ASF-CTDO and ASFΔRS-CTDO enhanced splicing of IgM-A3, whereas ASF/SF2 and ASFΔRS could not (Fig. 6A). This suggests that the RS domains of these proteins did not contribute to splicing under these conditions, which was not surprising since SR proteins exist naturally in nuclear extracts to perform this function. In stark contrast to what was found for IgM-A3 splicing in nuclear extracts, in reaction mixtures containing S100 and NF20-40 we observed a strong dependency on both the RS domain and the CTD of ASF-CTDO (Fig. 6B). Under conditions where neither ASF-CTDO nor ASFΔRS-CTDO activated splicing in S100 alone (Fig. 6B, lanes 1 to 3), the addition of an equal amount of NF20-40 to these reactions stimulated only those with ASF-CTDO (Fig. 6B, compare lane 5 with 4 and 6). Therefore, it seems that the very limited amount of SR proteins in NF20-40, which from Western blots we estimate to be similar to the amount detected in S100 (53) (data not shown), requires the addition of exogenous SR proteins with intact RS domains in order to activate splicing of the IgM-A3 substrate and that the CTDO is unable to perform this function. These data also suggest that the factor(s) in NF20-40 that cooperates with the CTDO is not SR proteins.
FIG. 6.
The RS domain in ASF-CTDO is not necessary for enhanced splicing of IgM-A3 in nuclear extract, but the RS domain ASF-CTDO is required for splicing of IgM-A3 in S100 plus NF20-40. (A) Standard in vitro splicing reaction mixtures were incubated in nuclear extract (NE) alone (lane 1) or in nuclear extract containing supplemental ASF/SF2 (lane 2), ASF-CTDO (lane 3), ASFΔRS (lane 4), or ASFΔRS-CTDO (lane 5) at a concentration of 200 nM. (B) Standard in vitro splicing reaction mixtures were incubated in S100 alone (lane 1) or in the presence of additional proteins and NF20-40 as indicated above the gel. All reaction mixtures were incubated for 80 min at 30°C. The migration of pre-mRNA and splicing products is indicated at the left.
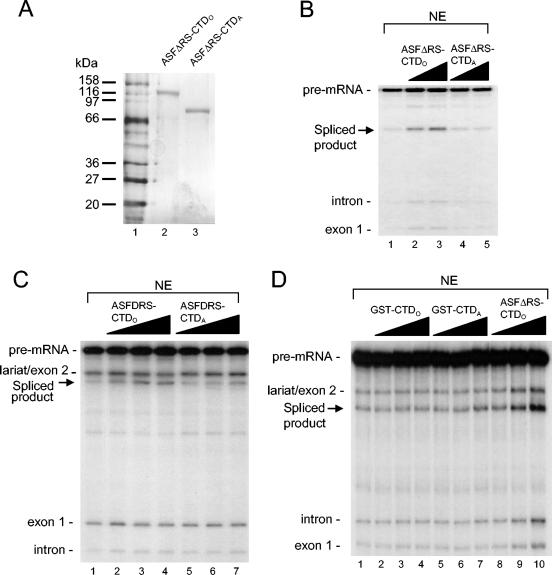
CTD phosphorylation is necessary for CTDO splicing activity.
The importance of CTD phosphorylation in pre-mRNA splicing is supported by the observation that RNAP IIO but not RNAP IIA enhances in vitro splicing (26). Since our ASF-CTDO fusion proteins were purified in a highly phosphorylated state, we wished to determine if CTD phosphorylation is necessary for ASF-CTDO splicing activity. However, since ASF/SF2 itself is highly phosphorylated in its RS domain, we chose to use ASFΔRS-CTD in these studies, so that dephosphorylation would affect only the CTD. To examine this, we dephosphorylated ASFΔRS-CTDO in vitro with CIP and repurified the resultant protein. A silver-stained SDS gel of the dephosphorylated protein, ASFΔRS-CTDA, shows the large mobility shift compared to that for the phosphorylated protein, ASFΔRS-CTDO (Fig. 7A).
FIG. 7.
