Abstract
Background
One potential use for the PR interval is as a biomarker of disease risk. We hypothesized that quantifying the shared genetic architectures of the PR interval and a set of clinical phenotypes would identify genetic mechanisms contributing to PR variability and identify diseases associated with a genetic predictor of PR variability.
Methods and Results
We used ECG measurements from the Atherosclerosis Risk in Communities (ARIC) study (n=6,731 subjects) and 63 genetically-modulated diseases from the Electronic Medical Records and Genomics (eMERGE) network (n=12,978). We measured pairwise genetic correlations (rG) between PR phenotypes (PR interval, PR segment, P wave duration) and each of the 63 phenotypes. The PR segment was genetically correlated with atrial fibrillation (AF) [rG=−0.88, p=0.0009]. An analysis of metabolic phenotypes in ARIC also showed that the P wave was genetically correlated with waist circumference [rG=0.47, p=0.02]. A genetically predicted PR interval phenotype based on 645,714 SNPs was associated with AF [OR=0.89 per standard deviation change, 95% CI (0.83–0.95), p=0.0006]. The differing pattern of associations among the PR phenotypes is consistent with analyses that show that the genetic correlation between the P wave and PR segment was not significantly different than 0 (rG=−0.03 [0.16]).
Conclusions
The genetic architecture of the PR interval comprises modulators of AF risk and obesity.
Keywords: ECG, genetic association, genetic epidemiology, atrial fibrillation
Journal Subject Terms: Electrophysiology, Genetic, Association Studies, Catheter Ablation and Implantable Cardioverter-Defibrillator
Introduction
The PR interval is an electrophysiological parameter derived from a cardiac electrocardiogram and measures the duration of conduction through the atrium and atrioventricular (AV) node. The PR interval comprises two components: the P wave, which primarily measures atrial conduction, and the PR segment, which primarily reflects AV nodal conduction. One potential use for the PR interval is as a biomarker for future disease risk. For instance, a prolonged PR interval is associated with an increased risk for atrial fibrillation (AF).1,2 If such associations are driven by heritable variation affecting both phenotypes, then a risk classifier based on genetic factors modulating the PR interval could be used to identify individuals at high risk for AF. Since heritable genetic risk is determined at birth, genetic classifiers can be evaluated at early time points, thereby enhancing early prevention and risk stratification strategies.
To date, a relatively small number of single nucleotide polymorphisms (SNPs) associated with the PR interval have been identified by genome wide association studies (GWAS)3–5, and they only account of a small portion of the underlying genetic variability. Hence, building and evaluating a robust genetic classifier for the PR interval based on known SNPs is not feasible. Newer genetic approaches, such as those based on generalized linear mixed models (GLMM) that measure the contribution of very large numbers of SNPs to a phenotype, can circumvent this limitation.6–8 Furthermore, these methods can also identify genetically related phenotypes across data sets by measuring genetic correlations based on additive genetics between pairs of phenotypes.8–10 A phenotype that is genetically correlated (i.e. has a non-zero genetic correlation) with the PR interval likely shares common physiological mechanisms and, potentially, can be predicted by a PR interval-based genetic classifier.
We used mixed modelling approaches to probe the additive genetic architecture of the PR interval based on the extent to which its architecture was shared by a set of clinically-recognized diseases. This approach identifies both clinical diagnoses genetically related to the PR interval and genetically-mediated disease mechanisms underlying PR variability. Specifically, we employed a discovery-oriented approach whereby we measured genetic correlations between PR interval phenotypes and a collection of clinical phenotypes. To ensure that associations are attributable to shared genetic risk factors and not environmental factors, we tested associations across populations: PR interval phenotypes were from the prospectively studied Atherosclerosis Risk in Communities (ARIC) cohort11 and clinical phenotypes were from the Electronic Medical Records and Genomics (eMERGE) network, a consortium of medical centers with observational electronic health records (EHR)-linked DNA biobanks data sets.12 We show distinct patterns of genetic disease associations among the PR phenotypes and that PR interval variability is driven by genetic factors associated with electrophysiological and metabolic phenotypes.
Materials and Methods
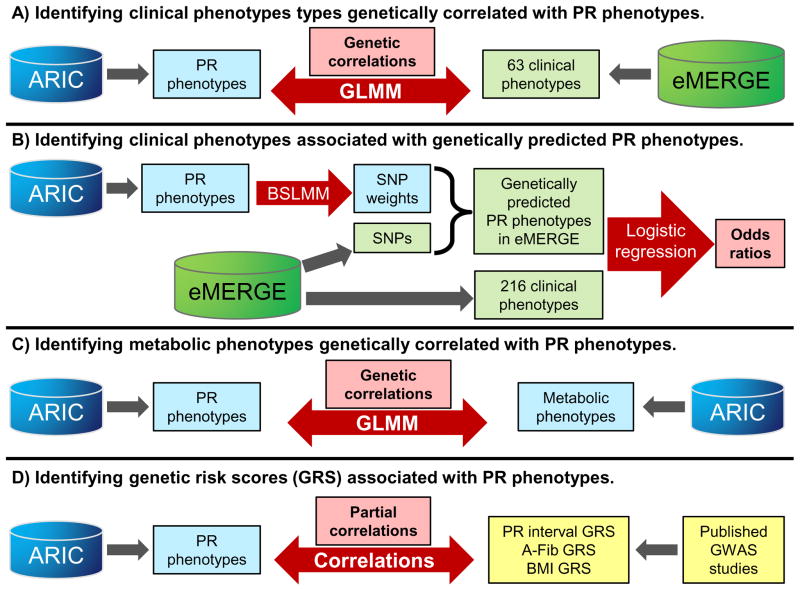
An overview of the analyses is presented in Figure 1.
