Abstract
Burkholderia thailandensis is a Gram-negative bacterium endemic to southeast Asia and northern Australia soils. Non-pathogenic, it is commonly used as a model organism for the related human pathogens Burkholderia mallei and Burkholderia pseudomallei. B. thailandensis is relatively easily genetically manipulated and a variety of robust genetic tools can be used in this organism. This unit describes protocols for conjugation, natural transformation, mini-Tn7 insertion, and allelic exchange in B. thailandensis.
Keywords: conjugation, transformation, allelic exchange, mutation, mini-Tn7
INTRODUCTION
Burkholderia thailandensis is a Gram-negative bacterium isolated from soil and water in southeast Asia and northern Australia. It is evolutionarily related to Burkholderia mallei and Burkholderia pseudomallei, which cause the diseases glanders and melioidosis, respectively. Lacking many virulence factors found in these human pathogens but otherwise sharing significant genomic similarity (Yu et al., 2006), B. thailandensis is utilized for comparative analyses or as an avirulent model organism. Indeed, B. thailandensis, like B. pseudomallei, can replicate in a variety of cell types (Haraga et al., 2008), utilize actin-based motility (Stevens et al., 2005), and induce the formation of multinucleate giant cells (Kespichayawattana et al., 2000; French et al., 2011). Moreover, its ease of genetic manipulation and the wide variety of genetic tools available for B. thailandensis has led to increased research in recent years, particularly in the study of interbacterial interactions (Anderson et al., 2012; Schwarz et al., 2010; Chandler et al., 2009). Protocols in this unit describe the basic genetic techniques used in B. thailandensis research, including the introduction of plasmids by conjugation (Basic Protocol 1), site-specific chromosomal insertion via Tn7 transposition (Basic Protocol 2), homologous recombination by natural transformation with linear DNA (Basic Protocol 3), antibiotic marker excision (Basic Protocol 4), and allelic exchange (Basic Protocol 5).
CAUTION: Burkholderia thailandensis is a Biosafety Level 1 (BSL-1) organism. Such organisms are not known to consistently cause disease in healthy adult humans, and are of minimal potential hazard to laboratory personnel and the environment. Standard microbiological practices should be followed when working with these organisms. See UNIT 1A.1 and other pertinent resources (APPENDIX 1B) for more information.
BASIC PROTOCOL 1 INTRODUCTION OF PLASMIDS BY CONJUGATION
Although Burkholderia thailandensis is naturally competent, the most effective way to introduce plasmids (particularly nonreplicative plasmids) is via conjugation. Donor Escherichia coli strain RHO3 (Δasd ΔaphA) is a 2,6-diaminopimelic acid (DAP) auxotroph and, following conjugation, is easily selected against by the exclusion of DAP from the selection medium (López et al., 2009). However, any donor E. coli strain that carries conjugation machinery may be used.
Materials
B. thailandensis from a frozen stock
Donor E. coli RHO3 strain(s) carrying the plasmid(s) to be transferred
Sterile swabs
Low salt LB (LSLB) plates supplemented with 200 μg/ml 2,6-diaminopimelic acid (DAP) and/or appropriate antibiotics (see Reagents and Solutions; Table 1)
Table 1.
Antibiotic and media supplement concentrations for use with B. thailandensis genetic tools.
| Supplement | Solvent | Stock conc.a | Final conc.b |
|---|---|---|---|
| 2,6-diaminopimelic acid (DAP) | 1 M NaOH | 100 mg/ml | 200 μg/ml |
| Chloramphenicol | Ethanol | 20 mg/ml | 20 μg/ml |
| Kanamycin | Water | 125 mg/ml | 250 μg/ml |
| Tetracyclinec | Ethanol | 10 mg/ml | 50 μg/ml |
| Ampicillind | Water | 100 mg/ml | 100 μg/ml |
| X-glucc,e | DMF or DMSO | 50 mg/ml | 50 μg/ml |
| Sucrose | Water | 50% (w/v) | 15% (w/v) |
| Rhamnosec | Water | 20% (w/v) | 0.2% (w/v) |
Most stock solutions should be filter-sterilized (0.22 μm filter) and stored at −20°C. Sucrose and rhamnose solutions may be stored at room temperature.
Final working concentration in liquid medium or agar plates. Media should be cooled to 55°C before the addition of antibiotics or supplements.
Light-sensitive. Stock solution and prepared media should be protected from light.
For use with E. coli only. B. thailandensis is resistant to ampicillin.
X-gluc, 5-bromo-4-chloro-3-indoxyl-beta-D-glucuronide
-
1
Streak B. thailandensis onto a LSLB plate and E. coli RHO3 carrying the plasmid(s) to be transferred on LSLB plate(s) supplemented with DAP and appropriate antibiotics.
-
2
Incubate overnight (B. thailandensis) or two days (E. coli RHO3) at 37°C.
-
3
Using a sterile swab, collect several large B. thailandensis colonies and thoroughly spread the bacteria onto approximately one half of a LSLB plate supplemented with DAP (no antibiotics).
The plate is spread evenly with a layer of bacteria, not streaked for isolated colonies.
