Abstract
We investigated the function of the histone H3/H4 chaperones anti-silencing function 1 (Asf1p) and chromatin assembly factor 1 (CAF-1) in global transcriptional regulation in budding yeast. Deletion of ASF1 or CAF-1 components led to global transcriptional misregulation, both activation and repression, of genes scattered throughout the 16 yeast chromosomes. To investigate direct effects on gene regulation, we developed an approach to destabilize Asf1p that results in its rapid degradation within minutes of transcriptional repression. Upon degradation of Asf1p, rapid global changes in gene expression occur without the requirement for passage through S phase or de novo protein synthesis. In particular, we demonstrate that the previously reported influence of Asf1p on histone gene expression is not a direct effect of loss of Asf1p. These data indicate that the histone chaperones CAF-1 and Asf1p regulate the gene expression of a broad array of genes in yeast and, in the case of Asf1p, this is likely to be due to a direct role in chromatin modulation during transcriptional regulation.
The packaging of the eukaryotic genome into the nucleoprotein structure known as chromatin has a profound influence on the activities of the genome (6, 15). The fundamental repeating unit of chromatin is the nucleosome, comprising ca. 147 bp of DNA wrapped around an octamer of core histones (20). The core histone octamer includes two molecules each of histones H3, H4, H2A, and H2B and is deposited onto the DNA in a stepwise process (23, 40). Histones H3 and H4 are deposited onto the DNA first, followed by histones H2A and H2B to complete the nucleosome.
The formation of chromatin is mediated by a class of proteins termed chromatin assembly factors (19, 40). Biochemical studies identified chromatin assembly factor 1 (CAF-1) as being required for the assembly of newly replicated DNA into chromatin in vitro (34). CAF-1 functions as a histone chaperone to deposit histones H3 and H4 onto newly replicated DNA (35). CAF-1 is a three subunit complex whose function and structure have been highly conserved through eukaryotic evolution (11-14, 42-44). CAF-1 may also assemble chromatin after DNA replication in vivo. For example, CAF-1 colocalizes with DNA replication foci (16), perhaps mediated via its interaction with the replication factor PCNA (32). In addition, inactivation of CAF-1 leads to defects in chromatin structure in human cells (28). However, CAF-1 is unlikely to be the only factor that assembles chromatin in the cell, since budding yeast lacking CAF-1 have no growth defects (13).
Anti-silencing function 1 (ASF1) is another highly conserved histone H3-H4 chaperone. ASF1 was found biochemically as a protein that facilitates CAF-1-mediated assembly of newly replicated DNA into chromatin in vitro (41). ASF1 may also function to assemble chromatin during DNA replication together with CAF-1 in vivo because ASF1 and CAF-1 coimmunoprecipitate (24, 31, 43), and CAF-1 and ASF1 copurify from human cells in a complex containing the form of histone H3 that is only assembled into chromatin during DNA replication (38). ASF1 also has functions that are independent of CAF-1. In the absence of CAF-1, ASF1 has no preference for assembling newly replicated DNA into chromatin, being fully able to assemble nonreplicating DNA (41). Unlike CAF-1 mutants, yeast lacking Asf1p have growth defects (41). Human ASF1 also exists in a CAF-1-independent complex, together with a histone variant, H3.3, that is specifically assembled into transcriptionally active genes (2, 38). Furthermore, genetic analyses have placed yeast Asf1p in a pathway independent from CAF-1 during transcriptional silencing (41). It is likely, therefore, that Asf1p functions to assemble chromatin at times other than after DNA replication. In yeast, Asf1p has an additional CAF-1-independent role in the cell during the disassembly of chromatin during transcriptional activation of the PHO5 and PHO8 genes (1).
It is widely accepted that changes in chromatin structure alter gene expression (10, 37). As such, defects in chromatin assembly or chromatin disassembly should influence gene expression. For example, yeast that lack Asf1p and CAF-1 have defects in transcriptional silencing (13, 17, 22, 25, 33, 41). Similarly, Drosophila heterozygous for ASF1 have defects in position effect variegation (26, 36). Transcriptional silencing and position effect variegation are specialized forms of transcriptional regulation that are mediated via heterochromatin formation. There is also evidence that ASF1 influences the expression of euchromatic genes. For example, yeast with ASF1 deleted misregulate the expression of histone genes (36), have an “Spt phenotype” that is indicative of a role in gene expression (4), and fail to induce the PHO5 and PHO8 genes (1). However, whether the influence of CAF-1 or Asf1p on transcriptional regulation is direct or indirect is unknown. Also, it is not clear how global or localized the roles of Asf1p and CAF-1 are on chromatin structure and, therefore, transcription.
Using transcriptional misregulation as a marker for altered chromatin structure, we have undertaken a genome-wide transcriptional analysis of yeast lacking Asf1p or CAF-1. This approach has demonstrated that Asf1p and CAF-1 have global and partially overlapping influences on chromatin structure and transcription. Furthermore, we have discovered that Asf1p most likely regulates gene expression in a direct manner that does not require passage through S phase or de novo protein synthesis.
MATERIALS AND METHODS
Yeast strains.
Yeast strains are presented in Table 1. All strains are W303-1 and were prepared for the present study, with the exception of LPY4180, which was kindly provided by Lorraine Pillus. Strains were made by using PCR-based gene modification (18). SRH001 was derived from JKT002 by using pFA6a-kanMX6-PGAL1-3HA (18). SRH007 was derived from LPY4180 by using pNKY51 (3). SRH015 was derived from SRH007 by using pFA6a-3HA-kanMX6 (18). SRH014 was derived from SRH015 by using pSVA13 (21) and pFA6a-kanMX6-PGAL1 (18). SRH016 was derived from SRH007 by using pFA6a-kanMX6 (18). All primer sequences used are available upon request.
