Summary
Natural tetracycline (TC) antibiotics were the first major class of therapeutics to earn the distinction of ‘broad-spectrum antibiotics’ and they have been used since the 1940s against a wide range of both Gram-positive and Gram-negative pathogens, mycoplasmas, intracellular chlamydiae, rickettsiae and protozoan parasites. The second generation of semisynthetic tetracyclines, such as minocycline and doxycycline, with improved antimicrobial potency, were introduced during the 1960s. Despite emerging resistance to TCs erupting during the 1980s, it was not until 2006, more than four decades later, that a third--generation TC, named tigecycline, was launched. In addition, two TC analogues, omadacycline and eravacycline, developed via (semi)synthetic and fully synthetic routes, respectively, are at present under clinical evaluation. Interestingly, despite very productive early work on the isolation of a Streptomyces aureofaciens mutant strain that produced 6-demethyl-7-chlortetracycline, the key intermediate in the production of second- and third-generation TCs, biosynthetic approaches in TC development have not been productive for more than 50 years. Relatively slow and tedious molecular biology approaches for the genetic manipulation of TC-producing actinobacteria, as well as an insufficient understanding of the enzymatic mechanisms involved in TC biosynthesis have significantly contributed to the low success of such biosynthetic engineering efforts. However, new opportunities in TC drug development have arisen thanks to a significant progress in the development of affordable and versatile biosynthetic engineering and synthetic biology approaches, and, importantly, to a much deeper understanding of TC biosynthesis, mostly gained over the last two decades.
Key words: antibiotics, biosynthesis, polyketides, polyketide synthase, tetracyclines, oxytetracycline, chlortetracycline, Streptomyces, Streptomyces rimosus, Streptomyces aureofaciens
Introduction
Tetracyclines (TCs) are polyketide natural products, which include a number of clinically important antibacterials, such as oxytetracycline (5-hydroxy-TC, OTC) and chlortetracycline (7-chlor-TC, CTC). TCs were the first major class of therapeutics to earn the distinction of ‘broad--spectrum antibiotics’, and they have been used since the 1940s against a wide range of both Gram-positive and Gram- -negative pathogens, mycoplasmas, intracellular chlamydiae, rickettsiae and protozoan parasites (1). The basic TC structure comprises four linearly fused six-membered carbon rings, to which different functional groups are attached (Fig. 1; 2). Clinically used TCs are characterized by a unique C2 amide functionality and one aromatic ring (D in Fig. 1), whereas other rings include saturated carbon centres (1, 3–5). The periphery of the TC molecule can be divided into two distinct regions (Fig. 1) designated the lower and upper peripheral regions, containing different chemical functional groups and substituents. The lower peripheral region, which extends from C10 to C4a, is involved in the interaction with the 30S ribosomal subunit via the oxygen functional groups on carbon atoms C10--C1, and the C2 amide functionality (2). It has been demonstrated that most modifications along the lower peripheral region of TC greatly decrease antimicrobial activity, whereas derivatives modified along the upper peripheral region, particularly on positions C7-C9, have demonstrated greater activity against many different biological targets (2, 4).
Fig. 1.
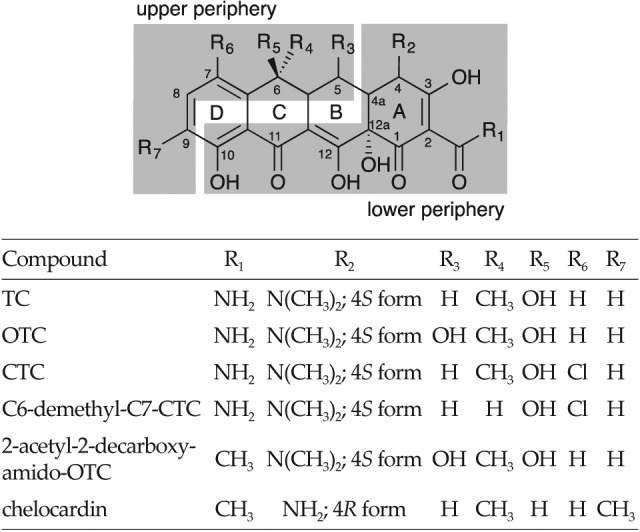
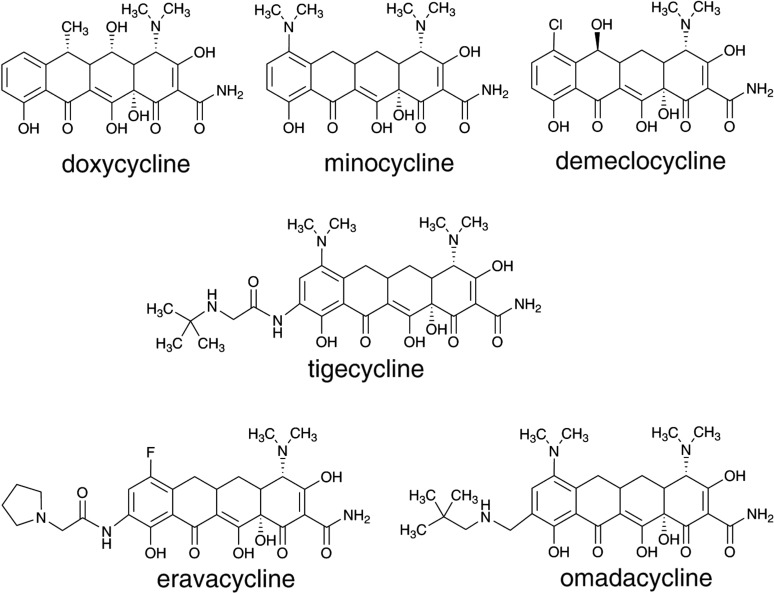
Natural tetracyclines produced by different Streptomyces species. The upper and the lower peripheral regions of the tetracycline backbone are shadowed (2)
Tetracyclines in Clinical Use
CTC, discovered in 1948 by Benjamin Duggar at the Lederle Laboratories (later known as American Cyanamid, Wayne, NJ, USA), and OTC, discovered in the early 1950s by Alexander Finlay at Pfizer (Groton, CT, USA), were the first members of the TC group (2). CTC was isolated from Streptomyces aureofaciens, and was named aureomycin because of the gold colouring of the bacterial culture on agar plates. OTC, on the other hand, is produced by Streptomyces rimosus and it was isolated in Terre Haute, IN, USA, and therefore called terramycin (2). Other naturally occurring TCs were identified later on, e.g. tetracycline from S. aureofaciens, S. rimosus and S. viridofaciens, and the important intermediate 6-demethyl-7-chlortetracycline (demeclocycline, 6DM-CTC), biosynthesized by a mutant strain of S. aureofaciens (3). 6DM-CTC is today one of the most important intermediates used at an industrial scale in semisynthetic processes for the production of second- and third-generation TCs (3, 6). TCs alone have been the fifth most prescribed antibiotics worldwide (7). Despite their availability as generic agents and the widespread occurrence of resistance by disease agents, TCs still have a relatively large market. For example, the semisynthetic TC doxycycline has an estimated market of over 250 million dollars (8).