-
4
Using the same technique, use a clean sterile swab to collect an equivalent amount of E. coli RHO3 and spread it on top of the area spread with B. thailandensis.
-
5
For triparental matings, repeat step 4 with the remaining donor E. coli strain.
-
5
To prepare control conjugations, use a sterile swab to collect fresh B. thailandensis and spread it alone onto a clean section (approximately one-quarter or less of the plate) of the conjugation plate.
-
6
Repeat step 5 with each donor E. coli strain.
-
7
Incubate the conjugation plate at 37°C for 4–6 h.
-
8
Using a sterile swab, collect bacteria from the conjugation (section of mixed B. thailandensis and E. coli) and spread onto approximately one-half of a LBLB plate supplemented with appropriate antibiotics (selection plate).
To select against the donor E. coli strain, the selection plate should not contain DAP.
-
9
Streak once for isolated colonies.
-
10
Using sterile swabs, collect bacteria from the control areas (sections of B. thailandensis and E. coli alone) of the conjugation plate and patch them onto clean sections of the selection plate.
-
11
Save the conjugation plate at room temperature overnight.
Conjugation will continue to occur. If no colonies are observed on the selection plate, selection (steps 8–12, omitting step 11) may be repeated once from this overnight conjugation.
-
12
Incubate the selection plate at 37°C overnight.
Depending on the efficiency of the mating, isolated colonies may be found in either the heavy-streaked area of the selection plate or in the area streaked for isolation (or both).
-
13
Observe the control conjugations on the selection plate. If growth is observed, the mating should be repeated following troubleshooting.
BASIC PROTOCOL 2 SITE-SPECIFIC CHROMOSOME INSERTION BY MINI-TN7 TRANSPOSITION
Unlike other transposons, Tn7 inserts site-specifically (and orientation-specifically) into Gram negative bacterial chromosomes at Tn7 attachment, or attTn7, sites, which are located downstream of glmS genes (Peters, 2014). Tn7-based vectors take advantage of this biology, allowing the stable insertion of DNA at a neutral chromosomal site. A mini-Tn7-based system developed in the Schweizer laboratory works robustly in B. thailandensis (Choi et al., 2005). In addition, B. thailandensis has two glmS genes and two corresponding attTn7 sites, each of which may be inserted with the same or different genetic material. Possible applications include mutant complementation, overexpression, introduction of gene expression reporters (lacZ, gfp), and fluorescent labeling (gfp, rfp) (Norris et al., 2010).
In this system, DNA of interest is cloned between the Tn7L and Tn7R recognition sequences on a mini-Tn7 suicide plasmid, which also contains an antibiotic resistance cassette. Together with a helper plasmid containing the required transposase, pTNS3 (Choi et al., 2008), the mini-Tn7 plasmid is introduced into B. thailandensis and the mini-Tn7 DNA (containing the DNA of interest and resistance cassette) is moved to an attTn7 site. Once introduced, insertions at attTn7 sites are stable and do not require antibiotic selection for maintenance (Choi et al., 2005). Moreover, many commonly-used mini-Tn7 plasmids feature Flp recombinase target (FRT) sequences that flank the antibiotic resistance cassette, allowing for subsequent marker excision, if desired (see Basic Protocol 4).
Materials
B. thailandensis from a frozen stock
Donor E. coli RHO3 carrying a mini-Tn7 plasmid (see Table 2)
Donor E. coli RHO3 carrying helper plasmid pTNS3 (see Table 2)
Sterile swabs
Low salt LB (LSLB) plates supplemented with DAP and/or appropriate antibiotics (see Reagents and Solutions; Table 1)
Sterile toothpicks or inoculating loops
Table 2.
Plasmids used routinely for genetic manipulation of B. thailandensis
| Plasmid | Description/features | Antibiotic resistance | Reference |
|---|---|---|---|
| pTNS3 | Helper plasmid for mini-Tn7, oriT, R6K ori | Amp | (Choi et al., 2008) |
| pUC18T-miniTn7T-Km | attTn7 site delivery plasmid with MCS | Kan (mini-Tn7), Amp (backbone) | (Choi et al., 2005) |
| miniTn7-kan-gfp | Delivers gfp to attTn7 site | Kan | (Norris et al., 2010) |
| mini-Tn7-kan-rfp | Delivers rfp to attTn7 site | Kan | (Norris et al., 2010) |
| pFlpe4 | Rham-inducible flp, TS ori | Kan | (Choi et al., 2008) |
| pFlpTet | Rham-inducible flp, TS ori | Tet | (Garcia et al., 2013) |
| pEXKm5 | Allelic exchange vector | Kan | (López et al., 2009) |
MCS, multiple cloning site; TS, temperature-sensitive; Rham, rhamnose; Amp, ampicillin; Kan, kanamycin; Tet, tetracycline
Perform a triparental mating with B. thailandensis, E. coli RHO3 carrying a mini-Tn7 plasmid, and E. coli RHO3 carrying the helper plasmid pTNS3 (see Basic Protocol 1).
-
Using aseptic technique, streak 4–8 transconjugant colonies from the conjugation selection plate onto a fresh LSLB plate supplemented with antibiotics. Streak for isolated colonies.
