Abstract
DASH is a microtubule- and kinetochore-associated complex required for proper chromosome segregation and bipolar attachment of sister chromatids on the mitotic spindle. We have undertaken a genetic and biochemical analysis of the DASH complex and uncovered a strong genetic interaction of DASH with the Ras/protein kinase A (PKA) pathway. Overexpression of PDE2 or deletion of RAS2 rescued the temperature sensitivity of ask1-3 mutants. Ras2 negatively regulates DASH through the PKA pathway. Constitutive PKA activity caused by mutation of the negative regulator BCY1 is toxic to DASH mutants such as ask1 and dam1. In addition, we have discovered two novel subunits of DASH, Hsk2 and Hsk3 (helper of Ask1), which are microproteins of fewer than 75 amino acids, as dosage suppressors of ask1 mutants. These are essential genes that colocalize with DASH components on spindles and kinetochores and are present in the DASH complex. Mutants in hsk3 arrest cells in mitosis with short spindles and broken spindle structures characteristic of other DASH mutants. Hsk3 is critical for the integrity of the DASH complex because in hsk3 mutants the association of Dam1, Duo1, Spc34, and Spc19 with Ask1 is greatly diminished. We propose that Hsk3 acts to incorporate Ask1 into the DASH complex.
Anaphase is the cell cycle period during mitosis in which chromosomes are segregated to daughter cells. The ability to accurately distribute replicated chromosomes is critical for the proper transfer of genetic material from one generation to the next. Mistakes in chromosome segregation can cause developmental defects, cancer, or death. Multiple layers of control ensure the fidelity of this process to prevent such mistakes.
The mitotic spindle is responsible for the physical movement of chromosomes during anaphase. The mitotic spindle is a bipolar microtubule-based structure that emanates from two organizing centers called centrosomes or spindle pole bodies (SPB) and connects to newly replicated chromosomes called sister chromatids (19, 51) through a proteinaceous structure on the centromere region of chromosomes called the kinetochore (4, 11, 32).
Sister chromatids are held together by a cohesion mechanism which must be eliminated in order for anaphase to occur. The elimination of cohesion is a highly regulated step that is catalyzed by a protease, separase (Esp1 in budding yeast), which is activated by the destruction of its inhibitor, securin (Pds1 in budding yeast) (10, 48).
Dissolution of cohesion and the resultant process of anaphase do not occur until all chromosomes form a bipolar attachment with the spindle and tension is generated. Cells have evolved the ability to detect the proper bipolar attachment of chromosomes. Each kinetochore generates an inhibitory signal through the spindle checkpoint pathway (2, 34) that stabilizes securin until the centromeres on sister chromatids are bound to the spindle in a bipolar fashion (35, 50). In budding yeast, at least six genes, MAD1 to -3, BUB1, BUB3, and MPS1 (14-16, 20, 28), are required for the spindle checkpoint to prevent anaphase.
How the spindle recognizes and binds to chromosomes is currently a topic of extensive research. It is generally thought that the kinetochore, which is composed of multiple protein complexes, captures microtubules and forms a stable attachment. How this capture is accomplished is not understood but is likely to involve a microtubule binding protein associated with the kinetochore or a kinetochore binding protein associated with the microtubule. In addition to forming a connection, cells must have a second activity that is capable of dissolving inappropriate connections such as those that form in a monopolar fashion whereby both sister kinetochores bind microtubules emanating from the same SPB. This is particularly important in budding yeast because centromere replication and maturation occur before the maturation of the newly synthesized SPB (1). Therefore, initially most of the sister kinetochores first attach in a monopolar fashion to the old SPB and need to be corrected, otherwise the two sister chromatids would be pulled into the same cell and undergo chromosome nondisjunction.
While the microtubule-kinetochore capture activity is unknown, two activities have been shown to participate in a regulatory pathway controlling attachment reversal. The first component is the Ipl1-Sli15 kinase complex of the Aurora-B-INCENP kinase superfamily (5, 27). Ipl1 localizes to the mitotic spindle, and in ipl1 mutants, the newly duplicated sister chromatids stay attached to the old SPB and segregate into the daughter bud (43).
The second activity implicated in monopolar attachment reversal is the DASH complex, also known as the Dam1-Duo1 complex. DASH was named for its components Dad1, Dad3, Dam1, Duo1, Ask1, Spc19, Spc34, and Hsk1/Dad2, each of which is coded for by an essential gene (9, 13, 18, 23, 29). Mutants in the DASH complex have multiple phenotypes, including monopolar attachment like ipl1 (23), chromosome loss, metaphase cell cycle arrest dependent upon the spindle checkpoint, sensitivity to hydroxyurea, and broken spindles (9, 18, 23-25, 29). All components of the DASH complex appear to associate with spindle microtubules, and Dam1 has been shown to bind purified microtubules in vitro (9). In addition to microtubule binding, after DNA replication, the DASH complex loads onto the kinetochore in a microtubule-dependent manner, where it presumably can act to regulate spindle-kinetochore interactions in response to the lack of tension (29).
Ipl1 has been shown to control Dam1 phosphorylation in vivo and in vitro (8, 29). Phospho-mimic mutants of Dam1 containing acid residues in place of Ipl1 phosphorylation sites are capable of weakly suppressing certain ipl1 temperature-sensitive alleles, suggesting the phosphorylation of Dam1 is a key function of Ipl1 (8). The prevailing model for function of this pathway is that Ipl1-Sli15 in some manner senses the absence of tension created by a monopolar attachment of sister chromatids and phosphorylates the DASH complex and other kinetochore components such as Ndc10 and Ndc80 to attenuate spindle-centromere attachment, thereby allowing bi-orientation to be established (41-43). It should be noted that no change in the activity of the Ipl1 kinase toward Dam1 in response to tension changes has ever been demonstrated, so the key tenet of this model remains untested. A third complex, Mtw1, is also required for bi-orientation and DASH localization, but its role in this pathway is unclear (36, 38).
In order to further investigate the regulation and composition of DASH, we carried out a systematic dosage suppressor screening of several DASH mutants. In this process, we found two new subunits of DASH as suppressors of ask1 mutants. These new subunits, Hsk2 and Hsk3, are microgenes and were missed as open reading frames (ORFs) by the Saccharomyces cerevisiae Genome Project. We show Hsk2 and Hsk3 are essential genes and associate with the DASH complex, binding to spindles and loading onto the kinetochore in a microtubule-dependent fashion. In addition to these subunits, we discovered that the Ras/protein kinase A (PKA) pathway acts antagonistically to DASH function during a normal cell cycle.
MATERIALS AND METHODS
Yeast strain and plasmid construction.
Yeast strains used in this study are isogenic with the W303-derived Y300 strain. The genotypes of the strains are given in Table 1. The BCY1 disruption was made by a PCR-based method and confirmed by the strain's inability to grow on a nonfermentable carbon source. The ras1::HIS3 mutation was generated by transforming the EcoRI-NaeI fragment of pras1::HIS3, and ras2::LEU2 was made by transforming the NcoI-HindIII fragment of pras2::LEU2 (26). Hemagglutinin (HA)-tagged Hsk2 and Hsk3 were also made by a PCR-based method (31).