Second-generation semisynthetic tetracyclines
The mode of action of TCs consists of their binding to the ribosome and inhibiting translation by preventing the binding of aminoacylated tRNA to the A site (6). Structural features within the TC nucleus and the application of organic synthetic reactions, investigated independently by Lederle and Pfizer, led to further modifications of their proprietary tetracycline scaffolds, with the goal of generating tetracycline antibiotics with improved pharmacokinetic properties, increased antimicrobial potency and decreased toxicity. Pfizer chemists modified the C ring of OTC (Fig. 1) to produce 6-deoxytetracycline (doxycycline, Fig. 2), an analogue with remarkable activity, stability and pharmacological efficacy, which was approved for use in 1967 by the USA Food and Drug Administration (FDA) (2). Doxycycline is still widely used today, not only for the treatment of bacterial infections such as pneumonia, acne, chlamydia infections, early lyme disease, cholera and syphilis, but also for chemoprophylaxis and treatment of malaria. Lederle´s scientists, on the other hand, intensively studied the biogenesis of CTC mutants in S. aureofaciens (2). Among others, they isolated the precursor 6DM-CTC (demeclocycline, Fig. 2), a TC scaffold that possessed unique C6 and C7 functional groups; further chemical reduction of demeclocycline resulted in the industrially important TC intermediate sancycline (6-demethyl-6-deoxytetracycline). Modifications of the aromatic D ring using semisynthetic approaches produced novel C7 and C9 derivatives of sancycline. Further semisynthetic modifications of sancycline resulted in an analogue possessing a 7-dimethylamino group, named minocycline (Minocin; Fig. 2), whose antibacterial and pharmacological activities against a broad range of bacterial pathogens were significantly improved, compared to the first-generation compounds and to doxycycline (2, 6, 9). This improvement can be explained by the fact that minocycline has a much higher affinity for the ribosome than TC and consequently inhibits in vitro translation more efficiently. Thus, minocycline has become one of the most widely used members of the TC group (2, 6).
Fig. 2.
Second and third generations of tetracyclines. Omadacycline and eravacycline are currently undergoing clinical evaluation
Third-generation (semi)synthetic tetracyclines
Due to the increasing incidence of bacterial resistance to the first and second generations of TCs, the clinical use of tetracyclines has significantly declined in most countries and in many instances they are no longer drugs of choice (3). Thus, in the late 1980s, the increasing prevalence of antibiotic resistance led pharmaceutical companies, including Wyeth (now Pfizer), to re-enter or to expand their antibiotic discovery programs. Wyeth launched a program to further chemically modify the minocycline scaffold, producing the first clinical example of a TC that is functionalized at the C9 of the D ring (Fig. 2) possessing an amide functionality with a glycine subunit attached (2, 6). In this way, they generated a series of new analogues for assay and evaluation, referred to as glycylcyclines. Many of the compounds were active against Gram--positive and Gram-negative bacteria and inhibited both, susceptible and resistant strains (expressing ribosomal protection or tetracycline efflux proteins), to a similar extent. Soon, their lead compound tigecycline (9-t-butylglycylamido-minocycline (Tygacyl), Fig. 2) entered clinical evaluation and was approved by the FDA for hospital use in 2006 (2). Tigecycline displays an affinity for the ribosome approx. 5-fold higher than that of minocycline (2, 6). This third-generation TC was the first new one to be introduced into the market in over 40 years. It was specifically designed to confront tetracycline-resistant mechanisms and today it is still clinically active within acceptable minimum inhibitory concentration (MIC) ranges against Gram--positive isolates obtained globally, including antibiotic- -resistant organisms, such as MRSA (methicillin-resistant Staphylococcus aureus), vancomycin-resistant enterococci, and penicillin-resistant S. pneumoniae (2, 6, 10, 11). However, due to its potent activity against some Gram-negative bacteria, it has also become a treatment of last resort against multidrug-resistant Gram-negative bacterial pathogens (12).
Tetracyclines in the pipeline
By applying transition metal-based semisynthetic chemistry to produce tetracycline derivatives, Paratek Pharmaceuticals (Boston, MA, USA) developed a 9-aminomethyl aromatic D ring group intermediate of minocycline that was subsequently modified to yield 9-alkylaminomethyl minocycline analogues with improved activity against a broad spectrum of tetracycline-susceptible and resistant bacteria (13, 14). One compound, now named omadacycline (PTK 0796; Fig. 2) was chosen for its superior activity, lack of toxicity and oral bioavailability, and it is now in phase III clinical trials (2, 6, 13, 14).
On the other hand, a team led by Myers, at Harvard University, developed a facile and versatile route to produce numerous novel TCs at positions that are not biosynthetically or semisynthetically amenable (15). This unique synthetic approach includes a conjunctive coupling of a BA ring fragment with a D ring precursor (C ring is formed in this process), thus generating the TC ring system. This pathway for tetracycline synthesis has been employed by Tetraphase Pharmaceuticals (Watertown, MA, USA), leading to the generation of a 7-fluorotetracycline derivative coded TP-434 (eravacycline, Fig. 2), containing 7-fluoro and 9-pyrrolidinoacetoamido modifications of D ring (16). Eravacycline was shown to have a much higher affinity for the ribosome than tetracycline; it consistently inhibits in vitro translation more efficiently than tetracycline. Eravacycline displays similar ribosome binding affinity to tigecycline (2, 6, 16–18). Eravacycline, developed by Tetraphase Pharmaceuticals, is now in phase III clinical trials (19).
Atypical tetracyclines
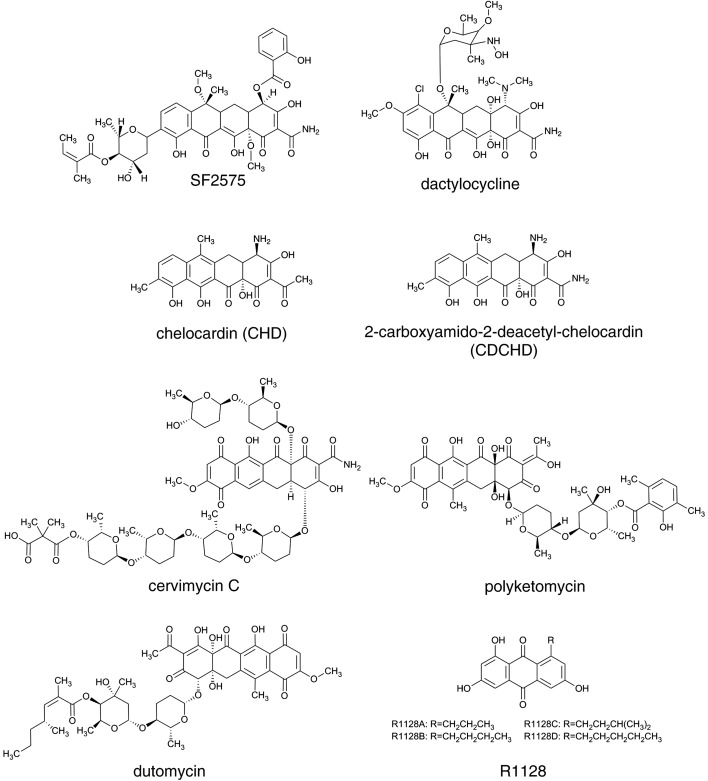
On the basis of their mode of action, or rather, on the basis of their ability to inhibit protein synthesis, TCs can be divided into two groups (20). Typical TCs, such as OTC, minocycline, doxycycline and tigecycline, comprise a group of effective protein synthesis inhibitors that bind to the 30S ribosomal subunit. On the other hand, a second class of TCs has been reported, such as chelocardin (CHD, Fig. 3) and anhydrotetracycline, which do not seem to be inhibitors of protein synthesis, at least not efficient ones, thus presenting an opportunity for the development of novel TCs with a different mode of action (20). TCs from the first group, the so-called typical or classic TCs, display a reversible bacteriostatic effect, while the atypical TCs from the second group lead to a strong bactericidal response, most likely by perturbing the organization of the bacterial cytoplasmic membrane (3, 20, 21). CHD, for example, is a natural polyketide produced by Amycolatopsis sulphurea (22), whereas other atypical TCs are either chemically synthesized as 6-thiatetracycline (23) or shunt products in OTC and CTC biosynthesis such as anhydrotetracycline, anhydrochlortetracycline and 4-epianhydrochlortetracycline. The mode of action of atypical TCs is still to be identified in order to facilitate the development of this group of antibiotics. Interestingly, during 1970s Abbott (Abbott Laboratories, Chicago, IL, USA) carried out a phase II clinical trial with CHD in collaboration with Dr. Josip Kajfeš hospital (Zagreb, Croatia) for the treatment of urinary tract infections (24). However, despite its potent activity against TC-resistant pathogens, CHD did not enter the clinic. In recent years, a team led by Petković has revisited the biosynthesis of this atypical TC analogue, produced by A. sulphurea (25). By applying biosynthetic engineering approaches, they have generated CHD analogues, some of which display potent antibacterial activity against multidrug-resistant pathogens, even against Gram-negative pathogens, which today represent a great threat in hospitals; they have thus demonstrated the great potential of atypical TCs (26).