Antibiotic included in the medium should correspond to the resistance cassette carried on the mini-Tn7 plasmid that is delivered to the B. thailandensis attTn7 site (located between the plasmid’s Tn7L and Tn7R sequences). Antibiotic resistance cassettes located elsewhere on the mini-Tn7 plasmid backbone will not transfer to B. thailandensis.
Incubate overnight at 37°C.
-
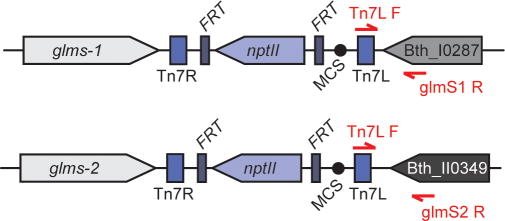
Test for attTn7 site insertions in each candidate using colony PCR (see Support Protocol 1). Two colony PCR reactions should be set up for each B. thailandensis candidate – one testing for insertion at the attTn7 site downstream of glmS-1 and a separate reaction to test for insertion at the site downstream of glmS-2 (Fig. 1) (Choi et al., 2005). Reaction 1 contains primers “Tn7L” (ATTAGCTTACGACGCTACACCC) and “glmS1” (GCGTTCGTCGTCCACTGGGA) and reaction 2 contains primers “Tn7L” and “glmS2” (TGTGAATGGTCAGACGCTGTTCG).
Occasionally, a single B. thailandensis colony will contain insertions at both attTn7 sites. Depending on the downstream application, this may or may not be acceptable.
Figure 1. Mini-Tn7 insertions at B. thailandensis E264 attTn7 sites.
The two attTn7 sites are located 3′ to glmS-1 (Bth_I0288/Bth_RS13655) on chromosome I and 3′ to glmS-2 (Bth_II0348/BTH_RS01805) on chromosome II. Primers (red arrows) that anneal to the Tn7L region of mini-Tn7 and either Bth_I0286 (Bth_RS13660) or Bth_II0349 (Bth_RS01810) are used to confirm attTn7 insertion (for primer sequences, see Basic Protocol 2). In this example based on insertion from pUC18T-miniTn7T-Km (Choi et al., 2005), nptII (conferring kanamycin-resistance) carried on mini-Tn7 is flanked by flp recombinase target (FRT) sequences for marker excision, leaving the multiple cloning site (MCS) intact.
BASIC PROTOCOL 3 HOMOLOGOUS RECOMBINATION BY NATURAL TRANSFORMATION WITH LINEAR DNA
Developed in the Manoil laboratory (Thongdee et al., 2008), this technique takes advantage of the fact that B. thailandensis becomes naturally competent (able to take up and incorporate exogenous DNA) when cultured in minimal glucose medium. By engineering specific DNA fragments, transformation with linear DNA is used to construct marked mutations. Using PCR and standard cloning methods, an antibiotic resistance marker is flanked by DNA homologous to the B. thailandensis genome so that integration of the linear DNA fragment replaces a specific chromosomal element with the resistance cassette. Alternatively, chromosomal DNA containing an antibiotic resistance marker may be used as the transformation substrate. In this way, the method can be used for both generating marked mutations (via transformation with PCR product), as well as for transferring mutations between strains (via transformation with whole chromosomal DNA).
Materials
B. thailandensis from a frozen stock
Sterile capped glass or plastic culture tubes
Low salt LB (LSLB) medium (see Reagents and Solutions)
Sterile sticks or inoculating loops
M63 minimal medium (see Reagents and Solutions)
Spectrophotometer and cuvettes
Microcentrifuge and plastic tubes
DNA containing an antibiotic resistance cassette that is flanked by >500 bp regions of homology to the B. thailandensis genome (may be PCR product or B. thailandensis chromosomal DNA). If subsequent antibiotic resistance marker removal is desired, the cassette should be flanked first by FRT sequences (see Basic Protocol 4).
Sterile cell spreader or glass beads
Low salt LB (LSLB) plates supplemented with appropriate antibiotics (see Reagents and Solutions; Table 1)
\
-
1
Inoculate several B. thailandensis colonies into 2 ml LSLB medium.
-
2
Incubate overnight with aeration at 37°C.
-
3
Dilute the overnight culture by adding 40 μl culture to 2 ml M63 minimal medium (1:50 dilution).
This may be scaled up depending on how many transformations are required. Adjust the resuspension volume accordingly in step 5 below.
-
4
Incubate with aeration at 37°C until OD600=0.5 (approximately 4–5 h).
-
5
Pellet culture in microcentrifuge (1 min at 15,000 × g), decant supernatant, and resuspend pellet in 100 μl fresh M63 (1/20th of the 2 ml culture volume).
-
6
Mix 50 μl cell suspension with 5 μl DNA (at least 100 ng) or water (no DNA) control.
Inclusion of a “no DNA” control with each set of transformations is important for data interpretation.
-
7
Incubate 30 min at room temperature with no agitation.
-
6
Add 2 ml fresh M63 minimal medium to each transformation and transfer each to a culture tube.
-
8
Incubate overnight with aeration at 37°C.
-
9
Pellet each culture as described in step 5, decant supernatant, and resuspend in 100 μl M63.
