Abstract
Mammalian NUMB is alternatively spliced generating four isoforms NUMB1-NUMB4 that can function as tumor suppressors. NUMB1-NUMB4 proteins, which normally determine how different cell types develop, are reduced in 21% of primary breast tumors. Our previous work has, however, indicated that two novel NUMB isoforms, NUMB5 and NUMB6 have the pro-oncogenic functions. Herein, we address a novel function of human NUMB isoform 6 (NUMB6) in promoting cancer cell migration and invasion. We found that NUMB6 induced expression of embryonic transcription factor Slug, which in turn actively repressed E-cadherin, prompting cells to undergo epithelial-mesenchymal transition (EMT). Low-metastatic breast cancer cells DB-7 stably expressing NUMB6, lost their epithelial phenotype, exhibited migratory and pro-invasive behavior, and ultimately elevated expression of mesenchymal markers. Among these markers, increased vimentin, β-catenin and fibronectin expression elicited metalloproteinase 9 (MMP9) production. Our results revealed that NUMB6-DB-7 cells have significantly increased level of Akt1 and Akt2 phosphorylation. Therefore, antagonizing Akt signaling using a chemical inhibitor LY294002, we found that NUMB6-induced Slug expression was reduced, and ultimately accompanied with decreased cell migration and invasion. In summary, this study identified a novel molecular determinant of breast cancer progression, uncovering a potential oncogenic role for the NUMB6 protein in cancer cell migration and invasion, coupled to the maintenance of mesenchymal-like cells.
Breast cancer is the most common neoplastic disease worldwide and the number one cause of cancer-related death among non-smoking women in the USA [Jemal et al., 2010; Dunn et al., 2010]. Tumor invasion and metastasis are the major causes of cancer mortality. Detection and treatment of metastatic breast cancer requires a better understanding of the mechanisms that cause breast tumor cells to become invasive. Treatment of metastatic breast cancer generally focuses on relieving symptoms and extending a patient’s life. To improve treatment outcomes and clinical decision-making, identification of novel pro-invasive protein targets is warranted. NUMB is an evolutionarily conserved gene that gives rise to 4 isoforms (NUMB1-NUMB4) involved in a variety of biochemical pathways associated with endocytosis, cell polarity and protein degradation [Pece et al., 2004; Colaluca et al., 2008; McGill et al., 2009; Gulino et al., 2010; Amson et al., 2011; Pece et al., 2011]. Considerable evidence implicates that NUMB1-NUMB4 can function as tumor suppressors [Pece et al., 2004; Rennstam et al., 2009; Pece et al., 2011]. For example, 21% of primary breast tumors are characterized by the loss or reduced expression of the NUMB protein, which normally functions to help determine how different cell types develop. Experiments indicate that mammalian Numb acts upstream of Notch action to inhibit Notch signaling, preventing the translocation of the Notch intracellular domain to the nucleus following ligand binding [Frise et al., 1996; McGill et al., 2009]. It is then anticipated, that loss of this negative regulation of NUMB over Notch signaling is an important factor in breast cancer carcinogenesis [Pece et al., 2004]. Interestingly, NUMB protein forms complex with TP53 and MDM2, thus protecting TP53 from its degradation. Therefore, loss of NUMB commonly observed in primary breast tumors, is associated with reduced TP53 levels and impaired apoptosis. Moreover, it has been demonstrated that reduced NUMB expression correlates with an increased risk of developing systemic metastases from primary breast tumors [Rennstam et al., 2009] and greatly impairs patients’ survival. Interestingly, recent work in endometrial cancer indicates that NUMB protein expression is not restricted to the cytoplasm and plasma membrane, but can localize in the nucleus which may implicate a novel insight of NUMB’s mechanism in cancer development [Wang et al., 2015]. Conversely, ectopic expression of NUMB in glioma does not exert its tumor suppressor function on cell proliferation or inducing cell differentiation [Euskirchen et al., 2011], suggesting rather a different role of NUMB in different neoplasms. Our laboratory reported that two novel human NUMB isoforms, NUMB5 and NUMB6 [Karaczyn et al., 2010], have the unique ability to induce Rho-GTPases, a family of proteins that govern cell migration and cancer metastasis [Raftopoulou et al., 2004; Vega et al., 2008]. This observation clearly suggested that NUMB5 and NUMB6 may exert their function in promoting cancer cell migration and invasion. Furthermore, it also advocates for an unexpected dual role of the NUMB proteins during cancer progression, depending on the isoform expression pattern.
Epithelial-mesenchymal transition (EMT) is a process of crucial importance in cancer metastasis, allowing cells to dissociate from the epithelial tissue and become motile [Thiery et al., 2006; Christfori, 2006; Micalizzi et al., 2009]. During this process, epithelial cells change their morphology to elongated phenotype, lose their polarity and cellular junctions, and show mesenchymal, fibroblast-like properties [Zavadil et al., 2008; Thiery et al., 2009; Kalluri et al., 2009]. Because of its clinical importance, an identification of reliable molecular markers and key mediators of EMT is warranted. One of the best-characterized markers of EMT is a tumor suppressor, E-cadherin (CDH1). It mediates calcium-dependent intercellular junctions, and its impaired function induces mesenchymal phenotypes. Decreased expression of E-cadherin is often correlated with advanced-stage carcinomas [Takeichi, 1993; Nollet et al., 1999; Handschuh et al., 1999; Kowalski et al., 2003; Jeanes et al., 2008]. The EMTs that occur both during tumor invasion is associated with the functional loss of E-cadherin. The molecular base of the E-cadherin downregulation during tumor progression in most carcinomas predominantly involves transcriptional repression. Several transcriptional repressors, which include Snail, Slug, SIP-1/ZEB-2, E12/E47, and Twist, have been characterized [Hajra et al., 1999; Cano et al., 2000]. They are found to interact with the proximal E-boxes of the E-cadherin promoter. Snail transcriptional repressors are the most widely studied effectors of EMT and CDH1 expression. Slug is a zinc finger protein that belongs to the Snail superfamily [De Herreros et al., 2010]. An inverse correlation between E-cadherin and Slug has been described in many different cell systems [Hajra et al., 2002; Come et al., 2006]. Ectopic expression of Snail and Slug in epithelial cells caused EMT by acquisition of migratory and invasive behaviors, which were also evident in breast cancer [Wu et al., 2010]. If considering Slug as a potent driver of EMT during cancer, several signaling pathways postulated to promote EMT including Akt [Kim et al., 2001; Irie et al., 2005].
In this study, we found that one of the novel NUMB isoforms, NUMB6, induces EMT in breast cancer cells, suggesting its function in the initiation of the metastatic cascade. We showed that NUMB6 overexpression acquired of both phenotypic and molecular changes typical of EMT in DB-7 breast epithelial cancer cells. NUMB6 promoted a spindle-like phenotype and significantly enhanced cell migration and invasion through extracellular matrix (ECM). These changes were associated with the inhibition of E-cadherin expression. Furthermore, we identified that NUMB6 induces expression of transcription factor Slug, mediating the promotion of EMT. This is the first demonstration of the distinct function of NUMB6 in breast tumor metastasis through the differential regulation of EMT-related genes. Because cells undergoing EMT are more invasive [Son et al., 2010] NUMB6 may be a potential therapeutic target.
MATERIALS AND METHODS
CELL CULTURE AND GENERATION OF STABLE CELL LINES
The DB-7 cells [Borowsky et al., 2005], provided by R. Friesel (Maine Medical Center Research Institute, Scarborough, ME), were grown in DMEM/F12, (Lonza, Princeton, NJ, USA) with 10% fetal bovine serum (Hyclone, Waltham, MA, USA), and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). For the generation of stable DB-7 cell lines expressing CMV-GFP constructs containing GFP alone, NUMB4-GFP, or NUMB6-GFP sequence inserts [Karaczyn et al., 2010], a selection was carried out in medium supplemented with 0.5 mg/ml of G418 sulfate (Calbiochem, Gibbstown, NJ, USA). After antibiotic selection, cells were maintained with 0.2 mg/ml of G418. Protein extractions were prepared 2 weeks after commencing drug selection.
CELL TREATMENT
A highly selective inhibitor, LY294002, used to block PI3 kinase-dependent Akt phosphorylation was purchased from Cell Signaling Technology (Danvers, MA, USA). For cell treatments 70 % confluent cells were incubated in fresh medium with 10 μM LY294002 alone or in combination for indicated times. Control cultures received the same volume of DMSO. All treatments were performed in duplicate or triplicate in each experiment.
TRANSIENT TRANSFECTION
Slug-specific siRNA oligos (predesigned siRNA) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Cells cultured in six-well plates were transfected with 2.5μM and 5μM siRNA duplexes using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA). Nonspecific siRNA duplexes were used as negative controls (Qiagen, Valencia, CA, USA). Cells were treated for 48 h to allow maximum knockdown.
COLLAGEN TYPE 1 GEL PREPARATION
For collagen sample preparation, the cells were trypsinized, added to culture media, counted and centrifuged (300 g, 3 min). Cells were resuspended in culture media at a concentration of 6x106 cells/ml. Collagen was prepared at a concentration of 1.6 mg/ml initially by neutralizing an acidic collagen solution (3.4 mg/ml, Collagen I, rat tail, BD Biosciences). To neutralize collagen a basic solution (5 N NaOH) was used: 4x PBS was added (one-quarter of the total volume) to make 1xPBS after neutralization, and culture media was added to adjust the collagen concentration. Sterile NaOH solution was carefully added to adjust the pH. Cells and culture media were added to the neutralized gel to achieve a final collagen concentration of 1.3 mg/ml and a cell concentration of 1x106 cells/mL (approximately 1200 cells/chamber).
