Abstract
Patients with human papillomavirus (HPV)-positive head and neck squamous cell carcinoma (HNSCC) have better responses to radiotherapy and higher overall survival rates than do patients with HPV-negative HNSCC, but the mechanisms underlying this phenomenon are unknown. P16 is used as a surrogate marker for HPV infection. Our goal was to examine the role of p16 in HPV-related favorable treatment outcomes and to investigate the mechanisms by which p16 may regulate radiosensitivity. HNSCC cells and xenografts (HPV/p16-positive and -negative) were used. P16-overexpressing and shRNA knockdown cells were generated, and the effect of p16 on radiosensitivity was determined by clonogenic cell survival and tumor growth delay assays. DNA double-strand breaks (DSBs) were assessed by immunofluorescence analysis of 53BP1 foci; DSB levels were determined by neutral comet assay; western blotting was used to evaluate protein changes; changes in protein half-life were tested with a cycloheximide assay; gene expression was examined by real-time polymerase chain reaction (PCR); and data from The Cancer Genome Atlas HNSCC project were analyzed. P16 overexpression led to downregulation of TRIP12, which in turn led to increased RNF168 levels, repressed DNA damage repair (DDR), increased 53BP1 foci, and enhanced radioresponsiveness. Inhibition of TRIP12 expression further led to radiosensitization, and overexpression of TRIP12 was associated with poor survival in patients with HPV-positive HNSCC. These findings reveal that p16 participates in radiosensitization through influencing DDR and support the rationale of blocking TRIP12 to improve radiotherapy outcomes.
Keywords: radiosensitivity, p16, TRIP12, human papillomavirus, head and neck cancer
INTRODUCTION
Patients with HPV-positive stage III–IV head and neck squamous cell carcinoma (HNSCC) are known to have a more favorable prognosis after radiation therapy (RT), with or without chemotherapy, than do those with HPV-negative stage III–IV HNSCC (5-year overall survival rates >80% versus 40%) (1,2). However, the mechanisms that underlie this phenomenon remain unknown. Clinically, p16 is used as a surrogate marker for HPV infection in HNSCC (2–4) because its expression is induced by HPV oncoprotein E7-related pRb inactivation and degradation (5,6). As a tumor suppressor, p16 has multiple biological functions, including cell cycle regulation (5); cancer cell RT sensitization (7–9); and radiation-induced senescence (10). Several studies have also reported that even in the absence of demonstrable HPV positivity, p16 positivity in patients with HNSCC is associated with improved survival (11). However, the specific role of p16 overexpression in the context of the radiosensitivity of HPV-related HNSCC is not clear.
Recently, HPV-positive HNSCC cells were found to sustain more RT-induced DNA damage than HPV-negative cells (9,12). In cervical cancer, the distribution of 53BP1 nuclear foci was similar to that of punctate HPV signals and p16 expression (13). In HPV-positive anal carcinoma, markedly enlarged 53BP1 foci were accompanied by dramatically increased RNF168 levels (14). As a DNA damage repair (DDR) mediator and marker of unrepaired double-strand breaks (DSBs) (15–20), 53BP1 is recruited to DNA damage sites and is regulated by RNF168 (21,22). These observations suggest that HPV influences the effectiveness of RT through RNF168-53BP1 pathway-regulated DDR. As a DDR-related protein (14,23–25) and a suppressor of RNF168, TRIP12 prevents excessive spreading of ubiquitinated chromatin at damaged chromosomes (14). However, whether TRIP12 is involved in the favorable prognosis of HPV-positive HNSCC and its relationship with p16 in this process remain unknown. Because p16 mutation and methylation are widely present in cancer (26–30), determining the role and mechanisms of p16 in radiation effects may reveal previously unidentified mediators of p16 and RT sensitization targets, leading to improved treatment outcomes.
In the current study, we used in vitro HNSCC cell models and in vivo HNSCC tumor xenografts to determine the role of p16 in HPV-positive RT sensitivity and explored the mechanisms of this effect. We found that p16 expression enhanced the RT responsiveness of HNSCC both in vitro and in vivo. In addition, p16 affected DDR via the TRIP12-RNF168-53BP1 cascade by suppressing TRIP12. We also verified that knockdown of TRIP12 rendered HNSCC cells sensitive to RT and delayed DDR. Finally, we found that in patients with HPV-positive head and neck cancer from the Cancer Genome Atlas, overexpression of TRIP12 was associated with poor outcome.