Figure 1.
Overview of approach. A) GLMMs were used to measure the pair-wise genetic correlations (rG) between each ARIC PR phenotype (PR interval, PR segment and P wave duration) and each genetic eMERGE phenotype. B) BSLMMs were used to compute SNP weights for each PR phenotype. These weights were used to compute genetically predicted PR phenotype values in eMERGE subjects. Logistic regression was used to test the association between the predicted PR phenotypes and eMERGE clinical phenotypes. C) GLMMs were used to compute rG between ARIC PR phenotypes and metabolic phenotypes. D) Partial correlation coefficients were calculated between ARIC PR phenotypes and genetic risk scores based on SNPs identified by prior GWAS studies.
Study populations
Analysis data sets
ARIC: The ARIC population was derived from 13,113 genotyped adult subjects and comprised 6,732 unrelated European ancestry (EA) subjects with normal ECGs.11 Genetic and phenotypic data were downloaded from dbGaP (phs000280.v3.p1). eMERGE: The eMERGE population comprised 12,978 unrelated EA adult subjects collected by the eMERGE Phase I Network [Vanderbilt University (VUMC), Marshfield Clinic, Northwestern University, Mayo Clinic and Group Health Research Institute], a consortium of medical centers using electronic health records as a tool for genomic research.13 Genetic data for the eMERGE network is available through dbGaP (phs000360.v2.p1).
Replication data sets
BioVU AF registry: The Vanderbilt Lone AF registry data set comprised 1,690 European ancestry patients between 18 and 65 years of age enrolled through Vanderbilt’s inpatient and outpatient services, as previously described, and had 1,022 AF cases and 668 control subjects.14 Of the cases, 220 have lone AF, 444 have paroxysmal AF and 259 have persistent AF. BioVU VESPA data set: The BioVU VESPA study population comprised 1,206 AF adult cases and 2,405 controls from VUMC’s collection of genotyped patients.15,16
All data sets were predominantly composed of self-reported whites so only EA subjects were evaluated, defined using STRUCTURE17 in conjunction with ancestry informative markers, with European ancestry defined as or >80% (eMERGE subjects) >90% (all other data sets) probability of being in the HapMap CEU cluster.
Genetic Data
ARIC: Genotype data were acquired on the Affymetrix 6.0 SNP array. Quality control steps for the ARIC data set followed the guidelines accompanying the dbGaP release and included removing SNPs with pre-identified chromosomal anomalies and with >5 discordant calls in replicate samples, and used a subset of unrelated subjects identified by the ARIC study. eMERGE: SNP genotype data were acquired on the Illumina Human660W-Quadv1_A. BioVU AF data set: Subjects were genotyped on the Illumina 610-quad Beadchip.14 BioVU VESPA: Subjects were genotyped on the Illumina HumanOmni1-Quad and HumanOmni5-Quad platforms. Quality control steps for the eMERGE and BioVU data sets used established protocols18 including filtering for a sample missingness rate<2.0%, a SNP missingness rate<2.0% and a SNP deviation from Hardy-Weinberg<0.001
All data sets were imputed to the October 2014 release of the 1000 Genomes cosmopolitan reference haplotypes. SNPs were pre-phased using SHAPEIT19 and imputed using IMPUTE2.20. The genetic correlation analyses used an intersection of the unimputed ARIC and imputed eMERGE data set and contained 503,404 SNPs with MAF>1.0%. The BSLMM analyses used an LD-reduced (r-square=0.9) set of SNPs with MAF>1.0% present on all platforms (n= 645,714 SNPs).
Phenotype data
The clinical phenotypes for the eMERGE and BioVU data sets were based on PheWAS Phecodes which are collections of related International Classification of Disease, Ninth revision, Clinical Modification (ICD9) diagnosis codes.21–24 There are over 1,600 defined Phecodes, described at http://PheWAScatalog.org. For each Phecode, cases are subjects with two or more instances of the phenotype appearing their medical record on two separate dates.23 Controls with no instances of the phenotype were randomly selected. There were 315 phenotypes with more than 500 cases in the eMERGE data set. Atrial fibrillation cases and controls were based on PheWAS code 427.21 (“Atrial Fibrillation”) which has been previously used in other genetic studies.22,23,25
ARIC phenotypes came from the GENEVA substudy (pht000114.v2.p1) and from ECG measurements taken at visit 1 (pht004071.v1.p1). Subjects with a baseline ECG diagnosis of atrial fibrillation, AV block other than first degree, Wolff-Parkinson-White, a non-sinus rhythm or a pacemaker were excluded. Subjects on AV nodal blocking drugs were also excluded. The PR interval was extracted from the ECG. The P wave duration was based on lead aVR and the PR segment duration was calculated as the difference between the PR segment and P wave duration. Subjects with a PR interval ≤80 or ≥320 ms were excluded, as were subjects with a P wave duration ≤50 or ≥140 ms. Phenotypes definitions for metabolic phenotypes were based on previously described thresholds for the ARIC data set26: elevated waist circumference (≥102 cm [men] or ≥88 cm [women]); insulin resistance (fasting glucose ≥100 mg/dl or history of diabetes); hypertension (systolic blood pressure ≥130 mm Hg, diastolic blood pressure ≥85 mm Hg or use of antihypertensive medications); elevated triglycerides (≥150 mg/dl or use of medications for elevated lipids); low HDL cholesterol (<40 mg/dl [men] or <50 mg/dl [women] or use of medications for elevated lipids); and metabolic syndrome (3 or more abnormal metabolic components). Subjects who were not fasting for >8 hours at their first visit were excluded from the analysis of metabolic phenotypes (n=150).