Phosphorylation of ASFΔRS-CTD is necessary for enhanced splicing activity in nuclear extract. (A) Silver-stained SDS-polyacrylamide gel of 50 ng of purified phosphorylated ASFΔRS-CTDO (lane 2) and dephosphorylated and repurified ASFΔRS-CTDA (lane 3). Protein molecular mass markers are shown in lane 1 (New England Biolabs; broad range, 7702S). (B) Addition of 15 (lane 2) or 30 nM (lane 3) ASFΔRS-CTDO stimulates splicing of IgM-A3 in nuclear extract (NE). Addition of 15 (lane 4) or 30 nM (lane 5) ASFΔRS-CTDA has no effect on splicing of IgM-A3. (C) Addition of ASFΔRS-CTDO (lanes 2 to 4) stimulates splicing of β-globin, but ASFΔRS-CTDA (lanes 5 to 7) has no effect on splicing. (D) Neither GST-CTDA (lanes 2 to 4) nor GST-CTDO (lanes 5 to 7) stimulates splicing of IgM-A3 under conditions where ASFΔRS-CTDO (lanes 8 to 10) enhances splicing. Reaction mixtures for panel D contained a titration of 5, 20, or 80 nM respective recombinant proteins and were incubated for 75 min at 30°C.
The splicing activity of ASFΔRS-CTDA was first tested with the IgM-A3 substrate by addition of increasing amounts of ASFΔRS-CTDA (Fig. 7B, lanes 4 and 5) or ASFΔRS-CTDO (Fig. 7B, lanes 2 and 3) to nuclear extract. ASFΔRS-CTDA was unable to activate splicing, whereas ASFΔRS-CTDO again produced a dose-dependent increase in splicing activity. We also tested splicing of the β-globin pre-mRNA in nuclear extract supplemented with either ASFΔRS-CTDO or ASFΔRS-CTDA, and again only ASFΔRS-CTDO produced increased splicing activity (Fig. 7C, lanes 2 to 4 compared to 5 to 7). Importantly, GST-CTD, either phosphorylated or dephosphorylated, had no detectable effect on the splicing of IgM-A3 at any concentration tested (Fig. 7D). These data together show that CTD phosphorylation is necessary for ASFΔRS-CTD-enhanced splicing in vitro and further strengthens the view that CTD fused to ASF/SF2 and CTD in its natural context function similarly.
DISCUSSION
Considerable evidence has established that RNAP II, and specifically the CTD, can play a positive role in the splicing of mRNA precursors. However, there has been very little progress in understanding how it actually functions in this process. This is due to the complexity both of the splicing reaction and of RNAP IIO itself. As we showed previously and confirmed here, and unlike the situation with capping and 3′ end formation, the isolated CTD does not function in standard splicing assays. We hypothesized that this might reflect the inability of the isolated CTD to be targeted to the substrate and/or splicing machinery, and the data presented here support that idea. Our findings are significant because they not only indicate that the CTD is sufficient to enhance splicing when targeted to the pre-mRNA but also provide a means for more readily analyzing CTD function in splicing. Below we discuss the evidence that the ASF-CTD fusion protein indeed recapitulates the behavior of RNAP II and the new insights it provides into how the CTD enhances splicing.
It is essential that the ASF-CTD fusion proteins function similarly to RNAP IIO if they are to provide insights into how the CTD functions in splicing. All the data presented here strongly support this. For example, our results showing that the fusion proteins function early in splicing are consistent with previous work in our laboratory which showed that RNAP IIO enhanced the splicing of several pre-mRNA substrates at an early step of spliceosome assembly. We also showed previously that CTD function could not be performed by excess SR proteins (26), which is consistent with our finding here that the CTD and RS domain have distinct functions. Additionally, in agreement with previous studies involving RNAP IIO and RNAP IIA (26), the CTD required phosphorylation in order to enhance splicing.
To understand the mechanism by which the CTD enhances splicing, it was important to determine where within the spliceosome assembly pathway CTDO performs this function. Previous evidence with RNAP IIO suggested a CTD-sensitive step during or prior to A complex formation (26). The results presented here are in agreement with this conclusion and extend our understanding by deemphasizing a CTDO role in ATP-independent E complex formation and more specifically implicate ATP-dependent A complex formation as a CTDO-sensitive step. At the snRNP level, E complex is characterized by the base pairing of the U1 snRNP to the 5′ splice site whereas the U2 snRNP is not yet base paired and is loosely associated with the complex (14). We conclude that the CTDO is most likely not involved in stabilizing the U1-5′ splice site interaction that occurs without ATP and is instead involved in the later events that occur in the presence of ATP, such as the base pairing of U2 to the branch site.