Fig. 3.
Selected aromatic polyketide metabolites displaying diverse biological activities. Their corresponding gene clusters present rich source of biosynthetic enzymes, which can be readily used in biosynthetic engineering approaches for the generation of new derivatives
Biosynthetic Pathway of Tetracycline Antibiotics
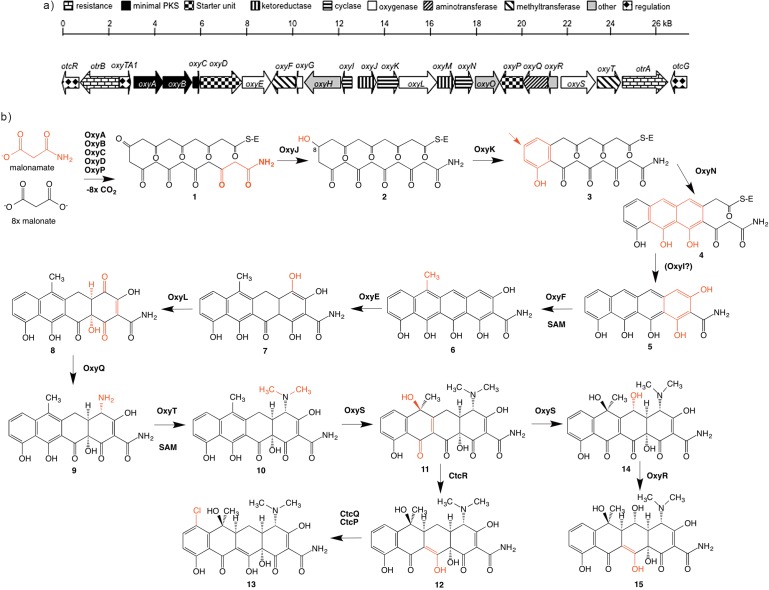
As described earlier, very productive studies on the biosynthesis of CTC, produced by S. aureofaciens, were carried out during the 1960s by McCormick et al. (27–35) at Lederle. They applied feeding experiments of the blocked mutants generated through random mutagenesis, which resulted in the production of different TC intermediates or shunt products (27–35). The entire biosynthetic gene clusters encoding CTC and OTC (Fig. 4a) were cloned in 1996 and 1989, respectively (36, 37). However, the complete DNA sequence of the gene cluster encoding for OTC biosynthesis in the wild type S. rimosus ATCC 10970 strain was only published in 2006 by Zhang et al. (38). A number of genes involved in the regulation of the OTC and CTC gene clusters have also been identified (39, 40). Important information on the biosynthesis of CTC, particularly related to the chlorination step in CTC biosynthesis, was contributed by Dairi et al. (41) and recently described in more detail by Zhu et al. (42). However, the most comprehensive studies related to the biosynthesis of TCs were carried out on the OTC biosynthetic pathway produced by S. rimosus. Therefore, this review predominantly focuses on the biosynthesis of OTC in S. rimosus.
Fig. 4.
Schematic presentation of: a) the gene cluster encoding for OTC biosynthesis from S. rimosus. The enzymatic activity of each putative gene is marked with different symbols and corresponds to the legend. The function of each gene product in the OTC gene cluster (in a) corresponds to the proposed OTC biosynthetic pathway (in b); and b) the biosynthetic pathway of oxytetracycline (OTC) and chlortetracycline (CTC), based on the biosynthetic gene clusters of OTC and CTC from S. rimosus and S. aureofaciens, respectively. The position on the tetracycline backbone of each proposed tetracycline precursor modified by the corresponding enzyme from the OTC (CTC) enzyme complex at each proposed biosynthetic step is shadowed. For colour version see: www.ftb.com.hr
Early studies on OTC and CTC biosynthesis were mostly carried out from the 1960s to the 1980s by research groups at Pfizer (Sandwich, UK), Lederle and Pliva (Zagreb, Croatia). Very productive work on the genetics of TC-producing strains, as well as on the biosynthesis of CTC and OTC was carried out by academic groups in the former Czechoslovakia and by Croatian researchers at the Faculty of Food Technology and Biotechnology at the University of Zagreb (43). Russian scientists from the All--Union Research Institute for Antibiotics in Moscow also contributed to the area of genetics of OTC production by S. rimosus; their work was mostly published in Russian journals (44). Early circular chromosomal linkage maps of three S. rimosus strains with nutritional markers and S. rimosus mutants, blocked in OTC production, were independently constructed by two research groups, as reported by Friend and Hopwood (45) in the UK, and by Alačević et al. (46–49) in Zagreb. Analysis of non-OTC- -producing mutants initially suggested the position of some loci responsible for the biosynthesis of OTC in the lower arc of the map. Interestingly, these studies were pointing to two locations of the genes involved in OTC biosynthesis on the physical map of the S. rimosus chromosome (46–49).
At the beginning of the 1980s, the Pfizer group published advanced genetic and biochemical studies based on S. rimosus mutants that were impaired in their OTC biosynthesis (50). However, following the advent of molecular biology approaches during the early 1980s, and very importantly, knowing that genes involved in secondary metabolite biosynthesis are most often clustered in a single location, the researchers at Pfizer managed to clone the entire gene cluster encoding for the biosynthesis of OTC (51, 52) (Fig. 4a). Subsequently, they managed to heterologously express the entire OTC gene cluster in Streptomyces lividans and Streptomyces albus, thus resulting in the production of OTC in heterologous hosts. This was a remarkable achievement (37, 53) and it was thus confirmed that the entire OTC biosynthetic pathway, of around 35 kb in size, is located in a single location on the S. rimosus chromosome. These efforts were later on followed by collaborative research between Hunter’s group in Glasgow (UK) (54) and a research group in Zagreb (Croatia) working at the pharmaceutical company Pliva (55, 56).
However, during the last decade, the most significant insight into OTC biosynthesis has been gained by Tang´s research group at the University of California, Los Angeles (USA) (57–63).
Initiation steps in TC biosynthesis
The polyketide skeleton of TCs is synthesized by type II minimal polyketide synthase (minimal PKS) genes (64, 65), consisting of ketosynthase α, ketosynthase β and acyl carrier protein (ACP), designated as OxyA, OxyB and OxyC in OTC biosynthesis, respectively (Fig. 4) (4). Although the exact mechanism of initiation in OTC biosynthesis is not well understood yet, the initial steps in OTC biosynthesis are carried out by a so-called minimal PKS consisting of two ketosynthase units KSα and KSβ, an ACP (OxyABC), an amidotransferase (AMT), OxyD (catalysing the transamination reaction of malonate to malonamate) and a putative acyltransferase (AT) homologue, OxyP (61) (Fig. 4). The function of the AT domain in OTC biosynthesis is not yet fully understood. Interestingly, inactivation of oxyP gene did not abolish the biosynthesis of OTC, but it did increase the proportion of 2-acetyl-2-decarboxyamido-oxytetracycline (ADOTC), compared to OTC (61) (Fig. 1).