Analyses
Linear mixed models (LMM) and generalized LMM (GLMM) estimate the additive-genetic variance or liability, respectively, attributable to a collection of common SNPs among unrelated individuals by modelling the genetic similarity between pairs of individuals as random effects.6,7,27 The linear mixed model is expressed as:
where y is phenotype vector, X is a vector of fixed effects (covariates and principal components) and ε is a vector of errors. The term gG is a vector of random polygenic effects, and AG is often referred to as the Genetic Relationship Matrix (GRM), with each element in the matrix defined by the equation (1/N) ΣNi=1 (xij−2pi)(xik−2pi) / 2pi(1−pi) where N in the number of SNPs analyzed, x is the genotype at that SNP (coded 0, 1 or 2) for individuals j and k, and p is the allele frequency. The variance components are estimated by a restricted maximum likelihood (REML) algorithm. These analyses used LMMs and GLMMs as implemented in the Genome-wide Complex Trait Analysis (GCTA) program.6,7,9,27,28 To ensure only unrelated subjects are analyzed, subjects with a genetic relatedness score>0.05 were excluded. Genetic liability estimates, adjusting for birth decade, sex and 20 principal components (PCs), were computed for each eMERGE PheWAS phenotype29 with >500 cases, and phenotypes with a genetic liability estimate p<0.05 (n=63) were used for the exploratory genetic correlation analyses (Supplementary Table 1).
A bivariate extension of the GLMM was used to undertake the exploratory genetic correlation analyses (Supplementary Table 1). Here, y is now comprised of pairs of phenotype vectors. For each pair of traits (t1 and t2), the bivariate GLMM estimates the genetic variance (σ2G) for the phenotypes and the genetic covariance between the phenotypes covg(G_t1,G_t2).9,10 The genetic covariance is a measure of how much pairs of traits change together based on the additive genetic effects from common SNPs. This model is most commonly applied to data from two different non-overlapping samples, where the trait (y) values are simply set to missing when not observed (e.g., for subjects in the study of trait t1, their t2 values are set to missing).9,10 The genetic correlation between pairs of traits is then defined as: rG = covg(G_t1,G_t2) / sqrt[(σ2Gt1)(σ2Gt2)]. This genetic correlation is a measure of the extent to which the additive genetic effects estimated from common SNPs are shared between a pair of traits. rG is computationally analogous to a Pearson’s correlation coefficient, and has a value of −1 to +1. Genetic correlations were computed between the ARIC PR phenotypes (PR interval, the P wave and PR segment) and each eMERGE PheWAS phenotype (n=63), adjusting for age, sex and 20 PCs. P-values for genetic correlations were determined using a likelihood ratio test comparing the bivariate GLMM to a model where the genetic correlation was fixed at 0. While standard errors are given for rG point estimates, the 95% confidence intervals surrounding these estimates under the assumption of asymptotic normality may fall outside the range of plausible values for rG. False discovery rate (FDR)-adjusted p-values (q-values) were determined using a Benjamini-Hochberg (B-H) adjustment. While not all pheWAS phenotype pairs are independent, the test statistics meet B-H criteria by the positive regression dependent criterion.30
Bayesian Sparse linear mixed modelling (BSLMM) was used to compute genetically predicted levels of PR phenotypes in the eMERGE and BioVU data sets. BSLMM employs a hybrid of GLMM and sparse regression models.31 In general, this method estimates the proportion of variance explained by a set of SNPs and the distribution of effect sizes for the SNPs and then jointly models the contribution of all SNPs to the phenotypic variance. The posterior SNP weights generated by this approach can be used in conjunction with SNP genotypes to compute a genetically predicted value for a phenotype. Each PR phenotype in the ARIC data set was first adjusted for age, gender and 3 PCs using linear regression. BSLMM was then used to generate SNP effect sizes (α and β) for the PR phenotype residuals. These effect sizes were then used to compute the genetically predicted value for a PR phenotype for an individual in the eMERGE and BioVU data sets using the equation:
where α is the small SNP effect, βγ is the large SNP effect.
Multivariable logistic regression adjusting for 3 PCs, age and sex was used to test the association between the predicted phenotype levels and the EMR PheWAS and AF phenotypes. The predicted phenotypes were set to have a standard deviation of 1, so ORs reflect risk per standard deviation increase in the predicted phenotype. A FDR-adjusted q-value<0.1 was considered significant.
Genetic risk scores based on either odds ratios (OR) or Beta coefficients for previously reported SNPs reaching genome-wide significance (p<5x10−8) were computed for the PR interval (n=9 SNPs), BMI (n=98) and atrial fibrillation (n=10 SNPs).5,14,32 The SNPs used to compute the GRS in each data set are shown in Supplementary Tables 2 and 3. A GRS was computed using previously the publish association statistics for each individual using the formulas:33
Only 8 of 9 SNPs for the PR GRS passed QC protocols and were used in the calculations. To ascertain whether the genetic risk scores are differentially associated with the PR phenotypes, partial correlation coefficients between each PR phenotype and each GRS were computed using PROC CORR (SAS) and adjusted for age and sex.
All quality control analyses and SNP association analyses were performed using PLINK v1.07.34 Genetic liability and correlation estimates were computed using the GCTA v1.24.27 BSLMM is part of the GEMMA v0.94.1 program package.31 All other analyses were performed using SAS v9.3 (SAS Institute, Cary, NC).