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| Y300 | MATatrp1-1 ura3-1 his3-11,3 leu2-3,112 ade2-1 can1-100 |
| Y1102 | As Y300 except Δask1::LEU2 pJBN81(pRS426-ASK1) |
| Y1103 | As Y300 except ask1-3 |
| Y1104 | As Y300 except ask1-2 |
| Y1113 | As Y300 except Δask1::LEU2 ASK1-9MYC-HIS3 |
| Y1173 | As Y300 except ASK1-TAP-TRP1 |
| Y1183 | As Y300 except DAM1-MYC-TRP1 |
| Y2293 | As Y300 except HSK2-3HA-TRP1 |
| Y2294 | As Y300 except Δask1::LEU2 ASK1-9MYC-HIS3 HSK2-3HA-TRP1 |
| Y2309 | As Y300 except Δras1::HIS3 ask1-3 |
| Y2310 | As Y300 except Δras2::LEU2 ask1-3 |
| Y2311 | As Y300 except Δras2::LEU2 ask1-2 |
| Y2313 | As Y300 except Δbcy1::TRP1 |
| Y2314 | As Y300 except Δbcy1::TRP1 ask1-3 |
| Y2315 | As Y300 except Δbcy1::TRP1 Δras2::LEU2 |
| Y2316 | As Y300 except Δbcy1::TRP1 Δras2::LEU2 ask1-3 |
| Y2317 | As Y300 except Δras2::LEU2 dam1-1 |
| Y2318 | As Y300 except Δbcy1::TRP1 dam1-1 |
| Y2319 | As Y300 except Δbcy1::TRP1 Δras2::LEU2 dam1-1 |
| Y2326 | As Y300 except dam1-1 |
| Y2327 | As Y300 except Δ duo1::duo1-1 LEU2 |
| Y2328 | As Y300 except Δras1::HIS3 |
| Y2329 | As Y300 except Δras2::LEU2 |
| Y2330 | As Y300 except Δras1::HIS3 dam1-1 |
| Y2331 | MATa/α trp1-1 ura3-1 his3-11 leu2-3,112 ade2-1 can1-100 Δhsk2::HIS3 |
| Y2332 | MATa/α trp1-1 ura3-1 his3-11 leu2-3,112 ade2-1 can1-100 Δhsk3::HIS3 |
| Y2333 | As Y300 except HSK3-3HA-TRP1 |
| Y2334 | As Y300 except cdc13-1 HSK3-3HA-TRP1 |
| Y2335 | As Y300 except Δask1::LEU2 ASK1-9MYC-HIS3 HSK3-3HA-TRP1 |
| Y2336 | As Y300 except DAM1-MYC-TRP1 HSK3-3HA-TRP1 |
| Y2337 | As Y300 except ASK1-TAP-TRP1 HSK3-3HA-TRP1 |
| Y2338 | As Y300 except Δhsk3::HIS3 pHSK3F2-CEN-URA3 |
| Y2339 | As Y300 except Δhsk3::HIS3 phsk3-16-CEN-TRP1 |
| Y2340 | As Y300 except Δhsk3::HIS3 phsk3-41-CEN-TRP1 |
| Y2341 | As Y300 except Δhsk3::HIS3 phsk3-44-CEN-TRP1 |
| Y2342 | As Y300 except Δhsk3::HIS3 pHSK3F2-CEN-URA3 ASK1-TAP-TRP1 |
| Y2343 | As Y300 except Δhsk3::HIS3 phsk3-16-CEN-TRP1 ASK1-TAP-TRP1 |
| Y2344 | As Y300 except Δhsk3::HIS3 phsk3-41-CEN-TRP1 ASK1-TAP-TRP1 |
| Y2345 | As Y300 except Δhsk3::HIS3 phsk3-44-CEN-TRP1 ASK1-TAP-TRP1 |
Suppressor screens.
For genomic library screens, a 2μm URA3 yeast genomic library (12) was used for transformation into the temperature-sensitive mutant strains. At least 104 transformants were spread on SC-Ura plates at room temperature for 8 h before shifting to the restrictive temperature. Surviving colonies were streaked at the restrictive temperature to confirm the suppression. Plasmids were then recovered and transformed back into the temperature-sensitive strains. To identify the genes within the genomic fragment that are responsible for the suppression, a randomly transposon-mutagenized collection of the plasmids was generated by the Genome Priming System (E7100S; BioLabs) method. The mutagenized plasmid collection was transformed back into the temperature-sensitive strain. The plasmids from the clones that failed to grow at restrictive temperature were recovered and analyzed.
For cDNA library screens, a CEN GAL URA3 yeast cDNA library was employed. Transformants were spread on SC-Ura-Gal plates at room temperature for 12 h before shifting to the restrictive temperature. The colonies were streaked at the restrictive temperature to confirm the suppression. Plasmids were then recovered and transformed back into the temperature-sensitive strains.
Construction of temperature-sensitive mutants of HSK3.
Temperature-sensitive alleles of HSK3 were generated by a random PCR mutagenesis method described previously (49). pHSK3F2-URA3 was used as the PCR template. The primers used for PCR were Hsk3-F2 containing a BamHI site (5′-CGGGATCCAAGGTCTTACTTATCAAGAAGCCATGGAGA-3′) and Hsk3-R1 containing an SpeI site (5′-GGACTAGTAACACAAACAATAAGGGTAATGGTAATAGT-3′). Mutagenic PCR was carried out with 1 μM each primer, 5 mM MgCl2, and 0.5 mM MnCl2. The PCR products were purified, digested with BamHI and SpeI, and then cloned into BamHI/SpeI-digested pRS414. Approximately 10,000 Escherichia coli transformants were obtained and transformed into a Δhsk3::HIS3 strain covered with pHSK3F2-URA3. The Trp+ transformants were replica plated to plates containing 0.1% 5-fluoroorotic acid (5-FOA) at 24 and 37°C to identify temperature-sensitive mutants. From 10 temperature-sensitive clones, plasmid DNA was recovered and transformed back to confirm the phenotype.
Cytological techniques.
Immunofluorescence microscopy was performed after formaldehyde fixation for 0.5 h and staining with antitubulin Yol1/34 (1:50 dilution) or anti-HA (1:100 dilution) antibodies.
Protein purification.
Protein extracts from a 3-liter yeast culture with an optical density of 1.2 were prepared with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-Cl, 5 mM EDTA, 0.5% Triton X-100), and TAP purification was carried out as described previously (29, 37). Briefly, Tris-Cl (pH 8.0) was added to the extracts to a final concentration of 10 mM and NP-40 was added to a final concentration of 0.1%. Then extracts were subjected to the two-step purification with immunoglobulin G-agarose (Sigma) and calmodulin beads (Stratagene). Purified proteins were precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (4 to 20% polyacrylamide), followed by Coomassie blue staining or Western blotting.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were conducted essentially as described previously (3, 52). Hsk2-HA and Hsk3-HA were immunoprecipitated with the monoclonal antibody 16B12. Primers for CEN3, CEN16, and an AT-rich region near PGK1 are the same as those described previously (33).