RESULTS
p16 sensitizes HNSCC cells and tumor xenografts to RT
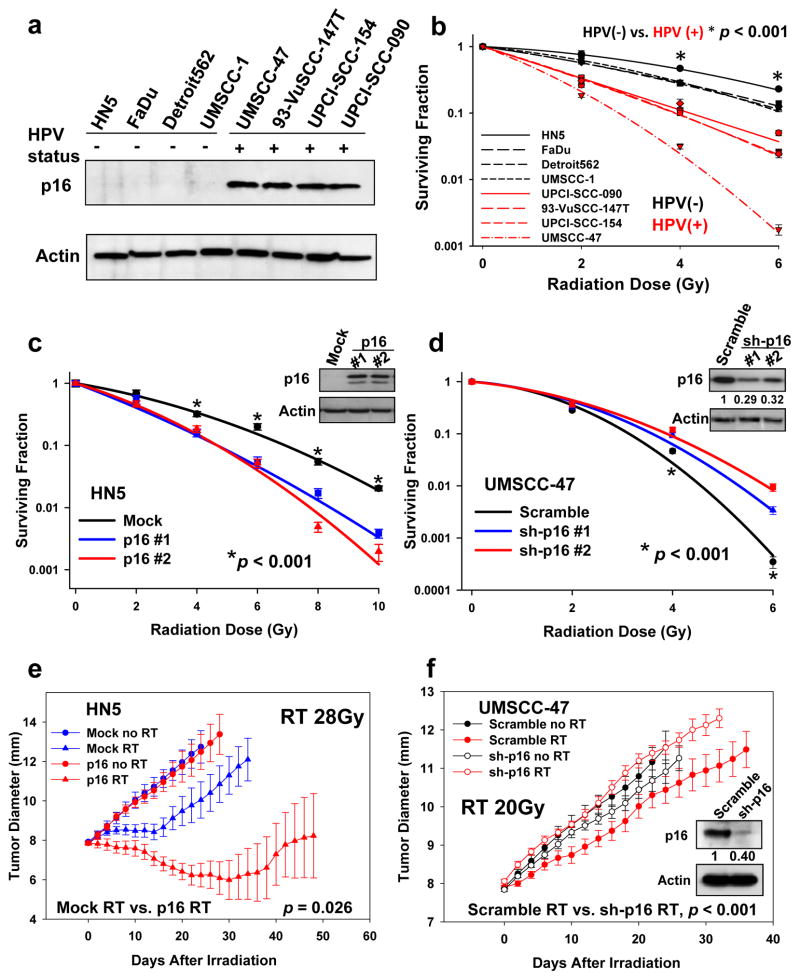
The HPV-negative head and neck cancer cell lines HN5, FaDu, UMSCC-1, and Detroit 562 and the HPV-positive head and neck cancer cell lines UMSCC-47, 93-VuSCC-147T, UPCI-SCC-154, and UPCI-SCC-090 were used (Fig. 1a). The treatment endpoint was clonogenic cell survival, which is the benchmark readout for in vitro RT sensitivity (31). As previously observed, HPV-positive HNSCC cells were more radiosensitive than HPV-negative cells (Fig. 1b). To investigate the role of p16 in HPV-positive RT enhancement, we generated p16-overexpressing cells (HN-5 and UMSCC-1) and p16 shRNA stable knockdown cells (UMSCC-47 and UPCI-SCC-154). Compared with scramble control cells, p16 overexpression in HPV-negative HN5 or UMSCC-1 cells led to significant radiosensitization (Fig. 1c and S1a), whereas knockdown of p16 rendered UMSCC-47 and UPCI-SCC-154 cells more radioresistant (Fig. 1d and S1b). To exclude the possibility that the above effects are actually due to the function of p53, we tested the radiosensitivity changes of the HPV-positive UMSCC-47 cells following siRNA inhibition of p53. We found that that following siRNA inhibition of p53, no significant change in radiosensitivity was observed in HPV-positive UMSCC-47 cells (Fig. S1c and d). This may due to the fact that p53 is inactivated by HPV E6 oncoprotein via several mechanisms (32–34). Because p16 is necessary for the survival of some HPV-positive cervical carcinoma cells (35), to exclude the possibility that loss of p16 may have led to slower growth of the HPV-positive HNSCC cells and subsequently to reduced sensitivity to RT, we assessed proliferation rates of HPV-positive UMSCC-47, UPCI-SCC-152, and UPCI-SCC-154 cells after ablation of p16 expression, using cell doubling time as an end point. Depletion of p16 expression had no influence on the proliferation rates of these HNSCC cell lines (Fig. S2a–c). The HPV-positive cervical carcinoma cell line SiHa, whose proliferation has been previously shown to be affected by p16 (35), served as a positive control (Fig. S2d). These observations indicated that p16 does not influence HNSCC cell radiosensitivity via cell growth rates.
Figure 1.
Effect of p16 on radiosensitivity. a, HPV and p16 status of the cell lines. b, HPV/p16-positive UPCI-SCC-154, UPCI-SCC-090, 93-VuSCC-147T and UMSCC-47 cells versus HPV/p16-negative HN-5, FaDu, UMSCC-1 and Detroit 562 cells. c and d, HN5 cells and UMSCC-47 cells treated with irradiation. Percentages of surviving cell colonies were normalized to mock or scramble vector control cells with no radiation. Values shown are the means + SE of three independent experiments. e, Xenografts generated by HN5 with p16-overexpressing cells. Growth delay during times of increasing diameter was significantly greater for HN5-p16 RT than for the Mock RT condition (unirradiated empty vector control) (p=0.026). f, Xenografts generated by UMSCC-47 cells with p16 knockdown. Growth delay was significantly greater (p<0.001) for Scramble RT than for sh-p16 RT at each diameter (9, 10, 11 etc. mm). Growth rate was significantly greater for sh-p16 RT (p<0.001). Each data point in the radiation-induced tumor growth delay curve represents means of 3 to 6 mice; bars, SE.
We further validated our findings in mice bearing HN5 and UMSCC-47 tumor xenografts by using an RT tumor growth delay assay (36). Clinically, HPV-positive tumors are more sensitive to RT than are HPV-negative tumors. Thus, in the current study, the HPV-positive UMSCC-47 tumors were irradiated for 5 days to a total dose of 20 Gy, and the HPV-negative HN5 tumors were irradiated for 7 days to a total dose of 28 Gy. In the HN5 tumors, forced expression of p16 had minimal effects on tumor growth in the absence of RT, but led to profound radiosensitization (Fig. 1e; red triangles vs. blue triangles for HN5, p<0.001). Conversely, inhibition of p16 expression in UMSCC-47 tumors led to radioresistance (Fig. 1f, red filled circle vs. red blank circle for UMSCC-47, p=0.002). Western blot analysis of tumor tissue lysates confirmed that p16 knockdown was retained throughout this tumor radiosensitivity study (Fig. 1f).
Together, these findings show that p16 expression in HNSCC cells can at least partially mediate favorable radiation response both in vitro and in tumor xenografts.
p16 does not regulate RT response through the CDK4/CDK6 pathway
One established function of p16 is the p16/cyclin D1/cdk4/pRb cell cycle regulatory cascade (5). To determine whether p16 regulates RT response through this pathway, we used the CDK4/CDK6-specific inhibitor PD0332991, which is being evaluated in phase II clinical trials (37). The PD0332991 dose that completely blocked CDK4/CDK6 activity was verified by western blot analysis (Fig. S3a). PD0332991 did not affect RT sensitivity in scrambled or p16-overexpressed HN5 cells (Fig. S3b), indicating that p16 does not regulate RT sensitivity through the p16/CDK4/cyclin D1/Rb pathway.