-
10
Using a sterilized cell spreader or glass beads, spread the cell suspension (100 μl) onto a LSLB plate supplemented with appropriate antibiotics (depending on the antibiotic resistance cassette included in transformed DNA).
-
11
Incubate plates ~20 h at 37°C.
Colonies should be visible on transformation plates, but not on “no DNA” control plates.
Check for the presence of the desired mutation in the resulting transformants by colony PCR (see Support Protocol 1).
BASIC PROTOCOL 4 FLP RECOMBINASE-MEDIATED MARKER EXCISION
Excision of antibiotic resistance markers allows markers to be recycled for subsequent genetic manipulations and provides antibiotic-sensitive bacteria for applications where this feature is desired. Flp recombinase mediates site-specific recombination between flp recombinase target (FRT) sequences and the Flp-FRT system has been adapted for many applications. Tools developed in the Schweizer laboratory allow an FRT-flanked antibiotic resistance marker to be excised from a B. thailandensis chromosome using a rhamnose-inducible flp contained on a temperature-sensitive plasmid (Choi et al., 2008). Plasmid pFlpe4 (or its Tet-resistant derivative, pFlpTet (Garcia et al., 2013)) replicates in Burkholderia at 30°C, but is lost at higher temperatures (42°C or 37°C).
Materials
B. thailandensis with a chromosomal FRT-flanked antibiotic resistance cassette
Donor E. coli RHO3 containing pFlpe4 or pFlpTet (see Table 2)
Sterile swabs
Low salt LB (LSLB) plates (see Reagents and Solutions)
LSLB plates supplemented with 200 μg/ml 2,6-diaminopimelic acid (DAP), 250 μg/ml kanamycin (Kan), 50 μg/ml tetracycline (Tet), and/or 0.2% rhamnose, where appropriate (see Reagents and Solutions; Table 1)
Sterile applicator sticks or inoculating loops
Perform a mating with B. thailandensis and E. coli RHO3 carrying pFlpe4 or pFlpTet according to Basic Protocol 1 with the following modification: incubate the conjugation at 30°C (instead of 37°C) for 6 h.
Select the mating on LSLB/Kan (for pFlpe4) or LSLB/Tet (for pFlpTet) plates (see Basic Protocol 1) and incubate the selection plate 48 h at 30°C.
To induce flp expression, streak 8–16 colonies for isolation onto LSLB/Kan/rhamnose (for pFlpe4) or LSLB/Tet/rhamnose (for pFlpTet) plates.
Incubate the plates 48 h at 30°C.
Sequentially patch 3–4 colonies from each plate/sector onto LSLB containing the antibiotic to which sensitivity is desired and either LSLB/Kan (for pFlpe4) or LSLB/Tet (for pFlpTet).
-
Incubate the plates 48 h at 30°C.
Clones should be sensitive to the antibiotic corresponding to the cassette removed and resistant to either Kan or Tet.
Using bacteria from the LSLB/Kan (for pFlpe4) or LSLB/Tet (for pFlpTet) plates, streak 4–5 antibiotic-sensitive clones onto LSLB.
Incubate the plates overnight at 37°C to induce loss of pFlpe4 or pFlpTet.
Streak colonies from the LSLB plates onto fresh LSLB plates.
Incubate the plates overnight at 37°C.
Patch 4–5 colonies from each clone onto LSLB plates and either LSLB/Kan (for pFlpe4) or LSLB/Tet (for pFlpTet) plates.
-
Incubate the plates 48 h at 30°C.
Growth should only be visible on LSLB plates, as colonies should be Kan- or Tet-sensitive after pFlpe4 or pFlpTet loss.
Confirm the loss of the antibiotic resistance marker by colony PCR (see Support Protocol 1).
BASIC PROTOCOL 5 ALLELIC EXCHANGE USING SACB COUNTERSELECTION
A common method for introducing unmarked chromosomal mutations, allelic exchange has been adapted for use in many bacterial species. This powerful method allows in-frame gene deletion, gene replacement, generation of point mutations, and introduction of sequences encoding epitope tags onto the bacterial chromosome. While several allele replacement systems exist for B. thailandensis, the pEXKm5-based system developed in the Schweizer laboratory is particularly robust, easy to use, and versatile (López et al., 2009).
In this protocol, a markerless mutation flanked by regions of homology (≥500 bp) is cloned into suicide vector pEXKm5. When introduced into B. thailandensis, the plasmid can integrate onto a chromosome via recombination at the homology regions. A kanamycin resistance cassette and gusA gene located on the pEXKm5 backbone allow selection and identification of colonies containing the cointegrated plasmid. Encoded by gusA, glucuronidase will cleave the indicator substrate X-Gluc, resulting in a blue color change and allowing for blue-white screening at several steps of the procedure. In the second stage of the protocol, subsequent loss of pEXKm5 from B. thailandensis by homologous recombination results in a mixture of bacteria that have either reverted to the wild-type genotype or that contain the desired mutation. Loss of pEXKm5 is achieved either through sacB counterselection (described here) or using I-SceI endonuclease-stimulated recombination (see Alternate Protocol 1). Located on pEXKm5, sacB encodes levansucrase, a Bacillus subtilis enzyme that is lethal for many Gram-negative bacteria when they are cultured in the presence of sucrose. Thus, B. thailandensis containing cointegrated pEXKm5 are sensitive to sucrose, while cells that have lost the plasmid by recombination (and which may contain the desired mutation) are rendered sucrose-resistant.