Nocodazole treatment of the cells was carried out as follows. Cells were arrested with α-factor for 3 h before being spun down and resuspended in YPD containing 15-μg/ml nocodazole and 1% dimethyl sulfoxide (DMSO). YPD containing 1% DMSO was used as a control. After 70 min of incubation at 32°C, cells were spun down for ChIP analysis.
RESULTS
A dosage suppressor screen of DASH mutants.
The DASH complex is one of the outer kinetochore protein complexes which are positioned intermediately between the spindle and the kinetohcore. It has been shown to possess spindle binding ability in vitro, and its localization at the kinetochore is dependent on an intact spindle structure and inner kinetochore proteins such as Ndc10 (13, 25, 29). In the ask1, dam1, or spc34 temperature-sensitive mutants, abnormal chromosome segregation and broken spindle structures are observed. Why these phenotypes occur is not at present clear. To gain further insight into the function of the DASH complex, we carried out a dosage suppressor screen for ask1-2 and ask1-3 mutants. We transformed either a 2μm yeast genomic library or a Gal cDNA library into the temperature-sensitive strain and selected for clones capable of colony formation at a nonpermissive temperature of 35 or 37°C. To ensure the suppression of temperature sensitivity is dependent on the resident plasmid, we recovered the plasmid and transformed it back into the parental strain. Clones that recapitulated suppression were chosen for further analysis, and 2μm clones were subject to transposon-mediated insertion mutagenesis to identify the gene responsible for suppression. The suppressors identified are listed in Table 2. Based on the annotated function, the suppressors could be divided into three different groups: regulators of the Ras/PKA pathway, DASH components, and novel proteins.
TABLE 2.
Dosage suppressors of mutants in DASH componentsa
| TS mutant | Genomic suppressor | cDNA suppressor |
|---|---|---|
| ask1-2 | ASK1 (9), DAD2/HSK1 (5), HSK2 (3), HSK3 (5) | ASK1 (5), DAD2/HSK1 (16), HSK2 (1), HSK3 (12) |
| ask1-3 | ASK1 (25), PDE2 (2), HSK3 (2) | ASK1 (18), PDE2 (4), RAS1 (1), HSK3 (1) |
The number of independent clones isolated is shown in parentheses.
Inactivation of the cAMP-dependent protein kinase (PKA) pathway suppresses the temperature-sensitive phenotype of DASH mutants.
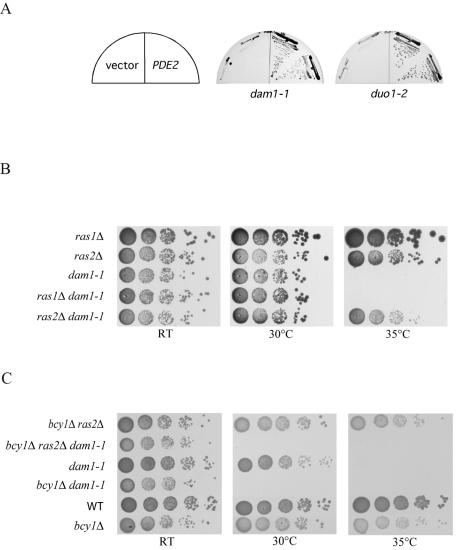
The phosphodiesterase gene PDE2 was identified as a strong suppressor of ask1-3 from both the genomic and Gal-cDNA libraries. PDE2 was also found to partially suppress dam1-1 and duo1-2 (see Fig. 2A). Moreover, the small G-protein gene RAS1 was isolated as a strong suppressor of ask1-3 from the Gal-cDNA library. In the cyclic AMP (cAMP)/PKA pathway, Ras1 and its paralogue Ras2 are the upstream GTPases that activate the PKA pathway by elevating the intracellular level of cAMP in response to glucose signaling (Fig. 1A). However, Pde2 is a phosphodiesterase which converts cAMP to AMP, thus inactivating the PKA pathway. How both a positive regulator of PKA, Ras1, and a negative regulator of PKA, Pde2, could be isolated from the same dosage screen was unclear. One possibility was that the Ras pathway was branched so that Ras1 has a PKA-independent function toward the DASH complex (Fig. 1A). If the PKA pathway exhibited negative feedback on Ras1 function, then down-regulating PKA by Pde2 overexpression might result in hyperactivation of Ras. A second possibility was that either Ras1 or Pde2 was acting in a dominant-negative manner to interfere with its normal function when overexpressed. If the first model was correct and Ras1 acted in a positive manner toward DASH, then deletion of Ras1 or Ras2 might exacerbate DASH temperature-sensitive mutants. To test this, we deleted either RAS1 or RAS2 in ask1-3 and dam1-1 mutant backgrounds and tested for growth at different temperatures. Deletion of RAS1 had no phenotype (Fig. 1B and 2B). In contrast to the possible enhanced TS phenotype, we found that deletion of RAS2 actually suppressed both ask1-3 and dam1-1 mutants (Fig. 1B and 2B).
FIG. 2.
The Ras-cAMP pathway genetically interacts with dam1-1 and duo1-2 mutants. (A) Overexpression of PDE2 rescued the temperature sensitivity of dam1-1 and duo1-2 mutants. Plates were streaked with dam1-1 mutant cells with vector alone or p2μ-PDE2 at 35°C. Plates were streaked with duo1-2 mutant cells with vector alone or p2μ-PDE2 at 37°C. (B) ras2 but not ras1 deletion suppresses the temperature lethality of dam1-1. dam1-1, dam1-1 ras2Δ, dam1-1 ras1Δ, ras1Δ, and ras2Δ cells were serially 10-fold diluted, and YPD plates were spotted with them at either room temperature (RT), 30°C, or 35°C (restrictive temperature) to assess their growth phenotypes. (C) bcy1 is required for the suppression of dam1-1 by ras2. Plates were spotted with 10-fold serial dilutions of strains of the indicated genotypes and incubated at room temperature, 30°C, or 35°C.
FIG. 1.