In contrast to the studies on the priming in OTC biosynthesis, Lešnik et al. (26) recently demonstrated that OxyP from the OTC gene cluster actually plays an essential role in the selection of an unusual malonamate starter unit when heterologously expressed in A. sulphurea. Heterologous expression of AT homologue oxyP gene, together with oxyD, resulted in the formation of a hybrid analogue of CHD, containing a carboxyamido group at C2 position, the 2-carboxyamido-2-deacetyl-chelocardin (CDCHD, Fig. 3). Thus, the hybrid structure CDCHD is efficiently produced by the recombinant strain of A. sulphurea only if both, AMT and AT, are heterologously expressed. Interestingly, in the biosynthesis of the tricyclic aromatic polyketide compound R1128 (Fig. 3), produced by Streptomyces sp. R1128 (66), Tang et al. (67) demonstrated that the OxyP homologue ZhuC, from the R1128 gene cluster, displays thioesterase activity. Therefore, these authors have suggested that AT in R1128 biosynthesis is in fact a proofreading enzyme, involved in the removal of inappropriate starter units from minimal PKS that might block the enzymatic complex. However, the function of the oxyP gene and the choice of starter unit in OTC biosynthesis are not yet fully understood.
Formation of the tetracycline backbone in OTC biosynthesis
The chemical structure of TCs implies that they are aromatic polyketides synthesized by a type II polyketide synthase (PKS) (64, 65). As discussed earlier, polyketide assembly from malonate-derived building blocks is achieved by iterative Claisen-like condensations, catalyzed by the minimal PKS, consisting of the ketosynthase (KSα), its cognate homologue KSβ (without an active site cysteine), also designated as chain length factor (CLF) (68), and a cognate ACP. The minimal PKS enzymes catalyze the iterative Claisen-like condensation of the starter unit, most likely malonamyl-CoA, and eight extender malonyl-CoA units, resulting in nascent amidated decaketide backbone (1 in Fig. 4b). It was proposed that malonyl- -CoA:ACP acyltransferase, which transfers the extender unit malonyl-CoA to ACP, can be shared with fatty acid biosynthesis (69). Initial folding of the growing polyketide chain is directed by a ketoreductase OxyJ, which catalyses the region-specific reduction of the keto group at C8 (chemical nomenclature in Fig. 1, and 2 in Fig. 4b) (54). Crucial biosynthetic steps of cyclisation and aromatization result in the formation of the basic tetracyclic structure of all TCs (Fig. 1). Key steps in closure and aromatisation of the first ring (D) (3 in Fig. 4b), catalysed by two-component cyclase/aromatase OxyK, are likely occurring rapidly with the C8 keto group reduction (54). Petković et al. (54) have demonstrated that in S. rimosus the cyclase/aromatase gene otcD1 (designated as oxyK by Zhang et al. (57), Fig. 4a) plays a key role in the formation of the first ring, considering that inactivation of otcD1 gene results in an aberrant cyclisation pattern of the nascent polyketide chain. Interestingly, inactivation of otcD1 also derailed the functionality of the minimal PKS complex, resulting in the formation of a number of compounds of various chain lengths consisting of 9, 15 or 17 carbon atoms (54). Remarkably, all these compounds contain a carboxyamido moiety, giving a strong indication that addition of the amino group does not happen after the formation of the basic TC backbone, a possibility that was considered in the early days. Subsequently, Zhang et al. (57) also confirmed the function of OxyK (OtcD1) as a cyclase/aromatase.
The formation and aromatisation of the first ring (D) is followed by the closure of the second ring (C), most likely catalysed by the mono-domain cyclase OxyN; the closure of the third ring (B) is believed to be spontaneous (4 in Fig. 4b) (58, 59). Last ring (A) formation, resulting in the first stable tetracyclic intermediate pretetramide (5 in Fig. 4b), was at first suggested to be catalysed by the third group of OxyI cyclases (58). However, it was later proposed that the closure of the final ring of OTC is spontaneous, because of the presence of the terminal amino group (58). To date, the function of the OxyI cyclase has not been elucidated; nevertheless, it is possible that OxyI still facilitates the formation of the last (A) ring (Fig. 4b).
It was originally proposed that the oxyH gene, present in the OTC gene cluster, is involved in the activation of the starter unit, being thus involved in the priming steps of OTC biosynthesis. OxyH homologues were also found in the CTC (42), mithramycin (70), SF2527 compound (71) and polyketomycin (72) gene clusters. The role of the acyl-CoA-ligase SsfL2, homologous to OxyH from the oxy gene cluster, putatively involved in the formation of the fourth ring of the TC-like compound SF2527 was discussed by Pickens et al. (71). Interestingly, the OxyH was demonstrated by gene-inactivation studies to be dispensable: therefore, its function is not yet understood (4). Considering that the cyclisation steps in TC biosynthesis occur between enzymatically catalysed reactions and spontaneous chemically-driven reactions, it is not surprising that the mechanism of cyclisation is not yet fully understood.
Based on the studies carried out using blocked mutants (27–35), the first stable OTC intermediate is likely the fully aromatised tetracyclic compound called pretetramide (5 in Fig. 4b), which then undergoes further processing, tailoring reactions of the fully formed TC backbone; this is also called post-PKS tailoring.
Tailoring reactions of the basic tetracycline backbone
A large enzyme complex designated as type II PKS, where a number of independent enzymes likely cooperate simultaneously, carries out the formation of OTC. Thus, when considering the biosynthetic pathway involved in the formation of TCs, it is probably not appropriate to define the biosynthetic steps as a number of ordered reactions. However, based on numerous OTC and CTC intermediates or shunt products isolated from blocked mutants of S. rimosus and S. aureofaciens, respectively (27–35), it is possible to speculate on the preferred order of enzymatic reactions involved in the modification of the early TC intermediate pretetramide (5 in Fig. 4b). Considering that the methyl group at carbon C6 is found in almost all early intermediates in the biosynthesis of OTC and CTC, when blocked mutants were analysed at Lederle (27–35), it was believed that methylation by methyl-transferase OxyF at the C6 position is one of the earliest steps in the tailoring reactions of the TC backbone (6 in Fig. 4b) (58). However, the isolation of the blocked S. aureofaciens mutant by Ryan (73), producing 6-demethyl- -CTC, confirms that methylation at C6 is not obligatory as the earliest step in the modification of the pretetramide. Hence, it is likely the most preferred route (30, 74). The C6 methylation is probably followed by a double hydroxylation of ring A at C4/C12a by oxygenase pairs OxyL in OxyE, whereas OxyE is believed to be an ancillary mono-oxygenase for OxyL with a nonessential but important role in improving its catalytic efficiency as a C4 hydroxylase (7 and 8 in Fig. 4b) (60). Hydroxylation at C4 is a prerequisite for the incorporation of an amino group at C4 by PLP-dependent aminotransferase OxyQ (9 in Fig. 4b), which is followed by dimethylation of the amino group by N,N-dimethyltransferases OxyT, to yield anhydrotetracycline (ATC) (10 in Fig. 4b) (59). Hydroxylation of ATC at C6 is catalysed by FAD-dependent monooxygenase OxyS (11 in Fig. 4b). Interestingly, OxyS has recently been found responsible also for the hydroxylation of C5 in the biosynthesis of OTC (14 in Fig. 4b) (63). An earlier observation by Perić-Concha et al. (55) that the deletion of the otcC gene (designated as oxyS by Zhang et al. (57)) resulting in the production of a tricyclic compound containing 17 carbon atoms was rather surprising. Perić-Concha et al. (55) demonstrated that even the enzyme involved in the late stages, the so-called post-PKS processing, can interact with minimal PKS, thus influencing the ability of the minimal PKS to make a polyketide of the correct chain length. As suggested by Wang et al. (63), the last step in OTC biosynthesis is believed to be the reduction of the C5a−C11a double bond by OxyR, to yield OTC (15 and 12 in Fig. 4b in CTC biosynthesis) (63). In the CTC biosynthesis, the halogenation as the final step occurs at the C7 position (13 in Fig. 4b). As expected, chlorination in the CTC biosynthetic pathway, a step that is not encoded in the OTC gene cluster, is catalysed by the flavin-dependent halogenase CtcP (13 in Fig. 4b) (42).
