Abstract
Rad6-mediated ubiquitylation of histone H2B at lysine 123 has been linked to transcriptional activation and the regulation of lysine methylation on histone H3. However, how Rad6 and H2B ubiquitylation contribute to the transcription and histone methylation processes is poorly understood. Here, we show that the Paf1 transcription elongation complex and the E3 ligase for Rad6, Bre1, mediate an association of Rad6 with the hyperphosphorylated (elongating) form of RNA polymerase II (Pol II). This association appears to be necessary for the transcriptional activities of Rad6, as deletion of various Paf1 complex members or Bre1 abolishes H2B ubiquitylation (ubH2B) and reduces the recruitment of Rad6 to the promoters and transcribed regions of active genes. Using the inducible GAL1 gene as a model, we find that the recruitment of Rad6 upon activation occurs rapidly and transiently across the gene and coincides precisely with the appearance of Pol II. Significantly, during GAL1 activation in an rtf1 deletion mutant, Rad6 accumulates at the promoter but is absent from the transcribed region. This fact suggests that Rad6 is recruited to promoters independently of the Paf1 complex but then requires this complex for entrance into the coding region of genes in a Pol II-associated manner. In support of a role for Rad6-dependent H2B ubiquitylation in transcription elongation, we find that ubH2B levels are dramatically reduced in strains bearing mutations of the Pol II C-terminal domain (CTD) and abolished by inactivation of Kin28, the serine 5 CTD kinase that promotes the transition from initiation to elongation. Furthermore, synthetic genetic array analysis reveals that the Rad6 complex interacts genetically with a number of known or suspected transcription elongation factors. Finally, we show that Saccharomyces cerevisiae mutants bearing defects in the pathway to H2B ubiquitylation display transcription elongation defects as assayed by 6-azauracil sensitivity. Collectively, our results indicate a role for Rad6 and H2B ubiquitylation during the elongation cycle of transcription and suggest a mechanism by which H3 methylation may be regulated.
Histone posttranslational modifications represent a major mechanism by which cells control the structure and function of chromatin (5, 22, 55). A diversity of histone modifications, such as acetylation, methylation, and ubiquitylation, are known to exist; significantly, many of these modifications have been linked to the regulation of gene activity. Although the precise mechanisms by which histone modifications contribute to the transcription process are not fully known, increasing evidence suggests that they work together in the form of a histone code to regulate the recruitment of chromatin-modulating factors (16, 23, 51, 54).
While much progress has been made on the mechanisms of transcriptional activation and repression, much less is known regarding how RNA polymerase II (Pol II) accesses DNA in chromatin and transcribes through it (2, 19, 29, 45, 46). Recently, a role for lysine-specific histone methylation in the elongation cycle of transcription has been uncovered. In the budding yeast Saccharomyces cerevisiae, the histone methyltransferases (HMTs) Set1 and Set2, which catalyze methylation of H3 lysines 4 (K4) and 36 (K36), respectively, have been found to be associated with the hyperphosphorylated (elongating) form of Pol II (17, 18). Those studies show that the recruitment of Set1 to Pol II is mediated by the Paf1 elongation complex and is dependent on Kin28, the TFIIH-associated kinase that phosphorylates serine 5 (Ser5) in the C-terminal heptapeptide repeat sequence (C-terminal domain; CTD) of Pol II (31, 42). In contrast, Set2 is recruited directly to the phosphorylated CTD through the actions of the Ctk1 kinase that mediates serine 2 (Ser2) CTD phosphorylation (35, 62). Given that Set1 is localized preferentially to the 5′ end of genes while Set2 is localized preferentially to the 3′ end of genes, distinct roles for these HMTs at different stages of the transcription elongation cycle have been proposed (18).
Recent studies have defined a trans-histone regulatory pathway involving the ubiquitylation of H2B as a prerequisite for the outcome of K4 and lysine 79 (K79) dimethylation, but not K36 dimethylation (9, 14, 43, 52). In yeast, Rad6 is the enzyme responsible for H2B ubiquitylation (ubH2B), and studies have shown that this enzyme is recruited to promoters to participate in the activation of several highly inducible yeast genes, most notably the GAL genes (13, 21, 24). Several of these studies have revealed that Rad6 is rapidly recruited to the upstream activation sequences of the GAL1 gene in a Gal4-dependent manner and that this association requires the E3 ligase Bre1. Importantly, recruitment of Rad6 leads to a concomitant ubiquitylation of H2B at the core promoter of GAL1, which is rapidly removed by the Ubp8 ubiquitin protease subunit of the Gcn5-containing histone acetyltransferase complex, SAGA (21). Given that the addition and subsequent removal of ubH2B are both required for GAL1 activation, the precise control of ubH2B levels at gene promoters is important for establishing proper transcription levels.
How ubH2B mediates its trans-histone effects on histone methylation is poorly understood. A paradox regarding this topic is how a relatively nonabundant, transient, and perhaps promoter-localized modification on H2B would regulate the global outcomes of K4 and K79 methylation, which are much more abundant and distributed throughout the genome. Starting with the notion that Rad6 and ubH2B might not be restricted just to promoter regions (24), coupled with the recent observation that the Paf1 transcription elongation complex regulates the outcome of K4 and K79 methylation (31, 42), we explored the possibility that Rad6 and ubH2B are associated with transcription elongation as well as with promoter activation.
In this report, we show that Rad6 is associated with the hyperphosphorylated form of Pol II and that this association depends on the Rtf1 and Paf1 components of the Paf1 complex and Bre1. Consistent with this observation, we find that the same Paf1 complex components are necessary for the ubiquitylation of H2B, in agreement with other reports (40, 61). However, we also find that while deletion of some Paf1 components nearly abolishes ubH2B and K4 and K79 dimethylation, others reduce ubH2B and K4 and K79 dimethylation levels in a tightly correlated manner that suggests a direct mechanistic link between the two types of histone modifications. Given that Rad6 interacts with the transcriptionally competent form of Pol II, we determined that Rad6 and ubH2B associate with both the promoters and coding regions of active genes. Significantly, using the inducible GAL1 gene as a model system, we found that Rad6 is transiently associated with the GAL1 gene at the onset of Pol II appearance but that the entrance of Rad6 into the body of this gene requires the Paf1 complex. Consistent with Rad6 and H2B ubiquitylation functioning in the elongation cycle of transcription, we demonstrate that Rad6-mediated ubH2B is reduced by Pol II CTD mutations and abolished by the inactivation of the Ser5 CTD kinase that promotes the initiation of elongation. In addition, we show that the Rad6 complex genetically interacts with other elongation factors and that yeast mutants bearing defects in the pathway to H2B ubiquitylation display sensitivity to drugs that affect elongation. Collectively, these studies link histone ubiquitylation with transcription elongation and suggest a mechanism for the trans-histone establishment of histone methylation patterns in the coding regions of genes.
MATERIALS AND METHODS
Yeast strains.