REPORTER GENE ASSAY
Luciferase reporter vector containing human E-cadherin promoter was obtained from Addgene, Cambridge, MA, USA (pGL2Basic-EcadK1 was a gift from Eric Fearon, Addgene plasmid # 19290) [Hajra et al., 1999]. All plasmids were cloned in the promoterless pGL3-Basic vector. Cells were transfected with 1 μg of each firefly luciferase reporter construct together with 0.5 μg Renilla plasmid as normalization reference for transfection efficiency. Transfections were performed with Lipofectamine 2000 (Life Technologies). Where indicated, cells were also co-transfected with Slug siRNA 24 hr prior to luciferase assay. Cells transfected with promoterless vector (pGL2-Basic) were used as control for background luciferase activity. Following transfection the cells were harvested in lysis buffer (Promega Corp., Madison, WI, USA) at designated time points. Luciferase units were calculated as luciferase activity presented as the mean of three individual experiments with triplication. The fold change was calculated by comparison with the promoterless pGL3-Basic.
QUANTITATIVE REAL-TIME PCR
Total RNAs extracted from cells were reverse-transcribed into cDNA by PowerScript cDNA synthesis strip (Clontech, Mountain View, CA, USA) with the standard protocol recommended by the manufacturer. Primer pairs for target genes such as Cdh1, and Mmp9 and Numb were designed base on the RefSeq (NIH) sequences by Primer3 (frodo.wi.mit.edu/primer3/input.htm). Primers for Slug, Zeb1, Zeb2, Sp1 and Twist were purchased from Santa Cruz Biotechnology. All real time PCRs are designed to follow a universal real time PCR thermocycle condition: 94 °C 2 min, 45 cycles of 94 °C 30 sec and 60 °C 30 sec with Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Specificity of the amplicons was checked on a 2.2% agarose gel (FlashGel System, Lonza, Allendale, NJ, USA) and by the dissociation curve to make sure there are no non-specific amplicons or formation of primer-dimer. The delta-delta-Ct approach was adopted for the relative expression calculations. Amplification efficiencies for all primers were checked against the housekeeping gene Rpl18 to ensure the signals were detected at the exponential phase and all primers have similar amplification efficiencies. Each test sample was run in triplicate.
ANTIBODIES
All antibodies used for Western blot and immunofluorescence analyses were obtained from commercial sources. Primary antibodies used were anti-E-cadherin (BD Bioscience, NJ, USA), anti-β-catenin (BD Bioscience), anti-active B-catenin (EMD Millipore, Billerica, MA, USA) anti-vimentin (Abcam, Cambridge, MA, USA), anti-fibronectin (Sigma-Aldrich, St. Louis, MO, USA), anti-GFP (Invitrogen, Waltham, MA, USA), anti-β-actin (Sigma-Aldrich), anti-Slug (Cell Signaling Technology, Danvers, MA, USA), anti-FSP-1 (NeoMarkers, Fremont, CA, USA), anti-phosphoAkt1 Ser473 (Cell Signaling Technology), anti-phosphoAkt2 Ser474 (Novus Biologicals Littleton, CO, USA), anti-total Akt1 (Cell Signaling Technology), anti-total Akt2 (Cell Signaling Technology), and rat polyclonal anti-MMP9 (EMD Millipore). β-actin protein levels were used as a control for adequacy of equal protein loading. Western blot membranes were developed using an appropriate Horseradish peroxidase (HRP) conjugated goat anti-mouse or rabbit IgG (Bio-Rad Laboratories, Hercules, CA, USA) or anti-rat antibody (Santa Cruz Biotechnology), and the ECL detection system (EMD Millipore). In the immunofluorescence detection secondary goat anti-mouse Alexa Fluor 488 IgG and goat anti-rabbit Alexa Fluor 546 IgG (Invitrogen) were used.
WESTERN BLOTTING AND IMMUNOFLUORESCENCE
Cells were grown in 6-well plates until 80 percent confluence, followed by harvest, protein isolation and quantification as previously described (12). Briefly, cells were lysed in Triton lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 10% (v/v) glycerol) cleared by centrifugation, and applied to Western blots. Protein concentration was determined using Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL, USA). Membranes were probed for GFP (Invitrogen), and β-actin (Sigma-Aldrich), followed by anti-mouse IgG horseradish peroxidase-linked whole antibody (Bio-Rad Laboratories) and detected using Luminata Crescendo Western HRP Substrate (Millipore, Billerica, MA, USA). In the immunofluorescence experiments, cells were seeded on 12-well plate until 70–90 percent confluence and treated as indicated in the experimental procedure. 24 hours later cells were rinsed with cold PBS buffer, fixed and permeabilized with 4% paraformaldehyde for 10 minutes, and rinsed again three times with PBS. Cells were blocked with 0.1% BSA and 1% goat serum in PBS buffer and incubated at room temperature for one hour. Primary antibodies were applied at 1:250 each in blocking solution and incubated at 4°C overnight. Cells were rinsed three times with PBS buffer and anti-mouse Alexa Fluor 488 IgG and goat anti-rabbit Alexa Fluor 546 IgG (Invitrogen) were applied at 1:1000. Images then were taken using an Axiovert 200 microscope Zeiss (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA) with 488 or 546 wavelength fluorescence filters and processed using MetaMorph software (Molecular Devices Inc., Sunnyvale, CA, USA).
ZYMOGRAPHIC ANALYSIS OF MMP-9 ACTIVITY
NUMB4-GFP, NUMB6-GFP or control GFP DB-7 cells were transfected with either ctrl siRNA or Slug siRNA at 70% confluence. Next day cell cultures were washed twice with PBS, and the medium was changed to serum free DMEM-F12 without supplements. After 24 h the conditioned medium was collected and centrifuged for 5 min. at 400 × g. A 1.500 μl supernatant was concentrated to <200 μl using a Microcon concentrator (Millipore, Billerica, MA, USA) at 6,500 × g at 4°C. Protein concentration was determined using a BCA assay (Thermo Scientific, Rockford, IL, USA), and 10 μg of total protein from each sample was electrophoresed on a 10% zymography gel containing 0.1% gelatin (Invitrogen). MMP activity was detected by using zymogram renaturing and developing buffer system from Invitrogen, according to the manufacturer’s protocol. Gels were stain with Coomassie Blue staining, destain and washed. Staining gels were air-dried in cellophane mounts and images were then captured and analyzed using ImageJ (NIH).
WOUND ASSAY
Cells were grown to confluence and formed a monolayer covering the surface of the entire plate. To inhibit mitosis of the cells and thus distinguish migration from proliferation 10 μg/ml of mitomycin C (Sigma-Aldrich) was incubated for 1 h at 37°C prior to the scratch assay; the cells were then washed twice with PBS. Before reaching confluence, the cells were kept in medium with appropriate siRNA-based treatments for 24 h before scratching. A linear scratch was made with 20ul sterile pipette tip across the diameter of the well and rinsed with warm PBS buffer to remove debris. Fresh media were added for continued drug treatment. For each well, at least three pictures were taken with the fluorescence microscope at a magnification of ×4. The images were taken following two time points, 0 and 24 h after scratch, during which intervals cell migration was closing the scratch.
TRANSWELL MIGRATION ASSAY
In addition to the wound assay, a cell migration was assessed using the transwell assay. 24-well inserts containing 8μm pore size polyethylene terephthalate-filters were assembled into transwell plates containing DMEM-F12 medium supplemented with 10% FBS as a chemoattractant. The cells were pretreated with 10 μg/ml mitomycin C for 1 h at 37°C; the cells were then washed twice with PBS. Transient siRNA Slug-transfected cells were harvested by trypsinization, and suspended in DMEM containing 0.2% bovine albumin. The cells were then spun down at 900 x RPM for 5 minutes, supernatant was removed and cells were resuspended in DMEM-F12-serum free medium. The cells were counted and added at a concentration 5.0 ×104 cell/0.9 ml to the transwell chamber. The assembled chambers were incubated for 20 h at 37°C, in which duration cells migrated toward 10% FBS into the lower chambers. At the end of the incubation, nonmigrating cells, which remained on the upper surface of the filter, were completely removed by swabbing with a cotton tip. The cells that have migrated through the membrane were fixed with cold 100% methanol for 3 minutes, stained with Hoechst 33342 (Life Technologies) for 10 minutes, and counted using fluorescence microscope. Experiments were repeated three times.
INVASION ASSAY IN MATRIGEL-COATED TRANSWELL CHAMBERS
The 24-well transwell units with 8um pore size polyethylene terephthalate-filters were coated with Matrigel basement membrane (BD Biosciences, Franklin Lake, NJ) just before the assay using FBS-free DMEM-F12 according to the manufacturer’s instructions. The chambers were assembled using freshly prepared Matrigel-coated filters and DMEM-F12 medium containing 10% FBS as a chemoattractant, gently placed in the lower compartment. Transient siRNA Slug transfected cells were harvested by trypsinization, and suspended in DMEM-F12 medium containing 0.2% bovine albumin. The cells were then spun down at 900 x RPM for 5 minutes, supernatant was removed and cells were resuspended in DMEM-F12-serum free medium. The cells were counted and added at a concentration 1.0 ×105 cell/0.9 ml to the invasion chamber containing a Matrigel-coated filter. The assembled chambers were incubated for 20 h at 37°C. At the end of the incubation, nonmigrating cells, which remained on the upper surface of the filter, were completely removed by swabbing with a cotton tip. The cells on the bottom surface of the filter were gently washed with PBS buffer, fixed with 100% cold methanol for 3 min and stained with Hoechst 33342 (Life Technologies) for 10 minutes. Cells migrated through Matrigel-coated membrane were counted. Results were reported as a fold of change compared with control cells. All experiments were carried out three times with three technical replicates each.