Regulation of tetracycline biosynthesis and genes involved in resistance
Environmental factors, such as carbon, nitrogen, inorganic phosphate, metal ion and other nutrient sources, cultivation condition parameters, such as pH and aeration (dO2), and cultivation methods in general, may affect the timing and the extent of TC production (43, 75, 76). Media containing high concentrations of starch as a carbon source are generally used in fermentation processes for the production of OTC with S. rimosus at the industrial level, although vegetable oil can also be used as a carbon source. It was shown that S. rimosus produces OTC abundantly when subjected to phosphate starvation (43, 75, 76). McDowall et al. (77) reported that the sequences of the promoter regions in the S. rimosus 4018 strain (now designated as DNA sequence encoding oxyI, oxyJ, oxyR, oxyS and oxyA genes (40)) contain tandem repeats that are similar to the DNA-binding sites of Streptomyces antibiotic regulatory protein (SARP) transcription activators. They suggested that OTC production by S. rimosus requires a SARP-like transcription factor that is either produced and/or activated when phosphate concentrations are low (77). Just recently, Yin et al. (40) identified a SARP-like regulator, OtcR, located immediately adjacent to the resistance gene otrB (Fig. 4a). OtcR acts as a positive pathway--specific activator of OTC biosynthesis, which leads to a significant increase in OTC production when overexpressed at the appropriate level. Based on amino acid sequence analyses, OtcR and Ctc11 (CtcB) from S. aureofaciens share 46% identity; furthermore, the predicted SARP-binding sites were also identified in the ctc cluster. Interestingly, Wang et al. (62) found that the SARP regulator, encoded by ctc11 (ctcB) from the CTC gene cluster (ctc) of S. aureofaciens (36), could activate the transcription of the oxy cluster in the heterologous host Streptomyces coelicolor (62). Since OTC and CTC are structurally highly similar antibiotics, it is reasonable to expect similar biosynthetic and regulatory mechanisms. The second regulatory element OtcG was identified by Lešnik et al. (39). OtcG belongs to the LAL (LuxR) family of transcriptional regulators and it is located on the other side of the otc gene cluster, in the vicinity of the otrA resistance gene (Fig. 4a). OtcG plays a conditionally positive role in OTC biosynthesis, considering that inactivation of the otcG gene in the S. rimosus 4018 strain reduces the production of OTC by more than 40%. However, overexpression of otcG by introducing a second copy of the gene under the strong constitutive promoter ermE* did not yield any statistically significant change in the production of OTC (39). To conclude, to date two regulatory proteins, OtcG and OtcR, have been identified in the gene cluster encoding OTC biosynthesis in the S. rimosus 4018 strain.
Interestingly, three genes encoding resistant determinants (otr), designated otrA, otrB and otrC, have been identified in S. rimosus (78, 79). OTC is a bacteriostatic antibiotic that inhibits bacterial growth by reversibly binding to the 30S ribosomal subunit and preventing the formation of the aminoacyl-tRNA–ribosome complex. OtrA protects the ribosome from translational arrest by TCs (80–82). The otrA, located in the right fringe of the OTC gene cluster, is a paralogue found in pathogens that cause tetracycline-resistant clinical infections (encoded by tetO and tetM). These proteins are similar to elongation factors and act by releasing tetracyclines from their inhibitory site on the ribosome (80). The second gene, otrB, encodes an integral membrane protein that is responsible for the efflux of OTC from the cell (80, 83, 84). Interestingly, this gene is regulated by another regulatory type protein, the putative MarR family repressor protein OxyTA1 (also designated in the past as OtrR (79)). The oxyTA1 has been identified in the OTC gene cluster, which is divergently transcribed from the resistance gene otrB (3). Thus, the overall topology of the otrB-oxyTA1 region mirrors that found in Tn10, with the expression of the tetracycline efflux pump being controlled by a divergent repressor (78, 83). Finally, the otrC gene, which is not part of the otc gene cluster, is only ensuring a lower level of resistance against OTC (85). Yu et al. (85) have presented evidence in their recent publication indicating that otrC encodes a protein with ATPase and multidrug efflux activity.
Future Directions in Biosynthetic Engineering of Tetracycline Analogues
As demonstrated by Charest et al. (15) and Sun et al. (17), who applied synthetic chemistry approaches, it is possible to envisage an entirely new synthetic route towards developing unique TC analogues, thus evading the established semisynthetic routes used in the development of the third generation of TCs. Similarly, biosynthetic engineering approaches also offer new routes. Early work at the Lederle laboratories resulted in the production/isolation of the extremely valuable TC precursor 6DM-CTC (27–37, 73). By applying random mutagenesis approaches, numerous tetracycline analogues have been isolated from the cultures of S. aureofaciens and S. rimosus mutant strains (reviewed by Vanĕk and Hoštálek in 1986 (86)). However, despite the early success by McCormick’s team at Lederle (27–35), and a much deeper understanding of TC biosynthesis gained over the last decades, a relatively limited number of new TC analogues have been reported to date in the literature. This is likely due to a general lack of interest in anti-infective drug development (87). In addition, relatively slow and tedious molecular biology approaches for the genetic manipulation of TC-producing actinobacteria have likely contributed to the relatively low success of the applied biosynthetic engineering efforts. However, affordable and versatile biosynthetic engineering and synthetic biology approaches, which have rapidly evolved in the recent years, will significantly facilitate any potential biosynthetic efforts directed towards producing new TC scaffolds. The reiterative nature of the type II PKS enzymatic machinery, catalyzing the biosynthesis of TCs, does not offer such diverse number of approaches to biosynthetic engineering routes compared to the type I PKS system, represented by the modular PKS systems involved in e.g. macrolactone biosynthesis (88). However, biosynthetic engineering efforts in TCs are not exhausted, and can be productively applied at the initial steps of TC biosynthesis (e.g. starter unit selection) as well as at the late stages of TC biosynthesis, the so-called post--PKS processing, steps that are often of crucial importance for the biological activity of these compounds (26, 62). A number of TC structures with a broad range of biological activities have been isolated (Fig. 3) and the corresponding gene clusters have been cloned, as recently exemplified by Wang et al. (62) and Lukežič et al. (25).
Rational biosynthetic engineering approaches represent at present a valuable complementary strategy to the existing semisynthetic approaches. Novel functional groups, identified in the scope of DNA sequencing strategies, can be introduced into the established TC scaffolds (e.g. CTC or OTC). However, the reverse approach, whereby functional group(s) found in clinically-used TC analogues, such as OTC or CTC, are introduced into novel TC scaffolds using biosynthetic engineering approaches, might as well be a valuable alternative. For example, the C2-carboxyamido moiety, found in typical tetracyclines such as OTC and CTC, is known to be an important structural feature for their bioactivity (2, 4). Applying the latter strategy, Lešnik et al. (26) introduced the C2-carboxyamido moiety found in typical tetracyclines into the CHD backbone. This was achieved through the expression of the amidotransferase OxyD, catalysing the transamination reaction of malonate to malonamate, and an acyltransferase homologue OxyP, both involved in the priming steps in the OTC pathway from S. rimosus in CHD-producing strain A. sulphurea. The recombinant strain constructed this way resulted in the efficient production of a new analogue with significantly improved antibacterial activity (26).