TABLE 1.
Yeast strains examined in this study
| Strain | Genotype |
|---|---|
| LPY4180 | MATabar1::LEU2 can1-100 his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 |
| JKT001 | MATaasf1::HIS5 bar1::LEU2 can1-100 his3-11 leu2-3,112 Δlys2 TELVIIL::URA3 trp1-1 ura3-1 |
| JKT002 | MATabar1::LEU2 can1-100 his3-11 leu2-3,112 Δlys2 TELVIIL::URA3 trp1-1 ura3-1 |
| RDY010 | MATabar1::LEU2 cac2::kanMX6 can1-100 his3-11 leu2-3,112 Δlys2 TELVIIL::URA3 trp1-1 ura3 |
| SRH001 | MATakanMX6-PGAL1-3HA-ASF1 bar1::LEU2 can1-100 his3-11 leu2-3,112 Δlys2 TELVIIL::URA3 trp1-1 ura3-1 |
| SRH007 | MATabar1::LEU2 can1-100 gal1::HISG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 |
| SRH014 | MATakanMX6-PGAL1-ASF1-3HA-PEST-HIS3 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 |
| SRH015 | MATaASF1-3HA-kanMX6 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 |
| SRH016 | MATaasf1::kanMX6 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 |
Serial dilution analysis.
Log-phase cultures were adjusted to 4 × 107 cells per ml, 10-fold serially diluted, and spotted onto the indicated plates with a pronged device.
Western analysis.
Total cell extracts were made as described previously (9). The Asf1 protein was detected by Western detection of the fused 3HA epitope with mouse antihemagglutinin (anti-HA; Covance MMS-101R, 1:1,000) and anti-mouse immunoglobulin G-peroxidase (Sigma A3682, 1:80,000). The addition of 3HA epitopes to the C terminus of Asf1p has no deleterious phenotypic consequences (data not shown). Tubulin was used as a loading control and was detected by using rat antitubulin (Serotec MCA785, 1:300) and anti-rat immunoglobulin G-peroxidase (Sigma A5795, 1:80,000).
Flow cytometry.
A total of 5 × 107 log-phase yeast cells were fixed in ethanol and stained with propidium iodide as described previously (41).
RNA preparation.
Total yeast RNA was harvested according to the hot acid phenol extraction protocol described previously (30), followed by clean up by using Qiagen's RNeasy minikit.
Reverse transcription-PCR (RT-PCR) analysis.
First-strand cDNA synthesis was performed with gene-specific 3′ primers and Stratagene RTase according to Stratagene's protocol. The subsequent PCR used 2 μl of the cDNA reaction and included a deoxynucleoside triphosphate mix containing [α-32P]dATP. The linear amplification range was empirically determined for each control primer set. Control primer sets were designed to amplify cDNAs whose RNA transcript numbers both do not change during the cell cycle, nor did they show any changes in the microarray experiments. CDC42 was used as the control for high-copy-number transcripts, and SWD2 was used as the control for low-copy-number transcripts. After amplification, PCR products were run on a 3% agarose gel and then dried. The dried gel was exposed to a phosphor screen, and the image was quantitated by using iQMac. Each experimental signal was normalized by the control signal in its reaction. All primer sequences are available upon request.
Microarray experiments.
RNA was isolated by using the hot acid phenol extraction method described previously (30). Biotinylated cRNA probes for use in Affymetrix GeneChips were prepared according to the supplier's protocol (Affymetrix, Santa Clara, Calif.). The cRNA was hybridized to oligonucleotide arrays (GeneChip YG-S98; Affymetrix) and scanned as described previously (45).
Data analysis.
Microarray data was analyzed by using Affymetrix and GeneSpring software. The data set was first filtered for noisy genes. A gene was considered noisy if it was given a change call in any of the three pairwise comparisons of the three wild-type (WT) t = 0 replicates. Of the remaining genes, those that were given change calls in the same direction with similar magnitudes in all three replicate time points were considered real changes in the present study. To determine whether the fold change between replicate measures was similar, the 95% confidence interval was calculated, and if the values throughout the entire interval still indicated a change, the measurements were considered consistent and similar. Using a strict fold cutoff was avoided because of that method's tendency to disproportionately include false positives among genes with low transcript numbers and exclude true positives among genes with high transcript numbers (5, 27). Protein functional class information was obtained from the MIPS database. Other chromatin-related array data were downloaded from yMGV database. The gene cluster tree was generated with Michael Eisen's Cluster and TreeView software. P values for the significance of the overlap between two sets of gene changes were calculated by using a single-sample chi-square test with Yates' correction. The chi-square test was used because it is a nonparametric test useful for classification situations (i.e., present or not present in the overlap set), whereas parametric tests (like the t test and the F-test) do not make sense in this situation, since they are used in value situations.
RESULTS
Global changes in gene expression upon loss of Asf1p or CAF-1.