STATISTICS
All assays were repeated at least three times in duplicate or triplicate. Statistical analysis was performed where appropriate using the Student’s t test. Differences with P < 0.05 (*, P < 0.05, **, P < 0.01) were considered to be statistically significant.
RESULTS
NUMB6 OVEREXPRESSION INDUCES MORPHOLOGICAL CHANGE TO THE EMT PHENOTYPE
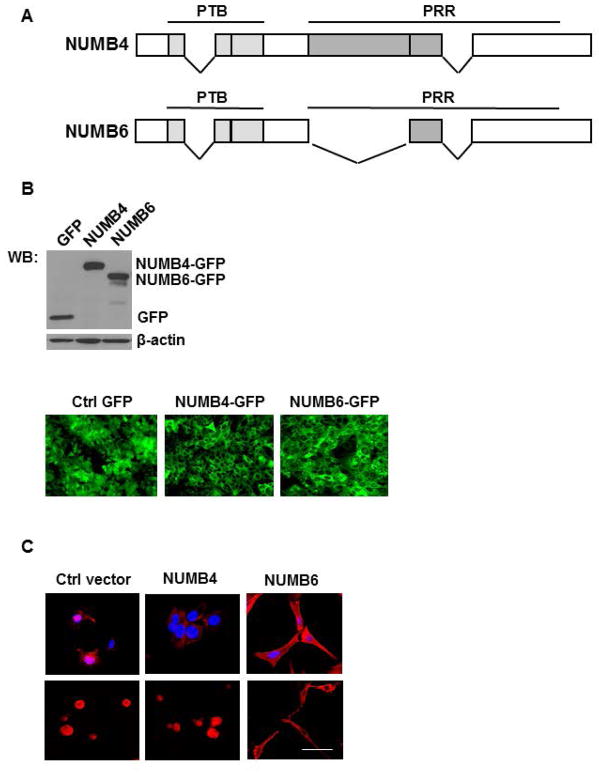
To directly investigate the involvement of NUMB6 in breast cancer cell migration and invasion, we generated a number of DB-7 cell lines stably expressing different NUMB isoforms constructs (Fig. 1A) including NUMB4-GFP and NUMB6-GFP along with the control GFP. The GFP positive detection in NUMB4-and NUMB-GFP tagged proteins was determined by Western blotting (Fig. 1B, upper panel) and immunofluorescence (Fig. 1B, lower panel). At first, we determined the basal transcript expression levels of all NUMB variants in the newly established cell lines. Supplementary Figure S1 shows the levels of mRNA expression of the various variants from Numb1 to Numb4 and Numb6 in control GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cell lines. mRNA level of Numb4 was elevated approximately 40-fold, and Numb6 70-fold in respective NUMB4-GFP and NUMB6-GFP transfected cell populations as compared to vector-GFP transfected control. Changes in mRNA expression of Numb1-Numb3 variants in NUMB4-GFP, or NUMB-GFP cell lines were minor as compared to control GFP group. Two different types of cell morphology were observed in normal growing conditions (Fig. 1C, upper panel) or when cells were cultured in 3D collagen type 1 substratum (Fig. 1C, lower panel). One type observed in control GFP, and NUMB4-GFP cells displayed the original epithelial morphology, while the other exhibited a fibroblast-like phenotype found in NUMB6-GFP cells. For example, Figure 1C shows the fibroblast-like phenotype induced by NUMB6, when the formation of stress fibers by F-actin expression was examined using rhodamine-conjugated phalloidin staining. Assembly of F-actin stress fibers was observed in NUMB-GFP overexpressing cells as compared to GFP and NUMB4-GFP cells. This data suggest that NUMB6-GFP cells acquired fibroblast-like morphology as they undergo EMT.
Fig. 1.
NUMB6 overexpression induces changes in DB-7 cell morphology. (A) Schematic representation of NUMB isoforms: isoform 4 (NUMB4) and isoform 6 (NUMB6). NUMB4 and 6 differ in respect to deletion within the proline-rich region (PRR) in NUMB6. PTB is the phospho-tyrosine binding domain. PRR is proline-rich region. (B) DB-7 cell lines stably expressing NUMB4-GFP, NUMB6-GFP or empty vector with GFP. NUMB4 or NUMB6 proteins expression was examined by Western blot (upper panel) and immunofluorescence (lower panel) measuring GFP staining. (C) NUMB6 induced EMT-like elongation in DB-7 cells. The representative fluorescence images of control GFP, NUMB4-GFP and NUMB6-GFP DB-7 cells cultured in the normal cell culture condition (upper panel) and in collagen type 1 substratum (lower panel). NUMB6 caused the redistribution of filamentous actin (F-actin) in DB-7 cells, as determined by F-actin red staining with rhodamine-phalloidin. Nuclei were stained DAPI in blue. Bars=15 μm.
NUMB6 OVEREXPRESSION INDUCES EMT-RELATED PROTEINS EXPRESSION
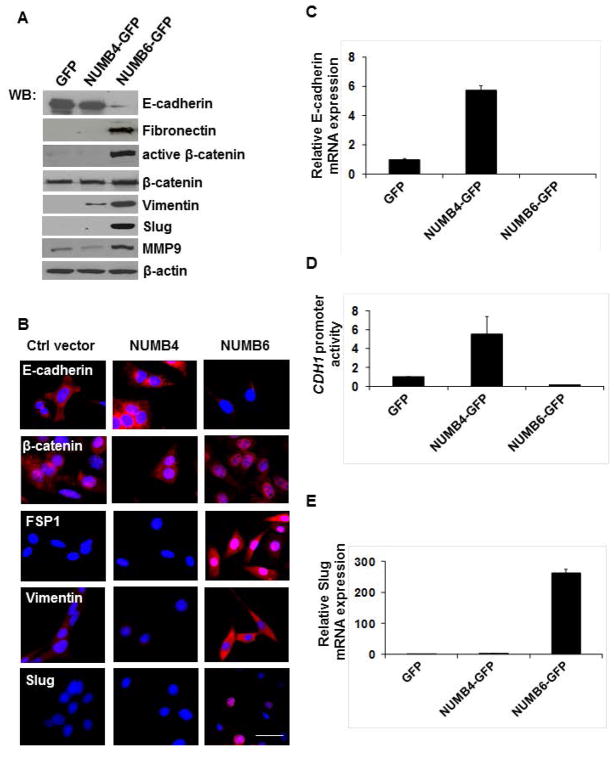
We surmised that this morphological change resulted from NUMB-GFP overexpression resembles the EMT phenotype; therefore we further examined the expression of well-known epithelial marker E-cadherin, and mesenchymal markers such as vimentin, fibronectin, β-catenin, fibroblast-specific protein1 (FSP-1, also called S100A4), and Slug in GFP, NUMB4-GFP, and NUMB-6 DB-7 cells. As shown in Figure 2A, Western blot analysis revealed that the expression of E-cadherin was significantly decreased, whereas that of vimentin, fibronectin, active-β-catenin, and Slug was dramatically increased in NUMB6-GFP DB-7 cells compared with NUMB4-GFP and vector control GFP DB-7 cells. Immunofluorescence showed that E-cadherin staining was restricted to GFP and NUMB4-GFP DB-7 cells, and a complete disappearance of E-cadherin positivity was observed in NUMB6-GFP DB-7 cells (Fig. 2B). In contrast to E-cadherin, proteins such as vimentin, and FSP-1 increased in cytoplasmic staining whereas Slug and β-catenin staining was observed in the nucleus of NUMB6-GFP cells as compared with NUMB4-GFP and control GFP DB-7 cells. These results demonstrate that the expression of EMT-associated markers is increased in a coordinated manner in NUMB6-GFP DB-7 cells. In light of the established role of Slug in the down-regulation of E-cadherin during epithelial-mesenchymal transition in development and cancer [Hajra et al., 2002; Come et al., 2006; De Herreros et al. 2010], we tested whether NUMB6 was capable in repressing CDH1 promoter activity and endogenous E-cadherin expression. Indeed, NUMB6-induced expression of Slug was strongly correlated with the absence of Cdh1 transcripts (Fig. 2C) and its promoter activity (Fig. 2D). These results imply that Slug is the more likely repressor of Cdh1 transcription in NUMB6-GFP DB-7 cells. Further, the mRNA expression of Slug (Fig. 2E), Zeb1, and Sp1 (Supplementary Fig. S2), key transcription factors that promote EMT, was greatly increased in NUMB6-GFP cells as compared with NUMB4-GFP, and control GFP cells, suggesting that NUMB6-GFP overexpression induces EMT in DB-7 cells. Interestingly, we found that Twist mRNA level was elevated in NUMB4-GFP, and NUMB6-GFP DB-7 cells implicating that those two variants share some common traits in the transcriptional regulation. We have previously shown an increased zymographic activity of MMP9 in breast cancer cell lines overexpressing NUMB6-GFP, including DB-7 cell line [Karaczyn et al., 2010]. MMP9 is a known target of Slug, and MMP9 upregulation correlates with increased tumor cell invasion and motility [Duffy et al., 2000; Radisky et al., 2010; Son et al., 2010]. Here we found that NUMB6 increases MMP9 protein (Fig. 2A) and mRNA expression (Supplementary Figure S3). Based on these findings, we speculate that NUMB6 induces MMP9 expression significantly contributing to EMT, most likely via Slug activation.
Fig. 2.