Our data have provided evidence that the phosphorylated CTD performs a function distinct from that of the RS domain. This is in fact consistent with our finding that the CTDO had no effect on E complex formation since SR proteins are important for the commitment of pre-mRNAs to splicing by stabilizing U1 binding to the 5′ splice site through the combined interactions between the RBD and pre-mRNA and RS domain and protein components of U1 snRNP (28, 32). This is additional evidence to support the theory that the CTDO is involved in ATP-dependent transition to the A complex by facilitating U2 base pairing to the branch site, although we can't rule out an involvement in later stages of B and C complexes. Although we never observed any effect of CTDO on E complex formation in our in vitro assays, it remains unclear if, in the presence of ATP, the CTDO might play a role in the initial interactions between the components of E complex and the pre-mRNA. This seems plausible since it has been demonstrated that ATP alters the composition of large snRNP/RNAP IIO complexes that form on short 5′ splice site RNAs (30). Notably, in these studies, ATP allowed for the incorporation of U2 into U1-containing complexes. In any event, our results were surprising because they suggest that the CTD, while functioning early in splicing, does not affect the earliest known step, i.e., E complex formation.
It remains an important question what CTD-binding proteins are necessary for CTD-enhanced splicing. However, our data suggest that different CTD-binding proteins have unique splicing functions, as illustrated by the strong nuclear protein requirement for CTD-activated splicing of IgM-A3, but not of β-globin or tat pre-mRNAs. Since CTDO did not show any effect on IgM-A3 splicing in S100 unless NF20-40 was added, CTDO-enhanced splicing of a subset of pre-mRNAs may be more strongly enhanced by cofactors that are absent from S100, whereas other pre-mRNAs may be less dependent on these factors due to specific strengths and weaknesses of a particular pre-mRNA. Therefore, the CTD may coordinate the actions of two or more CTDO-associated splicing regulators that have discrete functions. Accordingly, it is interesting that the CTD has also been shown to bind to the splicing factor PSF (17). In addition to linking CTDO to early spliceosomeal complex assembly (43), PSF has been shown to be required much later in the splicing reaction, during the second catalytic step (22). It is tempting to speculate that CTDO can, in a substrate-specific manner, remain associated with spliceosomes from prespliceosome formation through catalysis. In line with this possibility, RNAP IIO has been shown to form stable complexes with the late spliceosomal component U5 snRNP and the [U4/U6.U5] tri-snRNP (6, 57).
Our data confirm our previous suggestion that the isolated CTD cannot activate splicing in vitro. This contrasts with the ability of the CTD alone to function in in vitro capping (8, 27) and 3′ processing (25) and indicates that another property of RNAP IIO is necessary for CTDO-enhanced splicing. Fusion of the CTD to ASF/SF2, which normally enters spliceosomes, should localize the CTD closely with the pre-mRNA, perhaps in a manner analogous to that for a nascent transcript and the CTD of elongating RNAP IIO. Because we observed splicing enhancement by ASF-CTD, with or without the RS domain, we believe that it is most likely that the RNA binding activity of ASF/SF2 is in fact important for CTDO splicing activity, by providing an RNA targeting function. In possible contrast to our results, it has been reported previously that GST-CTD can enhance in vitro splicing, through the exon definition mode of splicing (62). We observed no requirement for having the exons in our substrate pre-mRNAs defined at both ends by splice sites. Perhaps the suggested exon definition role of the isolated CTD is bypassed by having the CTD fused to ASF/SF2, which itself is involved in identifying splice sites through interactions with U1 and U2AF (28, 32, 55, 59). Another difference is that our ASF-CTD was a highly phosphorylated protein preparation compared to the mostly unphosphorylated GST-CTD used by Zeng and Berget (62). Additionally, in our experiments, we never tested the high GST-CTD concentrations employed by these authors.