The genotypes and sources of strains used in this paper are shown in Table 1. For gene disruptions, the indicated gene was deleted by high efficiency transformation by using either a PCR product amplified from the KanMX plasmid pRS400 (4) or a PCR product amplified from genomic DNA of the target gene of interest, which had already been replaced by the KanMX gene (Research Genetics). Epitope tagging was performed by using plasmids and procedures as previously described (28). Briefly, the genomic copies of PAF1 or RAD6 were C-terminally tagged with the triple hemagglutinin (HA) epitope by transformation of a PCR product containing the KanMX gene as a marker along with genomic sequences flanking the stop codons for proper integration. All deletions and genomically tagged strains were confirmed by PCR analysis and/or Western blot analysis.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source |
|---|---|---|
| JR5-2A | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100, 〈pRS314-HTB1〉 | Recht and Osley (47a) |
| YCH002 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 bre1Δ::URA3 〈pRS314-HTB1〉 | Kao et al. (24) |
| YKH010 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 〈pRS314-HTB1〉 | Kao et al. (24) |
| YKH045 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UBI4::URA3 〈pRS314-Flag-HTB1〉 | Henry et al. (21) |
| YKH046 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UBI4::URA3 〈pRS314-Flag-htb1-K123R〉 | Henry et al. (21) |
| YCK001 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UBI4::URA3 rtf1Δ::KanMX 〈pRS314-Flag-HTB1〉 | This study |
| YCK003 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 GAPDH::HA-UBI4::URA3 bre1Δ::HIS3 〈pRS314-Flag-HTB1〉 | This study |
| YTX006 | MATaade2-1 can1-100 cyh2 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD6-3HA::KanMX | This study |
| YTX008 | MATaade2-1 can1-100 cyh2 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD6-ProA 7His::his3 paf1 3HA::KanMX | This study |
| YTX016 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 rtf1Δ::KanMX 〈pRS314-HTB1〉 | This study |
| YTX017 | MATahtb1-1 htb2-1 leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 RAD6-HA::his5 paf1Δ::URA3 〈pRS314-HTB1〉 | This study |
| YTX018 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 rtf1Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX019 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 paf1Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX020 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 ctr9Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX021 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 cdc73Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX022 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 leo1Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX023 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 set2Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YTX028 | MATaade2-1 can1-100 cyh2 his3-11,15 leu2-3,112 trp1-1 ura3-1 paf1 3HA::KanMX | This study |
| YTX031 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 (pZS145 HTA1-Flag-HTB1 CEN HIS3) kin28Δ::KanMX (pGK13-KIN28-HA) | This study |
| YTX032 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 (pZS145 HTA1-Flag-HTB1 CEN HIS3) kin28Δ::KanMX (pGK33-kin28-ts16-HA) | This study |
| YTX0037 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 ctk1Δ::KanMX (pZS145 HTA1-Flag-HTB1 CEN HIS3) | This study |
| YZS236 | MATaade2-1 can1-100 cyh2 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD6-ProA 7His::his3 | This study |
| YZS276 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 (pZS145 HTA1-Flag-HTB1 CEN HIS3) | Sun and Allis (52) |
| YZS277 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 (pZS146 HTA1-Flag-htb1-K123R CEN HIS3) | Sun and Allis (52) |
| YZS279 | MATahta1-htb1Δ::LEU2 hta2-htb2Δ leu2-3,-112 his3-11,-15 trp1-1 ura3-1 ade2-1 can1-100 rad6Δ::ura3 (pZS145 HTA1-Flag-HTB1 CEN HIS3) | Sun and Allis (52) |
| Z27 | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114] | Nonet et al. (44) |
| C3 | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (10 copies)] | Nonet et al. (44) |
| C23 | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (12 copies)] | Nonet et al. (44) |
| V26 | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (18 copies)] | Nonet et al. (44) |
| pY1WT(16) | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (16 copies)] | West and Corden (58) |
| pY1WT(12) | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (12 copies)] | West and Corden (58) |
| pY1WT(8) | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD (8 copies)] | West and Corden (58) |
| pY1A2(8)WT(7) | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD] | West and Corden (58) |
| pY1A5(5)WT(7) | MATα hisΔ200 ura3-52 leu2-3,-112 rpb1Δ187::HIS3 [pRP114ΔCTD] | West and Corden (58) |
Wild-type or KIN28 temperature-sensitive mutant strains were created in the H2B Flag-tagged strain YZS276 by using the plasmid-shuffling technique as previously described (7). Briefly, a wild-type KIN28 expression plasmid (created by cloning the KIN28 gene by PCR into an expression vector driven by the ADH1 promoter) was transformed into YZS276 by using the URA3 marker for selection. Following the replacement of the genomic KIN28 gene with KanMX, the wild-type KIN28-HA expression plasmid (pGK13), or the temperature-sensitive KIN28 allele (kin28-ts16-HA) expression plasmid (pGK33) was transformed into the KIN28-disrupted strain and selected for by using the TRP1 marker. These transformants were then replica plated onto 5-fluoroorotic acid-containing plates to select for the wild-type or mutated form of KIN28 (creating YTX031 or YTX032, respectively). As expected, the kin28-ts16 allele in the H2B-Flag background exhibited a slow growth phenotype at 25°C and was inviable at 37°C.
Generation of PAF1 and RTF1 expression constructs.
PAF1 and RTF1 open reading frames (ORFs) were PCR amplified from genomic DNA with a forward primer with a BamHI site added in front of the ATG codon and a reverse primer with a FLAG coding sequence inserted in front of the stop codon. Reverse primers for both ORFs have a SalI restriction site added after the stop codon. For PAF1 PCR amplification, the forward and reverse primer sequences used were 5′-GCGCGCGGATCCATGTCCAAAAAACAGGAATATATTGCACC-3′and 5′-GTACGCGTCGACCTACTTGTCATCGTCGTCCTTGTAGTCTTCTTCTTGTAAAGTTTCCTTTTCTTC-3′. For RTF1 PCR amplification, the forward and reverse primer sequences used were 5′-GCGCGGATCCATGTCTGATTTAGATGAGGATTTATTAGCC-3′ and 5′-GTACGCGTCGACCTACTTGTCATCGTCGTCCTTGTAGTCAAACTTAAGGTCAAATTTGATATC-3′. PAF1-Flag and RTF1-Flag PCR products were cloned into the PN823 yeast expression vector driven by the ADH1 promoter to produce PN823Paf1 and PN823Rtf1, respectively. Upon DNA sequencing, these fusion gene plasmids were transformed into the paf1Δ (YTX019) and rtf1Δ (YTX018) strains and selected for on synthetic complete-Ura plates.
Preparation and immunoblot analysis of yeast whole cell extracts and nuclei.
Yeast whole cell extracts (WCEs) and nuclei were prepared as previously described (62). Typically, 5 μl of WCE (∼50 μg of protein) or 3 to 5 μl of isolated nuclei (∼100 μg of protein) was loaded on sodium dodecyl sulfate (SDS)-polyacrylamide gels (8% gels for Pol II analysis and 15% gels for the analysis of histones or Rad6). Following electrophoresis and transfer to polyvinylidene difluoride membranes, samples were subjected to immunoblot analysis using procedures and detection reagents from Amersham Life Sciences. Rabbit anti-histone modification-specific antibodies were obtained from Upstate Biotechnology, Inc., and used at the following dilutions: 1:3,500 for anti-H3 dimethyllysine 36 (α-H3K36Me2), 1:25,000 for anti-H3 dimethyllysine 4 (α-H3K4Me2), 1:10,000 for anti-H3 dimethyllysine 79 (α-H3K79Me2), 1:10,000 for anti-H3 acetyllysine 14 (α-H3K14Ac), and 1:30,000 for anti-H4 acetylation (penta; α-H4Ac). Mouse monoclonal anti-FLAG antibody (M2; Sigma) was used at 1 μg/ml, and the mouse monoclonal anti-HA antibody (12CA5; Roche) was used at a 1:5,000 dilution. Anti-RNA Pol II CTD antibodies 8WG16 (unmodified CTD), H14 (serine 5 phosphorylation specific), and H5 (serine 2 phosphorylation specific) were purchased from Covance, Inc., and used at dilutions of 1:500, 1:50,000, and 1:750, respectively. The rabbit polyclonal antibodies to protein A (Sigma) and the C terminus of H3 (Abcam) were used at a 1:50,000 dilution and a 1:3,500 dilution, respectively. For detection of ubiquitylated H2B, cell pellets corresponding to approximately 106 cells from exponentially growing cultures at an optical density at 600 nm (OD600) of 1.0 were boiled directly in SDS-polyacrylamide gel electrophoresis loading buffer and examined by immunoblot analysis using the anti-Flag antibody.
Immunoprecipitations (IPs).