In order to identify genes whose transcription is altered upon loss of Asf1p or CAF-1, we performed global microarray analyses in yeast. In yeast, CAF-1 is encoded by three genes: CAC1, CAC2, and CAC3/MSI1. Deletion of any one of these genes yields the same phenotype as deleting all three genes (13). Therefore, we deleted CAC2 (Δcac2) to inactivate CAF-1. Yeast lacking Asf1p have a clear growth defect (41), making the comparison of transcriptional changes in asynchronous WT and asf1 mutant strains likely to yield false positives. Therefore, we isolated RNA from G2/M phase synchronized cultures of isogenic WT, ASF1-deleted (Δasf1), and CAC2-deleted (Δcac2) yeast strains (as confirmed by flow cytometry analysis; see Fig. S1 in the supplemental material), followed by analysis on Affymetrix open reading frame arrays. Transcription of 524 genes was significantly and reproducibly changed upon deletion of ASF1 in three independent experiments, whereas transcription of 252 genes was significantly and reproducibly changed upon deletion of CAC2 (see Table S1 in the supplemental material). An arbitrary cutoff of the fold change was not used for our analyses because of its tendency to overestimate gene changes among genes with low transcript numbers and underestimate gene changes among genes with high transcript numbers (5, 27). Instead, we used gene-specific cutoffs that were deemed significant by Wilcoxon's signed rank test (this is the test used by the Affymetrix software) in combination with noise filtering and only considering genes whose changes were reproducible over three independent experiments. It is likely that we have significantly underestimated the number of genes that are regulated by Asf1p and CAF-1 since our experiments were performed in rich growth conditions and only included one phase of the cell cycle. Of the 252 genes affected by loss of CAF-1, 186 of these are upregulated, whereas 66 are downregulated by loss of CAF-1. Similarly, of the 524 genes affected by loss of Asf1p, 376 are upregulated and 148 are downregulated by the loss of Asf1p.
To determine whether CAF-1 and Asf1p functions are restricted to specific regions of the genome, we examined the locations of the misregulated genes. The genes affected by deletion of ASF1 or CAC2 were dispersed over the 16 yeast chromosomes, with no clustering apparent (see Fig. S2 in the supplemental material). Also, we did not observe any telomere-proximal bias in the genes affected by loss of Asf1p or CAF-1, as was seen previously upon depletion of histone H4 (46). This result demonstrates that Asf1p and CAF-1 influence the chromatin structure throughout the entire yeast genome.
Next, we investigated whether genes that are misregulated by loss of Asf1p or CAF-1 are regulated by common pathways of transcriptional control. We failed to discover any enrichment in particular sequence-specific binding sites in the promoters of genes affected by loss of Asf1p or CAF-1, indicating that they are unlikely to be regulated by a common master regulator transcription factor. There was also no significant difference in the length of the transcripts of the genes that were affected by loss of Asf1p and CAF-1 compared to the average length for all transcripts from the genome, suggesting that they are not misregulated as a consequence of altered transcriptional elongation. Upon examination of the average transcript levels and frequency of transcription for the genes that were upregulated by loss of Asf1p and CAF-1, we did observe that these genes had transcript levels and transcription frequencies in WT cells that were significantly below the average transcript levels for the whole genome. This is consistent with loss of Asf1p and CAF-1 resulting in activation of genes that are repressed in WT cells.
Genes affected by loss of Asf1p or CAF-1 fall into specific functional classes and overlap significantly with genes misregulated by other perturbations to chromatin structure.
Next, we examined whether particular functional classes of genes were affected by loss of Asf1p or CAF-1. Genes involved in metabolism, energy production, and response to environmental stresses were over-represented upon deletion of ASF1 and CAC2 and genes involved in cell cycle control, protein fate, transcription, and membrane transport were underrepresented upon deletion of ASF1 and CAC2 (see Table S2 in the supplemental material). These results show that the overall metabolism of the cell is affected by loss of Asf1p or CAF-1.
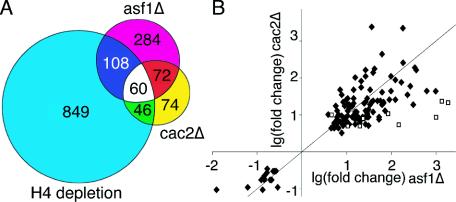
To investigate whether Asf1p and CAF-1 were influencing the same regions of the genome, we examined the overlap between the microarray results. Of the 524 genes affected by deletion of ASF1 and the 252 genes affected by deletion of CAC2, 115 genes are affected by deletion of both CAC2 and ASF1 (Fig. 1A). This overlap is highly significant (P value = 4.79 × 10−95), since only 21 genes would be expected to show up in this overlap if Asf1p and Cac2p affected independent gene sets. Strikingly, the direction and magnitude of the change in transcription of these 115 genes was always the same (Fig. 1B), indicating that these 115 genes are regulated by chromatin structure via a pathway that includes both Asf1p and CAF-1.
FIG. 1.
Comparison of the transcriptional changes caused by loss of Asf1p, CAF-1, and histone H4. (A) Venn diagram showing overlap of the genes whose transcription was affected significantly in all three independent microarray analyses upon deletion of CAC2, ASF1, or H4 depletion (using the data from 4 h after histone H4 depletion) (46). (B) The genes that are affected by deletion of ASF1 and CAC2 show changes of similar magnitudes in the same direction. The 115 gene expression changes common to Δasf1 and Δcac2 microarrays are plotted according to the log (base 2) of their fold change for Δasf1 (x axis) and Δcac2 (y axis). The diamonds (⧫) represent genes whose Δasf1 and Δcac2 change values are within each other's 95% confidence interval. The nine squares (□) represent genes whose Δasf1 and Δcac2 change values are outside of each other's 95% confidence interval.