The splicing-stimulatory activity of ASFΔRS-CTDO, like that of RNAP II, requires CTD phosphorylation, and therefore phosphorylated CTD heptads are likely to interact specifically with a component of the splicing machinery. Studies using yeast two-hybrid assays have revealed phospho-CTD-binding proteins that contain an RS domain, an RNA recognition motif, and a distinct CTD-binding domain (45, 61); however, the functional relevance of these proteins in splicing remains to be determined. The phosphorylated CTD has been shown to interact directly with splicing factors such as PSF (17) and the yeast splicing factor Prp40 (40). Nevertheless, the importance of these protein-protein interactions for CTD-enhanced splicing in our in vitro system requires further investigation.
CTD phosphorylation undergoes dynamic changes during transcription, and it is likely that this is critical for transcription-coupled RNA processing. For example, it has been shown that CTD Ser-5 phosphorylation is observable near the promoter and Ser-2 phosphorylation is seen throughout the gene (7). TFIIH kinase (16, 56) is involved in phosphorylation of Ser-5 during promoter escape, yielding a CTD that is competent for 5′ capping of the nascent transcript, and the transcription elongation factor P-TEFb (49, 65) is responsible for at least a fraction of the Ser-2 phosphorylation during the transcript elongation phase. The correlation between the timing of Ser-2 phosphorylation and the entry of elongating RNAP II into coding regions suggests the possibility that Ser-2 phosphorylation may be important for CTD-enhanced splicing. P-TEFb interacts with RNAP II and the HIV-1 Tat cofactor and general elongation factor, Tat-SF1 (19). Intriguingly, P-TEFb has also been shown to interact with snRNPs through a Tat-SF1-snRNP complex (20). It was demonstrated that this complex promotes transcription elongation and supports splicing in nuclear extracts depleted of snRNA. In yeast, the CUS2 protein was identified as a suppressor of a U2 snRNA mutation and, intriguingly, is homologous to Tat-SF1 (46, 60). The relationship between the Ser-2 kinase P-TEFb, RNAP IIO, Tat-SF1, and snRNPs (U2 in particular) is generally consistent with our suggestion that the CTDO may function during prespliceosomal A complex formation. The detection of P-TEFb, RNAP IIO, and snRNPs in large complexes formed on short 5′ splice-containing RNAs lends additional support for this hypothesis (30). Consistent with this model, purified ASF-CTDO used in our studies is phosphorylated on Ser-2, but we do not know if there is a Ser-2-specific splicing function because Ser-5 is also phosphorylated. Why purified ASF-CTDO expressed in insect cells maintains CTD phosphorylation at both Ser-2 and Ser-5 is unclear, since it should not be directly involved in transcription elongation. However, fusion of the CTD with a splicing factor such as ASF/SF2 may aid in localizing the CTD within regions of the nucleus that either contain CTD kinase activity or are deficient in CTD phosphatase activity.
In conclusion, our results strongly suggest a model in which the phosphorylated CTD is a functionally independent domain that, when targeted to the pre-mRNA, interacts with the splicing machinery to facilitate the formation of the prespliceosomal A complex. This is reflected in a significant decrease in the lag time typically seen in in vitro splicing reactions. The CTDO splicing function is distinct from that of the RS domain, consistent with evidence showing that CTDO must interact with protein domains different from those that interact with RS domains. For the future, this experimental system provides a means to examine not only splicing factors that functionally associate with the CTDO but also the role of specific heptads in splicing. Our results extend previous studies involving purified RNAP IIO and support the idea that the phosphorylated CTD can function as a general activator of splicing, provided it is properly recruited to the RNA substrate.
Acknowledgments
We thank N. Rao and C. Shin for assistance in the production and characterization of HeLa extracts and R. Reed for plasmids and technical advice.
This work was supported by NIH R37 GM48259 to J.L.M. S.M. was the recipient of NRSA F32 GM063322.