p16 regulates RT response by influencing DDR
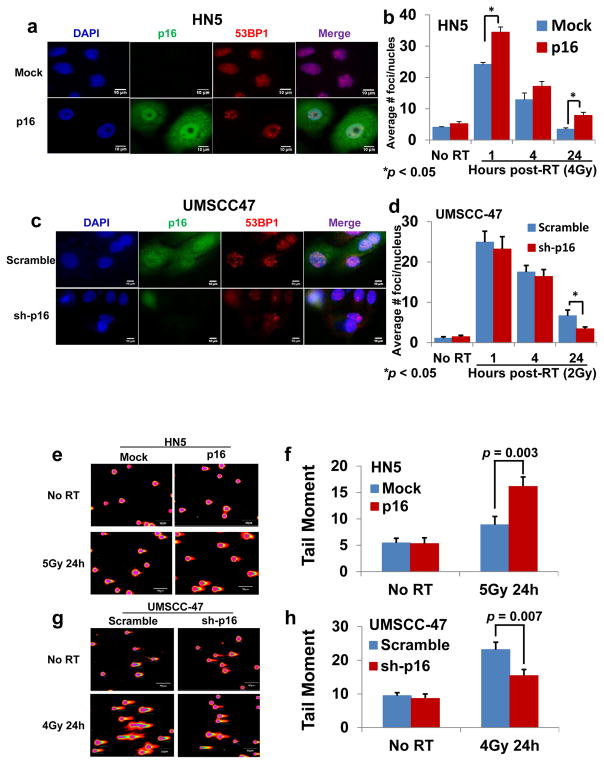
The persistence of 53BP1 foci indicates unrepaired DSBs (15–20). In p16-overexpressing HPV-negative HN5 cells, but not in mock control cells, 53BP1 foci persisted for 24 hours after 4 Gy γ-irradiation (Fig. 2a and b). Conversely, in scramble control HPV-positive UMSCC-47 cells, but not in p16-shRNA-knockdown cells, 53BP1 foci persisted for 24 hours after 2 Gy γ-irradiation (Fig. 2c and d). These findings indicate that cells that express p16 are less able to repair DNA DSBs.
Figure 2.
Effect of p16 on the kinetics of nuclear 53BP1 foci formation (a–d) and of DNA damage assessed by neutral comet assay (e–h). a and c, representative nuclei from 24 hours after radiation (RT). b and d, numbers of 53BP1 foci. e and g, representative cell nucleus fluorescent image. f and h, quantification of tail moment. Data are shown as means ± SE from three independent experiments.
To confirm this result, we used a neutral comet assay to measure levels of DSBs 24 hours after RT. P16-overexpressing HPV-negative HN5 cells exhibited a significant increase in the comet tail moment (percentage of DNA in the tail × tail length) (38) compared with the control mock cells (Fig. 2e and f). Conversely, the scramble control HPV-positive UMSCC-47 cells exhibited a significant increase in the comet tail moment compared with p16-shRNA-knockdown cells (Fig. 2g and h). The results of 53BP1 staining (Fig. 2a–d) and neutral comet assays (Fig. 2e–h) demonstrated that p16 is a negative regulator of RT-induced DDR in HNSCC cells.
p16 regulates the DDR-related TRIP12-RNF168-53BP1 pathway
RNF168 is a positive regulator of 53BP1 (21,22); in addition, in HPV-positive carcinoma, markedly enlarged 53BP1 foci have been shown to be accompanied by greatly increased RNF168 levels (14). On the basis of these findings, we investigated whether p16 regulates DDR through this DDR-related RNF168-53BP1 pathway. We found that p16 expression was associated with RNF168 protein levels (Fig. 3a and b, RNF168 antibody was verified in S4a). Interestingly, TRIP12, a negative regulator of RNF168 (14), was inversely related to p16 expression (Fig. 3a and b). This inverse association with p16 was observed in additional head and neck cancer cell lines (Fig. S4b and c) and in UMSCC-47 xenograft tumors (Fig. 3c). These previously unidentified observations suggest that p16 modulates the TRIP12-RNF168-53BP1 pathway. In this loop, TRIP12 regulates RNF168, causing changes in 53BP1 foci. Thus, suppression of TRIP12’s function may be a mechanism underlying p16-mediated RT sensitization.
Figure 3.
Effect of p16 on protein changes. a and b, cells were irradiated (RT) with a single 4-Gy dose. Samples were collected at 24 hours (h) after RT. c, western blots from UMSCC-47 xenografts. Numbers shown below the blots represent the relative density ratios of bands. HN5 Vector control (Mock), P16 overexpression (P16). The experiment shown was replicated for three times.
p16 regulates TRIP12 at the post-translational level
We found that the presence of p16 expression downregulated the expression of TRIP12 protein (Fig. 3a–c, S4b and c). To determine the link between p16 and TRIP12 expression, we examined the influence of p16 on TRIP12 mRNA expression by using a polymerase chain reaction (PCR) assay and the protein half-life of TRIP12 by using a standard protein-synthesis-inhibitor cycloheximide assay. We found that p16 overexpression appeared to reduce the half-life of TRIP12 protein (Fig. 4a, b and S5a) but did not reduce TRIP12 mRNA levels (Fig. S5c).
Figure 4.
Effect of p16 on TRIP12 half-life. a and b, HN5 cells were treated with cycloheximide (CHX; 50 μg/mL) for different durations as indicated. HN5 Vector control (Mock), P16 overexpression (P16). The experiment shown was replicated for three times.