Ethics Statement
The eMERGE study has been approved by the Institutional Review Board (IRB) at each site.12,15. Vanderbilt’s BioVU resource operates as nonhuman subjects research according to the provisions of 45 Code of Federal Regulations, part 46, with oversight by Vanderbilt’s Institutional Review Board (IRB), as previously described.15 IRB approval for the current study was obtained through Vanderbilt’s IRB.
Results
The ARIC population comprised 6,731 unrelated EA subjects with a normal ECG. Their median age was 54 years and 45% of subjects were males (Supplementary Table 4). Almost a quarter of subjects had three or more metabolic syndrome phenotypes. The eMERGE data set comprised 12,978 subjects, of which 48% were male, with an average of 44 clinical diagnoses per subject (Supplementary table 1).
Clinical phenotypes genetically correlated with PR phenotypes
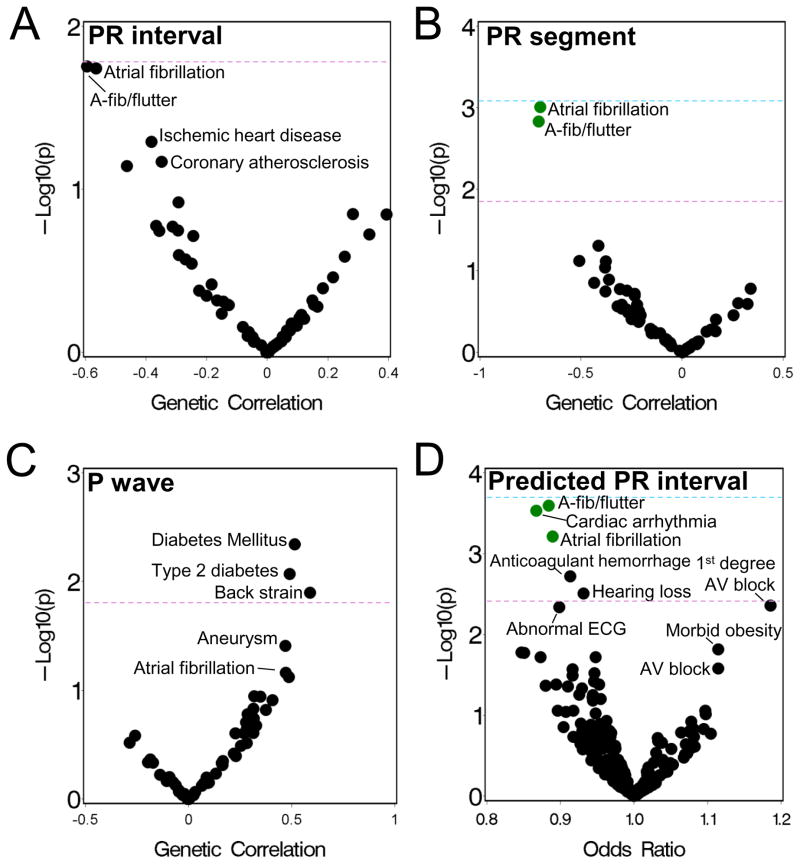
The estimated heritability explained by the SNPs for the PR interval in the ARIC data set was 0.23 (standard error 0.05) (Table 1). We measured the genetic correlation (rG) between the PR interval and 63 eMERGE phenotypes (listed in Supplementary Table 1). The strongest genetic correlations were with “atrial fibrillation/atrial flutter” [rG=−0.59, p=0.02] and AF [rG=−0.57, p=0.02], but were not significant (FDR q>0.1) after adjusting for multiple testing (Figure 2A and Table 2). (Characteristics of the AF cases and controls are shown in Supplementary Table 5).
Table 1.
Heritability/liability estimates for the ARIC cohort.
| Characteristic | Heritability/liability (s.e.)* |
|---|---|
| EKG parameters [mean (s.d.)] | |
| PR interval duration (ms) | 0.23 (0.05) |
| P wave duration (ms) | 0.19 (0.05) |
| PR segment duration (ms) | 0.18 (0.05) |
| Metabolic traits [n (%)]† | |
| Waist circumference | 0.14 (0.04) |
| Insulin Resistance | 0.05 (0.04) |
| Hypertension | 0.18 (0.04) |
| Triglycerides | 0.15 (0.04) |
| HDL cholesterol | 0.14 (0.04) |
| Metabolic syndrome | 0.21 (0.04) |
Heritability or liability estimates or metabolic traits and ECG phenotypes are based on genetic linear mixed models adjusting for age, sex and 20 principal components.
See Methods for metabolic trait definitions.
Figure 2.
Phenotypes genetically associated with PR phenotypes. (A–C) Genetic correlations between the (A) PR interval, (B) PR segment and (C) P wave duration, measured in ARIC subjects, and 63 phenotypes measured in eMERGE subjects, adjusted for age, gender and PCs. P-values are based on a likelihood ratio test comparing a full model to a model where the genetic correlation was fixed at 0. (D) Plot of p-value versus odds-ratio (OR) from logistic regression analyses of the association between a genetically predicted PR interval and 261 eMERGE phenotypes, adjusted for sex, gender and PCs. ORs represent the risk per standard deviation change in the predicted PR interval. For all graphs, each point represents a phenotype and green colored points have an FDR q<0.1. The purple dotted line denotes the expected false positive rate, and the blue line corresponds to a Bonferroni correction.
Table 2.