REFERENCES
- 1.Bauren, G., and L. Wieslander. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76:183-192. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 3.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2:754-765. [DOI] [PubMed] [Google Scholar]
- 4.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chabot, B., S. Bisotto, and M. Vincent. 1995. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res. 23:3206-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and γ-actin genes. Mol. Cell. Biol. 23:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. [DOI] [PubMed] [Google Scholar]
- 10.Corden, J. L., and M. Patturajan. 1997. A CTD function linking transcription to splicing. Trends Biochem. Sci. 22:413-416. [DOI] [PubMed] [Google Scholar]
- 11.Crispino, J. D., B. J. Blencowe, and P. A. Sharp. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265:1866-1869. [DOI] [PubMed] [Google Scholar]
- 12.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 13.Das, R., and R. Reed. 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5:779-787. [DOI] [PubMed] [Google Scholar]
- 15.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois, M. F., M. Vincent, M. Vigneron, J. Adamczewski, J. M. Egly, and O. Bensaude. 1997. Heat-shock inactivation of the TFIIH-associated kinase and change in the phosphorylation sites on the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 25:694-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929-933. [DOI] [PubMed] [Google Scholar]
- 21.Fu, X. D. 1993. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature 365:82-85. [DOI] [PubMed] [Google Scholar]
- 22.Gozani, O., J. G. Patton, and R. Reed. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13:3356-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 25.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 26.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 28.Jamison, S. F., Z. Pasman, J. Wang, C. Will, R. Luhrmann, J. L. Manley, and M. A. Garcia-Blanco. 1995. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 23:3260-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 30.Kameoka, S., P. Duque, and M. M. Konarska. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, E., L. Du, D. B. Bregman, and S. L. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 33.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4:1158-1171. [DOI] [PubMed] [Google Scholar]
- 35.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 36.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 37.Manley, J. L., and R. Tacke. 1996. SR proteins and splicing control. Genes Dev. 10:1569-1579. [DOI] [PubMed] [Google Scholar]
- 38.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 39.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 5:2534-2546. [DOI] [PubMed] [Google Scholar]
- 40.Morris, D. P., and A. L. Greenleaf. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935-39943. [DOI] [PubMed] [Google Scholar]
- 41.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25:1147-1149. [DOI] [PubMed] [Google Scholar]
- 43.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393-406. [DOI] [PubMed] [Google Scholar]
- 44.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 45.Patturajan, M., X. Wei, R. Berezney, and J. L. Corden. 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 18:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perriman, R., and M. Ares, Jr. 2000. ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 14:97-107. [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson, S. R., A. Dvir, C. W. Anderson, and W. S. Dynan. 1992. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 6:426-438. [DOI] [PubMed] [Google Scholar]
- 48.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 49.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 50.Reed, R. 1990. Protein composition of mammalian spliceosomes assembled in vitro. Proc. Natl. Acad. Sci. USA 87:8031-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp, P. A. 1994. Split genes and RNA splicing. Cell 77:805-815. [DOI] [PubMed] [Google Scholar]
- 52.Staknis, D., and R. Reed. 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 14:7670-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tacke, R., M. Tohyama, S. Ogawa, and J. L. Manley. 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93:139-148. [DOI] [PubMed] [Google Scholar]
- 55.Tange, T. O., and J. Kjems. 2001. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 312:649-662. [DOI] [PubMed] [Google Scholar]
- 56.Trigon, S., H. Serizawa, J. W. Conaway, R. C. Conaway, S. P. Jackson, and M. Morange. 1998. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 273:6769-6775. [DOI] [PubMed] [Google Scholar]
- 57.Vincent, M., P. Lauriault, M. F. Dubois, S. Lavoie, O. Bensaude, and B. Chabot. 1996. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 24:4649-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watakabe, A., K. Tanaka, and Y. Shimura. 1993. The role of exon sequences in splice site selection. Genes Dev. 7:407-418. [DOI] [PubMed] [Google Scholar]
- 59.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 60.Yan, D., R. Perriman, H. Igel, K. J. Howe, M. Neville, and M. Ares, Jr. 1998. CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol. Cell. Biol. 18:5000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, J., and J. L. Corden. 1991. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 266:2290-2296. [PubMed] [Google Scholar]
- 64.Zhang, J., and J. L. Corden. 1991. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 266:2297-2302. [PubMed] [Google Scholar]
- 65.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, J., A. Mayeda, and A. R. Krainer. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8:1351-1361. [DOI] [PubMed] [Google Scholar]
- 67.Zuo, P., and J. L. Manley. 1993. Functional domains of the human splicing factor ASF/SF2. EMBO J. 12:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zuo, P., and J. L. Manley. 1994. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc. Natl. Acad. Sci. USA 91:3363-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]