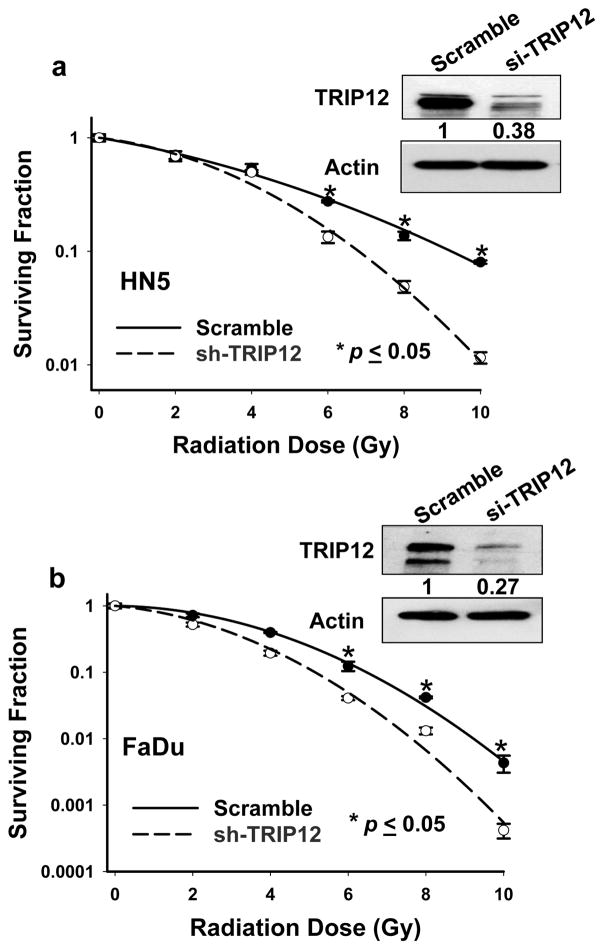
Silencing TRIP12 leads to enhanced radioresponsiveness through influencing DDR
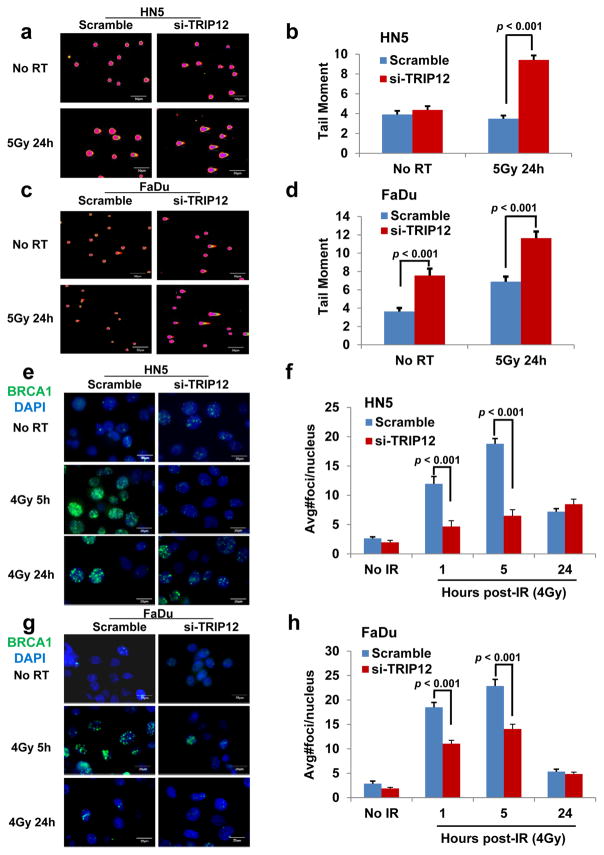
TRIP12 is known to influence the DDR (14,23–25). However, its role in regulating RT sensitivity and the mechanisms of this effect on DDR have not been established. We assessed the influence of TRIP12 on RT response in HNSCC HN5 and FaDu cells. Knockdown of Trip12 sensitized HN5 and FaDu cells to RT, which recapitulated the effect of p16 overexpression (Fig. 5a and b). To confirm this result, we used a neutral comet assay to measure levels of DSBs at 24 hours after RT. Trip12 knockdown in HN5 and FaDu cells led to significant increases in comet tail moment compared with the control scrambled cells (Fig. 6a–d). These findings indicate that cells with depleted TRIP12 expression have persistent DSBs and are less able to repair DNA DSBs. Because BRCA1 has an important role in DDR (39), especially in homologous recombination (HR)–mediated repair (40), we also examined BRCA foci after RT. We observed induction of BRCA1 foci at 1 and 5 hours after 4 Gy γ-irradiation in HN5 and FaDu scramble control cells, and UMSCC-47 p16 knockdown cells, but the levels of these foci were reduced in the TRIP12 knockdown HN5 and FaDu cells, and UMSCC-47 scramble control cells (Fig. 6e–h and S6a–c, suggesting that TRIP12 regulates DDR through influencing HR–mediated repair.
Figure 5.
Effect of TRIP12 on HN5 and FaDu cell radiosensitivity. a and b, percentages of surviving cell colonies were normalized to those of non-coding cells (scramble). Values shown are the means ± SE from three independent experiments.
Figure 6.
Effect of TRIP12 on DNA damage [assessed by neutral comet assay (a–d)] and on the kinetics of nuclear BRCA1 foci formation (e–h). a and c, representative cell nucleus fluorescent images. b and d, quantification of tail moment. e and g, representative nuclei from 24 hours after radiation (RT). f and h, numbers of BRCA1 foci. Shown are means ± SE from three independent experiments.
TRIP12 expression is associated with poor treatment outcome
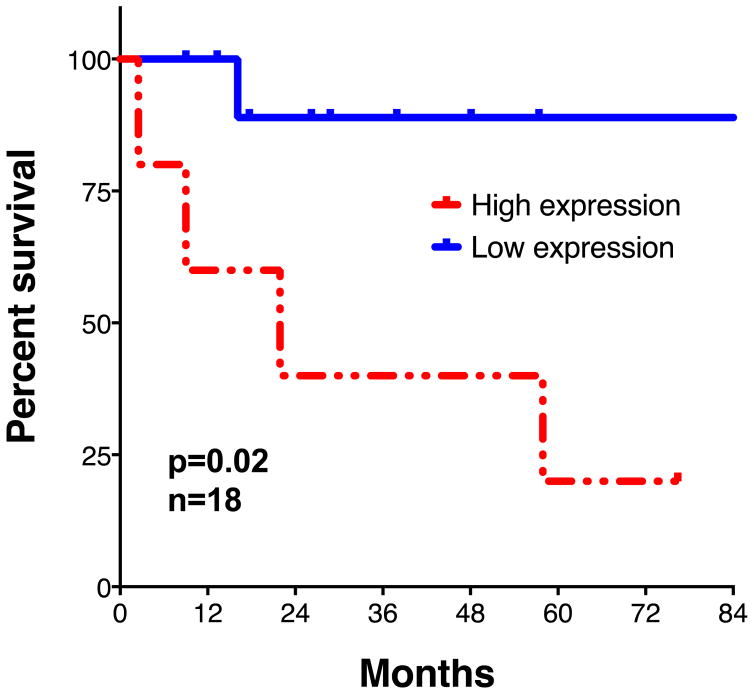
To investigate the influence of TRIP12 on clinical treatment outcomes, we examined potential correlations between TRIP12 expression status and overall survival times of patients with HPV-positive HNSCC in The Cancer Genome Atlas database. In this pilot study, outcomes were poorer for 18 patients with high-TRIP12-expressing tumors than for another 18 patients with low-TRIP12-expressing tumors (Fig. 7), indicating that the radioresistance conferred by TRIP12 observed in vitro may be associated with poor outcome in patients with HNSCC.