To determine whether the genes affected by loss of Asf1p and CAF-1 may represent a subset of genes that are hypersensitive to altered chromatin structure, we compared our microarray results to those generated by other chromatin modulations. This approach is likely to underestimate the overlap between our results and other microarray analyses due to the differences in growth media and experimental conditions (for example, we only examined changes in one cell cycle phase). If Asf1p and CAF-1 are chaperones for histones H3 and H4 in vivo, then we expected that the effect of their loss on transcription may be similar to depletion of histone H4 (46). We found significant overlap between the genes affected by deletion of ASF1 or CAC2 and depletion of H4 (P values = 5.63 × 10−42 and 8.18 × 10−45, respectively [Fig. 1A]). Significant overlap also existed between the genes affected by deletion of CAC2 and ASF1 and deletion of components of the SWI/SNF remodeling complex, deletion of histone deacetylases, treatment with the histone deacetylase inhibitor trichostatin A, and deletion of TAFs, both those specific to TFIID and those that are found in both TFIID and histone acetyltransferase complexes (see Table S3 in the supplemental material). In contrast, no significant overlap was observed between the genes affected by deletion of ASF1 or CAC2 and deletion of OPI1, HAC1, or IRE1, whose gene products are involved in the processes of phospholipid biosynthesis and the unfolded protein response (data not shown). These results demonstrate that a subset of the genome is highly sensitive to regulation by chromatin structure.
Generation of a rapidly degradable form of Asf1p.
Having identified many genes that are misregulated by loss of Asf1p and CAF-1, we wanted to determine whether these effects were direct or indirect. This was important because any perturbation to chromatin structure will have pleiotropic consequences on gene expression. The approach that we took was to generate repressible, destabilized versions of Asf1p and Cac2p. Replacement of the endogenous ASF1 and CAC2 promoters with the galactose-inducible and glucose-repressible pGAL1 promoter enabled rapid transcriptional shutoff. However, both the Asf1p and Cac2p were highly stable after transcriptional repression (Fig. 2A and data not shown). Because their half-lives were too long to permit us to look at the immediate transcriptional effects upon loss of Asf1p and CAF-1, we decided to destabilize the Asf1p and Cac2p proteins. We fused the PEST degradation sequence from Cln2p onto the C terminus of Asf1p and Cac2p to generate Asf1-Pest and Cac2-Pest. The Cln2p PEST domain leads to rapid protein turnover in a cell-cycle-independent manner (21). The Asf1-Pest protein was highly unstable even before transcriptional repression (Fig. 2B). The Asf1-Pest protein was not detectable 15 min after addition of glucose, indicating that its half-life is probably a matter of minutes (Fig. 2B). In contrast, the Cac2-Pest protein degraded in a biphasic manner upon transcriptional repression with a significant proportion remaining for more than 30 min (data not shown). We predict that this stable population of Cac2-Pest represents DNA-bound protein that is more resistant to degradation. For our purposes, the degradation of Cac2-Pest was too slow to determine immediate transcriptional effects upon loss of Cac2p and was not pursued further.
FIG. 2.
Generation of a degradable, repressible form of Asf1p. (A) Asf1p is a relatively stable protein. Strain SRH001 was grown in YEPR plus 1% galactose, followed by addition of glucose at t = 0 to repress transcription from the pGAL1 promoter. Total protein extracts were made at the indicated times after addition of glucose, and analyzed by Western blotting with the C-terminal 3HA epitope on Asf1p and tubulin as a loading control. (B) Fusion of a PEST domain destabilizes Asf1p, enabling rapid loss of Asf1p. Strain SRH014 was grown in the presence of galactose, allowing expression of ASF1. At t = 0, glucose was added to the medium to suppress the expression of ASF1, and protein degradation was monitored by taking samples at the indicated times after glucose addition. The asterisk indicates a major degradation product of the Asf1-Pest protein. (C) The Asf1-Pest protein is functional. Identical amounts of strains SRH015 (WT), SRH014 (Asf1-Pest), and SRH016 (asf1Δ) were 10-fold serially diluted onto plates containing either galactose or glucose carbon sources, as well as the indicated amounts of methyl methanesulfonate (MMS).
Next, it was important to demonstrate that the Asf1-Pest protein was fully functional. We tested whether the Asf1-Pest protein resulted in any of the known phenotypes of yeast with ASF1 deleted. In the presence of galactose, the Asf1-Pest strain was indistinguishable from WT for sensitivity to DNA-damaging agents (Fig. 2C), transcriptional silencing, growth rate, cell size, and cell cycle distribution (data not shown). In summary, the Asf1-Pest strain enables an extremely rapid switch from cells resembling WT to an asf1 mutant by switching the carbon source.
Transcriptional changes occur upon degradation of Asf1p prior to passage through the S phase.
In order to identify the immediate effects of loss of Asf1p on gene expression, we performed microarray analyses after transcriptional repression of Asf1-Pest. Yeast expressing the HA-tagged Asf1p protein from its own promoter (WT) and yeast expressing the Asf1-Pest protein from the pGAL1 promoter (Asf1-Pest) were synchronized in G1 phase by using alpha factor arrest. Glucose was added to repress Asf1-Pest upon release from G1 phase. The degradation of Asf1-Pest was confirmed by Western analysis (Fig. 3A), and the similar progression of both strains through the cell cycle was confirmed by flow cytometry analysis (Fig. 3B). RNA was isolated over the first cell cycle after removal of Asf1p and analyzed on Affymetrix open reading frame arrays. Figure 3C shows the red-green hierarchical cluster tree generated from the average of the gene changes observed in three independent experiments (Fig. 3C). Only genes that changed in the same direction in all three arrays are considered here, and “noisy” genes were filtered out. We are unlikely to have many false positives with these stringent conditions and are very likely to have underestimated the number of significant changes. We discovered 95 transcriptional changes in the first cell cycle after the loss of Asf1p, which represent the most direct transcriptional consequences of losing Asf1p (see Table S4 in the supplemental material). Many of these changes were confirmed by semiquantitative RT-PCR analysis (Fig. 3D and E and data not shown). Transcription of 59 genes increased upon loss of Asf1p, whereas transcription of 33 genes decreased. Three of the genes showed changes in both directions at different time points. Strikingly, many of the genes showed transcriptional changes in the first time point after loss of Asf1p, even before the majority of the cells had started going through S phase (Fig. 3B and C). This result suggests that changes in gene expression occur upon degradation of Asf1p independent of a role for Asf1p in chromatin assembly after DNA replication.