Materials
B. thailandensis from a frozen stock
Donor E. coli RHO3 containing pEXKm5 (see Table 2) that has been modified to contain the recombinant allele and homology regions
Sterile swabs
Low salt LB (LSLB) plates (see Reagents and Solutions)
LSLB plates supplemented with 200 μg/ml 2,6-diaminopimelic acid (DAP), 250 μg/ml kanamycin (Kan), and/or 50 μg/ml X-gluc, where appropriate (see Reagents and Solutions; Table 1)
Sterile applicator sticks or inoculating loops
YT plates supplemented with 15% sucrose and either 250 μg/ml Kan or 50 μg/ml X-gluc (see Reagents and Solutions; Table 1)
Sterile capped glass or plastic culture tubes
YT broth (see Reagents and Solutions)
Sterile cell spreader or glass beads
Perform a mating with B. thailandensis and E. coli RHO3 carrying the modified pEXKm5 plasmid (see Basic Protocol 1).
-
Select the mating on LSLB/Kan/X-gluc plates (see Basic Protocol 1) and incubate the selection plate overnight at 37°C.
Colonies with chromosomally-integrated pEXKm5 as the result of a single recombination event (called cointegrants or merodiploids) will be kanamycin-resistant and blue. GusA activity on X-gluc results in a subtle blue color (less pronounced than LacZ with X-Gal) that can be enhanced by incubation of the plate for several hours at 4°C.
Select 8–16 cointegrants and streak for isolated colonies onto LSLB/Kan/X-gluc plates.
Incubate the plates overnight at 37°C.
Check that the cointegrants contain pEXKm5 integrated at the correct chromosomal location (and determine orientation) using colony PCR and primers that anneal to the plasmid in combination with primers that anneal to the chromosome outside of the region used in homologous recombination.
Confirm the sucrose-sensitivity of cointegrants by patching each candidate sequentially onto a YT/Sucrose/Kan plate and a LSLB/Kan plate.
-
Incubate the plates 20–24 h at 30°C.
Ideal cointegrants should have heavy growth on the LSLB/Kan plate and little to no growth on the YT/Sucrose/Kan plate.
Select two sucrose-sensitive cointegrants for counterselection. Ideally, choose one clone that contains pEXKm5 integrated at the 5′ homology region and another clone that contains pEXKm5 integrated at the 3′ homology region (as determined by colony PCR in step 5).
Inoculate one colony into 1 ml YT broth.
Incubate with aeration for ≥4 h at 37°C.
-
Prepare serial dilutions from the YT culture and plate 100 μl of each dilution onto YT/Sucrose/X-gluc plates.
Which dilutions are plated depends on the incubation time of the YT culture (longer incubations will require plating more dilute cell suspension). To start, a greater number of dilutions are recommended (10−1, 10−2, 10−3, 10−4) to ensure that isolated colonies are observed.
-
Incubate for 48 h at 30°C.
A mixture of blue and white colonies may be observed. Bacteria that have undergone a second recombination event will “loop out” the pEXKm5 plasmid and lose it, resulting in colonies that are sucrose-resistant, kanamycin-sensitive, and white on X-gluc.
Select 8–16 white colonies and streak for isolated colonies onto LSLB plates.
Incubate overnight at 37°C.
Check for sensitivity to Kan by sequentially patching each candidate onto a LSLB/Kan plate and a LSLB plate.
-
Incubate overnight at 37°C.
Growth should only be observed on the LSLB plate.
For kanamycin-sensitive clones, use colony PCR (see Support Protocol 1) to determine the genotype (mutant or wild-type) of the colony.
SUPPORT PROTOCOL 1 COLONY PCR
B. thailandensis colonies that are candidates for introduced mutations (gene deletions, insertions, or replacements) or chromosomal insertions (ie. attTn7 site insertions) are routinely screened by PCR to determine whether or not they contain the desired genetic manipulation. PCR reactions can then be analyzed for their size or further digested with restriction enzymes, if necessary. Use of appropriate control reactions with each PCR is critical for data interpretation.
Materials
0.2 ml tubes
Sterile, deionized water
5× GoTaq® buffer (Promega), or alternative Taq buffer
10 mM dNTP mix
Primers, 10 μM each
Dimethyl sulfoxide (DMSO)
Go Taq® polymerase (Promega), or alternative Taq polymerase
B. thailandensis colonies for screening
Sterile toothpicks
Thermocycler
Restriction enzyme(s) and appropriate buffer (optional)
6× DNA gel loading dye
Agarose and 0.5× TAE buffer
Ethidium bromide or other DNA gel stain
Agarose gel electrophoresis apparatus
UV transilluminator
-
Prepare the following PCR reaction mixtures in 0.2 ml tubes:
water 14.3 μl 5× GoTaq® buffer 5.0 μl 10 mM dNTP mix 1.0 μl Forward primer, 10 μm 1.0 μl Reverse primer, 10 μm 1.0 μl DMSO 2.5 μl GoTaq® polymerase 0.2 μl -
Using a sterile toothpick, touch a single B. thailandensis colony and swirl in a PCR reaction tube.