Figure 7.
TRIP12 protein expression level and patient overall survival. Overall survival of patients with HPV-positive head and neck squamous carcinoma for tumors expressing different levels of TRIP12 protein expression in The Cancer Genome Atlas database (N=18).
DISCUSSION
In our study, p16 overexpression sensitized HPV-negative HN5 cells and tumor xenografts to radiation. Moreover, even in the presence of HPV, inhibition of p16 in UMSCC-47 cells and tumor xenografts led to radioresistance. Our in vitro findings are consistent with a recently reported in vitro study (9) and are validated in our tumor xenograft experiments. On the basis of these results, we conclude that p16 has a key role in HPV-related RT enhancement.
The tumor suppressor p16 has multiple biological functions and has been extensively studied. Even though it has been shown to lead to radiosensitization in several cell lines (7–9), its role in regulating radiosensitivity, particularly in the context of HPV-positive HNSCC, had not been elucidated, particularly in the context of DDR regulation. In the current study, we found that p16 enhanced the radiosensitivity of HNSCC by influencing DDR.
As an important DDR mediator and marker of unrepaired DSBs (15–20), 53BP1 is recruited to DNA damage sites and regulated by RNF168 (21,22). In HPV-positive cervical cancer, the distribution of 53BP1 foci is similar to that of p16 overexpression (12), and in HPV-positive anal carcinoma, markedly enlarged 53BP1 foci are accompanied by drastically increased RNF168 levels (13); however, the direct link between p16 and 53BP1-RNF168 was unclear. In the current study, we found that p16 overexpression not only caused persistence of 53BP1 foci 24 hours after RT but the protein levels of RNF168 were also modulated in a similar fashion. The converse was true in cells with inhibited p16. RNF168 is a histone E3 ubiquitin ligase with important positive roles in the DSB repair process, including avoiding collisions between transcription and repair intermediates (41), facilitating fusion of uncapped telomeres (42), and forming G1 nuclear bodies (43,44). However, RNF168 can amplify ubiquitin conjugates, generated by its own activity, that progressively spread away from the DSBs to undamaged chromatin and lead to uncontrolled amplification of chromatin ubiquitylation (21,45,46). TRIP12 is a known E3 ligase of ARF and a regulator of RNF168 with an important role in regulating DDR (14,23–25). By controlling the accumulation of RNF168, TRIP12 prevents excessive spreading of ubiquitinated chromatin at RNF168-damaged chromosomes (14). However, the mechanisms that regulate TRIP12 were unknown. In the current study, we found that TRIP12 was inversely related to p16 expression and consistent with RNF168-53BP1 and p16-induced DDR changes. Thus we concluded that mechanistically, p16 may affect DDR through the TRIP12-RNF168-53BP1 cascade and that suppression of TRIP12’s function is a mechanism underlying p16-mediated radiosensitization (Fig. 8). In terms of how p16 negatively controls TRIP12 protein levels, our results suggest that p16 may suppress TRIP12 through post-translational modification, although this will require confirmation in future studies. As for how p16 might affect the stability of TRIP12, p16 is not an ubiquitin E3 ligase and thus is unlikely to regulate TRIP12 degradation directly; rather, we believe intervening E3 ligases or deubiquitinases are involved. We are currently in the process of investigating this hypothesis.
Figure 8.
Working model of the regulation of radiosensitivity and DNA damage repair by p16.
P16 was recently found to regulate HR-mediated DDR in vitro by downregulating expression of cyclin D1 protein (9). E2F1 depletion has also been shown to downregulate cyclin D1 expression (47). In our study, we found that E2F1 gene expression was downregulated by TRIP12 knockdown (Table S1). Colllectively, these results suggest that p16 regulates HR-mediated DDR through p16-TRIP12-RNF168-53BP1 and p16-TRIP12-E2F1-cyclin D1 pathway loops and that TRIP12 is a previously unidentified key mediator of p16’s functions in both signaling cascades. TRIP12 has been reported to influence DDR (14,23–25). However, its role in regulating RT sensitivity has been unclear, and the mechanisms of this effect have not been established. In the current study, knockdown of TRIP12 rendered HNSCC cells sensitive to RT by influencing HR-mediated DDR, suggesting that TRIP12 is a target for RT sensitization.
Finally, we analyzed samples from The Cancer Genome Atlas database and found that high TRIP12 expression was associated with poor outcome in patients with HPV-positive HNSCC. Even though TRIP12 is known to participate in regulating DDR (14,23–25), the relationship of TRIP12 to clinical treatment outcomes had been unknown. Although limited by small numbers of patients, our findings indicate that TRIP12 may be a clinically relevant biomarker of radioresistance in HPV-positive disease and a potential treatment target for overcoming radioresistance.
In conclusion, we confirmed that p16 has a key role in HPV-related favorable treatment outcomes and that TRIP12 mediates p16-related radiation enhancement effects. P16 causes prolonged DDR and leads to RT sensitization. Knockdown of TRIP12 leads to an enhanced response to RT by influencing HR-mediated DDR. High levels of TRIP12 expression are associated with poor outcomes in patients with HPV-positive HNSCC. Because HPV infection is not a feature of most types of cancer, targeting TRIP12 may be a strategy for improving treatment outcomes in such patients.
MATERIALS AND METHODS
Cell lines
HN5, FaDu, UMSCC-1, Detroit 562, UMSCC-47, UPCI-SCC-154, and UPCI-SCC-090 cells were obtained from Dr. Jeffery Myers, The University of Texas MD Anderson Cancer Center, Houston, TX. 93-VuSCC-147T cells were obtained from Dr. Peter J. F. Snijders, UV University Medical Center, Amsterdam, The Netherlands. The identities of all cell lines were confirmed by genotyping (STR profiling) at MD Anderson’s Characterized Cell Line Core Facility (NCI CA016672). Other than UMSCC-47 cells, all cells were mycoplasma negative. HN5 and UMSCC-1 cells were maintained in DMEM/F-12 medium (Mediatech). FaDu, Detroit 562, UPCI-SCC-154 and UPCI-SCC-090 cells were maintained in MEM (Gibco). UMSCC-47 and 93-VuSCC-147T cells were maintained in DMEM (Gibco) supplemented with 2% MEM vitamins (Gibco), 1% sodium pyruvate (Lonza, Houston, TX, USA), and 1% non-essential amino acids (Gibco). All media were supplemented with 10% fetal bovine serum (Sigma) and 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco).