For IP experiments involving Rad6-3HA, 300 to 400 μl of WCE (containing 1.5 mg of protein) was diluted to a final volume of 750 μl by using extraction buffer and then incubated with 12.5 μl of pre-equilibrated α-HA affinity beads (A-2095; Sigma) for 2 h at 4°C. After three washes in extraction buffer, the bead-bound proteins were analyzed by immunoblot analysis using the antibodies indicated at the dilutions described above. For IPs involving phosphorylated CTD, WCEs at the same concentration and volume described above were incubated with H5 or H14 antibody (at a dilution of 1:400) for 3 h at 4°C. For immunoprecipitation, 12.5 μl of pre-equilibrated goat anti-mouse immunoglobulin M (IgM) agarose (ICN Biomedicals, Inc.) was added for another hour at 4°C and the bead-bound proteins were washed and analyzed. For studying the association between Rad6 and Paf1, WCEs (at the same concentration and volume as described above) from strains YTX008 and YZS236 were incubated with IgG-Sepharose (Amersham Biosciences) and the bead-bound proteins were washed and immunoblotted with anti-HA monoclonal antibody.
RT-PCR.
For reverse transcription (RT)-PCR analysis of Rad6 pathway genes, yeast strains were grown in 100 ml of yeast extract-peptone-dextrose (YPD) medium to an OD600 of 1.0. Total RNA was isolated by using a Clontech Nucleospin RNA purification kit, and 100 μg of total RNA was subjected to poly(A+) mRNA isolation by using a QIAGEN Oligotex mRNA kit. The resulting mRNA was used to synthesize cDNA by using M-MLV reverse transcriptase (Invitrogen) per the manufacturer's directions. The PCR consisted of 1/20 of the cDNA collected from the RT reaction and primer pairs specific to the indicated genes. For RT-PCR analysis of GAL1 RNA, yeast strains were grown to an OD600 of 0.5 in YPD medium, shifted to YP plus 2% raffinose, and induced with 2% galactose. Two micrograms of total RNA was used to synthesize cDNA by using an Omniscript reverse transcription kit (QIAGEN). Real-time PCRs were performed with 1/10 of the cDNA reactions by using a SYBR green master mix in an MJ Opticon PCR machine. Primer information is available upon request. Negative controls with no RT were also included.
Chromatin immunoprecipitation.
Strains containing Rad6 tagged with three copies of the HA epitope at its C terminus were fixed and immunoprecipitated with anti-HA antibodies as described by Kao et al. (24). Levels of ubH2B were determined by a chromatin double immunoprecipitation (ChDIP) method in strains containing a Flag-HTB1 or Flag-htb1-K123R gene and a constitutively expressed HA-UBI4 gene as previously described (21, 24). Pol II levels were determined by using the anti-RNA Pol II CTD antibody 8WG16 (Abcam ab817). Cells analyzed by chromatin immunoprecipitation were grown in YP medium containing 2% glucose or were washed and resuspended for 2 h in YP + 2% raffinose, and they were then shifted to YP + 2% galactose to induce the GAL genes as described by Henry et al. (21). Quantitative PCR in real time was carried out with a SYBR Green master mix and a 7000 Prism sequence detection system, both from ABI. Primer sequences are available upon request. The results represent the ratio of immunoprecipitated (IP) DNA to input DNA (Input) normalized to the IP/Input ratio from a telomere-associated region, TEL-V, or an ORF-free intergenic region on chromosome V, INT-V (24).
SGA technology.
Synthetic genetic array (SGA) analysis was carried out as previously described (53). An automated analysis of the results was carried out by procedures that will be described elsewhere (H. Ding et al., unpublished data).
RESULTS
Paf1 components selectively regulate H2B ubiquitylation.
Recent studies have shown that deletion of Rad6, in addition to deletion of the Rtf1 and Paf1 components of the Paf1 elongation complex, greatly reduces and/or abolishes the levels of H3 K4 and K79 methylation and H2B ubiquitylation (31, 40, 42, 61). To further address the extent to which the Paf1 complex regulates Rad6-mediated ubiquitylation of H2B, we examined the effects of deleting the Paf1 complex in a strain in which H2B was Flag tagged to monitor the presence of monoubiquitylated H2B. As expected, deletion of the RAD6 gene abolished all detectable levels of ubH2B as determined by immunoblot analysis using an anti-Flag antibody (Fig. 1A). In agreement with recently published reports (40, 61), we also found that deletion of genes encoding two Paf1 complex members, Rtf1 and Paf1, resulted in greatly reduced levels of ubH2B that correlated with the loss of H3 K4 and K79 dimethylation (Fig. 1A). Importantly, the loss of ubH2B in the rtf1 and paf1 deletion strains could be restored with ectopically expressed RTF1 and PAF1 (Fig. 1B). We additionally found that deletion of other Paf1 complex members, CTR9 and CDC73, also resulted in a significant reduction in the levels of ubH2B, while deletion of the LEO1 subunit had no effect (Fig. 1A). Interestingly, the reduction in ubH2B levels observed with the ctr9 and cdc73 deletion mutants was paralleled by a similar reduction in the levels of K4 and K79 dimethylation (Fig. 1A), suggesting that ubH2B levels are tightly correlated with the potential for K4 and K79 methylation.
FIG. 1.
The Paf1 elongation complex regulates H2B ubiquitylation. (A) RAD6 (YZS279), SET2 (YTX023), or the indicated Paf1 complex gene (YTX018-YTX022), was deleted from a yeast strain bearing a Flag-tagged copy of histone H2B (YZS276). Cell pellets derived from these strains were loaded directly on an SDS-polyacrylamide gel, and the levels of H2B ubiquitylation were determined by immunoblot analysis using an anti-Flag antibody. The presence of H2B and the monoubiquitylated form of H2B (ubH2B) are indicated. Independently, nuclei were prepared from these strains and analyzed by immunoblot analysis for the presence of H3 lysines 4 (K4), 36 (K36), and 79 (K79) dimethylation. Asterisks indicate N-terminal breakdown products of H3 that are typically observed. (B) The loss of H2B ubiquitylation in the rtf1Δ and paf1Δ strains can be restored. Cell pellets isolated from the WT (YZS276), rft1Δ (YTX018), or paf1Δ (YTX019) strain containing either the vector control plasmid or the appropriate Rtf1-Flag or Paf1-Flag expression plasmid were probed with the anti-Flag antibody. The locations of Rtf1 and Paf1 are indicated. It is noteworthy that Rtf1 shows a variety of N-terminal breakdown products that naturally occur during logarithmic growth.
The Paf1 complex, along with Bre1, mediates the association of Rad6 with the hyperphosphorylated form of RNA polymerase II.
To determine if the effects of the Paf1 complex on ubH2B levels are mediated through the ability of Rad6 to directly interact with this complex and possibly with Pol II, we genomically tagged Rad6 at the C terminus with a triple-HA epitope and immunoprecipitated Rad6-HA to examine its protein associations. Using antibodies that distinguish between the phosphorylated and unphosphorylated states of Pol II, we found by immunoblot analysis that both the Ser5- and Ser2-phosphorylated forms of Pol II copurified with Rad6-HA (Fig. 2A). Importantly, the unphosphorylated form of Pol II did not copurify with Rad6, indicating that Rad6 associates specifically with the elongating form of the polymerase (Fig. 2A). We confirmed this interaction in a reverse approach by first immunoprecipitating Pol II with the Ser5-phosphorylated-specific CTD antibody (H14), followed by immunoblotting for Rad6 with anti-HA antibodies (Fig. 2B). The Ser5 and Ser2 phospho-specific CTD antibodies were also used to confirm the presence of both forms of phosphorylated Pol II in the Rad6-containing immunoprecipitates (Fig. 2B). As a control, experiments in which the Ser5 phosphorylation-specific CTD antibody was excluded from the IPs showed that these bead-only immunoprecipitates did not contain Rad6-HA or Pol II (data not shown). Finally, we independently verified the Rad6-Pol II interaction in a different strain background in which Rad6 was genomically tagged at the C terminus with glutathione S-transferase and six-histidine (data not shown).
FIG. 2.