The Ras-cAMP pathway genetically interacts with ask1-3 mutants. (A) Genetic diagram of the Ras/PKA pathway. Regulatory interactions are represented by arrows for positive interactions and by bars for negative regulatory interactions. Question marks represent hypothetical interactions not previously documented in the literature that could explain the genetic interactions observed in our suppressor analysis. (B) ras2 deletion but not ras1 deletion rescues the temperature sensitivity of ask1-3 mutants. Cells of the indicated genotypes were serially 10-fold diluted, and YPD plates were spotted with them at either room temperature (RT), 30°C, or 35°C (restrictive temperature) to assess their growth phenotypes. (C) bcy1 deletion blocks the suppression by ras2 deletion and exacerbates the growth defect of ask1-3 mutants. Cells of the indicated genotypes were serially 10-fold diluted, and YPD plates were spotted with them at room temperature, 30°C, or 35°C. (D) ras2 deletion cannot suppress the temperature lethality of ask1-2 mutants. YPD plates were streaked with ask1-2, ask1-2 ras2Δ, and ras2Δ strains at room temperature or 37°C to assess their growth phenotypes. (E) ras2 deletion cannot bypass the lethality of ask1 deletion mutants. YPD or SC-5-FOA plates were streaked with ask1Δ, ask1Δ ras2Δ and ras2Δ containing pJBN81 (pRS426-ASK1).
There are three PKA genes (TPK1, -2, and -3) in budding yeast, and these genes are redundant because single- or double-deletion mutants are still viable while the triple-deletion mutant is dead. The fact that the DASH mutants are suppressed by Pde2 overproduction or Ras2 deletion suggests that the PKA pathway acts antagonistically with DASH, as hypothesized in the lower portion of Fig. 1A. If this is true, then hyperactivating PKA might exacerbate DASH mutants. To examine this, we made double mutants between DASH components and Δbcy1, which deletes a negative regulator of PKA, resulting in a high constitutive activity of PKA. Consistent with our hypothesis, the Δbcy1 ask1-3 double mutant could not survive at 30°C, a normally permissive temperature for both parental strains (Fig. 1C). The Δras2 Δbcy1 ask1-3 triple mutant behaved similarly to the Δbcy1 ask1-3 double mutant. Thus, the Δbcy1 phenotype was epistatic to the Δras2 phenotype with respect to ask1-3, indicating that RAS2 and BCY1 act in the same pathway to regulate DASH function as predicted. These results strongly argue that the PKA pathway acts antagonistically to the DASH complex, providing a link between Ras signaling and mitotic function.
We isolated RAS1 and PDE2 as a suppressor of ask1-3 but not ask1-2 mutants. To test whether the Ras/PKA pathway interacted specifically with ask1-3, we either transformed PDE2 or deleted RAS2 in the ask1-2 mutant background. We found that neither Pde2 overexpression (data not shown) nor RAS2 deletion rescued the temperature-sensitive phenotype of ask1-2 mutants (Fig. 1D), demonstrating that the suppression effect of down-regulating the Ras/PKA pathway is allelic specific. Consistent with this allele specificity, we found that Ras2 deficiency could not rescue the lethality of the ask1 deletion (Fig. 1E).
To examine whether the genetic interaction with the Ras/PKA pathway is specific for ASK1 or could also affect other DASH components, we overexpressed PDE2 in dam1-1 and duo1-2 mutants. Overexpression of PDE2 can rescue dam1-1 and duo1-2 viability at the nonpermissive temperature (Fig. 2A). We also found that Δras2 could rescue dam1-1 mutants (Fig. 2B), while deletion of bcy1 exacerbated the dam1-1 mutant's temperature sensitivity as it did for the ask1-3 mutant (Fig. 2C). Consistent with our previous results, the bcy1 deletion was epistatic to the ras2 deletion with respect to dam1-1 suppression. These results suggest that inhibition of the Ras/PKA pathway is capable of suppressing multiple defects in DASH function, indicating it may oppose the function of the entire complex.
Specific DASH proteins suppress the temperature-sensitive phenotypes of DASH mutants.
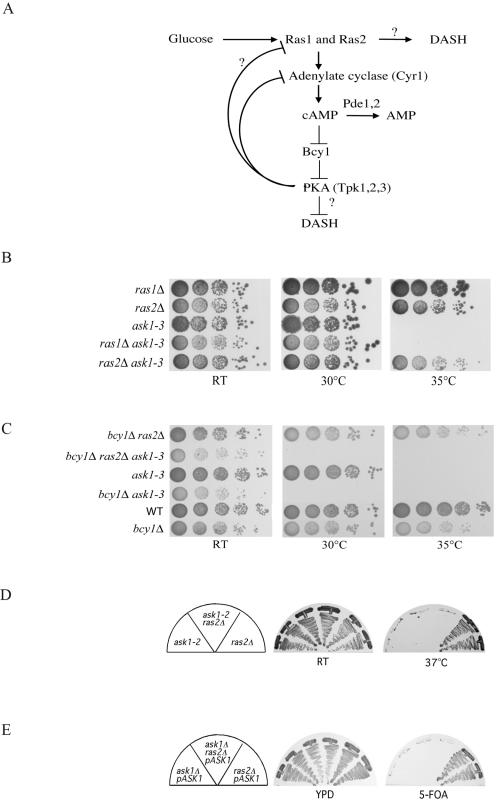
In addition to the known genes in the Ras/PKA pathway, we found that the known DASH component DAD2/HSK1 could suppress the temperature-sensitive phenotype of ask1-2 mutants (Table 2). Thus our screen identified functionally relevant proteins as dosage suppressors. In addition to these known proteins, we identified two novel proteins encoded by YDR320C-A and YKL138C-A and named their respective genes HSK2 and HSK3 (helper of Ask1). These two genes were missed by the Yeast Genome Project because of their extremely short coding sequences. The Hsk2 gene encodes a 72-amino-acid (aa) protein, and the Hsk3 gene encodes a 69-aa protein. However, we isolated each both as a genomic dosage suppressor and as a cDNA suppressor under GAL control, demonstrating that both are expressed sequences. To confirm that the observed suppression was due to the short coding regions, we deleted the N terminus of Hsk2 and Hsk3 and transformed it into ask1 mutants. Neither the truncated Hsk2 nor the truncated Hsk3 could suppress the lethality of ask1-2 at 37°C (Fig. 3A). We also cloned the precise Hsk2 and Hsk3 ORFs under the Gal promoter control and demonstrated that the coding region of Hsk2 and Hsk3 alone could rescue ask1-2 at the nonpermissive temperature (data not shown). To examine the phenotypes of Hsk2 and Hsk3 mutants, we replaced each gene with HIS3 in a diploid yeast strain. Sporulation of the resulting heterozygous diploids revealed 2:0 segregation for viability (Fig. 3B), and all viable colonies were His− (data not shown), indicating that both the Hsk2 and Hsk3 genes are essential genes. Consistent with their essentiality, both Hsk2 and Hsk3 are highly conserved among yeast species (Fig. 3C).
FIG. 3.
HSK2 and HSK3 are suppressors of ask1-2 mutants. (A) ask1-2 mutants containing either the complete ORF or an N-terminal truncation of HSK2 or HSK3 under Gal control were streaked onto YP galactose plates and incubated at room temperature (RT) or at 37°C, the nonpermissive temperature for ask1-2 mutants. The growth phenotype was examined after 3 days. The nucleotide (nt) positions of the deletions of the ORFs are diagrammed below. Nucleotide 1 refers to the A of the ATG start codon. (B) Hsk2 and Hsk3 are essential for cell growth. Tetrad dissection of the HSK2/Δ hsk2 and HSK3/Δhsk3 heterozygous diploid gave two viable and two dead spores. (C) Alignments of S. cerevisiae (Sc) Hsk2 and Hsk3 with their orthologues in Schizosaccharomyces pombe (SP) and Kluyveromyces lactis (Kl).