Plasmids and shRNA
CDKN2A cDNA was amplified by PCR from MB157 cells and cloned into the pLOC expression vector (Thermo Scientific). The human CDKN2A shRNAs were from Open Biosystems through MD Anderson’s shRNA and ORFeome Core (NCI CA016672): V2LHS_195839 (5′-TTCTTCCTCCGGTGCTGGC-3′), V3LHS_317755 (5′-GTCTGAGGGACCTTCCGCG-3′), and V3LHS_343118 (5′-CTTCTAGGAAGCGGCTGCT-3′).
siRNA transfection
P16-siRNA (GAUCAUCAGUCACCGAAGG, Thermo Scientific), Trip12-siRNA (GAACACAGAUGGUGCGAUA, Thermo Scientific), RNF168-siRNA (GACACUUUCUCCACAGAUA, Thermo Scientific) or TP53 (7157)-siRNA (GAAAUUUGCGUGUGGAGUA, Dharmacon) were transfected by electroporation (Nucleofector II, Amaxa) by using program T-001 with transfection reagent T (Lonza). Cells were collected 48 hours later for western blot analysis to validate transfection efficiency. Cells were irradiated 48 h after transfection.
Immunocytochemical analysis
Cells were plated on coverslips that were placed in 35-mm culture dishes. At specified time points after exposure to 4 Gy radiation, cells were fixed in 4% paraformaldehyde for 10 minutes at ambient temperature, briefly washed in phosphate-buffered saline (Mediatech), and placed in 70% ethanol overnight at 4ºC. Fixed cells were permeabilized with 0.1% Igepal for 20 minutes at ambient temperature, blocked in 2% bovine serum albumin (Sigma) for 1 h, and then incubated in anti-53BP1 (1:200, Cell Signaling) or anti-BRCA1 primary antibody (1:500, Santa Cruz) overnight at 4ºC. Cells were then washed four times with phosphate-buffered saline and then incubated for 1 hour in secondary anti-rabbit antibody conjugated to Cy3 (Jackson ImmunoResearch) for 53BP1 foci or in secondary anti-mouse antibody conjugated to FITC or Cy3 (Jackson ImmunoResearch) to visualize immunoreactivity. DNA was stained with 4′,6-diamidino-2-phenylindole (1:1000, Sigma). Immunoreactions were visualized with a Leica Microsystems microscope (Wetzlar, Germany), and foci were counted manually by using ImageJ software (rsbweb.nih.gov/ij/).
Clonogenic survival assay
Colony-forming ability was assayed as described previously (31). Briefly, exponentially growing cells were replated in specified numbers into six-well tissue culture dishes for each condition under investigation. Cells were exposed to a single dose of irradiation with a Mark I-68A 137Cs irradiator (JL Shepherd and Associates, San Fernando, CA) with the indicated doses and incubated for 10 days (HN5 and UMSCC-1), 14 days (FaDu and Detroit 562), 17 days (UMSCC-47 and 93-VuSCC-147T), or 20 days (UPCI-SCC-154 and UPCI-SCC-090). Colonies were stained with crystal violet and colonies consisting of more than 50 cells were counted. The survival fraction was calculated as (number of colonies/number of cells plated) irradiated / (number of colonies/number of cells plated) non-irradiated.
Neutral comet assay
DNA damage was assessed with a single-cell gel electrophoresis assay under neutral conditions with a CometAssay kit (Trevigen, 4250-050-K) according to the manufacturer’s protocol. Briefly, cells were harvested at 24 hours after 4 or 5 Gy γ-irradiation; mixed with agarose, which has a low melting point; and plated on the CometSlide. Cells were lysed overnight at 4°C, subjected to electrophoresis at 23 V for 1 hour under neutral conditions, and stained with SYBR Gold. The presence of comet tails was determined with a Leica fluorescence microscope. The tail moment (38) was calculated as: (percentage of the DNA in the tail) × (tail length), where the percentage of DNA in the tail and tail length were quantified with TriTek CometScore software (http://autocomet.com/index.php).
Western blot analysis
After irradiation, lysates were obtained at various time points and sonicated (QSonica) for 2 minutes in 0.4 M NaCl/20 mM HEPES buffer containing 1% Igepal, 0.1 mM ethylene glycol tetraacetic acid and ethylenediaminetetraacetic acid, 0.1 M dithiothreitol, 0.1 M phenylmethanesulfonylfluoride, and protease and phosphatase inhibitors (Sigma). Protein concentrations were measured with a DC protein assay kit (Bio-Rad, Hercules, CA, USA). Equalized proteins were subjected to electrophoresis at 60 mA in polyacrylamide pre-cast gels (Bio-Rad) and electrotransferred onto polyvinylidene fluoride membranes (Bio-Rad) at 4ºC for 1 hour at 100 V. Membranes were blocked with 5% milk (Bio-Rad). Antibodies were incubated overnight in 5% milk at various concentrations (P16, BD Bioscience, 1:5000; TRIP12, Santa Cruz, 1:1000; RNF168, Sigma, 1:1000; pRb, CST, 1:500; Rb, CST, 1:1000). Membranes were washed in Tris-buffered saline (Bio-Rad) with 0.1% Tween20 (Sigma) and incubated for 45 minutes at ambient temperature in 1:2000 anti-rabbit or anti-mouse IgG (GE Healthcare). Immunoreactions were visualized with the ECL2 system (Thermo Scientific) and then immediately exposed to autoradiographic film (Denville) for various periods. Images were scanned with an HP Scanjet 5550c and quantified with ImageJ software (http://rsbweb.nih.gov/ij/).