The Paf1 complex and Bre1 mediate Rad6 association with the hyperphosphorylated form of Pol II. (A) Whole cell extracts (WCEs) prepared from WT (W303) or genomically tagged Rad6 (Rad6-HA) cells (YTX006) were immunoprecipitated with anti-HA beads followed by immunoblotting with antibodies directed against unmodified CTD (8WG16; α-CTD), serine 2-phosphorylated CTD (H5; α-Ser2P), serine 5-phosphorylated CTD (H14; α-Ser5P), or α-HA. These WCEs were also examined by immunoblot analysis with the antibodies described above to monitor the presence of these proteins (Input). (B) Rad6 co-IPs with hyperphosphorylated RNA Pol II. WCEs from WT or Rad6-HA cells were immunoprecipitated with the α-Ser5P antibody followed by immunoblot analysis with α-HA, α-Ser5P, and α-Ser2P antibodies. (C) Rad6 interacts with Paf1. WCEs from either a Rad6-ProA-7Xhis-tagged strain (YZS236), a Paf1-HA tagged strain (YTX028), or a strain in which Rad6-ProA-7Xhis was double tagged with Paf1-HA (YTX008) were used for immunoprecipitation with IgG-Sepharose and then immunoblotted with an anti-protein A antiserum or the anti-HA antibody. WCEs were also probed with the anti-HA and anti-protein A antibodies to verify that the input levels of Rad6 and Paf1 were similar. (D) The Paf1 complex and Bre1 mediate the association of Rad6 to Pol II. PAF1, RTF1, or BRE1 (YTX016, YTX017, or YCH002, respectively) was deleted from an HA-tagged Rad6 strain (YKH010), which was then analyzed for Pol II association. WCEs from these strains were immunoprecipitated for Rad6 with anti-HA and then immunoblotted for phosphorylated Pol II with the α-Ser5P antibody H14. Similar results were obtained with the Ser2 CTD phosphorylation-specific antibody (data not shown). The input WCEs derived from these strains were also examined by immunoblot analysis to monitor the presence of Rad6 and Pol II. The asterisk indicates a nonspecific band that was observed.
To learn if Rad6 is also associated with the Paf1 elongation complex, we took advantage of a yeast strain in which Rad6 was C-terminally tagged with protein A and seven-His. Within this strain, Rtf1 or Paf1 was C-terminally tagged with 3× HA, and the resulting strains were used for co-IP experiments. As a control, we constructed isogenic strains in which only Rtf1 or Paf1 was HA tagged. Whole cell extracts were prepared, and then Rad6-protein A was immunoprecipitated with IgG-Sepharose followed by immunoblotting with anti-HA antibody. The results clearly show that Rtf1-HA and Paf1-HA are present in Rad6 immunoprecipitates, thereby confirming their association (Fig. 2C and data not shown).
We next sought to determine if the association of Rad6 with Pol II was dependent on the Paf1 complex as well as on other regulators of Rad6 function, such as Bre1. Bre1, the E3 ligase that directs Rad6 to ubiquitylate H2B, was recently shown to be a major determinant in the recruitment of Rad6 to gene promoters (24, 60). The RTF1, PAF1, or BRE1 gene was deleted from the Rad6-HA strain used in Fig. 2, and these strains were then employed in co-IP studies to examine the association of Rad6 with Ser5-phosphorylated Pol II. We found that deletion of BRE1 completely abolished the association of Rad6 with Pol II, whereas deletions of PAF1 and RTF1 severely reduced this association (Fig. 2D). The limited association of Pol II with Rad6 in the rtf1Δ and paf1Δ mutants is consistent with the observation that these deletions do not completely abolish ubH2B (Fig. 1A). Furthermore, we found that both Rad6-Pol II association and ubH2B levels were more severely compromised in an rtf1Δ mutant than in a paf1Δ mutant (see Fig. 1A and 2D). These data suggest that both Bre1 and the Paf1 complex regulate an association of Rad6 with Pol II that is important for ubiquitylation of H2B.
The Paf1 complex regulates a Pol II-dependent association of Rad6 with the transcribed region of genes.
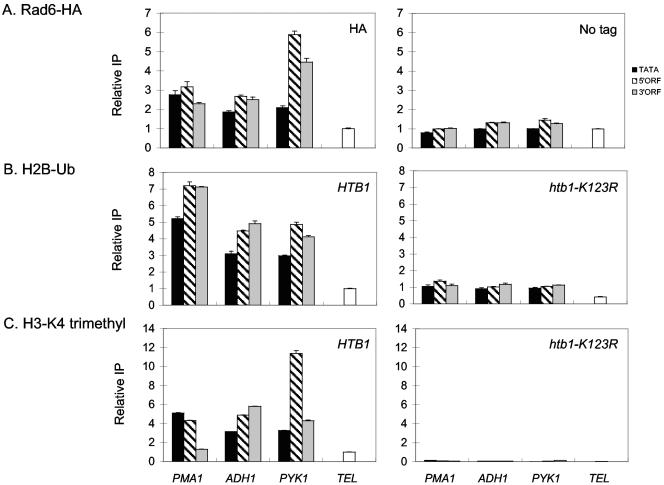
The finding that Rad6 associates with the phosphorylated form of Pol II suggests that Rad6 is involved in the process of transcription elongation and, thus, should be present at both the promoters and the transcribed regions of genes. Indeed, our previous studies showed that Rad6 associates across the body of the activated GAL1 gene (24). To determine if coding region association of Rad6 and ubH2B is a general property of active genes, we used the method of chromatin immunoprecipitation (ChIP) to measure the association of Rad6 and ubH2B with the promoters and ORFs of three highly transcribed constitutive genes, PMA1, ADH1, and PYK1 (Fig. 3). The results show that Rad6 is not restricted to promoters, as was previously reported (60, 61), but is present at the coding regions of all three genes (Fig. 3A). This association is independent of the epitope used to tag Rad6, as both Rad6-HA and Rad6-Flag show similar patterns of association at these genes (data not shown). Moreover, ubH2B is also found at both the promoters and ORFs of the same genes (Fig. 3B), supporting the view that the presence of this histone modification across the genes is a direct consequence of the extended association of Rad6 with the genes. In contrast, we find that H3 K4 trimethylation is more generally restricted to the 5′ region of the same genes, arguing that ubH2B performs other roles in addition to regulating K4 methylation (Fig. 3C).
FIG. 3.
Rad6 and ubH2B are associated with promoters and coding regions. Left panels: (A) ChIP was performed with anti-HA antibodies in a Rad6-HA strain (YKH010; WT); (B) ChDIP was performed with anti-Flag and anti-HA antibodies in a Flag-H2B strain (YKH045; HTB1) that contained HA-ubiquitin; and (C) ChIP was performed with anti-trimethyl H3 Lys 4 antibodies in strain Y131 (HTB1). Right panels: ChIP or ChDIP was performed in strains containing untagged Rad6 (JR5-2A; No tag), a Flag tagged version of the htb1-K123R allele (YKH046; htb1-K123R), or a strain with an htb1-K123R allele (Y133) as specificity controls. Purified DNA was analyzed by PCR in real time with primers corresponding to the core promoters and 5′ and 3′ ORF regions of the constitutively expressed PMA1, ADH1, and PYK1 genes. The relative IPs shown on the y axes represent the IP/Input ratios for each set of primers normalized to the IP/Input ratio for the INT-V region (A) or the TEL-V region (B and C), which were arbitrarily set as 1.0.
The Paf1- and Bre1-mediated interaction of Rad6 with elongating forms of Pol II suggests that both factors play a key role in the association of Rad6 with the coding region of transcriptionally active genes. To investigate this possibility, we used ChIP to measure the association of Rad6-HA with both the promoter and ORF of the GAL1 gene in wild-type (WT), rtf1Δ, and bre1Δ strains during growth in repressed (glucose), noninducing (raffinose) conditions and at 10 to 30 min intervals after induction with galactose (Fig. 4A). These measurements gave us a snapshot of the recruitment events that accompany gene activation. As previously reported (24), in the WT strain, Rad6 transiently associated with the GAL1 UAS after galactose induction, with peak association occurring 30 min after the shift, followed by a successive loss of Rad6 at the promoter over the next 60 min (data not shown). A similar temporal pattern of Rad6 association was observed at the GAL1 core promoter and also at two regions that define the beginning and end of the GAL1 ORF (Fig. 4A, left panels). Significantly, the appearance of Rad6 across the activated GAL1 gene coincided almost precisely with the appearance of Pol II across the gene, indicating that these two events are linked (Fig. 4A, left panels). However, while Rad6 association was transient, Pol II association was not transient, and the polymerase continued to accumulate over the GAL1 gene. These data suggest that the activity of Rad6 is associated with events linked to the earliest round(s) of activated transcription.