NUMB6 overexpression induces EMT-associated alterations in DB-7 cells. (A) Alterations of epithelial and mesenchymal markers expression in NUMB6-GFP DB-7 cells. Overexpression of NUMB6 reduces E-cadherin and increases expression of mesenchymal markers such as vimentin, fibronectin, and FSP-1, active β-catenin, Slug and MMP9. Protein lysates from DB-7 cells that stably expressed NUMB4-GFP, NUMB6-GFP, or vector alone with GFP were analyzed by immunoblot with antibodies against E-cadherin, fibronectin, vimentin, and FSP-1, active β-catenin, total β-catenin, Slug MMP-9 and β-actin. (B) Immunofluorescence study showing NUMB6 regulation of the expression and localization of EMT markers. NUMB4-GFP, NUMB6-GFP and GFP DB-7 cells were immunostained with E-cadherin, active β-catenin, FSP-1, vimentin, or Slug antibodies. Red is Alexa Fluor 594 anti-rabbit or anti-mouse. Nuclei were stained with DAPI. Bars=20um. (C) Numb6 regulation of E-cadherin transcription. NUMB6 caused loss of E-cadherin mRNA whereas NUMB4 increased it. The mRNA levels of E-cadherin were measured by quantitative RT-PCR analysis of total RNA extracted from NUMB4-GFP, NUMB6-GFP or control vector GFP DB-7 cells. The mRNA levels of E-cadherin are expressed relative to β-actin transcripts. Each experiment was performed in triplicate and repeated three times. Error bars represent standard error of the mean (SEM). (D) NUMB6 overexpression downregulated E-cadherin luciferase reporter activity. NUMB4-GFP, NUMB6-GFP or control vector GFP DB-7 cells were transfected with the E-cadherin-LUC reporter vector and Renilla constructs for 24 h before measuring luciferase activity. Cells were harvested after 24 h and lysates were assayed for luciferase activity. The relative luciferase activities are expressed in arbitrary units, and were normalized to co-transfected Renilla activity to control for transfection efficiency. Bars represent fold of change over control GFP DB7 cells. Each experiment was performed in triplicates. Error bars represent SEM. (E) NUMB6 overexpression induces Slug expression. The mRNA levels of Slug were measured by quantitative RT-PCR analysis of RNA extracted from DB-7 cells stably expressing NUMB4-GFP, NUMB6-GFP, or control vector GFP. The mRNA levels of Slug are expressed relative to β-actin transcripts. Each experiment was performed in triplicate. Error bars represent SEM.
NUMB6 INCREASES DB-7 CELL MIGRATION AND INVASION
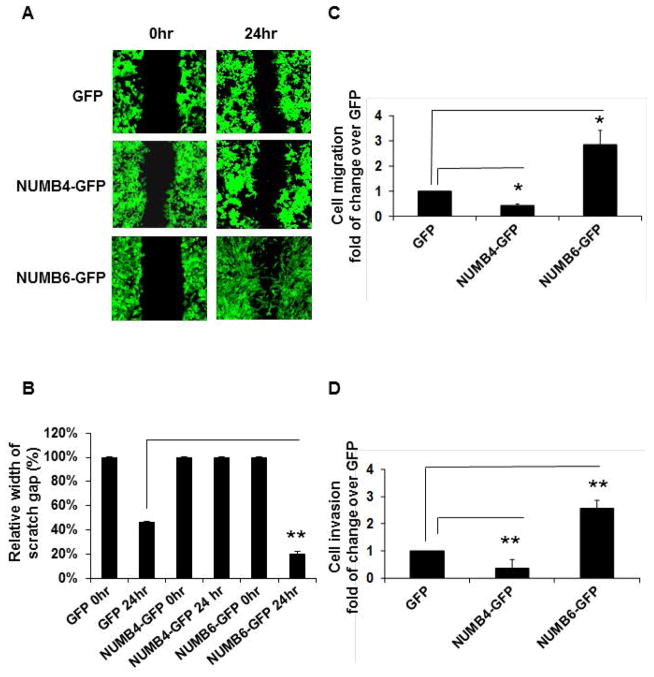
It has been demonstrated that cancer cells undergo EMT, thus acquiring the ability to migrate and metastasize [Pantel et al., 2004; Yang et al., 2008; Kalluri et al, 2009; Micalizzi et al. 2010]. Because the EMT phenotype induced by NUMB6-GFP overexpression might promote cancer cell migration and invasion, these features were examined. Therefore, we tested whether NUMB6 causes changes in cell migration and invasion as compared to NUMB4-GFP and control GFP DB-7 cells. We assessed migration by scratch and transwell migration assay, whereas cell invasiveness was evaluated using Matrigel invasion assay. Notably, we found that NUMB6-GFP overexpression significantly increased wound healing (Fig. 3A, B) and migration of DB-7 cells through the 8μm porous membrane (Fig. 3C) compared with control GFP, and NUMB4-GFP DB-7 cells. Furthermore, Figure 3D shows that approximately three times NUMB6-GFP cells were more invasive in the Matrigel invasion assay then NUMB4-GFP and control GFP cells. These results indicate that NUMB6 may positively regulate cancer cell progression. Finally, these results reveal that EMT phenotype induced by NUMB6 accelerates cancer cell migration and invasion in vitro.
Fig. 3.
NUMB6-induced EMT phenotype increases DB-7 cancer cell migration and invasion in vitro. (A) Wound healing migration assay of DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP. Cells as indicated were cultured until confluent and then a scratch wound was made using a 20ul pipette tip. Media were replaced and wounds were photographed at 20 hrs. (B) Graphic representation of migration differences between control GFP and NUMB4-GFP or NUMB6-GFP DB-7 cells. Error bars refer to SEM in pooled triplicate experiments. (C) Transwell migration assay of DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP. Cells as indicated were seeded in transwell containing 8μm PET membrane and incubated for 20 h. The migrated cells were stained and scored. Error bars represent SEM. (D) Matrigel invasion assay of DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP. Cells as indicated were plated in invasion chamber and incubated for 20 h. The transmigrated cells were stained and counted. The plotted data were averaged from three independent experiments and error bars represent SEM. Asterisks indicate significant differences between two groups.
NUMB6-INDUCED EMT PROTEINS EXPRESSION DEPENDS ON SLUG
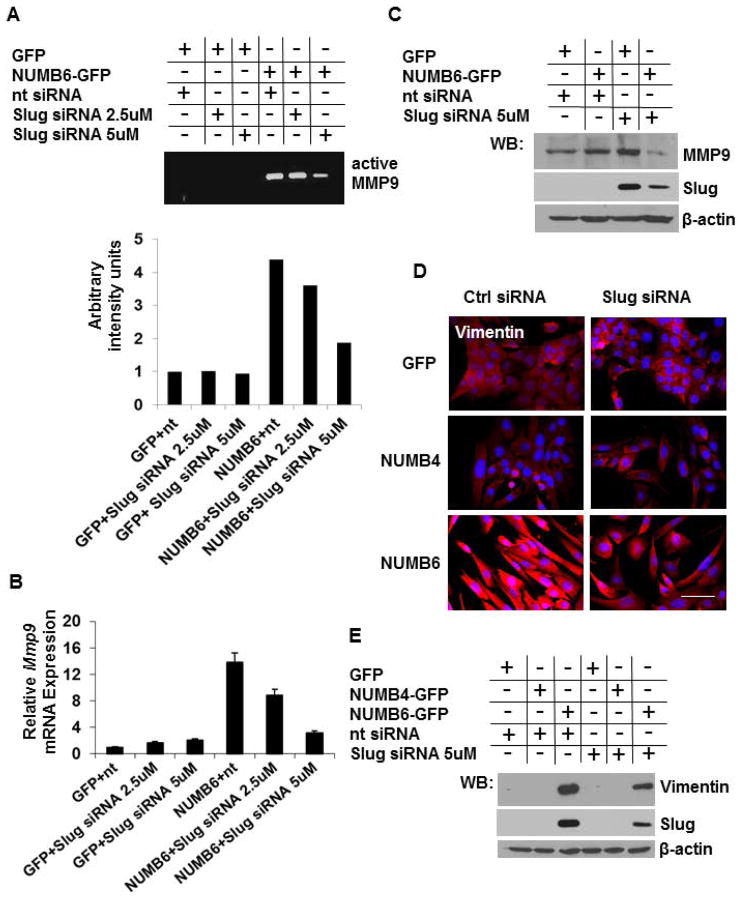
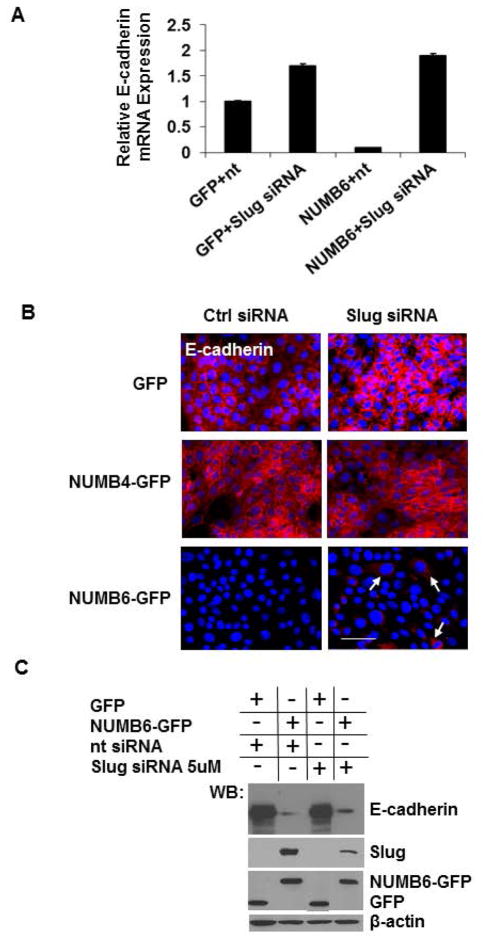
To investigate whether the loss of E-cadherin and gain of mesenchymal proteins were caused by increase of Slug expression, Slug knockdown was performed in NUMB6-GFP and in GFP control DB-7 cells. Slug specific siRNA was used to reduce the levels of Slug transcript in the NUMB-GFP and GFP DB-7 cells compared to those transfected with the control siRNA. NUMB6-induced Slug expression was decerased with increasing concentration of Slug siRNA, based on mRNA (Supplementary Fig. S4A) and protein (Supplementary Fig. S4B) expression analyses. Ultimately, an effective knockdown of Slug in NUMB6-GFP DB-7 cell was confirmed by immunofluorescence, when cell were treated with 5uM Slug siRNA (Supplementary Fig. S4C). We noted that the magnitude of Slug mRNA induction in NUMB6-GFP DB-7 cells transfected with control siRNA was lower than we observed earlier in Figure 2E, most likely due to transfection manipulation. To evaluate the effect of Slug knockdown on expression of EMT-related markers, we examined protein expression of MMP9 and vimentin in Slug-silenced cells. We measured MMP9 activity in conditioned media collected from NUMB6-GFP and GFP DB-7 cells transfected with Slug siRNA or control siRNA. A strong proteolytic activity was seen in NUMB6-GFP cells transfected with control siRNA; whereas little staining was detected in the Slug-inhibited cells using zymography analysis (Fig. 4A). Figure 4B and Figure 4C show that MMP9 mRNA and protein induction in NUMB6-GFP DB-7 cells was ablated by Slug knockdown. These results suggest that Slug knockdown impairs NUMB6-induced MMP9 activity and expression, which in turn reduces NUMB6-GFP DB-7 cell migratory and invasive potentials. Based on immunofluorescence (Fig. 4D) and Western blotting (Fig. 4E) analyses we found that an inhibition of Slug expression caused a pronounced decrease of vimentin levels in NUMB6-GFP compared to control GFP and NUMB4-GFP DB-7 cells. These data support the role of NUMB6, in part through its control of Slug expression, in promoting phenotypic changes consistent with an EMT process. Because Slug, among other transcription factors, regulates E-cadherin expression, decreased Slug transcriptional activity would restore E-cadherin protein production. Although, we observed an increase in E-cadherin mRNA level in Slug knockdown NUMB6-GFP cells (Fig. 5A), Western blot (Fig. 5B) and immunofluorescence (Fig. 5C) analyses revealed that E-cadherin protein expression was partially restored, implicating that either other transcription regulators might be involved in E-cadherin repression and/or rather posttranscriptional regulation plays a key part. This may suggest a hierarchical organized expression of EMT transcription factors through most likely directed activation, triggering an EMT process in NUMB6 DB-7 cells.