Hsk2 and Hsk3 are new components of the DASH complex.
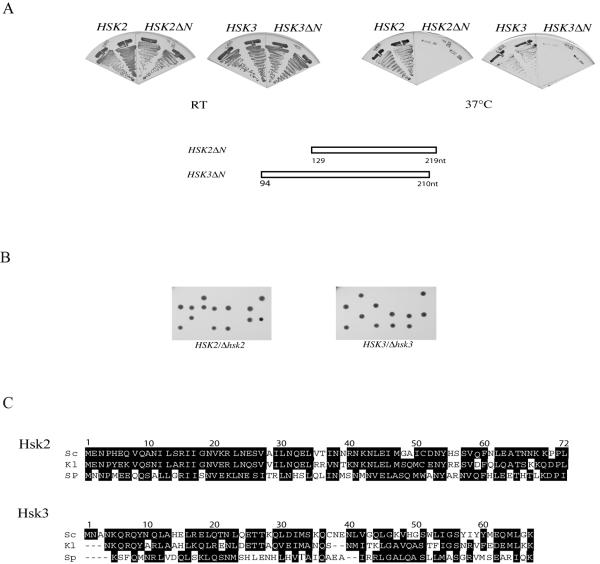
As both Ras/PKA and DASH components can suppress Ask1 mutants, we reasoned that the novel genes might function in one of those pathways. To characterize the function of Hsk2 and Hsk3 and determine in which pathway they function, we tagged the endogenous proteins with a triple-HA peptide and examined their localization. By indirect immunostaining, we showed Hsk2 and Hsk3 colocalized along with the mitotic spindle (Fig. 4A). This localization pattern is very similar to those of other components of the DASH complex, which are distributed along the mitotic spindle and kinetochores.
FIG. 4.
Hsk2 and Hsk3 are spindle and centromere-binding proteins. (A) Hsk2 and Hsk3 localize along the mitotic spindle and SPB. Cells carrying the integrated HSK2-HA or HSK3-HA genes were fixed for indirect immunostaining with anti-HA and antitubulin antibodies. (B) ChIP analysis of HSK2-HA and HSK3-HA. Binding of Hsk2 and Hsk3 to CEN3, CEN16, and a noncentromeric region, PGK1, was analyzed by ChIP with anti-Myc or anti-HA antibodies. (C) Hsk3-HA centromere binding is dependent on intact spindle structures. cdc13-1 HSK3-HA cells were α-factor arrested for 3 h at 25°C and released into YPD prewarmed to 32°C with or without nocodazole (Noc). Seventy minutes later, cells were collected for ChIP analysis. An untagged cdc13-1 strain was included as a control.
Since Hsk2 and Hsk3 have a similar localization pattern as DASH components, we suspected that Hsk2 and Hsk3 might also bind to kinetochores as DASH does. By ChIP, Hsk2 and Hsk3 were shown to bind to the centromere regions of chromosomes 3 and 16 (Fig. 4B). Neither Hsk2 nor Hsk3 showed appreciable binding to the PGK1 locus (Fig. 4B).
The loading of Hsk3 onto kinetochores is dependent on an intact spindle structure.
We previously showed by ChIP analysis that Ask1 bound to the centromere region of chromosomes in a microtubule-dependent manner (29). If Hsk3 is a DASH component, it should also show microtubule dependence for association with centromeric DNA. Therefore, we carried out a ChIP experiment in the presence or absence of the microtubule-depolymerizing reagent nocodazole. To eliminate cell cycle stage difference between the experimental and control samples, a cdc13-1 HSK3-HA strain was used in this experiment since cdc13-1 mutants show a pre-anaphase arrest similar to nocodazole arrest. The cells were synchronized in G1 by α-factor at 25°C and then released into prewarmed medium at 32°C in the absence or presence of nocodazole. Seventy minutes after release, cells were collected and fixed for ChIP analysis. We found the centromere binding of Hsk3 was decreased when cells progressed though S phase in the absence of intact spindle structures (Fig. 4C).
Hsk2 and Hsk3 associate with DASH proteins in vivo.
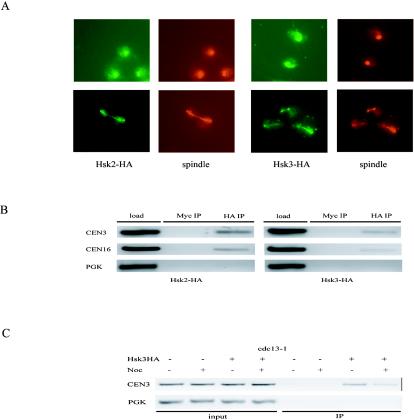
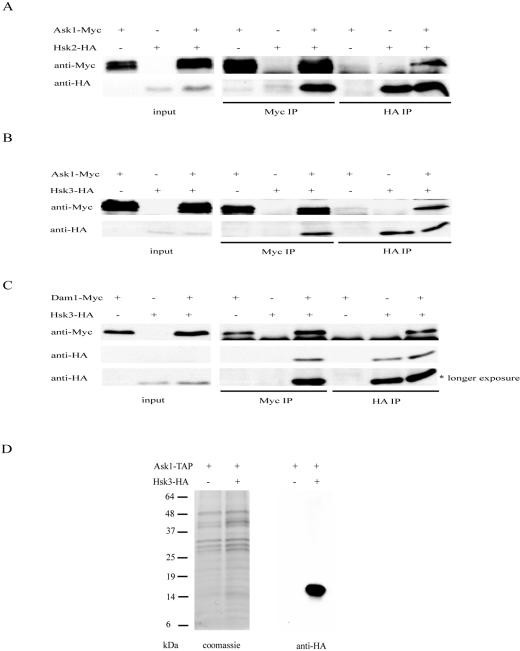
Based on their localization patterns and genetic interactions, we suspected that Hsk2 and Hsk3 are likely to be subunits of the DASH complex. To examine this directly, we tested whether Hsk2 and Hsk3 could bind DASH components by coimmunoprecipitation. Hsk2 was found to coimmunoprecipitate with Ask1 (Fig. 5A). In the reciprocal immunoprecipitation, Ask1 was also shown to coimmunoprecipitate with Hsk2 (Fig. 5A). This clearly demonstrates that Hsk2 interacts with Ask1. Hsk3 also interacts with Ask1 (Fig. 5B) and Dam1 (Fig. 5C), as shown by reciprocal coimmunoprecipitation.
FIG. 5.