FIG. 4.
The Paf1 complex is required for Rad6 association with the GAL1 coding region and for H2B ubiquitylation at the GAL1 gene. (A) ChIP was performed with anti-HA and anti-Pol II antibodies in a Rad6-HA containing WT strain YKH010 and rtf1Δ mutant YXT016 during growth either in glucose-containing medium (Glu), in 2% raffinose-containing medium (Raf), or at 10- to 30-min intervals after a shift to 2% galactose-containing medium. (B) ChIP was performed with the same antibodies in a Rad6-HA-containing bre1Δ mutant (YCH002) after growth in the medium as described for panel A. (C) Cells were shifted from YP + 2% glucose medium into YP + 2% galactose medium at 0 min in the Rad6-HA strains as described for panels A and B and in an untagged strain. ChIP was performed with anti-HA antibodies. ChDIP was performed with a Flag-H2B strain containing HA-ubiquitin under the same growth conditions with a WT strain (YKH045), an rtf1Δ (YCK001) or bre1Δ (YCK003) mutant, and a strain containing Flag-htb1-K123R (YKH046). Real-time PCR was performed and analyzed as described in the legend to Fig. 3, with primers that correspond to the GAL1 core promoter (TATA) and the 5′ or 3′ ORF region. The IP/Input data were normalized as described in the legend to Fig. 3. (D) GAL1 mRNA levels were measured for the WT strain YZS276 with or without rtf1Δ or bre1Δ deletion mutations. Strains were grown under the conditions described for panel A, samples were removed before and at 10- to 30-min intervals after galactose induction, and RT-PCR was performed on extracted RNA with primers against a region of the ORFs of the GAL1 gene and the ACT1 gene, which served as the internal control.
Next, we investigated the role of Bre1 and the Paf1 complex in Rad6 association with the activated GAL1 gene. Our previous work showed that Rad6 requires the Gal4 activator and the associated E3 ligase, Bre1, for its recruitment to the GAL1 UAS (24), and here we find that Bre1 is also required for the association of Rad6 with the core promoter and 5′ coding region (Fig. 4B). However, in striking contrast to the effects of a bre1Δ mutation, which significantly reduced the association of Rad6 with all regions analyzed, an rtf1Δ mutation resulted in a stable accumulation of Rad6 at both the UAS and TATA regions of the GAL1 promoter, with little to no turnover into the transcribed region (Fig. 4A, right panels, and data not shown). In addition, two constitutive genes, PMA1 and ADH1, also showed reduced levels of Rad6 in their coding regions in the absence of Rtf1 (data not shown), supporting the view that the Paf1 complex plays a role in releasing Rad6 from the promoter region into the body of a gene. Pol II also accumulated more slowly at the GAL1 gene in the rtf1Δ mutant (Fig. 4A, right panels), consistent with a role for the Paf1 complex in transcription elongation. While it is a formal possibility that the slow release of Pol II into the GAL1 coding region could account for the retention of Rad6 at the core promoter, our Rad6-Pol II interaction studies (Fig. 2), coupled with the finding that increased Pol II levels can be found across the GAL1 gene in the rtf1Δ mutant after activation (Fig. 4A), argue that the Paf1 complex plays a more direct role in the distribution of Rad6 by linking this enzyme to elongating Pol II. Collectively, the data suggest that Rad6 is associated with genes through two mechanisms: it is recruited to promoters through an activator- and Bre1-dependent mechanism and to coding regions through a Paf1- and Pol II-dependent mechanism (60) (see Fig. 7).
FIG. 7.
A model for how Rad6/H2B ubiquitylation becomes associated with the GAL1 gene and regulates H3 K4 methylation. (A) Based on recent studies and those herein, the recruitment of the Gal4 activator to the UAS in the promoter of the GAL1 gene results in the corecruitment of chromatin remodeling activities and transcriptional coregulators, such as Rad6/Bre1, SAGA, and mediator. Although Rad6/Bre1, SAGA, and mediator are depicted together, we do not suggest that they physically interact and have not indicated their ordered recruitment to the GAL1 promoter. In addition, with the exception of the nucleosome-free region at the upstream activation sequence, we do not accurately depict the nucleosome density of this promoter. Upon recruitment of Pol II and its associated general transcription factors (GTFs), the TFIIH-associated kinase, Kin28, phosphorylates the serine 5 (S5) position in the CTD to initiate transcription elongation. This phosphorylation results in the recruitment of transcription elongation factors to Pol II, such as the Paf1 complex, which in turn corecruits the Set1 HMT. (B) During or after the transcription initiation event and recruitment of the Paf1 complex, Rad6/Bre1 becomes associated with Pol II via the Paf1 complex in a mechanism we describe as a hand-off event. This handoff, or newly formed association of Rad6/Bre1 with Pol II, appears to be essential for the stimulation of Rad6's H2B-modifying activity, which, we propose, leads to localized nucleosome disruption around Pol II that is important for elongation. These disrupted nucleosomes may be those associated with the first or earliest rounds of new transcription, as Rad6 and ubH2B appear across the gene at the same time as Pol II's first appearance but occur transiently. The nucleosome disruption proposed to occur with ubH2B may also include the actions of the 19S proteasomal ATPases, which are recruited to chromatin in a H2B ubiquitylation-dependent manner and are important for H3 methylation (15). Given that Set1 (and perhaps Dot1; see text) is also associated with the Paf1 complex, we suggest that the localized disruption of nucleosomes caused by H2B ubiquitylation allows Set1 to access the core nucleosomes and histone tails for methylation of H3 on lysine 4. However, unlike histone methylation, H2B ubiquitylation is transient, and thus this modification's removal reverses the nucleosomal disruption that we propose to track with Pol II during gene transcription. (C and D) As Pol II moves and clears the promoter, S5 phosphorylation on the CTD is removed and followed by the recruitment of Ctk1, which phosphorylates the serine 2 (S2) position in the CTD and results in the direct association of Set2 with Pol II. Notably, Set1 is removed from the elongating Pol II complex shortly after initiation and loss of S5 phosphorylation; however, the Paf1 complex with Rad6/Bre1 remains associated throughout the length of the gene (Fig. 4; see references 26 and 49). Collectively, our results show that Rad6 travels with Pol II to mediate H2B ubiquitylation, a modification that appears important for the elongation cycle of newly activated transcription and the precise establishment of H3 methylation patterns.
The accumulation of Rad6 at the GAL1 promoter in the rtf1Δ mutant prompted us to ask if ubH2B also accumulated at the regulatory region in this strain. In striking contrast to Rad6, ubH2B was absent at the GAL1 promoter in the rtf1Δ mutant and, as expected, it was also absent at the 5′ORF because of the failure to accumulate Rad6 at that region (Fig. 4C). Since a bre1Δ mutation also abolishes H2B ubiquitylation globally and throughout the GAL1 gene (Fig. 4C) (24), this finding indicates that both factors are required for the activity of Rad6 towards H2B. In the absence of this activity, there is a reduced accumulation of GAL1 transcripts upon galactose induction (Fig. 4D). Coupled with the interaction data, these results suggest that the association of Rad6, via Bre1 and Paf1, to the phosphorylated form of Pol II is necessary for the activity of Rad6 in mediating H2B ubiquitylation on genes and the subsequent high levels of transcription. Recent results with the constitutively active PMA1 gene confirm this notion (60).
H2B ubiquitylation is regulated by the CTD and likely plays a role in transcription elongation.