Fig. 4.
Knockdown of Slug in NUMB6-GFP DB-7 cells modulates expression of MMP9 and vimentin. (A) NUMB6-induced MMP9 activity was decreased with Slug-siRNA treatment in DB-7 cells. MMP-9 activity was assessed by zymography in conditioned media collected after 24 hr post-transfection of NUMB4-GFP, NUMB-GFP or vector control GFP DB-7 cells with Slug-siRNA or control siRNA. (B) Slug knockdown reduces Mmp9 expression in NUMB6-GFP DB-7 cells. siRNA treatment with 2.5uM or 5uM Slug siRNA reduced Mmp9 mRNA levels. The mRNA levels of Mmp9 were measured by quantitative RT-PCR analysis of total RNA extracted from NUMB4-GFP, NUMB6-GFP or control vector GFP DB-7 cells treated with Slug-siRNA or control siRNA for 48hr. The mRNA levels of Mmp9 are expressed relative to β-actin transcripts. Each experiment was performed in triplicate and repeated three times. Error bars represent SEM. (C) Expression level of MMP9 protein was decreased in Slug-depleted NUMB6-GFP DB-7 cells. NUMB4-GFP, NUMB6-GFP and control vector GFP DB-7 cells were transfected with control siRNA or Slug 5uM siRNAs for 48hr. Expression levels of MMP9 and Slug were analyzed by immunoblotting. β-actin was used as loading control. (D) Decreased expression of vimentin resulted from Slug knockdown of NUMB6-GFP DB-7 cells was confirmed by immunofluorescence staining. Alexa-Fluor 594 conjugated anti-mouse IgG was used as a secondary antibody against vimentin (red). Blue is staining for nuclei. Bars=15um. (E) Western blot shows that vimentin protein was reduced in Slug-depleted NUMB6-GFP DB-7 cells. NUMB4-GFP, NUMB6-GFP and control vector GFP DB-7 cells were transfected with control siRNA or Slug 5μM siRNAs for 48hr. β-actin was used as loading control.
Fig. 5.
Slug knockdown partially restores E-cadherin expression in NUMB6-GFP DB-7 cells. Western blot analysis, real-time PCR and immunostaining assays were carried out to evaluate the expression of E-cadherin in DB-7 cells overexpressing control vector with GFP or NUMB6-GFP. (A) Control vector GFP and Numb-6 GFP DB-7 cells were transfected with control siRNA or Slug siRNA for 48 h and then E-cadherin levels were evaluated by qPCR. NUMB6-induced loss of E-cadherin mRNA expression was recovered when cells were treated with Slug siRNA. (B) Control GFP, NUMB4-GFP or NUMB6-GFP DB-7 cells were transfected with control siRNA or Slug siRNA, respectively. Immunostaining was performed in 48 hours later to assess the expression levels of E-cadherin; red is Alexa Fluor-546 for E-cadherin; nuclei were stained with DAPI. Bars=15um (C) Control GFP, or NUMB6-GFP DB-7 cells were transfected with control siRNA or Slug siRNA, and E-cadherin expression was evaluated by Western Blot.
SLUG KNOCKDOWN REDUCES NUMB6-INDUCED DB-7 CELL MIGRATION AND INVASION IN VITRO
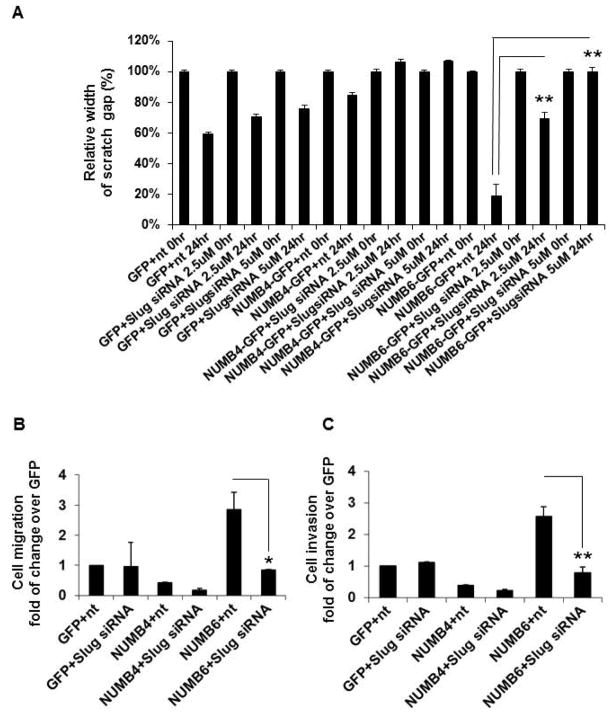
We then evaluated the effects of Slug knockdown on cell migration and invasion of the NUMB6-GFP, NUMB4-GFP and GFP DB-7 cells. As described in the earlier paragraph, cell migration was examined using scratch and transwell assays, whereas Matrigel assay was used for an assessment of cell invasion. Our data revealed that Slug knockdown reduced a width of the scratch gap in NUMB6-GFP cells as compared to GFP and Numb4-GFP DB-7 cells. Relative small 19% width of wound gap closed by migrating NUMB6-GFP DB-7 cells transfected with control siRNA was significantly enlarged to approximately 90% when NUMB6-GFP cells were transfected with Slug siRNA (Fig. 6A and Supplementary Fig. S5). Additionally, transwell assay confirmed, that after the same treatment schedule described above, a significantly reduced number of NUMB6-GFP migrated cells was detected after Slug siRNA transfection compared to those of control cells (Fig. 6B). Figure 6C shows that an invasion capability of NUMB6-DB-7 cells transmigrating through the ECM matrix was also significantly decreased as result of Slug knockdown. These results indicate that NUMB6 induces DB-7 cell migration and invasion via Slug activation.
Fig. 6.
Slug knockdown decreases NUMB6-induced DB-7 cell migration and invasion in vitro. NUMB4-GFP, NUMB6-GFP or control vector GFP DB-7 cells were treated with 2.5uM or 5 uM Slug siRNA or control siRNA; NUMB6-GFP DB-7 cell migration was reduced with Slug siRNA treatment assessed in scratch assay and transwell assay. (A) In scratch assay monolayers of NUMB4-GFP, NUMB6-GFP, and GFP DB-7 cells transfected with control siRNA or Slug siRNA were incubated at 37°C until confluence, then wounded with a uniform scratch, washed to remove cell debris, and cultured for indicated times in the completed DMEM-F12 medium. Images of cell cultures were captured at 0 and 24 h after scratching. (B) In transwell assay DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP were treated with control siRNA or Slug siRNA for 48hr. Cell were then trypsinized, counted and 1x104 cells were loaded into chamber inserts containing an 8-μm pore size membrane and migrated toward 10% FBS. The non-migrated cells were removed from the top of the membrane by cotton swap whereas migrated cells were fixed and stained with Hoechst 33342 after 20 h incubation at 37 °C. The migrated cells were counted. (C) Matrigel invasion assay of DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP treated with control siRNA or Slug siRNA for 48hr. Cell were trypsinized, counted and 1x104 cells were plated then in the invasion chamber and incubated at 37 °C for 20 h. The non-migrated cells were removed from the top of the membrane by cotton swap whereas migrated cells were fixed and stained with Hoechst 33342. The migrated cells were counted. Each experiment was performed in triplicate and repeated a minimum of three times. Error bars represent SEM. Asterisks indicate significant differences between two groups.