Genetic correlations (rG)* between PR components and atrial fibrillation.
| PR interval | P wave | PR segment | ||||
|---|---|---|---|---|---|---|
| Adjustment | rG (s.e.) | P-value | rG (s.e.) | P-value | rG (s.e.) | P-value |
| None | −0.57 (0.27) | 0.02 | 0.48 (0.29) | 0.07 | −0.88 (0.35) | 0.0009 |
| P wave adjusted† | −0.84 (0.34) | 0.001 | -- | -- | −0.84 (0.34) | 0.001 |
| PR segment adjusted† | 0.33 (0.28) | 0.22 | 0.32 (0.28) | 0.22 | -- | -- |
Genetic correlations between the PR component measured in ARIC subjects and AF measured in eMERGE. Genetic correlations were adjusted for age, sex and 20 PCs.
Additional covariates for ARIC subjects added to the model.
We next examined the P wave and the PR segment durations (Table 1), which comprise the PR interval. The point estimate of the genetic correlation between the PR segment and the PR interval [rG=0.89 (0.04)] was larger than that for the P wave and the PR interval [rG=0.49 (0.16)]. The genetic correlation between the P wave and the PR segment was not significantly different from zero (rG=−0.03 [0.16]). The PR segment showed a similar pattern of genetic correlations with the eMERGE phenotypes as the PR interval, with the exception that the genetic correlation with AF was significant after multiple testing correction [rG=−0.88 (95% CI: −1.6 to −0.19), p=0.0009, FDR q=0.047] (Table 2 and Figure 2B). For both the PR interval and PR segment, the AF correlation was negative indicating that genetic factors associated with a longer interval are associated with a decreased risk of AF. There were no significant genetic correlations with the P wave (Figure 2C). The most strongly genetically correlated phenotype was type 2 diabetes [rG=0.49, p=0.008, FDR q=0.26].
We examined the impact of adjusting for PR phenotypes on the genetic correlation between the PR interval duration and AF. Adjusting for the P wave duration minimally impacted the genetic correlation between the PR interval and AF [rG=−0.84, p=0.001] (Table 2). In contrast, adjusting for the PR segment further attenuated the P-wave-AF correlation [rG=0.33, p=0.22] (Table 2). Thus, the genetic signal in the PR interval that is associated with AF is most strongly captured by the PR segment.
Associations with a genetically predicted PR interval
The mixed models analyses indicate that a highly polygenic SNP-based genetic classifier could capture up to ~23% of the variability of the PR interval. We used BSLMM31 to construct a highly polygenic SNP classifier for the PR interval in then ARIC data set, and this was used to impute a genetically predicted PR interval for each subject in the eMERGE population. We then tested for an association between the predicted PR interval and 261 clinical phenotypes (with >250 cases and a genetic liability p<0.2).35 Significant associations were seen with arrhythmia phenotypes including AF [OR=0.89, 95% CI (0.83–0.95), p=0.0006, FDR q=0.04) (Table 3 and Figure 2D). Thus, a genetically predicted prolonged PR interval is associated with decreased AF risk. The magnitude of this association was modestly attenuated when adjusting for genetic risk scores based on significant GWAS SNP associations for the PR interval and AF or when adjusting for these SNPs (n=17) as covariates, though the p-value was no longer significant in the latter model (Table 3). While no associations with an opposite direction of effect were significant, the strongest associations were with first degree AV block, a diagnosis of a prolonged PR interval, and morbid obesity (Figure 2D). Analyses using a genetically predicted PR segment or P wave duration did not identify any significant associations, though the top associations for the PR segment were the same as those seen for the genetically predicted PR interval (Supplemental Figure 1).
Table 3.
Association between a genetically predicted PR interval and atrial fibrillation.
| Data set | Subjects | Cases/Controls | OR (95% CI)* | P-value |
|---|---|---|---|---|
| eMERGE | All AF cases | 1,547 / 3,128 | 0.89 (0.83–0.95) | 0.0006 |
| All AF cases, GRS adjusted† | 1,547 / 3,128 | 0.90 (0.83–0.98) | 0.02 | |
| All AF cases, SNP adjusted‡ | 1,547 / 3,128 | 0.90 (0.81–1.01) | 0.06 | |
| BioVU EHR set | All AF | 1,206 / 2,405 | 0.90 (0.85–0.98) | 0.01 |
| BioVU AF registry | All AF | 1,022 / 668 | 0.90 (0.81–0.99) | 0.03 |
| Lone AF | 220 / 668 | 0.87 (0.74–1.03) | 0.1 | |
| Paroxysmal | 444 / 668 | 0.90 (0.80–1.02) | 0.09 | |
| Persistent | 259 / 668 | 0.93 (0.80–1.08) | 0.34 |
The odds-ratio is per standard deviation increase in the genetically predicted PR interval. All association models are adjusted for age, gender and 3 PCs.
Adjusted for genetic risk scores for atrial fibrillation and the PR interval.
Adjusted for SNPs (n=17) previously associated with AF or the PR interval by GWAS.
Validating the atrial fibrillation association
To confirm the genetic correlation between the PR interval and AF, we tested the association between the genetically predicted PR interval and AF in two independent data sets. A second EHR-derived data set (1,206 AF cases and 2,405 controls) that used the same AF phenotype definition as the discovery set had a significant association [OR=0.90 (0.85–0.98), p=0.01] (Table 3). A comparable result was seen using subjects (1022 cases, 668 controls) from Vanderbilt’s AF registry [OR=0.90 (0.81–0.99), p=0.03] (Table 3). There was a similar magnitude and direction of effect when the results were stratified by AF subtypes (lone, paroxysmal and persistent AF) (Table 3).