The finding that the Paf1 complex mediates an association of Rad6 to the elongating form of Pol II as well as to the transcribed region of the GAL1 gene suggests a role for Rad6 and H2B ubiquitylation in the elongation cycle of transcription. Consistent with this view and as mentioned above, Pol II levels were reduced across the GAL1 gene in both rtf1Δ and bre1Δ mutants (Fig. 4A and B, right panels), and GAL1 transcripts accumulated with slower kinetics (Fig. 4D). Given that most elongation activities are coupled to CTD phosphorylation, we asked whether Rad6-mediated H2B ubiquitylation would also be dependent on this Pol II modification. Using the H2B-Flag tagged strain (see Fig. 1), we created a derivative in which the genomic KIN28 allele was deleted in the context of a URA3 plasmid expressing WT Kin28 protein. We then replaced the WT KIN28 gene with a temperature-sensitive KIN28 allele (kin28-ts16-HA) that is inactive at 37°C (11, 27). As expected, Ser5 phosphorylation was eliminated in the kin28-ts16-HA strain upon heat inactivation for 2 h, as shown by immunoblot analysis with the anti-Ser5 phosphorylation-specific antibody (Fig. 5A). Strikingly, thermal inactivation of Kin28 also resulted in a complete abolishment of H2B ubiquitylation (Fig. 5A). It is significant that thermal inactivation of this KIN28-ts allele does not affect the promoter occupancy of transcriptional initiation factors, including TBP, TFIIB, and Pol II (36), and does not result in a loss of Kin28, Pol II, or unmodified H2B protein levels (Fig. 5A) (42). In addition, reversible histone modifications, such as H3 and H4 acetylation and Ser2 CTD phosphorylation, are still present in this strain (Fig. 5A), indicating that the inactivation of this KIN28-ts allele (at least at the time point examined) has not yet fully abolished transcription. This idea is further supported by RT-PCR analysis, which shows that the mRNAs which code for proteins responsible for H2B ubiquitylation are abundant and present at similar levels to those found in the WT and nonshifted kin28-ts16-HA strains (Fig. 5B). Thus, the Kin28-dependent loss of ubH2B is correlated with the loss of Ser5 CTD phosphorylation rather than with the abolishment of transcription or the specific downregulation of genes important for H2B ubiquitylation. These data strongly suggest that H2B ubiquitylation is coupled to CTD phosphorylation, a result that is supported by the observation that a variety of CTD truncations and mutations also significantly reduce the levels of this modification (Fig. 5D).
FIG. 5.
H2B ubiquitylation is linked to events associated with transcription elongation. (A) KIN28 was deleted from the Flag-H2B described in Fig. 1 and replaced with either a WT copy or a temperature-sensitive allele of KIN28 (kin28-ts16) that was HA tagged at the C terminus (11). Similar to previous reports of the kin28-ts16 mutation, the H2B-Flag kin28-ts16-HA strain grew slowly at the permissive temperature and was inviable at 37°C (data not shown). The WT (YTX031) and kin28-ts16-HA H2B-Flag (YTX032) strains were grown at 25°C to 1 OD600 unit and then shifted to 37°C (or not shifted as the control) for 2 h prior to harvesting the cell pellets. A portion of these cell pellets were analyzed directly for the presence of ubiquitylated H2B as described for Fig. 1, and the rest were prepared as WCEs to examine the status of Pol II and Kin28 protein levels by using the indicated antibodies. (B) Expression levels of factors responsible for H2B ubiquitylation or ACT1 (used as the control) were not changed in the H2B-Flag kin28-ts16-HA strain after thermal inactivation for 2 h. Shown are reverse transcription PCRs with (+) or without (−) reverse transcriptase (RTase) before and after thermal inactivation. (C) A deletion of CTK1 was made in the H2B Flag-tagged strain to examine the requirement of H2B ubiquitylation for Ser2 CTD phosphorylation. H2B ubiquitylation and Pol II levels were determined as described above. (D) The CTD is required for H2B ubiquitylation. An expression plasmid containing a N-terminal Flag-tagged version of H2B was transformed into the indicated Pol II CTD truncation or mutation strain, and cell pellets were analyzed for H2B ubiquitylation as described for Fig. 1. The C3, C23, and V26 strains represent CTDs of Pol II that are ∼10, 12, and 18 repeats in length (44). The pY1A2(8)WT(7) and pY1A5(5)WT(7) strains represent mutations of the CTD that encompass a mixture of WT copies of the CTD that are in frame to CTD repeats that harbor alanine substitutions at either the Ser2 or Ser5 position (A2 or A5, respectively); parentheses indicate the repeat lengths associated with the WT or mutated CTD copies (58). These cell pellets were also analyzed for general H3 levels (for an additional loading control) by using an anti-C-terminal H3 polyclonal antibody.
We next examined if the Ser2 CTD kinase, Ctk1, would also affect H2B ubiquitylation. As expected, deletion of CTK1 resulted in the abolishment of Ser2 CTD phosphorylation without affecting the total levels of Pol II (Fig. 5B). However, we found that deletion of this factor in the Flag-H2B strain did not result in a significant decrease in ubH2B levels (Fig. 5B). This result suggests that Ser2 phosphorylation is not required to maintain the association of Rad6 with Pol II, which is consistent with earlier findings that deletion of CTK1 does not affect the levels of K4 and K79 dimethylation (62).
In an effort to further implicate the Rad6 complex in the process of transcriptional elongation, NatR strains harboring individual gene deletions of the components of the Rad6 complex (RAD6, BRE1, LGE1) were generated and crossed to a set of viable deletion strains selected for their involvement in gene expression, and the resulting double mutant strains were analyzed (data not shown). Similar sets of genetic interactions were obtained with all three gene deletions (Fig. 6A), and an enrichment of genetic interactions was obtained with a number of genes implicated in transcriptional elongation. For example, rad6Δ, bre1Δ, and lge1Δ deletions all resulted in growth defects when combined with deletions in genes encoding for several well-characterized transcriptional factors with direct roles in elongation, including DST1 (or TFIIS), SPT4, CTK1, and SET2 (1, 2, 19, 25, 34, 35, 37, 62). Interestingly, genetic interactions were also detected between the H2B ubiquitylation machinery and Rpb9, a nonessential subunit of Pol II whose activity is required for efficient elongation in vivo (3), as well as with components of Elongator, a histone acetyltransferase complex that copurifies with the elongating form of Pol II (32, 47, 59) (Fig. 6A and B). Growth defects were also apparent among rad6Δ, bre1Δ, lge1Δ mutants and nonessential components of the 19S (RPN4, RPN10, UBP6, SEM1) and 20S (PRE9) proteasome (57). The proteasome has not only been linked to transcriptional elongation (57), but directly to histone methylation and ubiquitylation as well (15). Finally, we find genetic interactions between RAD6/BRE1/LGE1 and the Swr1 chromatin-remodeling complex, which contains the histone H2A variant Htz1 (Fig. 6A). The Swr1 complex is required for the exchange of the canonical histone H2A with its variant counterpart, Htz1 (30, 33, 38), and was uncovered in several genetic screens by using nonessential transcriptional elongation factors (33). Since the components of the Rad6 complex genetically interact with so many factors impinging on elongation by Pol II, this finding provides strong evidence that the complex and its activity play a major role in transcriptional elongation.
FIG. 6.
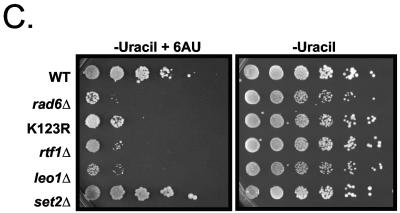
Genetic analysis of the Rad6/H2B ubiquitylation pathway reveals functional links to transcription elongation. (A) Synthetic genetic interactions of the Rad6 complex. SGA technology (53) was used to cross NatR strains harboring individual deletions of genes encoding for the Rad6 complex (rad6Δ, bre1Δ, and lge1Δ) with a transcription-targeted array of 384 Kanr deletion strains to create sets of NatR/Kanr haploid double mutants. Growth rates were assessed by automated image analysis of colony size, and lines in the diagram connect genes with synthetic genetic interactions. (B) Tetrad analysis documents the genetic interactions found between components of the Rad6 complex and other elongation-related factors identified by SGA. Genotyping of these tetrads revealed that the inviable or slowest growing colonies were the appropriate double mutants. (C) Analysis of strains carrying rad6Δ or H2B K123 tail mutations for 6-AU phenotypes. The WT Flag-H2B strain, the Flag-H2B K123R (K123R) mutant strain (YZS277), and strains with the indicated deletions made in the Flag-H2B background (see Fig. 1) were transformed with a plasmid bearing a copy of the URA3 gene (pRS316) and then plated as fivefold serial dilutions onto SD-uracil or SD-uracil containing 100 μg of 6-AU/ml. Similar results were observed by using 50 or 150 μg of 6-AU/ml, and deletions of BRE1 and LGE1 were also found to be sensitive to 6-AU (data not shown).