AKT ACTS UPSTREAM OF SLUG IN NUMB6-INDUCED CELL MIGRATION AND INVASION
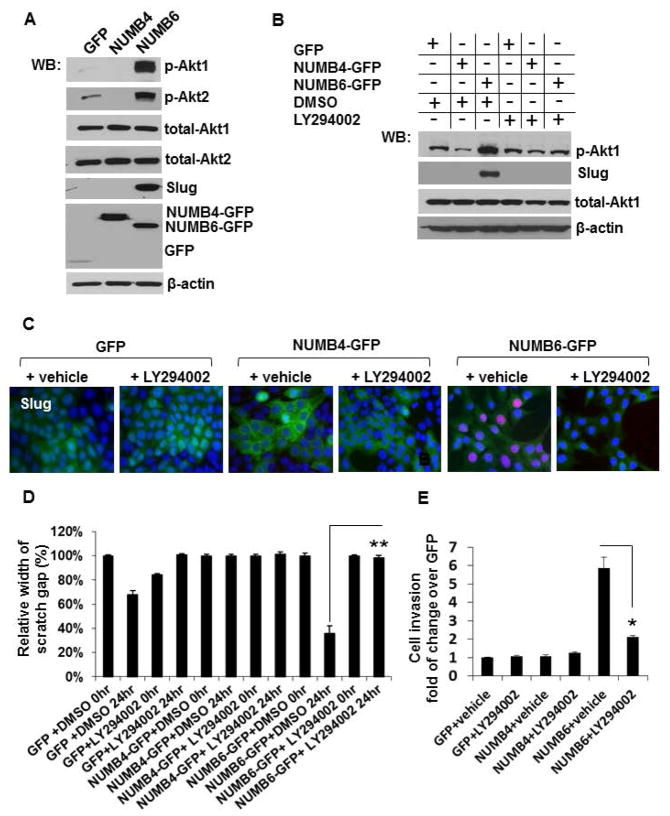
Considerable body of evidence implicates PI3K-Akt signaling in regulation of tumor cell invasion, growth, proliferation, survival, metabolism and apoptosis under different conditions [Dillon et al., 2010]. Aberrant activation of the PI3K-Akt pathway and genetic alterations of its components contribute to tumorigenesis [Bellacosa et al., 2010]. Notably, overexpression of Akt has been reported in 38% of invasive breast cancer samples [Zhou et al., 2004; Bose et al., 2006]. Our Western blot analysis revealed that NUMB6-DB-7 cells exhibit increased Akt phosphorylation, in both isoforms Akt1 and Akt2 (Fig. 7A). To establish, whether NUMB6 is linked to Slug via the PI3K-Akt signaling pathway, control GFP vector or NUMB6 overexpressing DB-7 cells were treated with the PI3K inhibitor (LY294002, 10 μM) for 24 hr. We found, that the pharmacological inhibitor efficiently suppressed the expression of Slug protein in cells displaying NUMB6-GFP overexpression (Fig. 7B). In addition, nuclear staining of Slug in NUMB6-GFP DB-7 cells was lost following LY294002 treatment (Fig. 7C). Our functional assays revealed that treatment of NUMB6-GFP DB-7 cells with LY294002 (Fig. 7D and Supplementary Fig. S6) inhibited wound healing in a scratch-wound assay that measures the amount of time for confluent cells to fill a wound in the monolayer. Similarly, Figure 7E shows that treatment of DB-7 cells overexpressing NUMB6 with LY294002 for 20 h led to a 3-fold decrease in the numbers of cells passing through the Matrigel-coated membrane. These results establish that NUMB6 enhances the invasive potential and mesenchymal properties of breast cancer cells via Akt activation, including induction of Slug (Fig. 9).
Fig. 7.
NUMB6 induces Slug expression via activating Akt signaling pathway. (A) Akt1 and Akt2 isoforms phosphorylation was evaluated in DB-7 cells stably expressing control GFP, NUMB4-GFP or NUMB6-GFP by Western blotting. Akt phosphorylation was detected with anti-phospho-AktS473 or AktS474 antibody specific for Akt1 or Akt2 isoform, respectively. Total level of Akt1, Akt2 was determined using anti-Akt1 or anti-Akt2 antibody. β-actin was used as loading controls. (B) Inhibition of Akt phosphorylation reduced Slug expression in NUMB6-GFP DB-7 cells. Western blot analyses were carried out to evaluate the expression of Slug in control vector GFP, NUMB4-GFP or NUMB6-GFP DB-7 cells untreated or treated with 1μM LY294002 inhibitor for 24 hr. NUMB6-induced Slug expression was ablated when cells were treated with LY294002. (C) Control GFP, NUMB4-GFP or NUMB6-GFP DB-7 cells were treated with DMSO or LY294002 for 24 hours. Immunostaining was performed to assess the expression levels of Slug; red is Alexa Fluor-546 for Slug; nuclei were stained with Hoechst 33342. (D) Inhibition of Akt signaling pathway decreased NUMB6-induced DB-7 cell migration and invasion in vitro. In scratch assay confluent monolayers of control GFP, NUMB4-GFP, or NUMB6-GFP DB-7 cells were wounded with a uniform scratch, washed to remove cell debris, and treated with DMSO or 1μM LY294002 inhibitor. Cells were cultured for indicated times in the completed DMEM-F12 medium. Images of cell cultures were captured at 0 and 24 h after scratching. (E) A Matrigel invasion assay was used to characterize FBS-induced invasion of DB-7 cells overexpressing GFP, NUMB4-GFP, or NUMB6-GFP treated with either a vehicle DMSO or LY294002. Cell migration through Matrigel coated membranes is expressed as percentage of the maximal migration induced by 10% FBS. Where indicated, suspended cells were treated with either control vehicle DMSO or 1μM LY294002, placed in migration chambers, and allowed to migrate to FBS for 20 hours. Error bars indicate SEM. Cell invasion for groups treated with LY294002 were compared with DMSO alone as determined using t test. Asterisks indicate significant differences between two groups.
DISCUSSION
The mammalian NUMB gene is alternatively spliced generating 4 isoforms NUMB1-NUMB4 with molecular weight ranging between 65 and 72 kDa. Our laboratory demonstrated that two novel NUMB isoforms, NUMB5 with molecular weight 55 kDa and NUMB6 with molecular weight 54 kDa differ from NUMB1-NUMB4, in structure and function. For example, NUMB5 and NUMB6 are lacking entire proline-rich region and can interact with proteins involved in the regulation of various cellular processes such as organization of the actin cytoskeleton and cell adhesion [Karaczyn et al., 2010]. Interestingly, the presence of NUMB5 and NUMB6 among other novel human NUMB isoforms, NUMB7, 8, and 9 has been recently reported in placental tissues [Haider et al., 2011]. Because we have previously reported that NUMB5 and NUMB6 are expressed in different grades of human breast tumor, and their expression linearly correlated with tumor progression it is anticipated that NUMB5 and NUMB6 have a prometastatic function. Indeed, in this study, we have shown that human NUMB isoform6, NUMB6, originally described by our laboratory in neural progenitors [Karaczyn et al., 2010], becomes involved in epithelial-to-mesenchymal transition (EMT) of breast cancer cells, a crucial step in tumor invasion. We hypothesized that because of its involvement in EMT, NUMB6 gains a prometastatic function related to increased cell migration and invasion. EMT results in phenotypic changes that are accompanied by an increased cellular motility [Pantel et al., 2004; Kalluri et al., 2009] and production of proteolytic enzymes resulting in the disruption of cell–cell adhesions, which are mediated by E-cadherin [Takeichi, 1993]. We found that NUMB6 induces expression of embryonic transcription factor Slug, which in turn actively represses tumor suppressor E-cadherin, prompting cells to acquire an invasive phenotype. This cellular transition was anticipated as we previously demonstrated in neural progenitors that NUMB6 alters expression of proteins involved in reorganization of the actin-cytoskeleton network, a process crucial for cell movement. It is strongly believed that Slug is a key mediator of EMT being overexpressed in numerous cancers [Hajra et al., 2002], including breast cancer. Slug also reported to be more relevant for generating breast cancer cells with cancer stem cell phenotype [Phillips et al., 2014]. In our work the siRNA-mediated knockdown of Slug in NUMB6-DB-7 cells revealed that NUMB6-induced EMT promotes expression of pro-invasive proteins such as vimentin. Increased vimentin expression reflects cytoskeleton remodeling and strongly suggests impaired cell adhesion. Therefore, we speculate that NUMB6-mediated downregulation of E- cadherin contributes to the invasive property of DB-7 cells characterized by high expression of mesenchymal markers, such as vimentin, Slug, Zeb1/2. This strongly agrees with the notion that during cancer metastasis, EMT is initiated and mesenchymal markers are induced. Further, in this study, we found that EMT was also accompanied by significant increase of metalloproteinase 9 (MMP9) expression, a proteolytic enzyme of the invasive property of tumor cells by degrading the extracellular matrix (ECM) [Pantel et al., 2004; Micalizzi et al., 2010; Radisky et al., 2010]. We observed decreased expression and activity of matrix metalloproteinase-9 (MMP9) following Slug knockdown in DB-7 cells overexpressing NUMB6, implicating that production of MMP9 may be the primary cause of NUMB6-trigerred invasiveness. This MMP9-mediated ECM remodeling process is necessary for tumor cells to cross tissue boundaries. We conclude that DB-7 cells stably overexpressing NUMB6 exhibit not only a complete loss of E-cadherin expression but also a capacity to induce proteolytic enzymes, indicating that a cellular program of epithelial cell phenotype is severely disrupted, and therefore the cells become more malignant. Since an increased cellular motility plays a significant role in the invasive properties of cancer cells, both wound and transwell assays revealed that NUMB6-DB7 cells migration was increased, which were further related to EMT and activation of MMP9. In vitro invasion assay unveiled that cell phenotypic alterations due to NUMB6-induced EMT resulted in accelerated DB-7 cell invasion. However, in is unclear if this phenotypic switch can be completely reversed by knockdown of Slug exclusively, which implicates that co-knockdown of other Snail genes might be required. In most solid tumors, the function of epithelial E-cadherin is altered due to various genetic and epigenetic mechanisms, including activation of Akt signaling pathway. This in turn, activates signaling molecules that promote tumor-cell migration, invasion and dissemination [Thiery et al., 2006; Zavadil et al., 2008]. We found that DB-7 cells overexpressing NUMB6 exhibit high level of phosphorylated Akt1 and Akt2. Our approach with chemical inhibition of PIK3/Akt signaling pathway revealed that NUMB6 upregulates Slug followed by Akt activation. We postulate that NUMB6-mediated activation of the Akt pathway also promotes production of MMP9, which in turn causes cleavage of E-cadherin, leading to the disruption of cell–cell contacts. We anticipate that because activation of Akt pathway is required for cancer cell survival, DB-7 cells are likely to become resistant to anti-cancer drug treatments.