FIG. 3.
Immediate transcriptional changes occur upon degradation of Asf1p. Strains SRH015 (WT) and SRH014 (Asf1-Pest) were grown in galactose medium and arrested with alpha factor. At time zero, the cells were released into glucose media and samples for microarray analysis were taken at the indicated times. (A) Western blot showing Asf1p levels in both the Asf1-Pest and WT strains throughout the time course. (B) DNA content of yeast at each time point. (C) Hierarchical clustering of genes showing changed expression profiles in Asf1-Pest compared to WT in all three independent microarray time courses. (D and E) RT-PCR confirmation of transcriptional changes following degradation of Asf1. The histograms show the average of four independent experiments and the error bars represent the 95% confidence interval for the measurement. (D) RT-PCR of SRL3 shows increased transcription in Asf1-Pest over WT in time points 60, 75, and 150, which is what was observed in the microarrays. (E) RT-PCR of HYR1 shows increased transcription in Asf1-Pest over WT in all time points after t = 0, the same as what was observed in the microarrays.
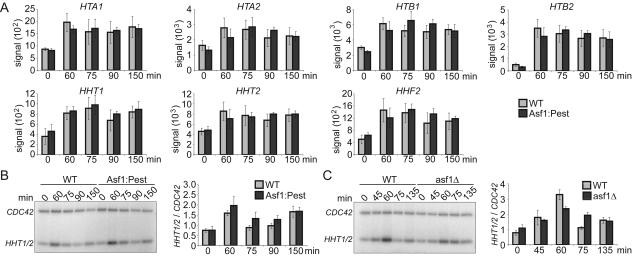
Changes in histone gene expression is not a direct effect of loss of Asf1p.
It has been reported previously that deletion of ASF1 leads to transcriptional misregulation of the histone genes (36). It was important to rule out the possibility that Asf1p may be affecting histone gene expression directly; otherwise, all of the transcriptional changes that occur upon degradation of Asf1p could be indirect effects of altered histone levels. We found that transcription of the yeast core histone genes showed no significant difference in transcription levels in the first cell cycle after degradation of Asf1-Pest by either microarray or semiquantitative RT-PCR analyses (Fig. 4A and B). However, we were able to confirm the previously reported effects of deletion of ASF1 on histone gene expression (Fig. 4C). These results demonstrate that the changes in histone expression that are observed in yeast deleted for ASF1 are indirect consequences of loss of Asf1p. As such, this finding suggests that Asf1p has a more direct effect on transcription of at least 95 genes than acting through control of histone transcription.
FIG. 4.
Absence of direct changes to histone gene expression upon loss of Asf1p. (A) Histograms of changes in transcription of the histone encoding genes HTA1, HTA2, HTB1, HTB2, HHT1, HHT2, and HHF2 as determined by microarray analyses after degradation of Asf1p. Times on the x axis represent time after transcriptional repression of ASF1 in strain SRH014 (Asf1-Pest), as shown in the experiment in Fig. 3. The average and 95% confidence intervals are shown for three independent experiments. (B) RT-PCR analysis showing lack of immediate changes in histone H3 gene expression after degradation of Asf1p. RNA harvested from the time courses described in Fig. 3 was subjected to quantitative RT-PCR analysis with primers to the indicated genes. The fold change was calculated by first normalizing the experimental signals to the CDC42 control and then dividing the Asf1-Pest signal by the WT signal. The error bars represent the 95% confidence interval for the changes observed over four experiments. (C) RT-PCR analysis showing significant changes in histone H3 gene expression in strains deleted for ASF1. Cultures of strain SRH015 (WT) and SRH016 (Δasf1) were arrested by addition of alpha factor, and samples were taken for RT-PCR analysis at the indicated times after release from alpha factor. A representative RT-PCR analysis is shown on the left, and the quantitation of four independent experiments is shown on the right, as described above.
Transcriptional changes occur upon degradation of Asf1p when passage through S phase is prevented.
To determine whether Asf1p can influence gene expression independent of passage through S phase, we inhibited DNA replication. To achieve this, our first approach was to release G1-arrested cells that had been grown in galactose into 200 mM hydroxyurea (HU) at the time of glucose addition. This amount of HU blocks all DNA synthesis (8). Flow cytometry analysis demonstrated that cells released from the G1 arrest into HU remained with a G1 DNA content (Fig. 5A). We confirmed that the transcriptional changes that occur upon Asf1-Pest degradation occur even when the cells are prevented from entering S phase by HU (Fig. 5B to E). We chose to examine two of the genes, SRL3 and HYR1, whose transcription was significantly deregulated at the first time point after degradation of Asf1 (Fig. 3D and E). For both SRL3 and HYR1, release from alpha factor arrest resulted in transcriptional repression in the WT strain, and this repression was significantly reduced upon degradation of Asf1-Pest in the presence or absence of HU (Fig. 5). The identical results were obtained when we repeated the experiment with the temperature-sensitive cdc6-1 allele to prevent DNA replication instead of using HU (data not shown).