A number of non-commercial TC analogues of bacterial or fungal origin are known, whose biosynthetic pathways have been studied, such as SF2575 produced by Streptomyces sp. SF2575 (71), dactylocycline produced by Dactylosporangium sp. SC14051 (62), and CHD produced by A. sulphurea (25). Other structurally related tetracyclic secondary metabolites reported in the literature and produced by actinobacteria include cervimycin C produced by Streptomyces tendae HKI-179 (89), polyketomycin produced by Streptomyces diastatochromogenes Tü6028 (72) and dutomycin, isolated from the culture of Streptomyces sp. 1725 (90) (Fig. 3). As exemplified by Wang et al. (62), rapidly expanding genomics data, available in the recent years thanks to significantly improved and affordable DNA sequencing technologies, will undoubtedly result in the identification of numerous novel gene clusters encoding for the production of TC scaffolds.
Conclusion
Despite the much deeper understanding gained over the last two decades on tetracycline biosynthesis, biosynthetic approaches to the development of TC antibiotics have not been productive. However, recent development of rapid and powerful molecular biology tools for the manipulation of TC pathways in antibiotic-producing strains, and rapidly expanding genomics data available today will undoubtedly augment the capacity of the biosynthetic engineering and semisynthetic efforts applied for the development of novel TC antibiotics.
Acknowledgement
Professor Marija Alačević passed away in Zagreb (Croatia) on February 25, 2015. She significantly contributed to the scientific field of the genetics of Streptomyces, which has been internationally recognized over the years as the ‘Zagreb School of Streptomyces Genetics’. In the scope of this review article, we particularly thank Marija Alačević for her essential role over many years in stimulating the research on the genetics of Streptomyces rimosus.
References
- 1.Chopra I, Hawkey PM, Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992;29:245–77. 10.1093/jac/29.3.245 [DOI] [PubMed] [Google Scholar]
- 2.Nelson ML, Levy SB. The history of the tetracyclines. Ann N Y Acad Sci. 2011;1241:17–32. 10.1111/j.1749-6632.2011.06354.x [DOI] [PubMed] [Google Scholar]
- 3.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickens LB, Tang Y. Oxytetracycline biosynthesis. J Biol Chem. 2010;285:27509–15. 10.1074/jbc.R110.130419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67:419–31. 10.1007/s00018-009-0172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. Tetracycline antibiotics and resistance mechanisms. Biol Chem. 2014;395:559–75. 10.1515/hsz-2013-0292 [DOI] [PubMed] [Google Scholar]
- 7.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–50. 10.1016/S1473-3099(14)70780-7 [DOI] [PubMed] [Google Scholar]
- 8.Top 100 drugs for Q4 2013 by sales, Drugsite Trust, Auckland, New Zealand; 2014. Available from: http://www.drugs.com/stats/top100/sales.
- 9.Church RFR, Schaub RE, Weiss MJ. Synthesis of 7-dimethylamino-6-demethyl-6-deoxytetracycline (minocycline) via 9-ni--tro-6-demethyl-6-deoxytetracycline. J Org Chem. 1971;36:723–5. 10.1021/jo00804a025 [DOI] [PubMed] [Google Scholar]
- 10.Schafer JJ, Goff DA. Establishing the role of tigecycline in an era of antimicrobial resistance. Expert Rev Anti Infect Ther. 2008;6:557–67. 10.1586/14787210.6.5.557 [DOI] [PubMed] [Google Scholar]
- 11.Sum PE, Lee VJ, Testa RT, Hlavka JJ, Ellestad GA, Bloom JD, et al. Glycylcyclines. 1. A new generation of potent antibacterial agents through modification of 9-aminotetracyclines. J Med Chem. 1994;37:184–8. 10.1021/jm00027a023 [DOI] [PubMed] [Google Scholar]
- 12.Huttner B, Jones M, Rubin MA, Neuhauser MM, Gundlapalli A, Samore M. Drugs of last resort? The use of polymyxins and tigecycline at US Veterans Affairs medical centers, 2005–2010. PLoS One. 2012;7:e36649. 10.1371/journal.pone.0036649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draper MP, Weir S, Macone A, Donatelli J, Trieber CA, Tanaka SK, et al. Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob Agents Chemother. 2014;58:1279–83. 10.1128/AAC.01066-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macone AB, Caruso BK, Leahy RG, Donatelli J, Weir S, Draper MP, et al. In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob Agents Chemother. 2014;58:1127–35. 10.1128/AAC.01242-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charest MG, Lerner CD, Brubaker JD, Siegel DR, Myers AG. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science. 2005;308:395–8. 10.1126/science.1109755 [DOI] [PubMed] [Google Scholar]
- 16.Clark RB, Hunt DK, He M, Achorn C, Chen CL, Deng Y, et al. Fluorocyclines. 2. Optimization of the C-9 side-chain for antibacterial activity and oral efficacy. J Med Chem. 2012;55:606–22. 10.1021/jm201467r [DOI] [PubMed] [Google Scholar]
- 17.Sun C, Wang Q, Brubaker JD, Wright PM, Lerner CD, Noson K, et al. A robust platform for the synthesis of new tetracycline antibiotics. J Am Chem Soc. 2008;130:17913–27. 10.1021/ja806629e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao XY, Hunt DK, Zhou J, Clark RB, Dunwoody N, Fyfe C, et al. Fluorocyclines. 1. 7-fluoro-9-pyrrolidinoacetamido-6-demethyl-6-deoxytetracycline: a potent, broad spectrum antibacterial agent. J Med Chem. 2012;55:597–605. 10.1021/jm201465w [DOI] [PubMed] [Google Scholar]
- 19.Tetraphase announces zop-line results from IGNITE2 phase 3 clinical trial of eravacycline in cUTI, Tetraphase Pharmaceuticals. Watertown, MA, USA; 2015. Available from: http://ir.tphase.com/releasedetail.cfm?Release ID=930613.
- 20.Oliva B, Gordon G, McNicholas P, Ellestad G, Chopra I. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob Agents Chemother. 1992;36:913–9. 10.1128/AAC.36.5.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen B, Noller HF, Daubresse G, Oliva B, Misulovin Z, Rothstein DM, et al. Molecular basis of tetracycline action: identification of analogs whose primary target is not the bacterial ribosome. Antimicrob Agents Chemother. 1991;35:2306–11. 10.1128/AAC.35.11.2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver TJ, Sinclair AC. Antibiotic M-319. US patent 3155582A. 1964.
- 23.Rogalski W, Kirchlechner R, Seubert J, Gottschlich R, Steinigeweg R, Bergmann R, et al. Tetracyclic compounds. US patent 4024272A. 1977.