We conclude that NUMB6 overexpression supports multiple metastatic traits in breast cancer, including enhanced cell migration and invasion. Mechanistically, this pathway involves NUMB6 activation of Akt signaling and transcriptional program regulation via Slug induction, resulting in change of cell morphology form epithelial to fibroblast-like. Altogether, these data suggest a pro-invasive role of deregulated NUMB6 in breast cancer, promoting cell migration and invasion when antagonizing E-cadherin-dependent suppression of EMT. Although NUMB isoforms have been proposed to exert potentially overlapping functions in cell fate specifications [Verdi et al., 1999], our results show that these molecules have instead entirely distinct functions in cancer, with NUMB4 suppressing and NUMB6 promoting tumor cell migration and invasion.
In summary, this study identified a novel molecular determinant of breast cancer progression, uncovering a potential prometastatic role for the NUMB6 protein in tumor cell migration and invasion coupled to the maintenance of mesenchymal-like cells characteristics. Together gene expression analysis and cellular assays demonstrated that NUMB6 overexpression in breast cancer cells caused the acquisition of a mesenchymal phenotype and cellular changes that are required for tumor invasion and metastasis [Thiery et al., 2009].
Supplementary Material
Supplementary Fig. S1. mRNA detection of Numb1-Numb4 and Numb6 in control GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cell lines. RT-PCR analysis was used to determine mRNA relative expression levels of Numb1-Numb4 and Numb6 in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells. Numb1-Numb4 and Numb6 variants were amplified by RT-PCR and resolved on 1.5% agarose gel. Numb variants were then identified according to their unique amplicon size and quantitated by densitometry Y-axis depicts mRNA expression as a fold of change over GFP control groups. Results were normalized to β-actin housekeeping gene. Error bars represent SEM.
Supplementary Fig. S2. NUMB6 overexpression upregulates EMT-related transcription factors expression. The mRNA levels of EMT-inducing transcription factors including Twist, Zeb1, Zeb2, and Sp1 were measured by quantitative RT-PCR analysis of RNA extracted from DB-7 cells stable overexpressing NUMB4-GFP, NUMB6-GFP or control vector GFP. The mRNA levels of Twist, Zeb1, Zeb2, and Sp1 are expressed relative to β-actin transcripts. Each experiment was performed in triplicate. Error bars represent SEM.
Supplementary Fig. S3. NUMB6 overexpression increases Mmp9 mRNA. Mmp9 expression was measured by quantitative RT-PCR analysis of total RNA extracted from DB-7 cells stable overexpressing NUMB4-GFP, NUMB6-GFP or control vector GFP. Relative Mmp9 mRNA expression is normalized to β-actin transcripts. Each experiment was performed in triplicate. Error bars represent SEM.
Supplementary Fig. S4. NUMB6-induced Slug expression is attenuated by Slug specific siRNA. siRNA-mediated Slug knockdown was performed in GFP control and NUMB6-GFP DB-7 cells. siRNA treatment with 2.5uM or 5uM Slug siRNA effectively reduced Slug mRNA levels in NUMB6 DB-7 cells (A) The mRNA levels of Slug were measured by quantitative RT-PCR analysis of total RNA extracted from NUMB6-GFP or control vector GFP DB-7 cells treated with Slug-siRNA or control siRNA for 48hr. The mRNA levels of Slug are expressed relative to β-actin transcripts. Each experiment was performed in triplicate and repeated three times. Error bars represent SEM. (B) Expression of Slug on protein level was significantly decreased in Slug-depleted NUMB6-GFP DB-7 cells when 5uM Slug specific siRNA was used. NUMB6-GFP and control vector GFP DB-7 cells were transfected with control siRNA or Slug 5uM siRNAs for 48hr. Expression levels of Slug were analyzed by immunoblotting. β-actin was used as loading control. (C) Immunofluorescence depicts siRNA-mediated loss of Slug nuclear staining in NUMB6-GFP cells. NUMB6-GFP and GFP DB-7 cells were transfected with control siRNA or Slug specific siRNA at the 5uM concentration. After 48 hours cells were fixed, immunostained with Slug antibody (red). Secondary antibody Alexa Fluor-594 was used. Nuclei were stained with DAPI. Bars=20um.
Supplementary Fig. S5. NUMB6-induced DB-7 cell migration is reduced by Slug knockdown. Representative images of wound healing assay in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells transfected with control siRNA or Slug specific siRNA at the concentration of 2.5uM and 5uM. Cells as indicated were cultured until confluent and then a scratch wound was made using a 20ul pipette tip. Media were replaced and wounds were photographed at 20 hrs.
Supplementary Fig. S6. NUMB6-induced DB-7 cell migration is attenuated by inhibition of Akt signaling. Representative images of wound healing assay in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells treated with a vehicle DMSO or PI3K specific inhibitor LY294002 (10uM). Cells as indicated were cultured until confluent, and then a scratch wound was made using a 20ul pipette tip. Cell monolayers were washed with PBS, medium was replaced with DMSO or 10 μM LY294002, and wounds were photographed at 20 hrs.
Fig. 8.
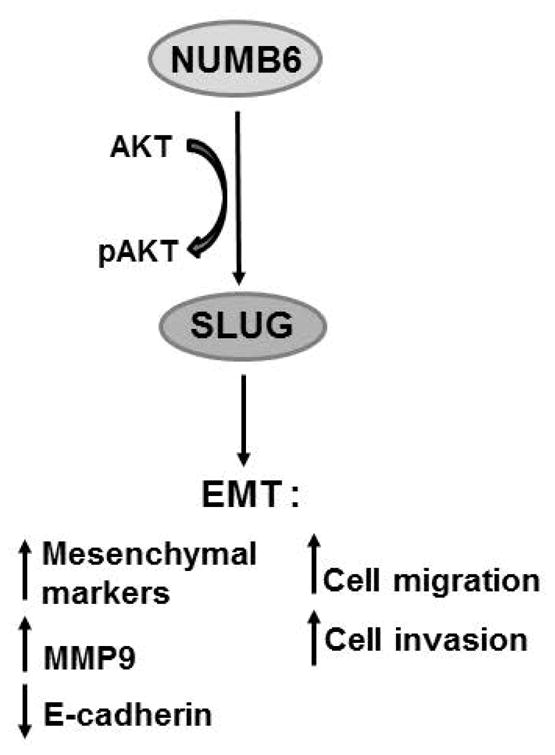
Schematic model of NUMB6-induced EMT in the regulation of cancer cell migration and invasion. The interaction of NUMB6 with Akt signaling pathway by unknown mechanism mediates induction of Slug transcription factor, ultimately leading to EMT. We found that EMT activation by NUMB6 overexpression triggered DB-7 cells migratory and invasive behavior. Because Akt pathway has been shown to affect cancer cell migration and invasion, we showed that chemical inhibition of the Akt phosphorylation abolished Slug expression in NUMB6 DB-7 cells, which in turn displayed reduced migration and invasion in vitro.
Acknowledgments
This work was supported by a pilot grant from the Maine Cancer Foundation.
In this project Axiovert 200 IF microscope Zeiss (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA) was used at the Protein, Nucleic Acid Analysis and Cell Imaging Core facility supported by grant number P30GM103392, Phase III COBRE in Vascular Biology (R. Friesel, P.I.), a grant supported by the National Institute of General Medical Sciences. The qPCR Bio-Rad cycler was used for real-time PCR analyses at the Progenitor Cell Analysis Core supported by grant number P30GM103465, COBRE in Stem Cell Biology and Regenerative Medicine (D. Wojchowski, P.I.), a grant support by the National Institute of General Medical Sciences.
All authors are in agreement with submitting and publishing this manuscript in your journal and also declare no competing interests.