Toothpick does not need to contain visible material from the bacterial colony.
Repeat step 2 for remaining reactions.
-
Place tubes in thermocycler and run the following program, where Tanneal = (Primer Tm) – (4–5°C):
Temperature Time Cycles
95°C 2 min 1
95°C 30 sec Tanneal 30 sec 30 72°C 1 min/kb expected product
72°C 5 min 1
If necessary, digest the PCR product with the appropriate restriction enzyme(s).
Mix 2 μl DNA loading dye with 2–10 μl PCR product and run on an agarose gel.
Analyze the gel by UV transillumination for presence/absence of PCR product, PCR product size, or size of digested PCR products.
REAGENTS AND SOLUTIONS
Low salt LB broth (or agar)
10 g tryptone
5 g yeast extract
5 g NaCl
1 L distilled, deionized water
Autoclave
Store broth at room temperature for 3 months
For plates: add 15 g agar to broth mixture before autoclaving. Cool to 55°C and add antibiotics or other supplements, if necessary (see Table 1). Pour cooled agar into sterile plastic petri dishes. Dry plates at room temperature overnight and store at 4°C for 6 months.
M63 minimal medium
-
5× M63 salts solution
15 g KH2PO4
35 g K2HPO4
10 g (NH4)2SO4
2.5 ml 1 mg/ml FeSO4 ·7H2O
pH to 7.0 with KOH
distilled, deionized water to 1 L final volume
Autoclave
Store at room temperature for 12 months
-
1× M63 broth
400 ml distilled, deionized water
Autoclave
Cool to 55°C and add the following:
100 ml 5× M63 salts solution
5 ml 20% (w/v) glucose (or alternate carbon source), filter sterilized
0.5 ml 1 M MgSO4, sterile
Store at room temperature for 3 months
YT broth
10 g tryptone
10 g yeast extract
1 L distilled, deionized water
Autoclave
Store broth at room temperature for 3 months
YT agar with 15% sucrose
10 g tryptone
10 g yeast extract
15 g agar
700 mL distilled, deionized water
Autoclave
Cool to 55°C and add 300 ml 50% (w/v) filter sterilized sucrose. Add antibiotics or other supplements, if necessary (see Table 1). Pour cooled agar into sterile plastic petri dishes. Dry plates at room temperature overnight and store at 4°C for 2–3 weeks.
COMMENTARY
Background Information
Occupying overlapping niches in tropical soils and water, B. thailandensis and the human pathogen B. pseudomallei are very closely related. Thus, most genetic tools developed for use in B. pseudomallei are also compatible with B. thailandensis. As a model organism, B. thailandensis offers greater flexibility in antibiotic marker use and ease of genetic manipulation, as, unlike B. pseudomallei, it is not a CDC Select Agent nor does it require BSL-3 containment.
Genome sequences of several B. thailandensis strains have been completed and numerous draft genome sequences are available on NCBI and other databases. Like B. pseudomallei, B. thailandensis has two circular chromosomes that, in reference strain E264, are 3.8 MB and 2.9 MB in size and are GC-rich (approximately 68% GC) (Yu et al., 2006). Phylogenetic analysis using 16S rRNA and housekeeping gene sequences suggests that B. thailandensis diverged from B. pseudomallei and Burkholderia mallei approximately 47 million years ago (Yu et al., 2006). Horizontal gene transfer appears to be a driver of B. pseudomallei and B. thailandensis diversity, as strains of both species contain numerous genomic islands (Yu et al., 2006; Tuanyok et al., 2008).
Robust genetic tools, including those described in this unit, have accelerated B. thailandensis research, particularly the ability to generate in-frame unmarked mutations using allelic exchange. Although the use of sacB counterselection was initially thought to be problematic in B. pseudomallei and related species due to the presence of endogenous sacB genes (Holden et al., 2004), modifications have allowed the system to function effectively. A modified sacB gene that includes a Burkholderia leader sequence (Hamad et al., 2009) and optimized culture conditions (Logue et al., 2009) enabled this method to function efficiently in B. thailandensis and B. pseudomallei (López et al., 2009). While other counterselectable markers have been used for Burkholderia spp., including pheS (Barrett et al., 2008), the pEXKm5-based sacB system offers several advantages (López et al., 2009). This vector enables counterselection on rich medium, supplies a useful gusA marker to screen cointegrants and resolved cointegrants, and provides versatility with the inclusion of an alternative I-SceI based system.
An alternative to sacB counterselection described in Basic Protocol 4 is the use of homing endonuclease I-SceI to stimulate recombination in pEXKm5 cointegrants. The 18 bp recognition sequence of I-SceI does not occur in any bacterial genomes sequenced to date, but is included on the pEXKm5 backbone. Generation of a double-stranded DNA break by I-SceI cleavage leads to an increased rate of intramolecular recombination and can be exploited to promote cointegrant resolution (Pósfai et al., 1999). The current tools available for I-SceI use with pEXKm5 in B. pseudomallei are incompatible with B. thailandensis, as I-sceI is encoded on a plasmid conferring zeocin resistance in this system (López et al., 2009). Although B. thailandensis is intrinsically zeocin-resistant, simple cloning to alter the resistance cassette should allow this approach to be applied to B. thailandensis as well, further expanding the genetic tool repertoire for this organism.