To further investigate the potential involvement of Rad6 and ubH2B in transcription elongation, we examined strains containing a deletion of RAD6 or BRE1, as well as a H2B tail mutation that prevents ubiquitylation (htb1-K123R) in a phenotypic plate assay involving 6-azauracil (6-AU). This drug, which reduces the nucleotide pools for Pol II elongation, results in a slow-growth phenotype that is further affected upon deletion of factors involved in the transcription elongation process. As a control and for comparison, we also examined the deletions of various Paf1 complex members and SET2 (rtf1Δ, leo1Δ, and set2Δ). Under control (synthetic defined medium without uracil [SD-uracil]) conditions, we found that rad6Δ and leo1Δ mutants showed a subtle slow-growth phenotype, whereas the htb1-K123R mutant and strains with an rtf1Δ or set2Δ deletion did not (Fig. 6C). Strains with deletions of other Paf1 complex members (paf1Δ, cdc73Δ, and ctr9Δ) were extremely slow growing and therefore could not be analyzed with confidence in this assay (data not shown). Interestingly, we observed a significant sensitivity to 6-AU in strains with a rad6Δ, bre1Δ, or htb1-K123R mutation (Fig. 6C and data not shown). In comparison, and consistent with previous studies (6, 12, 34, 50), this sensitivity was also witnessed with deletion mutants of the Paf1 complex (Fig. 6C). In contrast, deletion of SET2 resulted in modest resistance to 6-AU, a result that has been previously seen (B. D. Strahl and A. Greenleaf, submitted for publication; D. Stillman, personal communication). As Rad6 and H2B ubiquitylation at K123 are required for H3 K4 and K79 methylation, it was possible that the 6-AU phenotypes observed with the rad6Δ, bre1Δ, and htb1-K123R mutants were a direct result of the loss of elongation-related histone methylation events. However, we note that mutations of K4 and K79, singly or in combination to arginine, do not result in 6-AU phenotypes, whereas mutations at K36 do (Strahl and Greenleaf, submitted). Collectively, these data support the notion that H2B ubiquitylation has a direct role in the elongation process, which appears to be distinguishable from its effects on methylation of H3 at K4 and K79.
DISCUSSION
Recent evidence has revealed an important role for Rad6 and H2B ubiquitylation both in the transcription activation process and in the establishment of histone methylation at H3 K4 and K79 in S. cerevisiae (63). In light of this finding, we sought to determine the mechanism behind this regulation in an effort to understand the role(s) of Rad6 in gene regulation. In this report, we show that Rad6 interacts with the hyperphosphorylated form of Pol II and that this interaction is established through the actions of the Paf1 complex and Bre1. We also show that Rad6 and H2B ubiquitylation are found in the transcribed regions of a number of constitutively active and galactose inducible genes, a result which is consistent with a role for Rad6 as an elongation factor. This idea is supported by the results of SGA analysis, which show that Rad6 components genetically interact with known elongation factors, and from the finding that strains carrying either a RAD6 or BRE1 deletion or a H2B-K123R tail mutation are sensitive to 6-AU. Furthermore, the requirement for Kin28 and the CTD of Pol II in H2B ubiquitylation underscores the fact that the activity of Rad6 is dependent on the initiating events of elongation. This result is also consistent with our finding that the Paf1 complex is required for the efficient transfer of Rad6 into the transcribed regions of genes, which occurs temporally with the initial appearance of Pol II.
A novel role for Rad6 in transcription elongation.
Recent efforts have revealed an increasingly prominent role for histone-modifying activities in transcript elongation. While a number of studies suggest a role for histone acetylation in this process, more recent evidence links the actions of ATP-dependent chromatin remodeling activities and histone methyltransferases directly to this event. Specifically, the Isw1 ATPase has been found to associate with H3 K4 methylation and to act as a checkpoint regulator of transcription initiation (39, 48). Likewise, the recently identified ATPase Swr1 was shown to remodel nucleosomes to incorporate the histone variant Htz1, which has been connected to elongation events as well (30, 33, 38). Independently, studies have shown that the Set1 and Set2 HMTs both associate with Pol II during transcription elongation (17, 18). While Set1 association is dependent on the Ser5 CTD kinase activity of Kin28, Set2 association is dependent on the Ser2 CTD kinase activity of Ctk1.
Our results show that similar to Set1 and Set2, Rad6 is associated with the elongating form of Pol II (see Fig. 2). However, unlike the two Set proteins, which have been shown to selectively associate with distinct gene regions, we find that Rad6, as well as its associated H2B modification, is present broadly over the promoter and coding regions of transcriptionally active genes. This result is likely explained by the fact that Rad6 has an initial role in the activation of gene transcription (perhaps in the initiation event of elongation) but then participates in the later stages of elongation as well. This explanation would be consistent with the observation that a wide variety of elongation factors, including members of the Paf1 complex, are present at both the promoter and transcribed region of genes and that some of these elongation factors, such as Isw1, have been demonstrated to have distinct roles at different stages of the transcription elongation cycle (39, 49).
Similar to Set1, Rad6 is also associated with Pol II via the Paf1 complex (see Fig. 2). Strikingly, both enzymes appear to require the action of Kin28, suggesting a general requirement of Ser5 phosphorylation on the CTD for the recruitment of the Paf1 complex and its associated factors. In contrast, however, the major fraction of Set1 is thought to exit the polymerase after the loss of Ser5 phosphorylation, which is generally restricted to the promoter and 5′ regions of genes (17, 18). We take these data to suggest that the Paf1 complex is initially recruited, along with a variety of associated factors, such as Set1, Rad6 and possibly Dot1, in a Ser5 CTD phosphorylation-dependent manner, but that the long-term association of the Paf1 complex and some of the Paf1-associated factors are not CTD phosphorylation-dependent (see Fig. 7). This idea is consistent with results showing that the Paf1 complex is stably associated throughout the promoters and transcribed regions of genes (26, 49). In addition, the deletion of Ctk1, which presumably results in a significant loss of CTD phosphorylation in the body of genes, does not affect H2B ubiquitylation levels (Fig. 5C), which suggests that Paf1 and Rad6 association across genes is not disrupted.
As mentioned above, our results show that in addition to Rad6 being present at promoters, it is also recruited to the coding region of genes. This finding is in contrast to recent studies showing that Rad6 is exclusively recruited to promoter regions (60, 61). While we are unable to explain previous results regarding Rad6 localization, the differences in the findings may lie in the choice of the tagging method used (TAP versus HA or Flag tag) and in the fact that Rad6 levels on genes are generally quite low and, thus, may be more readily detected by using a real-time PCR assay.
Intriguingly, we find by using the GAL1 gene that Rad6 is still recruited to the promoter region of this gene in an rtf1Δ mutant but shows almost no association with this region in a bre1Δ mutant. Moreover, in the absence of Rtf1, Rad6 stably accumulates at the GAL1 promoter and fails to significantly accumulate in the GAL1 ORF (Fig. 4). This result suggests that Rad6's association with promoters and coding regions occurs in two steps, via distinct but interrelated mechanisms (see Fig. 7). First, Rad6 is recruited to a promoter by a mechanism that is Paf1 complex independent, e.g., through its association with a gene-specific activator and Bre1. Next, Rad6 that has accumulated at the promoter is handed off to Pol II phosphorylated on Ser5 of the CTD through its association with the Paf1 complex. This handoff to elongating Pol II serves two functions: it activates the ubiquitin conjugating activity of Rad6 towards H2B, and it provides a mechanism for Rad6 to enter the coding region. The E3 ligase Bre1 appears to function at both steps and thus may bridge the interaction of Rad6 with both activators and the Paf1 complex and promote H2B ubiquitylation. By this model, the turnover of Rad6 that has been observed at the GAL1 promoter would be due primarily to the activity of the Paf1 complex in handing off Rad6 from the Gal4 activator to Pol II.