PR components and metabolic syndrome phenotypes
Other than AF, the strongest genetic correlations for the PR phenotypes were with metabolic phenotypes (diabetes, obesity). Epidemiological studies have also shown that P wave duration is positively associated with metabolic syndrome phenotypes.26 We measured the genetic correlations between each PR interval component and metabolic phenotypes in the ARIC subjects. The PR interval and PR segment were not genetically correlated with any metabolic phenotype (Table 4). The P wave was positively genetically correlated with waist circumference [rG=0.47, p=0.03].
Table 4.
Genetic correlations (rG)* between PR components and metabolic syndrome phenotypes in ARIC.
| PR interval | P wave | PR segment | ||||
|---|---|---|---|---|---|---|
| Metabolic phenotype | rG (s.e.) | P-value | rG (s.e.) | P-value | rG (s.e.) | P-value |
| Waist circumference | 0.16 (0.19) | 0.42 | 0.47 (0.21) | 0.03 | −0.06 (0.22) | 0.80 |
| Insulin resistance | −0.21 (0.30) | 0.47 | −0.14 (0.32) | 0.66 | −0.21 (0.33) | 0.52 |
| Hypertension | −0.10 (0.17) | 0.56 | 0.23 (0.19) | 0.22 | −0.23 (0.19) | 0.22 |
| Triglycerides | −0.17 (0.18) | 0.34 | −0.14 (0.20) | 0.46 | −0.11 (0.20) | 0.57 |
| HDL cholesterol | −0.15 (0.19) | 0.44 | −0.15 (0.21) | 0.62 | −0.11 (0.22) | 0.31 |
| Metabolic syndrome | −0.05 (0.16) | 0.76 | −0.06 (0.17) | 0.72 | −0.03 (0.17) | 0.66 |
Analyses are adjusted for age, sex and 20 PCs.
Associations between PR components and genetic risk scores
Finally, we examined whether there was a differential association between genetic risk scores based on known genetic modulators of the PR interval, atrial fibrillation and weight (measured by body mass index [BMI]), and the PR phenotypes. The PR GRS was significantly linearly correlated with each PR phenotype, and had the largest linear correlations with PR interval and PR segment (Table 5). The AF GRS was weakly correlated with the P wave duration (partial r=0.024, p=0.049), while the BMI GRS was correlated with both the PR interval (partial r=0.035, p=0.004) and P wave (partial r=0.048, p<0.001) (Table 5).
Table 5.
Partial correlation coefficients (r)* between PR interval, AF and BMI genetic risk scores and PR phenotypes.
| PR GRS | AF GRS | BMI GRS | ||||
|---|---|---|---|---|---|---|
| Phenotype | Partial r | p-value | Partial r | p-value | Partial r | p-value |
| PR interval | 0.17 | <.0001 | 0.014 | 0.25 | 0.04 | 0.004 |
| P wave duration | 0.07 | <.0001 | 0.024 | 0.049 | 0.05 | <0.001 |
| PR segment | 0.14 | <.0001 | 0.002 | 0.87 | 0.01 | 0.36 |
Correlations are adjusted for age and sex.
Discussion
We employed a discovery-oriented approach to identify clinical phenotypes modulated by genetic factors that also modulate the PR interval. We found that AF risk was genetically correlated with the PR interval, and this association was also observed using a highly polygenic risk score derived from the PR interval. We also observed genetic correlations with metabolic phenotypes including measures of adiposity. Thus, the genetic architecture underlying PR interval variability is driven, in part, by SNP variation that predisposes to AF risk and SNP variation which modulates body mass. Our analyses also found that the constitutive components of the PR interval (the PR segment and the P wave) were associated with different phenotypes and further characterizing their individual genetic architectures may enable the development of better genetic risk prediction tools.
While the PR interval is a genetically modulated measure of cardiac conduction, relatively few SNPs associated with this phenotype have been identified.3–5 This paucity is not unexpected, as the genetic variability underlying many complex phenotypes is driven by numerous SNPs with small effect sizes that are difficult to detect by GWAS. We used modelling approaches which analyze the contributions of large number of SNPs to broadly characterize the genetic architecture of the PR interval. We found that common SNP variation accounted for at least 23% of phenotypic variability in the PR interval, indicating that much of the additive heritability of PR interval is currently hidden. When we examined the individual constituents of the PR interval, we found that the genetic correlation between PR segment and P wave durations was not significantly different for zero, suggesting that they differing genetic architectures. This observation is consistent with GWAS studies which have found that these intervals are associated with different SNPs.36 To further characterize the genetic architectures of the PR phenotypes, we examined their genetic correlations with a large number of clinical phenotypes.
The individual PR phenotypes were not uniformly genetically correlated with the same clinical phenotypes. The most significant association was between the PR interval and PR segment and AF. The genetic correlation was negative, indicating that a genetically prolonged PR interval is associated with decreased risk of AF. This finding was not anticipated, as epidemiological studies have frequently observed that a prolonged PR interval is associated with an increased risk of AF.1,2 This epidemiological association is attributed, in part, to prolongation in the PR interval due to acquired structural changes to the atrium that manifest as slowed atrial conduction and lead to increased atrial arrythmogenicity.37 Indeed, the PR interval duration increases with age, cardiac diseases38 and metabolic phenotypes such as obesity and hypertension.39–41 These increases are most pronounced for the P wave.26 These epidemiological associations are consistent with the trends in the genetic correlations that we observed when analyzing P wave duration. The P wave was most strongly genetically correlated with metabolic phenotypes including waist circumference and type 2 diabetes and a genetically predicted P wave duration was most strongly associated with a diagnosis of obesity. While not significant, the genetic correlation between the P wave and AF was positive, suggesting that a prolonged P wave duration is associated with an increased risk of AF. In turn, these results indicate that there are genetic factors, such as those that modify BMI, which prolong the PR interval by affecting the P wave and which increase the risk of AF.
