 The development of ligation conditions suitable for highly hydrophobic peptide segments allowed the first chemical synthesis of the head-to-tail cyclized protein AS-48.
The development of ligation conditions suitable for highly hydrophobic peptide segments allowed the first chemical synthesis of the head-to-tail cyclized protein AS-48.
Abstract
An antibacterial cyclic AS-48 protein was chemically synthesized by α-ketoacid–hydroxylamine (KAHA) ligation. Initial challenges associated with the exceptionally hydrophobic segments arising from the amphiphilic nature of the protein were resolved by the development of bespoke reaction conditions for hydrophobic segments, using hexafluoroisopropanol (HFIP) as a co-solvent. The synthetic protein displays similar biological activity and properties to those of the native protein. To support the current understanding of its antibacterial mode of action, we demonstrate the ability of AS-48 to be incorporated into synthetic multilamellar vesicles (MLVs).
Introduction
AS-48 is a cyclic antibacterial protein produced by Enterococcus faecalis.1,2 At 70 residues it is, together with uberolysin, the largest member of the class of circular bacteriocines and its structure and bactericidal mechanism have been extensively studied.3–7 It consists of five amphipathic α-helices and adopts a saposin fold. Owing to its cyclic nature it is exceptionally thermodynamically stable and shows an increased resistance to proteolytic degradation.8,9 It is active against a wide range of pathogenic bacteria – including L. monocytogenes and B. cereus – and has been proposed as a biopreservative and as a treatment for acne vulgaris.2,10 These unique properties and the cyclic nature of AS-48 make it a formidable target for chemical synthesis.11–20
During the preparation of this manuscript, Tam published a chemoenzymatic synthesis of cyclic bacteriocins21 – including AS-48 – by butelase-mediated cyclization22–24 of linear unprotected peptide precursors. Although an elegant approach, it required both a folded cyclization precursor and an enzyme that is not broadly available.25 Tam also reported that preparation of this protein by chemical synthesis using native chemical ligation was unsuccessful due the low solubility of the extremely hydrophobic peptide segments, prompting us to disclose our own work on the chemical synthesis of this cyclic protein.
In this report, we document the chemical synthesis of AS-48 by α-ketoacid–hydroxylamine (KAHA) ligation, an amide-forming ligation reaction that is particularly well-suited for assembling hydrophobic peptides and proteins. KAHA ligation relies on the chemoselective reaction of α-ketoacids with hydroxylamines and tolerates both organic and aqueous solvents.26,27 We have developed cyclic hydroxylamines that form serine or homoserine residues upon ligation.28–30 The requisite peptide α-ketoacids and hydroxylamines can be conveniently prepared by Fmoc–SPPS and we have applied this reaction to the synthesis of numerous linear proteins as well as small cyclic peptides.31,32 The homoserine-forming variant, which uses (S)-5-oxaproline as the ligation partner, has the unique property of forming depsipeptides as the primary ligation products, a feature that can greatly aid in the preparation of hydrophobic sequences.
Results and discussion
Preliminary studies
In preliminary studies on the synthesis of AS-48 we identified the hydrophobicity of the peptide segments as the main obstacle. In the first attempt to synthesize AS-48 1–44 by Fmoc–SPPS, after 19 residues it became impossible to analyze the peptide by RP-HPLC. Only by employing Liu’s removable poly-arginine solubilizing tag33 at a glycine residue were we able to isolate the segment.
Design
In order to reduce the synthetic overhead, we desired to develop a synthesis strategy unconstrained by the need for additional steps and non-standard manipulations of the peptide sequences. This required careful selection of the ligation sites to adjust the hydrophobicity of each segment while avoiding mutations possibly detrimental to biological activity. The synthesis of a cyclic peptide facilitates the selection of non-conventional strategies, as any amino acid pair can serve as a potential cyclization site.