Critical Parameters and Troubleshooting
Introduction of plasmids by conjugation
All conjugations should utilize B. thailandensis freshly-cultured from a frozen stock. Bacteria stored at 4°C may have decreased viability and repeated serial passages of B. thailandensis may allow mutation accumulation. Growth of control conjugations on the antibiotic selection plate most likely suggests that the wrong antibiotic was used for selection or that the wrong bacterial strain was used in the mating. Occasionally pinpoint colonies on the selection plate will be observed for these control conjugations (as well as for the true conjugation), especially if the plate was incubated for >24 h. These are spontaneous antibiotic-resistant mutants and are not a concern, provided that true transconjugant colonies (larger in size) are easily identified on the selection plate. Finally, frequently-used plasmids should be periodically re-transformed into fresh E. coli RHO3 cells because, although rare, reversion to DAP-independence has been observed for some strains.
Site-specific chromosome insertion by mini-Tn7 transposition
Difficulty obtaining antibiotic-resistant transconjugants after conjugations with mini-Tn7 plasmids may indicate that the helper plasmid pTNS3 was omitted from the mating or the wrong antibiotic was used in the selection plate. Antibiotic-resistant colonies that lack attTn7 site insertions (determined by colony PCR) can occur and likely represent spontaneous mutants or, if the mini-Tn7 plasmid contained sequences present in the B. thailandensis genome, bacteria that have integrated the whole plasmid into their genome. The frequency of this occurrence increases as the size (bp) of homologous B. thailandensis sequence present on the mini-Tn7 plasmid increases. Usually, simply screening additional colonies by PCR will identify bacteria with the desired attTn7 site insertion.
Homologous recombination by transformation with linear DNA
When designing PCR products for linear DNA transformation, at least 500 bp of homologous DNA flanking the antibiotic resistance cassette must be included. For increased transformation efficiencies, 800–1000 bp may be used. For preparation of the bacteria to be transformed, it is not critical for the M63 culture to reach OD600=0.5 exactly. A range of mid-late log phase culture densities (OD600=0.3–0.8) can result in successful transformations, provided that sufficient numbers of cells are used. Transformant colonies may not be visible after overnight incubation, but should be visible after 20–24 h at 37°C. Spontaneous antibiotic-resistant mutants will also arise on the “no DNA” control plate, particularly after extended incubation. All plates should be checked periodically between 18–24 h to ensure that transformant colonies can be distinguished from spontaneous mutants. Incubation times >24 h are not recommended as spontaneous mutants will most likely occur and will be difficult to distinguish from transformants.
Flp recombinase-mediated marker excision
If clones are not found that are sensitive to the desired antibiotic, screen additional colonies by repeating steps 5–6 of the protocol. Alternatively (or additionally), colonies obtained in step 4 may be re-streaked onto LSLB/Kan/rhamnose or LSLB/Tet/rhamnose to re-induce Flp production and then screened for marker excision by testing antibiotic sensitivity.
Allelic exchange using sacB counterselection
It is important to ensure that the cointegrants chosen for counterselection start out sucrose-sensitive, as spontaneous sacB mutations occur at a relatively high frequency and will negatively impact counterselection. If all tested cointegrants appear sucrose-resistant, test additional clones or simply repeat the conjugation and try again with fresh conintegrants. YT/Sucrose plates may only be stored at 4°C for 2–3 weeks. Likely due to degradation of the sucrose, prolonged storage will result in plates that are less effective in counterselection. Freshly-prepared YT/Sucrose plates are recommended for each round of allelic exchange.
In theory, homologous recombination at the 5′ and 3′ homology regions of pEXKm5 should occur with equal frequency. Thus, counterselection should yield a 50:50 ratio of clones that contain the desired mutation (having recombined at the 5′ side during cointegration and the 3′ side during counterselection, or vice versa) and those that reverted to the wild-type sequence (having recombined at the same side for both cointegration and counterselection). However, in practice this is not always the case and screening additional “loopout” candidates may be necessary to identify the desired mutant. Initiating counterselection with both cointegrants that have 5′-integrated plasmid as well as those that have 3′-integrated plasmid is often useful in overcoming this problem.
Colony PCR
If no products or nonspecific products of incorrect sizes are observed after PCR, several parameters of the reaction, including DMSO concentration and annealing temperature, may be adjusted. Use of an annealing temperature gradient may be helpful. If reaction optimization is unsuccessful, primer redesign may be necessary, paying careful attention to GC content and melting temperature.
All protocols
Particularly during sacB-mediated allelic exchange (but possible during any genetic manipulation), spontaneous biofilm-deficient B. thailandensis mutants can arise. Mutations should be complemented and marked mutations should be transferred to a fresh genetic background (see Basic Protocol 3) to ensure that phenotypes are not due to secondary mutations.