The epidemiological association between PR interval and AF is U-shaped, as a short PR interval is also associated with increased AF risk.42–45 Hence, our observation that a genetically shorter PR interval and PR segment is associated with an increased AF risk suggest the inverse association is genetically mediated, and that a short PR interval represents an accumulation of PR-shortening genetic variants, some of which also predispose to AF risk. Our results also suggest that the genetic mechanisms modulating the PR interval duration modulate AF risk in different directions. Thus, the genetic risk relationships between AF and each PR phenotype should be evaluated individually to better define this association. Another approach to examining the U-shaped relationship between the PR interval and AF is to employ non-linear statistical models. However, we believe that ascribing the non-linear association to the individual effects of the PR phenotypes is biologically more plausible than non-linear additive genetic effects underlying the PR interval. Our findings also indicate there is opportunity for more discovery. For instance, we found that a genetic risk score comprised known AF SNPs more strongly reflected the genetic risk associated with the P wave, as compared to the PR segment. Thus, identifying and evaluating additional SNP variants associated with the PR segment may reveal additional genetic mechanisms contributing to AF risk.
A significant genetic correlation between a pair of phenotypes suggests that they are modulated by a common set of genetic factors. Hence, a genetic predictor derived from one phenotype should associate with the other phenotype, provided that that predictor is able to capture a sufficient portion of the underlying genetic architecture of the first phenotype. We used BSLMM, which models phenotypes based on large numbers of SNP, to compute genetically predicted PR intervals in three data sets. This genetically predicted PR interval was associated with AF risk in each data set, and the direction was consistent with that observed with the genetic correlations analyses. As larger sample sizes become available and new polygenic modelling techniques are developed, it may be possible to develop a PR interval-derived genetic classifier which can robustly predict AF risk and can offer sufficient lead time to maximize the benefit of intervention strategies.
There are several limitations to this study. We used phenotypes derived from EHR data sets, which often lack rigid phenotype definitions and can have incomplete ascertainment. Incomplete ascertainment and phenotype misclassification can attenuate associations. In support of the validity of our EHR AF phenotype, we note that it has been used for several genetic studies and has been shown to replicate known SNP associations.22,23,25 It is possible that the genetic correlations we observed are spurious and are caused by SNPs simultaneously tagging disparate causative genetic variants that impact the phenotypes through distinct mechanisms.46 However, all of our genetic correlations are supported by epidemiological observations, so this is unlikely for the phenotypes we identified. Our AF cases also had more comorbidities as compared to our controls, which could inflate genetic correlation estimates for risk factors related to the metabolic syndrome. We also did not have sufficient individuals of other ancestries to evaluate and validate our findings in these other racial groups.
In conclusion, we used mixed models to characterize the genetic architecture of the PR interval. We found that SNP variants which predispose to AF and elevated body mass, modulate the PR interval and that these variants differentially influence the P wave and PR segment durations. Future GWAS studies should examine the constitutive PR phenotypes separately in order to more fully define the genetic modulators of the PR interval. Furthermore, focusing on genetic variation underlying the PR segment may identify novel AF genetic risk factors and mechanisms, which may lead to better AF risk prediction models.47 Finally, a portion of the genetic predisposition towards AF is driven by genetic factors for metabolic risk factors including obesity, highlighting the continued need for aggressive risk modification and treatment for these predisposing conditions.
Supplementary Material
Clinical Perspective.
Biomarkers which predict disease risk enable risk stratification and disease prevention. Since many biomarkers and diseases are modulated by underlying genetic risk, it is possible to associate them based on this shared genetic risk. Importantly, these genetic associations can be assessed across different datasets, as long as all subjects have genotypic data, and the approach can be used to study relationships between potential biomarkers and disease. Here, we measured genetic correlations, a measure of genetic association, between a potential biomarker, the PR interval (and its individual components, the P wave and the PR segment), and 63 electronic health record (EHR) disease phenotypes. The ECG phenotypes were analyzed in the Atherosclerosis Risk in Communities (ARIC) cohort, and the EHR phenotypes in the Electronic Medical Records and Genomics (eMERGE) network. We found that a genetically predicted PR interval was associated with atrial fibrillation (AF) risk, consistent with previous epidemiological studies, but with an opposite direction of association. The individual components had different genetic architectures, were not correlated with each other, and AF risk was predominantly associated with genetically-determined PR segment. This study establishes that the shared genetic architectures of clinical phenotypes like AF and putative biomarkers like the PR and its components can identify epidemiological associations, validate the biomarkers, and point to disease mechanisms.
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions.
Sources of Funding: This work was supported by a career development award from the Vanderbilt Faculty Research Scholars Fund (JDM), American Heart Association (15MCPRP25620006 and 16FTF30130005) (JDM), PGRN (P50 GM115305), R01 LM010685, K23 HL127704 (MBS) and R01 HL092217 (Darbar). BioVU is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. The eMERGE Network is funded by NHGRI and NIGMS through the following grants: U01-HG-004610 (Group Health Cooperative/University of Washington); U01-HG-004608 (Marshfield Clinic Research Foundation and VUMC); U01-HG-04599 (Mayo Clinic); U01-HG-004609 (Northwestern University); U01-HG-006378 and U01-HG-04603; U01-HG-004438 (CIDR) and U01-HG-004424 (the Broad Institute) serving as Genotyping Centers. ARIC is supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Funding for GENEVA was provided by NHGRI grant U01HG004402 (E. Boerwinkle).
Footnotes
Disclosures: None.