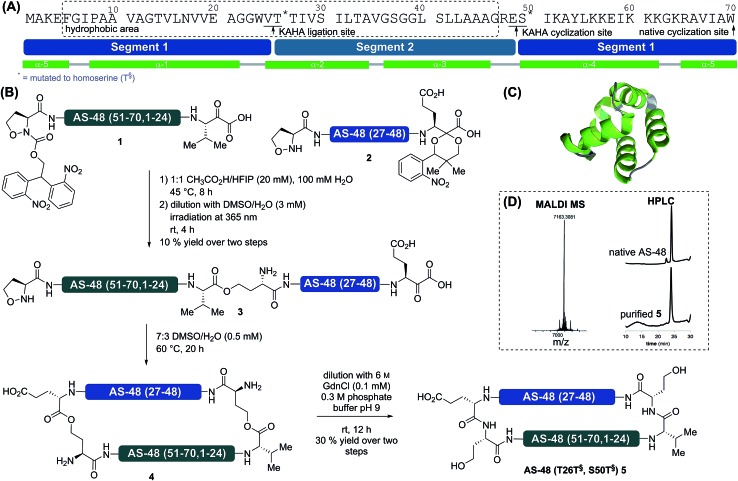
Based on several considerations, including the predicted hydrophobicity of the segments, the secondary structure of the cyclization site, and the desire to place the two non-canonical homoserine residues at positions that would not disturb the biological activity, we selected V25T26 as the first ligation site and E49S50 as the cyclization site (Scheme 1). In order to minimize handling and purification of the hydrophobic segment and linear peptide, we sought to utilize photolabile protecting groups for the 5-oxaproline in segment 2 and for the α-ketoacid in segment 1. This would allow the release of the linear cyclization precursor 3 immediately after ligation. For the α-ketoacid we used our recently reported protecting group, which is introduced as a linker attached to a solid support.34 To avoid the formation of diastereomers on the (S)-5-oxaproline residue, we used the achiral photolabile protecting group 2,2-bis(2-nitrophenyl)ethoxycarbonyl (di-NPEOC).35
Scheme 1. Synthesis of AS-48 by KAHA ligation of two segments. (A) Sequence of AS-48 with division into two peptide segments. The dashed box indicates an area of mainly hydrophobic residues. Alpha helices are depicted in green, loop and coil regions in gray. The asterisks (*) indicate residues mutated to Hse in the synthesis by KAHA ligation. (B) Synthesis by a two-segment strategy with photoprotected 5-oxaproline segment 2 and photoprotected α-ketoacid segment 1. (C) Three dimensional structure of native AS-48. (D) MALDI mass spectrum and analytical HPLC traces at 220 nm of purified cyclic AS-48 5 and comparison with HPLC trace of native AS-48.
Protein synthesis
We prepared both segments 1 and 2 by Fmoc–SPPS and subjected them to our standard ligation conditions (15 mM, 9 : 1 DMSO/H2O, 0.1 M oxalic acid, 60 °C). Possibly due to the extremely hydrophobic nature of the segments, we could not observe any conversion. Variation of the solvent ratio and switching the organic solvent component to N-methylpyrrolidinone (NMP) did not improve the outcome; increasing the temperature to 95 °C led to decomposition.
We speculated that the steric bulk of the valine α-ketoacid might hamper the reactivity, and resynthesized segment 1, replacing valine with leucine α-ketoacid at the C-terminus. As this ligation also failed we excluded steric repulsion as the likely cause for the lack of reactivity and considered aggregation or the formation of other perturbing structures. As both hexafluoroisopropanol (HFIP) and acetic acid have a pronounced effect on protein secondary and tertiary structure, we attempted the ligation in 1 : 1 HFIP/CH3CO2H.36,37 Gratifyingly, we observed almost complete conversion after 8 h. The reaction proceeded sufficiently fast at lower temperatures than usually employed for KAHA ligation (45 °C compared to 60 °C).
Using this purely organic solvent mixture increases the applicability of the KAHA ligation, especially for exceptionally hydrophobic peptide segments. Using these optimized conditions for hydrophobic segments, 1 and 2 were ligated in 1 : 1 HFIP/CH3CO2H at 45 °C for 8 h. After dilution with DMSO/H2O the reaction mixture was irradiated with 365 nm UV light from a handheld lamp to remove both photoprotecting groups in a one-pot fashion. Purification by preparative RP-HPLC afforded 3 in 10% yield over three steps. This rather low yield is attributed to the lower recovery of hydrophobic and amphiphilic peptides on RP-HPLC. The linear cyclization precursor 3 was soluble in acidic media and the standard KAHA ligation conditions (7 : 3 DMSO/H2O, 0.5 mM, 60 °C) were employed for the cyclization. As the retention time of the cyclized product 4 was identical to that of the starting material, the conversion was monitored by mass spectrometry (loss of CO2 – 44 Da). After 20 h, conversion was complete and the mixture was diluted with 0.3 M pH 9 buffer containing 6 M guanidine hydrochloride to induce the O-to-N acyl shifts. One rearrangement proceeded quickly38 and moderately increased the retention time by two minutes; the second acyl shift proceeded more slowly and increased the retention time of the final product 5 by an additional eight minutes. This might indicate the formation of a secondary or tertiary structure. Purification by RP HPLC afforded 1.0 mg (30% yield) of pure AS-48 (T26T§, S50T§) 5.
Biological activity and folding
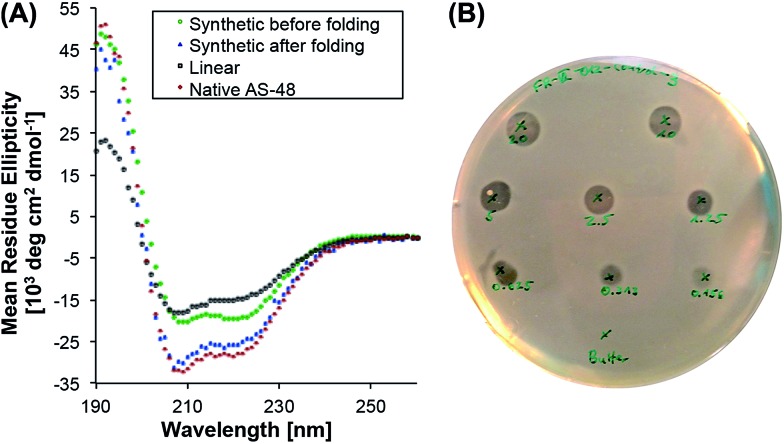
To evaluate its biological activity, we tested the synthetic protein in a spot on lawn assay against Listeria innocua. To our surprise, the minimum inhibitory concentration (MIC) was significantly higher (>10 µM) than the reported value8 (0.5 µM) for the natural protein.39 Although the synthetic AS-48 contains two mutations (Thr26Hse and Ser50Hse) – and it is known that the biological activity can be altered by single mutations8,40,41 (e.g. Trp24Ala) – we were surprised by the reduced activity and suspected misfolding as the reason.