A mechanism for the establishment of K4 and K79 methylation.
Because previous studies suggested that Rad6 is selectively recruited to the promoter regions of genes, a paradox arose as to how H2B ubiquitylation at a promoter is capable of establishing long-range K4 and K79 methylation marks throughout the rest of the gene and beyond. Models to explain this phenomenon include a wedge model, in which transient promoter H2B ubiquitylation results in a long-range disruption of higher-order condensed chromatin structure around an entire locus that makes the histones accessible to the appropriate HMTs (20). While one function of ubH2B may be to disrupt higher-order chromatin structure, we find that Rad6 and H2B ubiquitylation both track throughout the promoter and body of genes and that ubH2B levels are intimately coupled to K4 and K79 methylation levels (see Fig. 1 and 3). Given these results and the observation that Rad6 and ubH2B occur transiently over the activated GAL1 gene at the onset of Pol II appearance (Fig. 4), we suggest a model whereby Pol II- and Paf1-associated Rad6 ubiquitylates H2B while moving with the elongating polymerase (Fig. 7). In this model, ubH2B is closely associated with the nucleosomes being disrupted for Pol II elongation. We suggest that transient Pol II-associated H2B ubiquitylation results in the placement of a bulky moiety on chromatin that leads to a momentary disruption of nucleosomes, making H3 briefly accessible to Set1 and Dot1, which are thought to also track with Pol II through their association with the Paf1 complex. This disruption may also include the actions of the 19S proteasome, which has been shown to link H2B ubiquitylation and H3 methylation (15). While this idea may explain the regulation of histone methylation on H3, we suggest that the transient disruption of nucleosomes by ubH2B may have a more direct role in elongation, based on the fact that the H2B-K123R tail mutation results in a 6-AU phenotype whereas H3-K4R and H3-K79R mutations do not (Fig. 6C and data not shown). We speculate that the elongation and Pol II-coupled disruption of nucleosomes by ubH2B may be linked with the process of H2A/H2B dimer removal by FACT and that the disruption of chromatin may be caused by other elongation-associated ATP-dependent remodeling activities, such as Isw1 and Swr1. Future work will be required to uncover whether an association exists between H2B ubiquitylation and other mechanisms of chromatin disruption that occur during elongation.
A pioneering role for Rad6 and H2B ubiquitylation?
From our ChIP results (shown in Fig. 4), we find that Rad6 associates across the activated GAL1 gene in a timing that matches Pol II; however, unlike Pol II, which continues to accumulate over the gene, Rad6 rapidly turns over (Fig. 4A). This transient association of Rad6 at the early stages of newly activated transcription suggests a role for Rad6 and its modification in the early round(s) of transcript elongation by RNA polymerase II. Recently, it has been suggested that there is a pioneer round of transcription for newly transcribed genes in which Pol II is coupled to specialized chromatin modification and disruption (45). This round is postulated to result in a disrupted chromatin template that is permissive for the first and future rounds of gene transcription within a given cell cycle. Thus, it is intriguing to speculate that the early and transient association of Rad6 with the GAL1 gene may indicate that Rad6-mediated H2B ubiquitylation is part of such a pioneer polymerase. Given this possibility, we wonder whether the K4 and K79 methylation events instigated by Rad6/H2B ubiquitylation, which have recently been proposed to serve as a “memory of recent transcription” (42), would act as a memory system by maintaining the disrupted state of chromatin created during the pioneering round of gene transcription.
In summary, our results reveal a physical and functional interaction between Rad6, the Paf1 complex, and Pol II that points to a novel and unexpected role for histone ubiquitylation in the transcription elongation process. The requirement of Kin28 and the CTD for H2B ubiquitylation, along with the SGA and 6-AU analysis of the Rad6 complex, provide strong evidence that Rad6 and H2B ubiquitylation are functionally linked to this event. However, future investigation will be required to determine the specific functions of Rad6 and ubH2B in the elongation process and why these events happen transiently and in conjunction with the earliest appearances of Pol II in activated transcription. Given the conserved nature of these enzymes and their modifications in eukaryotes, we propose that the function of H2B ubiquitylation in transcription elongation and its regulation of histone methylation will be highly conserved. Finally, we note that H3 K4 and K79 dimethylation levels are also thought to be global in yeast and not necessarily restricted to the Pol II-transcribed regions of the genome (8, 10, 41, 56). Given the requirement for the Paf1 complex and the CTD to mediate H2B ubiquitylation and these histone methylation marks, it is intriguing to speculate that the Pol II CTD and the Paf1 complex play a much broader role in chromatin regulation than previously realized.
.
Acknowledgments
This work is supported by NIH grants GM068088 (B.D.S.) and GM40118 (M.A.O.). B.D.S. is a Pew Scholar in the Biomedical Sciences.
We thank E. Schiebel for providing the C-terminal tagging constructs, M. Solomon for providing the Kin28 expression plasmids, and J. Corden and R. Young for providing the CTD mutation strains. We also thank B.D.S. and M.A.O. lab members for their technical advice, strain construction, and comments on the manuscript. We are grateful to J. Lieb and Y. Zhang for very helpful comments on the manuscript.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, K. M., and C. M. Kane. 2003. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 19:543-550. [DOI] [PubMed] [Google Scholar]
- 3.Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272:14747-14754. [DOI] [PubMed] [Google Scholar]
- 4.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. [DOI] [PubMed] [Google Scholar]
- 7.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 8.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 10.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165-170. [DOI] [PubMed] [Google Scholar]
- 11.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 14.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 15.Ezhkova, E., and W. P. Tansey. 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol. Cell 13:435-442. [DOI] [PubMed] [Google Scholar]
- 16.Fischle, W., Y. Wang, and C. D. Allis. 2003. Binary switches and modification cassettes in histone biology and beyond. Nature 425:475-479. [DOI] [PubMed] [Google Scholar]
- 17.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 18.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 19.Hartzog, G. A. 2003. Transcription elongation by RNA polymerase II. Curr. Opin. Genet. Dev. 13:119-126. [DOI] [PubMed] [Google Scholar]
- 20.Henry, K. W., and S. L. Berger. 2002. Trans-tail histone modifications: wedge or bridge? Nat. Struct. Biol. 9:565-566. [DOI] [PubMed] [Google Scholar]
- 21.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C.-F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iizuka, M., and M. M. Smith. 2003. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 13:154-160. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Kao, C.-F., C. Hillyer, T. Tsukuda, K. Henry, S. Berger, and M. A. Osley. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 18:184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 29.Kobor, M., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 30.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 32.Krogan, N. J., and J. F. Greenblatt. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:8203-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 34.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 37.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 39.Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot, and J. Mellor. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425-435. [DOI] [PubMed] [Google Scholar]
- 40.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 41.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 43.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 44.Nonet, M., D. Sweetser, and R. A. Young. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50:909-915. [DOI] [PubMed] [Google Scholar]
- 45.Orphanides, G., and D. Reinberg. 2000. RNA polymerase II elongation through chromatin. Nature 407:471-475. [DOI] [PubMed] [Google Scholar]
- 46.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 47.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 47a.Recht, J., and M. A. Osley. 1999. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 18:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 49.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 52.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 53.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 54.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 55.van Holde, K. E. 1989. Histone modifications, p. 111-148. Springer, New York, N.Y.
- 56.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 57.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 58.West, M. L., and J. L. Corden. 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]
- 60.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]
- 61.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 62.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y. 2003. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 17:2733-2740. [DOI] [PubMed] [Google Scholar]