Hsk2 and Hsk3 are components of the DASH complex. (A) Hsk2 associates with Ask1. (Top) Extracts from ASK1-MYC, HSK2-HA, and ASK1-MYC HSK2-HA strains were immunoprecipitated (IP) with anti-HA or anti-Myc antibodies and probed with either anti-HA or anti-Myc as indicated. Input represents 2% of the total. (B) Hsk3 associates with Ask1. (Top) Extracts from ASK1-MYC, HSK3-HA, and ASK1-MYC HSK3-HA strains were immunoprecipitated with anti-HA or anti-Myc antibodies and probed with either anti-HA or anti-Myc as indicated. (C) Hsk3 associates with Dam1. (Top) Extracts from DAM1-MYC HSK3-HA strains were immunoprecipitated and probed with either anti-Myc or anti-HA antibodies. (Bottom) A longer exposure of the anti-HA Western blot. (D) Hsk3 is a component of the DASH complex. TAP-tagged purification of the DASH complex from ASK1-TAP and ASK1-TAP HSK3-HA strains was resolved by 4 to 20% polyacrylamide SDS-PAGE and stained with Coomassie blue or Western blotted with anti-HA antibodies.
Previous studies failed to identify Hsk3 in the DASH complex. This might be due to its small size, 69 aa, or the absence of HSK3 in the annotated yeast protein database. To further confirm that Hsk3 is in the DASH complex, we epitope tagged HSK3 with HA in the ASK1-TAP strain. By TAP-tagged purification of Ask1, we purified the whole DASH complex as described previously (29) and found that Hsk3-HA was present within the DASH complex by immunoblotting analysis (Fig. 5D). One interesting note is that both Hsk2 and Hsk3 immunoprecipitate only one of the three different migrating forms of Ask1. We had previously shown that Ask1 mobility shifts were in part due to Cdk phosphorylation (30). This may indicate that the complex is more stable during interphase when Cdk1 is active and is consistent with our previous observation that Cdk phosphorylation positively regulates Ask1 function.
Hsk3 mutants display a metaphase-arrested phenotype.
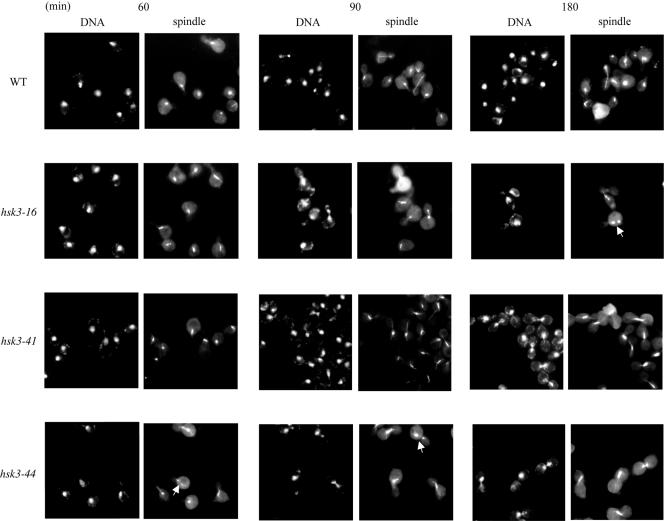
To better understand the role of Hsk3, we generated temperature-sensitive alleles by PCR mutagenesis. A library of PCR-mutagenized HSK3 genes was cloned into pRS414 in the HSK3 deletion strain, which contains wild-type HSK3 on a URA3 CEN plasmid. Clones that allowed growth on 5-FOA at 25°C but not at 37°C were selected. Three temperature-sensitive alleles were identified and characterized. Since the DASH complex is a mediator of spindle-kinetochore interaction and also contributes to spindle integrity, we examined the integrity of spindle and cell cycle progression phenotypes in the temperature-sensitive mutants. Cells were α-factor arrested for 3 h before release into the nonpermissive temperature. Samples were collected after 60, 90, and 180 min at the restrictive temperature and fixed for spindle staining. In contrast to the wild type, all the TS mutants arrested in metaphase with short spindles and DNA in the bud necks (Fig. 6). This was due to arrest as opposed to delayed entry or progression through the cell cycle as cell cycle progressions for the wild type and hsk3 mutants were identical at the 60- and 90-min time points (Fig. 6). We also observed broken spindle structures in hsk3-16 and hsk3-44 mutants, as indicated by the arrowheads in the figure. Given that certain alleles of DASH components give a metaphase arrest and display broken spindle structures like ask1 mutants, we take this as further support that Hsk3 is a bona fide component of the DASH complex.
FIG. 6.
HSK3 is required for mitotic progression and spindle integrity. hsk3-16, hsk3-41, and hsk3-44 mutants were α-factor arrested for 3 h at 25°C and released into YPD at 37°C. Cells were collected for DNA and spindle staining at 60, 90, and 180 min after release from the α-factor block. The white arrows show examples of broken spindles. WT, wild type.
Hsk3 is required for integrity of the DASH complex.
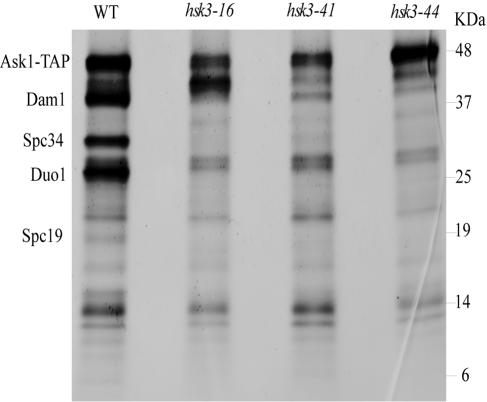
HSK3 overproduction can suppress DASH mutants and is required genetically for DASH function in vivo. We sought to investigate the role of Hsk3 in DASH biochemically. To do this, we purified DASH complexes by TAP-Ask1 affinity purification. Purification of DASH and analysis of Coomassie staining protein can easily identify the large subunits of the complex Ask1, Dam1, Duo1, Spc34, and Spc19 from wild-type cells as previously shown (29). In hsk3-44 and hsk3-41 mutants, we see a significant reduction in the amount of Spc34, Dam1, Duo1, and Spc19 associated with Ask1 (Fig. 7). hsk3-16 mutants show a similar phenotype with a significant reduction of Spc34, Dam1, Duo1, and Spc19; however, there is a significant increase in the abundance of a protein that runs between Ask1 and Dam1. Western blotting revealed this band to be Ask1. Thus the slower-migrating form of Ask1 might be its nonphosphorylated form, which has a faster mobility (29), or it could be a proteolytic fragment of the TAP-Ask1 protein. The identity of this Ask1 species will require further investigation; however, it is clear that Hsk3 plays an important role in maintaining the association of Ask1 with multiple components of the DASH complex and may be directly responsible for recruitment of Ask1 into the complex.
FIG. 7.
The integrity of the DASH complex depends upon Hsk3. TAP tag purification of the DASH complex from ASK1-TAP and ASK1-TAP hsk3-16, ASK1-TAP hsk3-41, and ASK1-TAP hsk3-44 mutant strains was resolved by 4 to 20% polyacrylamide SDS-PAGE, and the strain was stained with Coomassie blue. WT, wild type.