References
- Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J, Chaloin O, Hoebeke J, Marine JC, Di Fiore PP, Telerman A. Reciprocal repression between P53 and TCTP. Nat Med. 2011;18:91–9. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Larue L. PI3K/AKT pathway and the epithelial-mesenchymal transition. In: Tikhonenko AT, editor. Cancer Genome and Tumor Microenvironment. Springer Science&Business Media, LCC; 2010. pp. 11–32. [Google Scholar]
- Borowsky AD, Namba R, Young LJ, Hunter KW, Hodgson JG, Tepper CG, McGoldrick ET, Muller WJ, Cardiff RD, Gregg JP. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- Bose S, Sindhu C, Mirocha JM, Bose N. The Akt pathway in human breast cancer: a tissue-array-based analysis. Modern Pathol. 2006;19:238–245. doi: 10.1038/modpathol.3800525. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Christfori C. New signals from an invasive front. Nature. 2006;41:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Come C, Magnino F, Bibeau F, De Santa Barbara P, Becker KF. Snail and slug play distinct roles during breast carcinoma progression. Clin Cancer Res. 2006;12:5395–5402. doi: 10.1158/1078-0432.CCR-06-0478. [DOI] [PubMed] [Google Scholar]
- De Herreros AG, Peiró S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia. 2010;15:135–147. doi: 10.1007/s10911-010-9179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon RL, Muller WJ. Distinct biological roles for the Akt family in mammary tumor progression. Cancer Res. 2010;70:4260–4264. doi: 10.1158/0008-5472.CAN-10-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Maguire TM, Hill A, McDermott E, O’Higgins N. Metalloproteinases: role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000;2:252–7. doi: 10.1186/bcr65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn BK, Agurs-Collins T, Browne D, Lubet R, Johnson KA. Health disparities in breast cancer: biology meets socioeconomic status. Breast Cancer Res Treat. 2010;121:281–292. doi: 10.1007/s10549-010-0827-x. [DOI] [PubMed] [Google Scholar]
- Euskirchen P, Skaftnesmo KO, Huszthy PC, Brekka N, Bjerkvig R, Jacobs AH, Miletic H. NUMB does not impair growth and differentiation status of experimental gliomas. Exp Cell Res. 2011;317:2864–2873. doi: 10.1016/j.yexcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Frise EJ, Knoblich A, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- Haider M, Qiu Q, Bani-Yaghoub M, Tsang BK, Gruslin A. Characterization and role of NUMB in the human extravillous trophopblast. Placenta. 2011;32:441–449. doi: 10.1016/j.placenta.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Hajra KM, Ji X, Fearon ER. Extinction of E-cadherin expression in breast cancer via a dominant repression pathway acting on proximal promoter elements. Oncogene. 1999;18:7274–7279. doi: 10.1038/sj.onc.1203336. [DOI] [PubMed] [Google Scholar]
- Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- Handschuh G, Candidus S, Luber B, Reich U, Schott C, Oswald S, Becke H, Hutzler P, Birchmeier W, Hofler H, Becker KF. Tumour-associated E-cadherin mutations alter cellular morphology, decrease cellular adhesion and increase cellular motility. Oncogene. 1999;18:4301–4312. doi: 10.1038/sj.onc.1202790. [DOI] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial–mesenchymal transition. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. Journal of Clinical Investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaczyn A, Bani-Yaghoub M, Tremblay R, Kubu C, Cowling R, Adams T, Prudovsky I, Spicer D, Friesel R, Vary C, Verdi JM. Two novel human NUMB isoforms provide a potential link between development and cancer. Neur Dev. 2010;5:31–46. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim S, Koh H, Yoon SO, Chung AS, Cho KS, Chung J. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. Faseb J. 2001;15:1953–62. doi: 10.1096/fj.01-0198com. [DOI] [PubMed] [Google Scholar]
- Kowalski PJ, Rubin MA, Kleer CG. E-cadherin expression in primary carcinomas of the breast and it distant metastases. Breast Cancer Res. 2003;5:R217–R222. doi: 10.1186/bcr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of notch1. J Biol Chem. 2009;284:26427–438. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micalizzi DS, Ford HL. Epithelial-mesenchymal transition in development and cancer. Future Oncol. 2009;5:1129–43. doi: 10.2217/fon.09.94. [DOI] [PubMed] [Google Scholar]
- Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet F, Berx G, van Roy F. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol Cell Biol Res Commun. 1999;2:77–85. doi: 10.1006/mcbr.1999.0155. [DOI] [PubMed] [Google Scholar]
- Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Confalonieri S, Romano P, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochem Biophys Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley SH, Perou, Gill G, Gupta PB, Kuperwasser C. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J Mammary Gland Neoplasia. 2010;15(2):201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Rennstam K, McMichael N, Berglund P, Honeth G, Hegardt C, Ryden L, Luts L, Bendahl PO, Hedenfalk I. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res Treat. 2009;122:315–324. doi: 10.1007/s10549-009-0568-x. [DOI] [PubMed] [Google Scholar]
- Son H, Moon A. Epithelial-mesenchymal Transition and Cell Invasion. Toxicol Res. 2010;26:245–252. doi: 10.5487/TR.2010.26.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Vega FM, Ridley AJ. Rho GTPases in cancer biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Verdi JM, Bashirullah A, Goldhawk DE, Kubu CJ, Jamali M, Meakin SO, Lipshitz HD. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci USA. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cui T, Feng W, Li H, Hu L. Role of Numb expression and nuclear translocation in endometrial cancer. Oncology Lett. 2015;9:1531–1536. doi: 10.3892/ol.2015.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YD, Zhou BHP. Snail more than EMT. Cell Adhesion & Migration. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-Mesenchymal Transition. Cancer Res. 2008;68:9574–9577. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tan M, Hawthorne VS, Klos K, Lan KH, Yang Y, Yang W, Smith TL, Shi D, Yu D. Activation of the Akt/Mammalian Target of Rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancer. Clin Cancer Res. 2004;10:6779–6788. doi: 10.1158/1078-0432.CCR-04-0112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. mRNA detection of Numb1-Numb4 and Numb6 in control GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cell lines. RT-PCR analysis was used to determine mRNA relative expression levels of Numb1-Numb4 and Numb6 in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells. Numb1-Numb4 and Numb6 variants were amplified by RT-PCR and resolved on 1.5% agarose gel. Numb variants were then identified according to their unique amplicon size and quantitated by densitometry Y-axis depicts mRNA expression as a fold of change over GFP control groups. Results were normalized to β-actin housekeeping gene. Error bars represent SEM.
Supplementary Fig. S2. NUMB6 overexpression upregulates EMT-related transcription factors expression. The mRNA levels of EMT-inducing transcription factors including Twist, Zeb1, Zeb2, and Sp1 were measured by quantitative RT-PCR analysis of RNA extracted from DB-7 cells stable overexpressing NUMB4-GFP, NUMB6-GFP or control vector GFP. The mRNA levels of Twist, Zeb1, Zeb2, and Sp1 are expressed relative to β-actin transcripts. Each experiment was performed in triplicate. Error bars represent SEM.
Supplementary Fig. S3. NUMB6 overexpression increases Mmp9 mRNA. Mmp9 expression was measured by quantitative RT-PCR analysis of total RNA extracted from DB-7 cells stable overexpressing NUMB4-GFP, NUMB6-GFP or control vector GFP. Relative Mmp9 mRNA expression is normalized to β-actin transcripts. Each experiment was performed in triplicate. Error bars represent SEM.
Supplementary Fig. S4. NUMB6-induced Slug expression is attenuated by Slug specific siRNA. siRNA-mediated Slug knockdown was performed in GFP control and NUMB6-GFP DB-7 cells. siRNA treatment with 2.5uM or 5uM Slug siRNA effectively reduced Slug mRNA levels in NUMB6 DB-7 cells (A) The mRNA levels of Slug were measured by quantitative RT-PCR analysis of total RNA extracted from NUMB6-GFP or control vector GFP DB-7 cells treated with Slug-siRNA or control siRNA for 48hr. The mRNA levels of Slug are expressed relative to β-actin transcripts. Each experiment was performed in triplicate and repeated three times. Error bars represent SEM. (B) Expression of Slug on protein level was significantly decreased in Slug-depleted NUMB6-GFP DB-7 cells when 5uM Slug specific siRNA was used. NUMB6-GFP and control vector GFP DB-7 cells were transfected with control siRNA or Slug 5uM siRNAs for 48hr. Expression levels of Slug were analyzed by immunoblotting. β-actin was used as loading control. (C) Immunofluorescence depicts siRNA-mediated loss of Slug nuclear staining in NUMB6-GFP cells. NUMB6-GFP and GFP DB-7 cells were transfected with control siRNA or Slug specific siRNA at the 5uM concentration. After 48 hours cells were fixed, immunostained with Slug antibody (red). Secondary antibody Alexa Fluor-594 was used. Nuclei were stained with DAPI. Bars=20um.
Supplementary Fig. S5. NUMB6-induced DB-7 cell migration is reduced by Slug knockdown. Representative images of wound healing assay in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells transfected with control siRNA or Slug specific siRNA at the concentration of 2.5uM and 5uM. Cells as indicated were cultured until confluent and then a scratch wound was made using a 20ul pipette tip. Media were replaced and wounds were photographed at 20 hrs.
Supplementary Fig. S6. NUMB6-induced DB-7 cell migration is attenuated by inhibition of Akt signaling. Representative images of wound healing assay in GFP, NUMB4-GFP, and NUMB6-GFP DB-7 cells treated with a vehicle DMSO or PI3K specific inhibitor LY294002 (10uM). Cells as indicated were cultured until confluent, and then a scratch wound was made using a 20ul pipette tip. Cell monolayers were washed with PBS, medium was replaced with DMSO or 10 μM LY294002, and wounds were photographed at 20 hrs.