- 24.Molnar V, Matković Z, Tambić T, Kozma C. Clinico-pharmacological investigation of chelocardine in patients suffering from urinary tract infection. Lijec Vjesn. 1977;99:560–2. [in Croatian] [PubMed] [Google Scholar]
- 25.Lukežič T, Lešnik U, Podgoršek A, Horvat J, Polak T, Šala M, et al. Identification of the chelocardin biosynthetic gene cluster from Amycolatopsis sulphurea: a platform for producing novel tetracycline antibiotics. Microbiology. 2013;159:2524–32. 10.1099/mic.0.070995-0 [DOI] [PubMed] [Google Scholar]
- 26.Lešnik U, Lukežič T, Podgoršek A, Horvat J, Polak T, Šala M, et al. Construction of a new class of tetracycline lead structures with potent antibacterial activity through biosynthetic engineering. Angew Chem Int Ed Engl. 2015;54:3937–40. 10.1002/anie.201411028 [DOI] [PubMed] [Google Scholar]
- 27.McCormick JRD, Jensen ER. Biosynthesis of the tetracyclines. XII. Anhydrodemethylchlortetracycline from a mutant of Streptomyces aureofaciens. J Am Chem Soc. 1969;91:206. 10.1021/ja01029a046 [DOI] [PubMed] [Google Scholar]
- 28.McCormick JRD, Jensen ER. Biosynthesis of tetracyclines. X. Protetrone. J Am Chem Soc. 1968;90:7126–7. 10.1021/ja01027a045 [DOI] [PubMed] [Google Scholar]
- 29.McCormick JRD, Jensen ER. Biosynthesis of the tetracyclines. VIII. Characterization of 4-hydroxy-6-methylpretetramid. J Am Chem Soc. 1965;87:1794–5. 10.1021/ja01086a034 [DOI] [PubMed] [Google Scholar]
- 30.McCormick JRD, Jensen ER, Arnold N, Corey HS, Joachim UH, Johnson S, et al. Biosynthesis of tetracyclines. XI. Methylanthrone analog of protetrone. J Am Chem Soc. 1968;90:7127–9. 10.1021/ja01027a046 [DOI] [PubMed] [Google Scholar]
- 31.McCormick JRD, Jensen ER, Johnson S, Sjolander NO. Biosynthesis of the tetracyclines. IX. 4-Aminodedimethylaminoanhydrodemethylchlortetracycline from a mutant of Streptomyces aureofaciens. J Am Chem Soc. 1968;90:2201–2. 10.1021/ja01010a063 [DOI] [PubMed] [Google Scholar]
- 32.McCormick JRD, Joachim UH, Jensen ER, Johnson S, Sjolander NO. Biosynthesis of the Tetracyclines. VII. 4-hydroxy-6- -methylpretetramid, an intermediate accumulated by a blocked mutant of Streptomyces aureofaciens. J Am Chem Soc. 1965;87:1793–4. 10.1021/ja01086a033 [DOI] [PubMed] [Google Scholar]
- 33.McCormick JRD, Sjolander NO, Johnson S, Doerschuk AP. Biosynthesis of tetracyclines. II. Simple, defined media for growth of Streptomyces aureofaciens and elaboration of 7-chlorotetracycline. J Bacteriol. 1959;77:475–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick JRD, Johnson S, Sjolander NO. Biosynthesis of the tetracyclines. V. Naphthacenic precursors. J Am Chem Soc. 1963;85:1692–4. 10.1021/ja00894a037 [DOI] [Google Scholar]
- 35.McCormick JRD, Miller PA, Johnson S, Arnold N, Sjolander NO. Biosynthesis of the tetracyclines. IV. Biological rehydration of the 5a,6-anhydrotetracyclines. J Am Chem Soc. 1962;84:3023–5. 10.1021/ja00874a040 [DOI] [Google Scholar]
- 36.Ryan MJ, Lotvin JA, Strathy N, Fantini SE. Cloning of the biosynthetic pathway for chlortetracycline and tetracycline formation and cosmids useful therein. US patent 5589385A. 1996.
- 37.Binnie C, Warren M, Butler MJ. Cloning and heterologous expression in Streptomyces lividans of Streptomyces rimosus genes involved in oxytetracycline biosynthesis. J Bacteriol. 1989;171:887–95. 10.1128/jb.171.2.887-895.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Ames BD, Tsai SC, Tang Y. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol. 2006;72:2573–80. 10.1128/AEM.72.4.2573-2580.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lešnik U, Gormand A, Magdevska V, Fujs Š, Raspor P, Hunter I, et al. Regulatory elements in tetracycline-encoding gene clusters: the otcG gene positively regulates the production of oxytetracycline in Streptomyces rimosus. Food Technol Biotechnol. 2009;47:323–30. [Google Scholar]
- 40.Yin S, Wang W, Wang X, Zhu Y, Jia X, Li S, et al. Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb Cell Fact. 2015;14:46. 10.1186/s12934-015-0231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci Biotechnol Biochem. 1995;59:1099–106. 10.1271/bbb.59.1099 [DOI] [PubMed] [Google Scholar]
- 42.Zhu T, Cheng X, Liu Y, Deng Z, You D. Deciphering and engineering of the final step halogenase for improved chlortetracycline biosynthesis in industrial Streptomyces aureofaciens. Metab Eng. 2013;19:69–78. 10.1016/j.ymben.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 43.Vanĕk Z, Hoštálek Z, Blumauerová M, Mikulík K, Podojil M, Bĕhal V, et al. The biosynthesis of tetracycline. Pure Appl Chem. 1973;34:463–86. 10.1351/pac197334030463 [DOI] [PubMed] [Google Scholar]
- 44.Mindlin SZ, Alikhanian SI, Vladimirov AV, Mikhailova GR. A new hybrid strain of an oxytetracycline-producing organism, Streptomyces rimosus. Appl Microbiol. 1961;9:349–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friend EJ, Hopwood DA. The linkage map of Streptomyces rimosus. J Gen Microbiol. 1971;68:187–97. 10.1099/00221287-68-2-187 [DOI] [PubMed] [Google Scholar]
- 46.Alačević M, Strašek-Vešligaj M, Sermonti G. The circular linkage map of Streptomyces rimosus. Microbiology. 1973;77:173–85. 10.1099/00221287-77-1-173 [DOI] [Google Scholar]
- 47.Alačević M. Genetics of tetracycline-producing streptomycetes. Proceedings of the First International Symposium on the Genetics of Industrial Microorganisms (GIM), Prague, Czechoslovakia; 1970. pp. 59–70. [Google Scholar]
- 48.Alačević M. Recent advances in Streptomyces rimosus genetics. In: McDonald KD, editor. Proceedings of the Second International Symposium on the Genetics of Industrial Microorganisms, Sheffield, United Kingdom; London, UK: Academic Press; 1974. pp. 513–9. [Google Scholar]
- 49.Pigac J, Alačević M. Mapping of oxytetracycline genes in Streptomyces rimosus. Period Biol. 1979;81:575–82. [Google Scholar]
- 50.Rhodes PM, Winskill N, Friend EJ, Warren M. Biochemical and genetic characterization of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. Microbiology. 1981;124:329–38. 10.1099/00221287-124-2-329 [DOI] [Google Scholar]
- 51.Butler MJ, Binnie C, Hunter IS, Sugden DA, Warren M. Genetic manipulation of the oxytetracycline biosynthetic pathway genes. Dev Ind Microbiol. 1990;31:41–50. [Google Scholar]
- 52.Butler MJ, Friend EJ, Hunter IS, Kaczmarek FS, Sugden DA, Warren M. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus. Mol Gen Genet. 1989;215:231–8. 10.1007/BF00339722 [DOI] [PubMed] [Google Scholar]
- 53.McDowall KJ, Doyle D, Butler MJ, Binnie C, Warren M, Hunter IS. Molecular genetics of oxytetracycline production by Streptomyces rimosus. In: Baumberg S, Krügel H, Noock D, editors. Proceedings of the International Symposium on the Genetics and Product Formation in Streptomyces, Erfurt, Germany; New York, NY, USA: Plenum Press; 1990. pp. 105–16. [Google Scholar]
- 54.Petković H, Thamchaipenet A, Zhou LH, Hranueli D, Raspor P, Waterman PG, et al. Disruption of an aromatase/cyclase from the oxytetracycline gene cluster of Streptomyces rimosus results in production of novel polyketides with shorter chain lengths. J Biol Chem. 1999;274:32829–34. [PubMed] [Google Scholar]
- 55.Perić-Concha N, Borovička B, Long PF, Hranueli D, Waterman PG, Hunter IS. Ablation of the otcC gene encoding a post-polyketide hydroxylase from the oxytetracyline biosynthetic pathway in Streptomyces rimosus results in novel polyketides with altered chain length. J Biol Chem. 2005;280:37455–60. 10.1074/jbc.M503191200 [DOI] [PubMed] [Google Scholar]