Anticipated Results
Conjugations (Basic Protocol 1) are efficient in B. thailandensis and should result in numerous antibiotic-resistant transconjugants. Triparental matings are generally less efficient, but still should yield at least 5–30 colonies. Basic Protocol 2 should result in B. thailandensis colonies having an insertion at an attTn7 site. Transformation efficiency with linear DNA (Basic Protocol 3) depends on the DNA used for transformation and is expected to be approximately 2–600 transformants/ng DNA/109 CFU for chromosomal DNA and 0.2–1600 transformants/ng DNA/109 CFU for PCR fragments (Thongdee et al., 2008). Transformations usually result in 3–50 transformant colonies per plate. Marker excision frequencies (via Flp recombinase) are expected to range from 75–96% (Choi et al., 2008), but are often observed at lower rates. Allelic exchange (Basic Protocol 5) should yield B. thailandensis colonies that have the desired chromosomal mutation, insertion, or replacement.
Time Considerations
B. thailandensis colonies should be visible on plates after overnight growth at 37°C, although slightly longer incubation (20–24 h) may be required for plates containing antibiotics. Two days of growth are required for B. thailandensis plates incubated at 30°C or for E. coli RHO3 strains incubated at 37°C. With the exception of colony PCR, which will take several hours, all of the protocols described in this unit are completed over multiple days. Conjugation and mini-Tn7 insertion can be completed in 4 days (including streaking the strains from frozen stocks) and transformation with linear DNA will take 4 days to complete (5 days including streaking B. thailandensis from a frozen stock). Removal of an antibiotic resistance cassette by flp-recombination can be completed in 11 days. Allelic exchange using sacB counterselection can be completed 11 days and, although more time may be required if additional screening steps are required (for example, screening for sucrose-sensitive colonies).
Acknowledgments
Research reported in this unit was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number K22 AI118949. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
INTERNET RESOURCES
The Burkholderia Genome Database contains a comprehensive collection of sequences, annotations, and comparative tools for completed and draft genomes within the Burkholderia genus, including B. thailandensis.
http://tools.nwrce.org/tn_mutants/
A useful tool for B. thailandensis researchers, the laboratory of Colin Manoil has generated a near-saturation transposon mutant library in B. thailandensis E264, consisting of 87,000 unique mutants (7.5 mutants per gene) (Gallagher et al., 2013). The Transposon Mutant Library Browser displays available mutants and provides links for information on how to request mutants.
LITERATURE CITED
- Anderson MS, Garcia EC, Cotter PA. The Burkholderia bcpAIOB genes define unique classes of Two-Partner secretion and contact dependent growth inhibition systems. PLoS Genetics. 2012;8:e1002877. doi: 10.1371/journal.pgen.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AR, Kang Y, Inamasu KS, Son MS, Vukovich JM, Hoang TT. Genetic tools for allelic replacement in Burkholderia species. Applied and Environmental Microbiology. 2008;74:4498–4508. doi: 10.1128/AEM.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JR, Duerkop BA, Hinz A, West TE, Herman JP, Churchill MEA, Skerrett SJ, Greenberg EP. Mutational analysis of Burkholderia thailandensis quorum sensing and self-aggregation. Journal of Bacteriology. 2009;191:5901–5909. doi: 10.1128/JB.00591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. A Tn7-based broad-range bacterial cloning and expression system. Nature Methods. 2005;2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Applied and Environmental Microbiology. 2008;74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CT, Toesca I, Wu TH, Teslaa T, Beaty SM, Wong W, Liu M, Chiou PY, Teitell MA, Miller JF. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10295–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Patrapuvich R, Weiss E, Brittnacher M, Manoil C. Sequence-defined transposon mutant library of Burkholderia thailandensis. mBio. 2013;4:e00604–13. doi: 10.1128/mBio.00604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Molecular Microbiology. 2013;89:1213–1225. doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad MA, Zajdowicz SL, Holmes RK, Voskuil MI. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene. 2009;430:123–131. doi: 10.1016/j.gene.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infection and Immunity. 2008;76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infection and Immunity. 2000;68:5377–5384. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue CA, Peak IRA, Beacham IR. Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. Journal of Microbiological Methods. 2009;76:320–323. doi: 10.1016/j.mimet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- López CM, Rholl DA, Trunck LS, Schweizer HP. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Applied and Environmental Microbiology. 2009;75:6496. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris MH, Kang Y, Wilcox B, Hoang TT. Stable, site-specific fluorescent tagging constructs optimized for Burkholderia species. Applied and Environmental Microbiology. 2010;76:7635–7640. doi: 10.1128/AEM.01188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE. Tn7. Microbiology Spectrum. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0010-2014. MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Research. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathogens. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Ulrich RL, Taylor LA, Wood MW, Deshazer D, Stevens MP, Galyov EE. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. Journal of Bacteriology. 2005;187:7857–7862. doi: 10.1128/JB.187.22.7857-7862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Applied and Environmental Microbiology. 2008;74:2985–2989. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, et al. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 2008;9:566. doi: 10.1186/1471-2164-9-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, et al. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiology. 2006;6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]