FIG. 5.
Passage though S phase is not required for changes in gene expression that occur upon Asf1p degradation. Cultures of strains SRH015 (WT) and SRH014 (Asf1-Pest) were arrested in G1 as in Fig. 3 and then released into glucose-containing medium with or without 200 mM HU. Samples were taken before release from G1 (t = 0), 45 min after release into HU or 45 min after release into medium containing no HU. (A) Flow cytometry analysis of DNA content by propidium iodide staining. (B) RT-PCR analysis of SRL3 gene expression. SWD2 was included as an internal control. (C) RT-PCR analysis of HYR1 gene expression. CDC42 was included as an internal control. (D) Quantitation of RT-PCR analysis of SRL3 for four independent experiments, as described in Fig. 4B. (E) Quantitation of RT-PCR analysis of HYR1 as described above.
As a second approach to demonstrate that Asf1p-mediated changes in gene expression are independent of passage through S phase, we maintained cells in G1 phase after degradation of Asf1-Pest. Flow cytometry analysis of samples taken between 0 and 120 min after glucose addition demonstrated that the transcriptional changes that occur upon degradation of Asf1-Pest were not a consequence of loss of G1 arrest (Fig. 6A). Again, we examined two of the genes, HYR1 and NCE103, whose transcription was significantly deregulated at the first time point after degradation of Asf1 (Fig. 3E; see also Table S4 in the supplemental material). It is apparent that HYR1 and NCE103 transcript levels decrease with increasing time after addition of glucose in the WT strain but decreased to a much lesser degree upon degradation of Asf1-Pest (Fig. 6B to E). Taken together, these experiments using genetic, pharmacological, and temporal means of preventing S-phase entry demonstrate that transcriptional changes mediated by Asf1p are independent of its role during DNA replication-coupled chromatin assembly.
FIG. 6.
Changes in gene expression occur upon Asf1p degradation when cells are maintained in the G1 phase. Cultures of strains SRH015 (WT) and SRH014 (Asf1-Pest) were arrested in G1 as in Fig. 3, followed by the addition of glucose (at t = 0). Samples were taken before and after addition of glucose. (A) Flow cytometry analysis of DNA content by propidium iodide staining. (B) RT-PCR analysis of HYR1 gene expression. CDC42 was included as an internal control. (C) RT-PCR analysis of NCE103 gene expression. CDC42 was included as an internal control. (D) Quantitation of RT-PCR analysis of HYR1, performed as described in Fig. 4B. (E) Quantitation of RT-PCR analysis of NCE103, performed as described in Fig. 4B.
Transcriptional changes occur upon degradation of Asf1p independent of de novo protein synthesis.
To address whether Asf1p-mediated transcriptional control was an indirect consequence of degradation of Asf1p, we examined whether de novo protein synthesis was required for the Asf1p-mediated changes. The approach was to arrest WT and Asf1-Pest strains growing in galactose in G1 phase, followed by addition of cycloheximide to prevent de novo protein synthesis at the same time as addition of glucose to repress expression of Asf1-Pest. We then performed microarray analyses before and 60 min after cycloheximide addition to identify transcriptional changes. We identified genes that had changed significantly upon degradation of Asf1-Pest in the presence of cycloheximide (see Table S5 in the supplemental material), and these changes included 56 of those identified by degradation of Asf1-Pest in the absence of cycloheximide. Presumably, not all 95 genes that were transcriptionally misregulated in the first cell cycle after degradation of Asf1-Pest were detected in the cycloheximide experiment because we were only examining changes during the G1 phase of the cell cycle, whereas many of the 95 changes were not apparent until later times during the cell cycle. These results demonstrate that transcriptional changes that occur upon degradation of Asf1-Pest do not require de novo protein synthesis and are therefore likely due to direct affects of Asf1p on gene expression.
DISCUSSION
CAF-1 and Asf1p are global transcriptional regulators.
CAF-1 and Asf1p influence the expression of hundreds of yeast genes distributed over the entire yeast genome, suggesting that their function is not limited to the assembly of specialized chromatin structures such as heterochromatin. The common themes among genes regulated by Asf1p and CAF-1 appear to be that (i) they belong to particular functional classes, such as genes involved in the stress response and metabolism and (ii) they share significant overlaps with genes influenced by other perturbations to chromatin structure. There are two possible explanations for this. First, it is possible that altered gene expression is an indirect consequence of loss of Asf1p or CAF-1, in that the cell may respond to changes in chromatin structure by activating certain stress and metabolic responses. Second, it is possible that these changes are direct and that the genes involved in stress and metabolic responses may be particularly highly regulated by chromatin structure.
Although most of the changes in gene expression that occur in cells lacking Asf1p and CAF-1 were increases, there were also many decreases. The increased gene expression upon loss of Asf1p or CAF-1 is consistent with these genes being assembled into repressive chromatin structures by Asf1p and CAF-1. It is possible that the decreases in gene expression that were observed in yeast deleted for Asf1p or CAF-1 were due to indirect transcriptional effects. However, this is unlikely to be the case for all of the transcriptional decreases that we observed. For example, upon examining changes of gene expression that occurred immediately after degradation of Asf1p in the presence of cycloheximide (which prevents de novo protein synthesis and therefore indirect transcriptional effects), we still observed many decreases in gene expression compared to normal cells. Furthermore, about half of the genes that were affected by CAF-1 loss were also affected by Asf1p loss, always by the same magnitude and in the same direction of change, and roughly one-quarter of these changes were decreases upon loss of Asf1p or CAF-1. This overlap in the identity of the genes affected and the identical manner in which they were affected by independent loss of Asf1p and CAF-1 is not consistent with their being indirect effects. Similarly, depletion of histone H4 resulted in both increases and decreases in gene expression (46). It would appear that chromatin assembly may serve to either repress or activate gene expression in a gene-specific manner. Precedent for this exists at the MMTV and Xenopus vitellogenin B1 promoters where positioned nucleosomes facilitate transcription (29, 39).