- 56.Hranueli D, Perić N, Petković H, Biuković G, Toman Z, Pigac J, et al. Novel hybrid polyketide compounds produced by genetic engineering of the oxytetracycline biosynthetic pathway. Food Technol Biotechnol. 1999;37:117–25. [Google Scholar]
- 57.Zhang W, Ames BD, Tsai SC, Tang Y. Engineered biosynthesis of a novel amidated polyketide, using the malonamyl-specific initiation module from the oxytetracycline polyketide synthase. Appl Environ Microbiol. 2006;72:2573–80. 10.1128/AEM.72.4.2573-2580.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, Watanabe K, Wang CC, Tang Y. Investigation of early tailoring reactions in the oxytetracycline biosynthetic pathway. J Biol Chem. 2007;282:25717–25. 10.1074/jbc.M703437200 [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Watanabe K, Cai X, Jung ME, Tang Y, Zhan J. Identifying the minimal enzymes required for anhydrotetracycline biosynthesis. J Am Chem Soc. 2008;130:6068–9. 10.1021/ja800951e [DOI] [PubMed] [Google Scholar]
- 60.Wang P, Zhang W, Zhan J, Tang Y. Identification of OxyE as an ancillary oxygenase during tetracycline biosynthesis. ChemBioChem. 2009;10:1544–50. 10.1002/cbic.200900122 [DOI] [PubMed] [Google Scholar]
- 61.Wang P, Gao X, Chooi YH, Deng Z, Tang Y. Genetic characterization of enzymes involved in the priming steps of oxytetracycline biosynthesis in Streptomyces rimosus. Microbiology. 2011;157:2401–9. 10.1099/mic.0.048439-0 [DOI] [PubMed] [Google Scholar]
- 62.Wang P, Kim W, Pickens LB, Gao X, Tang Y. Heterologous expression and manipulation of three tetracycline biosynthetic pathways. Angew Chem Int Ed Engl. 2012;51:11136–40. 10.1002/anie.201205426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang P, Bashiri G, Gao X, Sawaya MR, Tang Y. Uncovering the enzymes that catalyze the final steps in oxytetracycline biosynthesis. J Am Chem Soc. 2013;135:7138–41. 10.1021/ja403516u [DOI] [PubMed] [Google Scholar]
- 64.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–98. 10.1021/cr960034i [DOI] [PubMed] [Google Scholar]
- 65.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–90. 10.1039/B507395M [DOI] [PubMed] [Google Scholar]
- 66.Hori Y, Abe Y, Ezaki M, Goto T, Okuhara M, Kohsaka M. R1128 substances, novel non-steroidal estrogen-receptor antagonists produced by a Streptomyces. I. Taxonomy, fermentation, isolation and biological properties. J Antibiot. 1993;46:1055–62. 10.7164/antibiotics.46.1055 [DOI] [PubMed] [Google Scholar]
- 67.Tang Y, Koppisch AT, Khosla C. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry. 2004;43:9546–55. 10.1021/bi049157k [DOI] [PubMed] [Google Scholar]
- 68.Das A, Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009;42:631–9. 10.1021/ar8002249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Revill WP, Bibb MJ, Hopwood DA. Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J Bacteriol. 1995;177:3946–52. 10.1128/jb.177.14.3946-3952.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prado L, Lombó F, Braña AF, Méndez C, Rohr J, Salas JA. Analysis of two chromosomal regions adjacent to genes for a type II polyketide synthase involved in the biosynthesis of the antitumor polyketide mithramycin in Streptomyces argillaceus. Mol Gen Genet. 1999;261:216–25. 10.1007/s004380050960 [DOI] [PubMed] [Google Scholar]
- 71.Pickens LB, Kim W, Wang P, Zhou H, Watanabe K, Gomi S, et al. Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J Am Chem Soc. 2009;131:17677–89. 10.1021/ja907852c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daum M, Peintner I, Linnenbrink A, Frerich A, Weber M, Paululat T, et al. Organisation of the biosynthetic gene cluster and tailoring enzymes in the biosynthesis of the tetracyclic quinone glycoside antibiotic polyketomycin. ChemBioChem. 2009;10:1073–83. 10.1002/cbic.200800823 [DOI] [PubMed] [Google Scholar]
- 73.Ryan MJ. Strain for the production of 6-demethyltetracycline, method for producing the strain and vector for use in the method. US patent 5965429A. 1999.
- 74.Vanĕk Z, Cudlín J, Blumauerová M, Hoštálek Z. How many genes are required for the synthesis of chlortetracycline? Folia Microbiol (Praha). 1971;16:225–40. 10.1007/BF02884211 [DOI] [PubMed] [Google Scholar]
- 75.Martin JF, Demain AL. Control of antibiotic biosynthesis. Microbiol Rev. 1980;44:230–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bĕhal V. The tetracycline fermentation and its regulation. Crit Rev Biotechnol. 1987;5:275–318. 10.3109/07388558709079475 [DOI] [Google Scholar]
- 77.McDowall KJ, Thamchaipenet A, Hunter IS. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J Bacteriol. 1999;181:3025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bĕhal V, Hunter IS. Tetracyclines. In: Vining LC, Stuttard C, editors. Genetics and biochemistry of antibiotic production. Boston, MA, USA: Butterworth-Heinemann; 1995. pp. 359–84. http://dx.doi.org/ 10.1016/B978-0-7506-9095-9.50022-8 [DOI] [Google Scholar]
- 79.Petković H, Cullum J, Hranueli D, Hunter IS, Perić-Concha N, Pigac J, et al. Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol Mol Biol Rev. 2006;70:704–28. 10.1128/MMBR.00004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohnuki T, Katoh T, Imanaka T, Aiba S. Molecular cloning of tetracycline resistance genes from Streptomyces rimosus in Streptomyces griseus and characterization of the cloned genes. J Bacteriol. 1985;161:1010–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doyle D, Butler MJ, Hunter IS. Molecular analysis of the oxytetracycline resistance gene, otrA, from Streptomyces rimosus. Heredity. 1988;61:305. [DOI] [PubMed] [Google Scholar]
- 82.Doyle D, McDowall KJ, Butler MJ, Hunter IS. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol Microbiol. 1991;5:2923–33. 10.1111/j.1365-2958.1991.tb01852.x [DOI] [PubMed] [Google Scholar]
- 83.McMurry LM, Levy SB. Revised sequence of OtrB (tet347) tetracycline efflux protein from Streptomyces rimosus. Antimicrob Agents Chemother. 1998;42:3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reynes JP, Calmels T, Drocourt D, Tiraby G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. J Gen Microbiol. 1988;134:585–98. 10.1099/00221287-134-3-585 [DOI] [PubMed] [Google Scholar]
- 85.Yu L, Yan X, Wang L, Chu J, Zhuang Y, Zhang S, et al. Molecular cloning and functional characterization of an ATP-binding cassette transporter OtrC from Streptomyces rimosus. BMC Biotechnol. 2012;12:52. 10.1186/1472-6750-12-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vanĕk Z, Hoštálek Z. Overproduction of microbial metabolites: strain improvement and process control strategies. Boston, MA, USA: Butterworths; 1986. [Google Scholar]
- 87.Brandenburg K, Schürholz T. Lack of new antiinfective agents: passing into the pre-antibiotic age? World J Biol Chem. 2015;6:71–7. 10.4331/wjbc.v6.i3.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu W, Qiao K, Tang Y. Structural analysis of protein-protein interactions in type I polyketide synthases. Crit Rev Biochem Mol Biol. 2013;48:98–122. 10.3109/10409238.2012.745476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herold K, Xu Z, Gollmick FA, Gräfe U, Hertweck C. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Org Biomol Chem. 2004;2:2411–4. 10.1039/B409221J [DOI] [PubMed] [Google Scholar]
- 90.Xuan LJ, Xu SH, Zhang HL, Xu YM, Chen MQ. Dutomycin, a new anthracycline antibiotic from Streptomyces. J Antibiot. 1992;45:1974–6. 10.7164/antibiotics.45.1974 [DOI] [PubMed] [Google Scholar]