Protein half-life detection
The half-life of TRIP12 was tested by using a standard protein-synthesis inhibitor cycloheximide assay as previously described (48). Briefly, control cells or cells overexpressing p16 were treated with 50 μg/mL cycloheximide for the indicated times before lysis, and the expression levels of TRIP12, p16, and glyceraldehyde 3-phosphate dehydrogenase were detected by western blot analysis.
RNA isolation and real-time PCR with reverse transcription
Total RNA was isolated with a mirVana RNA Isolation Kit (Ambion) and then reverse-transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA was used for quantitative PCR with TaqMan Gene Expression Assays (Applied Biosystems), and data were normalized to an endogenous control (glyceraldehyde 3-phosphate dehydrogenase). Real-time PCR and data collection were done with a CFX96 instrument (Bio-Rad).
Tumor radiosensitivity study
Animal experiments were done as previously described (36), in accordance with a protocol approved by the Institutional Animal Care and Use Committee of MD Anderson. Mice were euthanized when they met the institutional euthanasia criteria for tumor size and overall health condition. Tumors were introduced by intramuscular injection of 1–3 × 106 control (mock) or p16-overexpressing HN5 tumor cells or p16-depleted (shp16) or control (scramble) UMSCC-47 tumor cells into the hind legs of 3- to 4-month-old NCR nu/nu mice. Three mutually orthogonal tumor diameters were measured every other day, and mean values were used to quantify tumor regrowth after treatment when tumors reached 8.0 mm (range 7.7–8.2 mm) in diameter. Mice were randomly assigned to groups (using flipping group numbers, e.g., #1- empty vector no irradiation; #2- empty vector irradiation; #3- gene changes no irradiation; #4- gene changes plus irradiation) without blinding and treated or not treated with fractionated RT. Fractionated radiation (2 Gy per fraction, twice daily for 7 or 5 consecutive days) was delivered to the tumor-bearing limbs of mice by using an irradiator (Co-V, Theratron 780; MDS Nordion, Ottawa, Ontario, Canada) with a cobalt-60 source (field size, 10 × 10 cm; source axis distance, 64.9 cm), at a dose rate of 0.955 Gy/min. During irradiation, unanesthetized mice were mechanically immobilized in a jig so that the tumor was exposed in the radiation field and the animal’s body was shielded from radiation exposure.
Clinical data analysis
Samples from patients with HPV-positive HNSCC were identified from The Cancer Genome Atlas HNSCC project by using the cBioPortal for Cancer Genomics (49,50). This resource includes data on TRIP12 mRNA expression and survival outcomes for 18 patients with HPV-positive HNSCC.
Statistical analysis
Each experiment was repeated three times or more. Unless otherwise noted, data are presented as mean ± standard error of the mean (SEM), and Student’s t tests (unpaired, unequal variance) were used to compare two groups of independent samples for in vitro radiosensitivity, 53BP1 foci, and comet tail moment. Differences in tumor growth delay (Fig. 1e and 1f) were tested with nonparametric bootstrapping of the measured diameters. Delay was estimated as different times to a fixed diameter from linear fits of the two growth curves (Fig. 1e: Mock RT vs. HN5-p16 RT; Fig. 1f: Scramble RT vs. sh-p16 RT). Data were confined to times of increasing diameter; these were the times of increasing diameter in Figure 1e, as opposed to all data in Figure 1f. Differences in tumor growth rates (Fig. 1f) were tested by first confirming that the increase in tumor diameter was linear in time (coefficient of t2 in fit was not significant), then by fitting a linear model that included a parameter that distinguished between the slopes of the two lines. The p value of this parameter was the test of significance of difference in growth rates. For clinical analysis, the upper tertile of TRIP12 expression was used as a cutoff between high and low expression. Group comparisons were done with the Kaplan-Meier method and log-rank tests. Statistical analyses of the clinical data were done with Graph Pad Prism (v6.0), whereas STATA 11 (College Station TX) was used to analyze animal study data. p<0.05 was considered statistically significant in all analyses.
Supplementary Material
Acknowledgments
Financial support: This work was supported by grants R01CA168485 and R01CA181029 from the National Cancer Institute (REM and KKA) and (LM) respectively, the Gilbert H. Fletcher Chair (KKA), Cancer Center Support Grant (P30 NCI CA016672), Individual Investigator Research Award RP150293 from the Cancer Prevention Research Institute of Texas (HDS), an MD Anderson Cancer Head and Cancer SPORE Career Development Award (HDS), and a Center for Radiation Oncology Research Development Award (HDS). The content is solely the responsibility of the authors and does not represent the official views of any grant-awarding agency.