DISCUSSION
In this study, we have undertaken a genetic analysis of the DASH complex to explore the pathways in which it functions and to learn more about the complex itself. These genetic suppressor screens have identified new components of DASH essential for its function as well as a previously unappreciated interaction with the Ras/PKA pathway in cells.
The Ras pathway in budding yeast consists of two homologous proteins, Ras1 and Ras2, which share some redundancy as the Δras1 Δras2 double mutant is lethal (26). This lethality is due to an inability to activate the PKA kinase family, Tpk1, -2, and -3, because deletion of the inhibitor of PKA, BCY1, suppresses this lethality (44). Ras proteins normally act to inhibit Bcy1 through the stimulation of cAMP synthesis. While there is redundancy between Ras1 and Ras2, there are also differences. Ras1 and Ras2 show different patterns of transcriptional regulation (7) and different genetic interactions (46, 47). Our studies have revealed additional differences. Deletion of RAS2 but not RAS1 was capable of suppressing ask1-3 and dam1-1 mutants. This might indicate that Ras2 is the dominant member of the pathway as the remaining Ras1 protein is insufficient to provide sufficient PKA activity to antagonize DASH function. However, this explanation does not explain the fact that overproduction of Ras1 also suppresses ask1-3 and dam1-1 mutants. The most likely explanation for this apparently contradictory observation is that Ras1 and Ras2 have different roles in the cell and that Ras1 overproduction interferes with Ras2 function to mimic the ras2 deletion mutant.
A key question is how the PKA pathway antagonizes DASH function. Since turning down the PKA pathway suppresses multiple mutants in the DASH complex (ask1, dam1, and duo1) and increasing PKA activity exacerbates the temperature sensitivity of several DASH mutants, it is unlikely to be affecting the activity or stability of a single-mutant protein in the complex. In addition, there is some specificity in the suppression in that reducing PKA activity suppresses some ask1 alleles but not others and fails to suppress the null allele. Using this information, we can envision two models to explain these genetic interactions. The first model is that the PKA pathway inhibits a pathway redundant with one of the essential functions for DASH. The essential functions of DASH are not clear, but there could be as many as three different essential functions: control of microtubule stability, kinetochore capture by microtubules, and the ability to reverse monopolar microtubule-kinetochore binding in the absence of tension. If different alleles of DASH mutants primarily affected distinct essential functions, only those alleles affecting the function compensated for by activation of the redundant pathway would be suppressed. Ras has been previously implicated in mitotic control through regulation of Cdc20 and APC (6, 22). In addition, a recent study showed it could have a role in budding in yeast cytokinesis (39) and stimulation of the Ras/PKA pathway was also reported to repress the expression of G1 cyclins and coregulated genes (45). Perturbing any of these processes could affect cell cycle transitions that indirectly have consequences for DASH function. A hypothetical example of how a redundant PKA-regulated pathway might suppress is by reducing the severity of the monopolar spindle attachment problem. One of the unique cell biological problems faced by budding yeast is that newly replicated centromeres attach primarily to the old SPB because the new SPB matures after centromere duplication and kinetochore maturation. Thus, there are many more monopolar attachments that must be reversed than would be expected if both SPB were active prior to kinetochore maturation. If loss of Ras2 and lowering of PKA activity delayed the timing of replication of centromeres, delayed the maturation of kinetochores relative to SPB maturation, or sped up the duplication and maturation of the new SPB, the number of mono-attached sister chromatids might be decreased and contribute to the suppression of DASH mutants defective in the correction of monopolar attachment reversal.
A second model is that PKA directly regulates DASH function. In this model, the allele specificity could arise by suppression of an allelic series with the most severe alleles failing to be suppressed by loss of RAS2 and down-regulation of the PKA pathway. If direct regulation by PKA occurs, then it would be anticipated that DASH components should be substrates for PKA phosphorylation. We examined this in vitro by treating purified DASH components with PKA and labeled ATP and observed phosphorylation of several DASH components (data not shown). While it would be necessary to prove this phosphorylation has in vivo significance with respect to PKA regulation of DASH, the in vitro phosphorylation observation does lend support to the second model.
Two important questions remain. (i) Why would PKA and glucose signaling regulate DASH? (ii) How is this regulation achieved? It is unclear why nutrient availability should directly affect the activity of mitotic regulators such as DASH, but PKA has been linked to other mitotic events (6, 17, 22, 40) with no clear biological rationale. It is possible that the cell is willing to accept higher rates of mitotic infidelity under conditions of rich nutrient availability in order to duplicate more rapidly and outcompete other consumers of nutrients in their ecosystem. Further investigations will be required before we truly understand the rationale for this link.
How the regulation of DASH by PKA is achieved is still not understood. If it is by direct phosphorylation, identifying the sites of phosphorylation and mutating those sites will be critical for demonstrating this. It is clear that components of DASH are phosphorylated by both Ipl1 (8, 29) and Cdk1 (30), and several phosphorylation sites have been identified (8, 30). The consensus phosphorylation site for PKA is R-R-X-S/T-B, where B is hydrophobic. The suggested consensus site for Ipl1 is R/K-X-S/T-[ILV]. Thus there is some potential overlap between these kinases. Analysis of the consensus PKA phosphorylation sites in DASH members identified three potential PKA phosphorylation sites that were also shown to be phosphorylated in vivo by Ipl1: Ask1 S200 (RKRKISLLLQQ), Spc34 T199 (NQRRKTIFVED), and Dam1 S257 (KLRRKSILHTI) (phospho-amino acids are underlined) (8). Thus PKA and Ipl1 may phosphorylate the same sites. It has been hypothesized that in the presence of monopolar attachement, Ipl1 phosphorylates DASH on Dam1 (and possibly other DASH components) and negatively regulates DASH to reduce its affinity for the kinetochore, causing a reversal of binding and providing an additional opportunity for bipolar attachment of the sister kinetochores. If PKA could constitutively phosphorylate the same inhibitory sites as Ipl1, PKA would negatively regulate DASH, providing a possible mechanism for the exacerbation of DASH mutants by the PKA pathway. The fact that they happen to share overlapping specificities could be completely coincidental and normally of no real biological consequence if phosphorylation occurs at low levels in wild-type cells. Only in a mutant with compromised DASH function might this situation become deleterious.
Our genetic analyses also identified two new components of DASH, YDR320C-A and YKL138C-A, named HSK2 and HSK3 (helper of Ask1) with respect to their suppressor function toward ask1 mutants. These two genes were not annotated by the Saccharomyces cerevisiae Genome Project due to their small size. Based on several lines of evidence, we have concluded that they are components of the DASH complex. First, upon overproduction they suppress ask1 mutants. Second, they are essential genes, like all other DASH components. Third, they display microtubule association as well as kinetochore localization by ChIP analysis, like other DASH components. Fourth, they physically associate with multiple members of the DASH complex, as measured by reciprocal immunoprecipitation. Finally, conditional mutants of Hsk3 display phenotypes at the nonpermissive temperature, such as G2 arrest and broken spindles characteristic of other DASH mutants. While this work was in progress, Cheeseman et al. also identified Hsk2 as a DASH component by immunoprecipitation (8). Thus, we conclude that both Hsk2 and Hsk3 are bona fide components of DASH.