Our data indicate that Asf1p and CAF-1 do function together in vivo at many regions of the genome. The 115 genes whose expression was affected in the same manner by deletion of either ASF1 or CAC2 are likely to be genes that are assembled into chromatin via a pathway requiring both Asf1p and CAF-1, presumably after DNA replication. This is consistent with the copurification of Asf1p and CAF-1 from cells (24, 31, 38, 43) and their functional codependence in vitro during assembly of newly replicated DNA into chromatin. The remaining genes that were affected by loss of CAF-1, but not by loss of Asf1p, may either be indirect transcriptional effects or may represent genes that are assembled into chromatin by CAF-1 in an Asf1p-independent manner.
Asf1p is likely to be a direct regulator of transcription.
By generating an experimental system that enabled degradation of all Asf1p in the cell within minutes, we have gained evidence to strongly support a likely direct role for Asf1p in transcriptional regulation. By direct, we mean to propose that Asf1 is likely to function in the vicinity of its transcriptional targets during transcriptional regulation. Asf1 is not formally a transcription factor, since it does not possess sequence-specific DNA binding properties, but rather it is likely to influence chromatin structure of the promoter and/or open reading frame of its transcriptional targets, as was shown recently at the PHO5 and PHO8 genes (1).
The influence of Asf1p on the expression of many genes does not require the passage of the cell through S phase and therefore is not an indirect effect of Asf1p assembling chromatin after DNA replication. Furthermore, the role of Asf1p in gene expression does not require de novo protein synthesis because transcriptional changes still occurred upon degradation of Asf1p in the presence of cycloheximide. Interestingly, transcriptional misregulation of the apparent direct targets of Asf1 that are misregulated in the first cell cycle after Asf1 degradation is not responsible for the known growth phenotypes of asf1 mutants, since we have found that these phenotypes do not appear until at least 24 h after degradation of Asf1p and are therefore due to indirect transcriptional consequences of loss of Asf1p (28a).
The possibility that Asf1p is exerting its immediate effects on gene expression indirectly through influencing histone gene expression (36) or histone stability have also been ruled out in the present study and by another study (7), respectively. For example, we have found that histone RNA levels do not differ from the WT in the first cell cycle after Asf1p degradation. In contrast, histone RNA levels are different from WT levels for cells from which ASF1 has been deleted (36), and this may account for many of the indirect transcriptional effects of deleting ASF1 that we observe. At the protein level, neither the amount nor the stability of histone proteins is affected by deletion of ASF1 (1a) (7). It is formally possible that Asf1p may exert its effect on gene expression indirectly, for example, via its ability to stabilize an unknown protein that is required for transcription and, upon degradation of Asf1p, this putative protein may degrade and affect transcription. However, we favor a more direct role for Asf1p in gene expression via a role in assembling and disassembling chromatin in a dynamic and gene-specific manner during transcriptional regulation. In favor of this model, we have recently shown that Asf1p is required for activation of the PHO5 and PHO8 yeast genes via its ability to disassemble nucleosomes from the promoter regions of these genes during transcriptional induction (1).
Physical and genetic interactions between Asf1p and transcription factors also place Asf1p at the site of transcription. For example, Asf1p interacts with the yeast Bdf1 and Bdf2 proteins that contain the bromodomains of TAFII250 of higher eukaryotes (4). Similarly, Drosophila ASF1 interacts with Brahma, the Drosophila counterpart of SWI/SNF (26). Furthermore, Drosophila ASF1 is localized to the transcriptionally active intergenic bands of polytene chromosomes (43). The definitive experiment to show a direct role of Asf1p in gene regulation would be to localize Asf1p to its transcriptional targets by chromatin immunoprecipitation. However, despite trying seven different epitope tags, we have not been able to chromatin immunoprecipitate Asf1p, perhaps due to dynamic interactions between Asf1p and DNA. Taken together, with all of the circumstantial evidence above, we feel that demonstrating transcriptional changes within 30 min of degradation of Asf1p in the presence of replication and protein synthesis inhibitors is fairly strong evidence for a direct role in transcriptional regulation.
We predict that, in addition to its role in replication-dependent chromatin assembly with CAF-1, Asf1p has two CAF-1-independent and replication-independent functions: one during assembly and the other during disassembly. Simplistically, we propose that the genes that were upregulated in the first cell cycle after degradation of Asf1p are normally assembled into transcriptionally repressive chromatin structures by Asf1p. This could be via Asf1p-mediated exchange of histones bearing posttranslational modifications that foster transcription for unmodified transcriptionally repressive histones or via the reassembly of naked regions of DNA into chromatin. In support of this later idea, we recently showed that nucleosomes are reassembled onto a naked region of the PHO5 promoter during transcriptional repression (1). Although Asf1p was not required for reassembly at the PHO5 promoter, it is possible that Asf1p mediates reassembly at other genes during transcriptional repression. Similarly, we propose that the genes that were downregulated in the first cell cycle after degradation of Asf1p in yeast are normally subject to Asf1p-mediated formation of active chromatin structures. This could either be via Asf1-mediated exchange of unmodified histones for histones bearing modifications that foster transcriptional activation or via Asf1p-mediated chromatin disassembly during gene activation, as is the case for the PHO5 and PHO8 genes (1). In higher eukaryotes, the existence of ASF1 in a protein complex with the histone variant H3.3 that is assembled into actively transcribing regions of the genome strongly supports such a replication-independent assembly function of Asf1 (38). Future studies will define how the various functions of Asf1p are regulated and coordinated during transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Melissa Adkins, Jeffrey Linger, Beth Tamburini, and Nicholas White for critical reading of the manuscript. We acknowledge David Bentley and Mark Longtine for plasmids and Lorraine Pillus for strains. We are highly grateful to Larry Hunter for advice with the statistical analyses of the microarray results and to the University of Colorado Cancer Center Affymetrix microarray and flow cytometry core facilities for assistance on this project.