We thank Dr. Jeffery Myers and Dr. Peter J. F. Snijders for kindly sharing their cell lines.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Ang KK, Sturgis EM. Human papillomavirus as a marker of the natural history and response to therapy of head and neck squamous cell carcinoma. Semin Radiat Oncol. 2012;22:128–142. doi: 10.1016/j.semradonc.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Naggar AK, Westra WH. p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: a guide for interpretative relevance and consistency. Head Neck. 2011;34:459–461. doi: 10.1002/hed.21974. [DOI] [PubMed] [Google Scholar]
- 4.Klussmann JP, Gültekin E, Weissenborn SJ, Wieland U, Dries V, Dienes HP, et al. Expression of p16 protein identifies a distinctentity of tonsillar carcinomas associated with human papillomavirus. Am J Pathol. 2003;162:747–753. doi: 10.1016/S0002-9440(10)63871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall M, Bates S, Peters G. Evidence for different modes of action of cyclin-dependent kinase inhibitors: p15 and p16 bind to kinases, p21 and p27 bind to cyclins. Oncogene. 1995;11:1581–1588. [PubMed] [Google Scholar]
- 6.Li W, Thompson CH, Cossart YE, O’Brien CJ, McNeil EB, Scolyer RA, et al. The expression of key cell cycle markers and presence of human papillomavirus in squamous cell carcinoma of the tonsil. Head Neck. 2004;26:1–9. doi: 10.1002/hed.10335. [DOI] [PubMed] [Google Scholar]
- 7.Matsumura Y, Yamagishi N, Miyakoshi J, Imamura S, Takebe H. Increase in radiation sensitivity of human malignant melanoma cells by expression of wild-type p16 gene. Cancer Lett. 1997;115:91–96. doi: 10.1016/s0304-3835(97)04714-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee AW, Li JH, Shi W, Li A, Ng E, Liu TJ, et al. p16 gene therapy: a potentially efficacious modality for nasopharyngeal carcinoma. Mol Cancer Ther. 2003;2:961–969. [PubMed] [Google Scholar]
- 9.Dok R, Kalev P, Van Limbergen EJ, Asbagh LA, Vázquez I, Hauben E, et al. p16INK4a impairs homologous recombination-mediated DNA repair in human papillomavirus-positive head and neck tumors. Cancer Res. 2014;74:1739–1751. doi: 10.1158/0008-5472.CAN-13-2479. [DOI] [PubMed] [Google Scholar]
- 10.Mirzayans R, Andrais B, Hansen G, Murray D. Role of p16(INK4A) in replicative senescence and DNA damage-induced premature senescence in p53-deficient human cells. Bio Chem Res Int. 2012;2012:951574. doi: 10.1155/2012/951574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis JS, Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, et al. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34:1088–1096. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieckmann T, Tribius S, Grob TJ, Meyer F, Busch CJ, Petersen C, et al. HNSCC cell lines positive for HPV and P16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother Oncol. 2013;107:242–246. doi: 10.1016/j.radonc.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda K, Miura S, Kurashige T, Suzuki K, Kondo H, Ihara M, et al. Significance of p53-binding protein 1 nuclear foci in uterine cervical lesions: endogenous DNA double strand breaks and genomic instability during carcinogenesis. Histopathology. 2011;59:441–451. doi: 10.1111/j.1365-2559.2011.03963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grøfte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 15.DiTullio RA, Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, et al. DNA damage-induced G(2)-M checkpoint activation by histone H2AX and53BP1. Nat Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 18.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage signaling pathways. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Bohgaki M, Bohgaki T, El Ghamrasni S, Srikumar T, Maire G, Panier S, et al. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proc Natl Acad Sci. 2013;110:20982–20987. doi: 10.1073/pnas.1320302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collado M, Serrano M. The TRIP from ULF to ARF. Cancer Cell. 2010;17:317–318. doi: 10.1016/j.ccr.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W. Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Mol Cell. 2013;51:46–56. doi: 10.1016/j.molcel.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velimezi G, Liontos M, Vougas K, Roumeliotis T, Bartkova J, Sideridou M, et al. Functional interplay between the DNA-damage-response kinase ATM and ARF tumour suppressor protein in human cancer. Nat Cell Biol. 2013;15:967–977. doi: 10.1038/ncb2795. [DOI] [PubMed] [Google Scholar]
- 26.Smith-Sørensen B, Hovig E. CDKN2A (p16INK4A) somatic and germline mutations. Hum Mutat. 1996;7:294–303. doi: 10.1002/(SICI)1098-1004(1996)7:4<294::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 27.Xu R, Wang F, Wu L, Wang J, Lu C. A systematic review of hypermethylation of p16 gene in esophageal cancer. Cancer Biomark. 2013;13:215–226. doi: 10.3233/CBM-130355. [DOI] [PubMed] [Google Scholar]
- 28.Lou-Qian Z, Rong Y, Ming L, Xin Y, Feng J, Lin X. The prognostic value of epigenetic silencing of p16 gene in NSCLC patients: a systematic review and meta-analysis. PLoS One. 2013;8:e54970. doi: 10.1371/journal.pone.0054970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Tang L, Xie R, Nie W, Chen L, Guan X. p16 promoter hypermethylation is associated with increased breast cancer risk. Mol Med Rep. 2012;6:904–908. doi: 10.3892/mmr.2012.1001. [DOI] [PubMed] [Google Scholar]
- 30.Bihl MP, Foerster A, Lugli A, Zlobec I. Characterization of CDKN2A(p16) methylation and impact in colorectal cancer: systematic analysis using pyrosequencing. J Transl Med. 2012;10:173. doi: 10.1186/1479-5876-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Raju U, Milas L, Molkentine D, Zhang Z, Yang P, et al. Huachansu, containing cardiac glycosides, enhances radiosensitivity of human lung cancer cells. Anticancer Res. 2011;31:2141–2148. [PubMed] [Google Scholar]
- 32.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 33.Moody CA1, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 34.Li C1, Johnson DE. Liberation of functional p53 by proteasome inhibition in human papilloma virus-positive head and neck squamous cell carcinoma cells promotes apoptosis and cell cycle arrest. Cell Cycle. 2013;12:923–34. doi: 10.4161/cc.23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci. 2013;110:16175–16180. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Mason KA, Ang KK, Buchholz T, Valdecanas D, Mathur A, et al. MK-4827, a PARP-1/-2 inhibitor, strongly enhances response of human lung and breast cancer xenografts to radiation. Invest New Drugs. 2012;30:2113–2120. doi: 10.1007/s10637-011-9770-x. [DOI] [PubMed] [Google Scholar]
- 37.Dickson MA, Tap WD, Keohan ML, D’Angelo SP, Gounder MM, Antonescu CR, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31:2024–2028. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer E, Recknagel RD, Fiedler U, Wollweber L, Bock C, Greulich KO. The distribution of the tail moments in single cell gel electrophoresis (comet assay) obeys a chi-square (chi2) not a gaussian distribution. Mutat Res. 1998;398:101–110. doi: 10.1016/s0027-5107(97)00246-7. [DOI] [PubMed] [Google Scholar]
- 39.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 40.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peuscher MH, Jacobs JJ. DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat Cell Biol. 2011;13:1139–1145. doi: 10.1038/ncb2326. [DOI] [PubMed] [Google Scholar]
- 43.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;93:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 45.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol. 2011;31:118–126. doi: 10.1128/MCB.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates an ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 47.Yang ZH, Zhou CL, Zhu H, Li JH, He CD. A functional SNP in the MDM2 promoter mediates E2F1 affinity to modulate cyclin D1 expression in tumor cell proliferation. Asian Pac J Cancer Prev. 2014;15:3817–3823. doi: 10.7314/apjcp.2014.15.8.3817. [DOI] [PubMed] [Google Scholar]
- 48.Chou TF, Deshaies RJ. Quantitative cell-based protein degradation assays to identify and classify drugs that target the ubiquitin-proteasome system. J Biol Chem. 2011;286:16546–16554. doi: 10.1074/jbc.M110.215319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.