The function of individual components within the DASH complex is not known. An interaction map for DASH components was reported, but no interaction between Ask1 and other DASH components was observed (21, 41). Given the strong suppression of ask1 mutants by Hsk3 overproduction, we hypothesize that Hsk3 may be required to allow Ask1 to bind to components of DASH. This idea is supported by the analysis of Ask1-associated DASH components in hsk3 mutants. In three different temperature-sensitive alleles of HSK3, Ask1 associations of several of the large DASH proteins that can be detected by Coomassie staining, Dam1, Duo1, Spc34, and Spc19, were significantly reduced. Therefore, Hsk3 is required for the integrity of the DASH complex and may be the missing link in the interaction map that recruits Ask1 into the DASH complex in vivo.
In summary, this study has identified new components and new regulatory pathways that regulate the DASH complex to carry out its essential function in yeast. The identification of these new subunits should aid in the reconstitution of the DASH complex for future biochemical determination of its function in promoting proper chromosome segregation.
Acknowledgments
We thank members of the Elledge laboratory for helpful discussions.
This work was supported in part by a National Institutes of Health grant (GM44664) to S.J.E. S.J.E. is an investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Adams, I. R., and J. V. Kilmartin. 2000. Spindle pole bodies duplication: a model for centrosome duplication? Trends Cell Biol. 10:329-335. [DOI] [PubMed] [Google Scholar]
- 2.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: restriction of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 4.Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, J. R., and G. D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454-459. [DOI] [PubMed] [Google Scholar]
- 6.Bolte, M., P. Dieckhoff, C. Krause, G. H. Braus, and S. Irniger. 2003. Synergistic inhibition of APC/C by glucose and activated Ras proteins can be mediated by each of the Tpk1-3 proteins in Saccharomyces cerevisiae. Microbiology 149:1205-1216. [DOI] [PubMed] [Google Scholar]
- 7.Breviario, D., A. G. Hinnebusch, and R. Dhar. 1988. Multiple regulatory mechanisms control the expression of the RAS1 and RAS2 genes of Saccharomyces cerevisiae. EMBO J. 7:1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheeseman, I. M., S. Anderson, M. Jwa, E. M. Green, J. Kang, J. R. Yates III, S. M. Chan, D. Grubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the aurora kinase Ipl1p. Cell 111:163-172. [DOI] [PubMed] [Google Scholar]
- 9.Cheeseman, I. M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J. R. Yates, T. C. Huffaker, D. G. Drubin, and G. Barnes. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067-1076. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 12.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enquist-Newman, M., I. M. Cheeseman, D. Van Goor, D. G. Drubin, P. B. Meluh, and G. Barnes. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12:2601-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farr, K. A., and M. A. Hoyt. 1998. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 18:2738-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwick, K. G., and A. W. Murray. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131:709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwick, K. G., E. Weiss, F. C. Luca, M. Winey, and A. W. Murray. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science 273:953-956. [DOI] [PubMed] [Google Scholar]
- 17.Heo, S. J., K. Tatebayashi, and H. Ikeda. 1999. The budding yeast cohesin gene SCC1/MCD1/RHC21 genetically interacts with PKA, CDK and APC. Curr. Genet. 36:329-338. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann, C., I. M. Cheeseman, B. L. Goode, K. L. McDonald, G. Barnes, and D. G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyt, M. A., and J. R. Geiser. 1996. Genetic analysis of the mitotic spindle. Annu. Rev. Genet. 30:7-33. [DOI] [PubMed] [Google Scholar]
- 20.Hoyt, M. A., L. Totis, and B. T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66:507-517. [DOI] [PubMed] [Google Scholar]
- 21.Ikeuchi, A., Y. Sasaki, Y. Kawarasaki, and T. Yamane. 2003. Exhaustive identification of interaction domains using a high-throughput method based on two-hybrid screening and PCR-convergence: molecular dissection of a kinetochore subunit Spc34p. Nucleic Acids Res. 31:6953-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irniger, S., M. Baumer, and G. H. Braus. 2000. Glucose and ras activity influence the ubiquitin ligases APC/C and SCF in Saccharomyces cerevisiae. Genetics 154:1509-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janke, C., J. Ortiz, T. U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, M. H., J. B. Bachant, A. R. Castillo, T. H. Giddings, and M. Winey. 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell 10:2377-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, M. H., X. He, T. H. Giddings, and M. Winey. 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. USA 98:13675-13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kataoka, T., S. Powers, C. McGill, O. Fasano, J. Strathern, J. Broach, and M. Wigler. 1984. Genetic analysis of yeast RAS1 and RAS2 genes. Cell 37:437-445. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. H., J. S. Kang, and C. S. Chan. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, R., and A. W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell 66:519-537. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., J. Bachant, A. Alcasabas, Y. Wang, J. Qin, and S. J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, Y., and S. J. Elledge. 2003. The DASH complex component Ask1 is a cell cycle-regulated Cdk substrate in Saccharomyces cerevisiae. Cell Cycle 2:78-83. [PubMed] [Google Scholar]
- 31.Longtime, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.McAinsh, A. D., J. D. Tytell, and P. K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519-539. [DOI] [PubMed] [Google Scholar]
- 33.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vitro cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signaling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 35.Pangilinan, F., and F. Spencer. 1996. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol. Biol. Cell 7:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinsky, B. A., S. Y. Tatsutani, K. A. Collins, and S. Biggins. 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5:735-745. [DOI] [PubMed] [Google Scholar]
- 37.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 38.Scharfenberger, M., J. Ortiz, N. Grau, C. Janke, E. Schiebel, and J. Lechner. 2003. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 22:6584-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneper, L., A. Krauss, R. Miyamoto, S. Fang, and J. R. Broach. 2004. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 3:108-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Searle, J. S., K. L. Schollaert, B. J. Wilkins, and Y. Sanchez. 2004. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 6:138-145. [DOI] [PubMed] [Google Scholar]
- 41.Shang, C., T. R. Hazbun, I. M. Cheeseman, J. Aranda, S. Fields, D. G. Drubin, and G. Barnes. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell 14:3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka, T. U. 2002. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, T. U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M. J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 8:317-329. [DOI] [PubMed] [Google Scholar]
- 44.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 45.Tokiwa, G., M. Tyers, T. Volpe, and B. Futcher. 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371:342-345. [DOI] [PubMed] [Google Scholar]
- 46.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 47.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 48.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 49.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y., and D. J. Burke. 1995. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6838-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winey, M., and E. T. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:23-27. [DOI] [PubMed] [Google Scholar]
- 52.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3089-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]