References
- 1.Cheng S, Keyes MJ, Larson MG, McCabe EL, Newton-Cheh C, Levy D, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA. 2009;301:2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng M, Lu X, Huang J, Zhang S, Gu D. Electrocardiographic PR Prolongation and Atrial Fibrillation Risk: A Meta-Analysis of Prospective Cohort Studies. J Cardiovasc Electrophysiol. 2014 doi: 10.1111/jce.12539. [DOI] [PubMed] [Google Scholar]
- 3.Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler AM, Yin X, Evans DS, Nalls MA, Smith EN, Tanaka T, et al. Novel loci associated with PR interval in a genome-wide association study of 10 African American cohorts. Circ Cardiovasc Genet. 2012;5:639–646. doi: 10.1161/CIRCGENETICS.112.963991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8:e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 12.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowton E, Field JR, Wang S, Schildcrout JS, Van Driest SL, Delaney JT, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6:234cm3. doi: 10.1126/scitranslmed.3008604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 20.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie MD, Denny JC, Crawford DC, Ramirez AH, Weiner JB, Pulley JM, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Israel RA. The International Classification of Disease. Two hundred years of development. Public Health Rep. 1978;93:150–152. [PMC free article] [PubMed] [Google Scholar]
- 25.Weeke P, Denny JC, Basterache L, Shaffer C, Bowton E, Ingram C, et al. Examining rare and low-frequency genetic variants previously associated with lone or familial forms of atrial fibrillation in an electronic medical record system: a cautionary note. Circ Cardiovasc Genet. 2015;8:58–63. doi: 10.1161/CIRCGENETICS.114.000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnani JW, Lopez FL, Soliman EZ, Maclehose RF, Crow RS, Alonso A. P wave indices, obesity, and the metabolic syndrome: the atherosclerosis risk in communities study. Obesity (Silver Spring) 2012;20:666–672. doi: 10.1038/oby.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Methods Mol Biol. 2013;1019:215–236. doi: 10.1007/978-1-62703-447-0_9. [DOI] [PubMed] [Google Scholar]
- 29.Mosley JD, Witte JS, Larkin EK, Bastarache L, Shaffer CM, Karnes JH, et al. Identifying genetically driven clinical phenotypes using linear mixed models. Nat Commun. 2016;7:11433. doi: 10.1038/ncomms11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 31.Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9:e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verweij N, Mateo Leach I, van den Boogaard M, van Veldhuisen DJ, Christoffels VM, et al. LifeLines Cohort Study. Genetic determinants of P wave duration and PR segment. Circ Cardiovasc Genet. 2014;7:475–481. doi: 10.1161/CIRCGENETICS.113.000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soliman EZ, Cammarata M, Li Y. Explaining the inconsistent associations of PR interval with mortality: the role of P-duration contribution to the length of PR interval. Heart Rhythm. 2014;11:93–98. doi: 10.1016/j.hrthm.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Alonso A, Soliman EZ, Chen LY, Bluemke DA, Heckbert SR. Association of blood pressure and aortic distensibility with P wave indices and PR interval: the multi-ethnic study of atherosclerosis (MESA) J Electrocardiol. 2013;46:359.e1–6. doi: 10.1016/j.jelectrocard.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnani JW, Johnson VM, Sullivan LM, Gorodeski EZ, Schnabel RB, Lubitz SA, et al. P wave duration and risk of longitudinal atrial fibrillation in persons ≥ 60 years old (from the Framingham Heart Study) Am J Cardiol. 2011;107:917–921. e1. doi: 10.1016/j.amjcard.2010.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnani JW, Gorodeski EZ, Johnson VM, Sullivan LM, Hamburg NM, Benjamin EJ, et al. P wave duration is associated with cardiovascular and all-cause mortality outcomes: the National Health and Nutrition Examination Survey. Heart Rhythm. 2011;8:93–100. doi: 10.1016/j.hrthm.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnani JW, Zhu L, Lopez F, Pencina MJ, Agarwal SK, Soliman EZ, et al. P-wave indices and atrial fibrillation: cross-cohort assessments from the Framingham Heart Study (FHS) and Atherosclerosis Risk in Communities (ARIC) study. Am Heart J. 2015;169:53–61. e1. doi: 10.1016/j.ahj.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magnani JW, Wang N, Nelson KP, Connelly S, Deo R, Rodondi N, et al. Electrocardiographic PR interval and adverse outcomes in older adults: the Health, Aging, and Body Composition study. Circ Arrhythm Electrophysiol. 2013;6:84–90. doi: 10.1161/CIRCEP.112.975342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soliman EZ, Prineas RJ, Case LD, Zhang Z, Goff DC. Ethnic distribution of ECG predictors of atrial fibrillation and its impact on understanding the ethnic distribution of ischemic stroke in the Atherosclerosis Risk in Communities (ARIC) study. Stroke. 2009;40:1204–1211. doi: 10.1161/STROKEAHA.108.534735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen JB, Pietersen A, Graff C, Lind B, Struijk JJ, Olesen MS, et al. Risk of atrial fibrillation as a function of the electrocardiographic PR interval: results from the Copenhagen ECG Study. Heart Rhythm. 2013;10:1249–1256. doi: 10.1016/j.hrthm.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, et al. Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE-AF consortium. J Am Heart Assoc. 2013;2:e000102. doi: 10.1161/JAHA.112.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gianola D, de los Campos G, Toro MA, Naya H, Schön C-C, Sorensen D. Do Molecular Markers Inform About Pleiotropy? Genetics. 2015;201:23–29. doi: 10.1534/genetics.115.179978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J. 2013;34:2243–2251. doi: 10.1093/eurheartj/eht033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.