J.K.T. is a Leukemia and Lymphoma Society Scholar. This study was supported by a grant from the National Institutes of Health (GM64475) to J.K.T. and supported in part by research grant FY02-201 from the March of Dimes Birth Defects Foundation.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 1a.Adkins, M. W., and J. K. Tyler. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem., in press. [DOI] [PubMed]
- 2.Ahmad, K., and S. Henikoff. 2002. Epigenetic consequences of nucleosome dynamics. Cell 111:281-284. [DOI] [PubMed] [Google Scholar]
- 3.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chimura, T., T. Kuzuhara, and M. Horikoshi. 2002. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 99:9334-9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claverie, J. M. 1999. Computational methods for the identification of differential and coordinated gene expression. Hum. Mol. Genet. 8:1821-1832. [DOI] [PubMed] [Google Scholar]
- 6.Ehrenhofer-Murray, A. E. 2004. Chromatin dynamics at DNA replication, transcription, and repair. Eur. J. Biochem. 271:2335-2349. [DOI] [PubMed] [Google Scholar]
- 7.Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in Saccharomyces cerevisiae. Cell 115:537-549. [DOI] [PubMed] [Google Scholar]
- 8.Hartwell, L. H. 1976. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol. 104:803-817. [DOI] [PubMed] [Google Scholar]
- 9.Horvath, A., and H. Riezman. 1994. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 10:1305-1310. [DOI] [PubMed] [Google Scholar]
- 10.Jones, K. A., and J. T. Kadonaga. 2000. Exploring the transcription-chromatin interface. Genes Dev. 14:1992-1996. [PubMed] [Google Scholar]
- 11.Kamakaka, R. T., M. Bulger, P. D. Kaufman, B. Stillman, and J. T. Kadonaga. 1996. Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor 1. Mol. Cell. Biol. 16:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufman, P. D., R. Kobayashi, N. Kessler, and B. Stillman. 1995. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81:1105-1114. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 14.Kaya, H., K. I. Shibahara, K. I. Taoka, M. Iwabuchi, B. Stillman, and T. Araki. 2001. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104:131-142. [DOI] [PubMed] [Google Scholar]
- 15.Khorasanizadeh, S. 2004. The nucleosome: from genomic organization to genomic regulation. Cell 116:259-272. [DOI] [PubMed] [Google Scholar]
- 16.Krude, T. 1995. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res. 220:304-311. [DOI] [PubMed] [Google Scholar]
- 17.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 18.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 19.Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677:3-11. [DOI] [PubMed] [Google Scholar]
- 20.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 21.Mateus, C., and S. V. Avery. 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16:1313-1323. [DOI] [PubMed] [Google Scholar]
- 22.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 24.Mello, J. A., H. H. Sillje, D. M. Roche, D. B. Kirschner, E. A. Nigg, and G. Almouzni. 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monson, E. K., D. de Bruin, and V. A. Zakian. 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. USA 94:13081-13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodeling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutch, D. M., A. Berger, R. Mansourian, A. Rytz, and M. A. Roberts. 2002. The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data. BMC Bioinformatics 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nabatiyan, A., and T. Krude. 2004. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Biol. 24:2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Ramey, C. J., S. Howar, M. Adkins, J. Linger, J. Spicer, and J. K. Tyler. 2004. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 24:10313-10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schild, C., F. X. Claret, W. Wahli, and A. P. Wolffe. 1993. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 12:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 32.Shibahara, K., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 33.Singer, M. S., A. Kahana, A. J. Wolf, L. L. Meisinger, S. E. Peterson, C. Goggin, M. Mahowald, and D. E. Gottschling. 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150:613-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, S., and B. Stillman. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15-25. [DOI] [PubMed] [Google Scholar]
- 35.Smith, S., and B. Stillman. 1991. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svejstrup, J. Q. 2004. The RNA polymerase II transcription cycle: cycling through chromatin. Biochim. Biophys. Acta 1677:64-73. [DOI] [PubMed] [Google Scholar]
- 38.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 39.Truss, M., R. Candau, S. Chavez, and M. Beato. 1995. Transcriptional control by steroid hormones: the role of chromatin. Ciba Found. Symp. 191:7-23. [DOI] [PubMed] [Google Scholar]
- 40.Tyler, J. K. 2002. Chromatin assembly. Eur. J. Biochem. 269:2268-2274. [DOI] [PubMed] [Google Scholar]
- 41.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 42.Tyler, J. K., M. Bulger, R. T. Kamakaka, R. Kobayashi, and J. T. Kadonaga. 1996. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 16:6149-6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Biol. Cell 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1996. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95-104. [DOI] [PubMed] [Google Scholar]
- 45.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 46.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402:418-421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.