To probe the protein folding, we recorded the circular dichroism spectrum of freshly dissolved 5 (T26T§, S50T§) in 10 mM pH 3 phosphate buffer and compared it to the spectrum of the expressed protein (Fig. 1). Even though the synthetic cyclic AS-48 5 showed a spectrum typical for α-helical peptides, it differed from the spectrum of the native AS-48 in intensity and more importantly in the relative intensities of the minima at 208 and 220 nm, suggesting different or incomplete folding. After storing the synthetic protein for one month at 4 °C in pH 3 buffer, we remeasured the spectrum and noticed that it had become essentially identical to that of the native protein. Apparently, the synthetic AS-48 (T26T§, S50T§) slowly adopted the correct fold upon storage. We used this folded sample to evaluate its activity against L. innocua and were pleased to find that the activity had significantly increased (MIC 0.5 µM) and compared well with our experimental results and the reported value for the natural protein.
Fig. 1. (A) Circular dichroism spectra of synthetic linear, cyclic unfolded and cyclic folded AS-48 and native AS-48 at pH 3. (B) Agar well diffusion antibacterial assay (concentrations: 20.0–0.156 µM) of synthetic AS-48 with L. innocua as the indicator strain.
Membrane incorporation
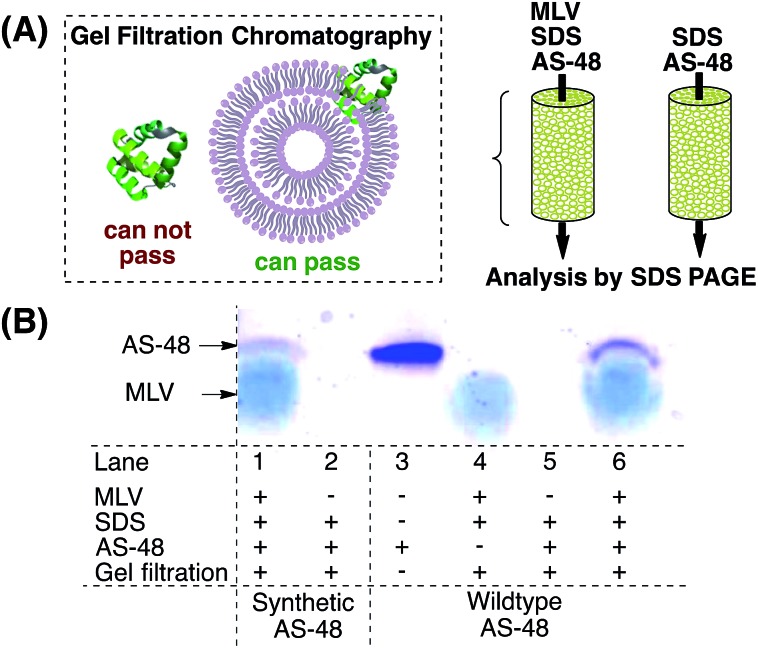
It is currently believed that AS-48 operates by either direct incorporation into the membrane42 or by accumulation at the membrane surface, leading to molecular electroporation.6 It was shown by lipid cosedimentation assays3 that AS-48 interacts with liposomes and can permeate membrane vesicles.43 To complement this data we used multilamellar vesicles (MLVs) to mimic the (bacterial) membrane (Scheme 2).44 AS-48 (10 µM in pH 3 phosphate buffer) was incubated with MLVs and SDS45 for 1 h, then passed through the gel filtration resin. The filtrate was analyzed by SDS PAGE.
Scheme 2. Incorporation of AS-48 into multilamellar vesicles. (A) After incubation of 10 µM AS-48 with 6 mg mL–1 MLV, gel filtration through Sepharose CL-4B resin prevents free AS-48 from passing through the column and only vesicle-bound AS-48 can pass through. (B) Analysis of the filtrate by SDS PAGE stained with Coomassie Brilliant Blue shows vesicle-bound AS-48 (lane 1 vs. 2 for synthetic AS-48 and lane 5 vs. 6 for native AS-48). The broad band captioned MLV arises from the remaining multilamellar vesicles (lane 4).
Both the synthetic and native protein were incorporated into the MLV and were detected by SDS PAGE. In accordance with previous reports, the bands of synthetic and native AS-48 appeared at a lower molecular weight than expected, presumably due to their cyclic structure. This experiment indicates that AS-48 is able to be incorporated into membranes without the need for specific receptors or other active mechanisms and supports the currently assumed bactericidal mechanism of action.
Conclusions
In summary, we have reported the chemical synthesis of a biologically active variant of AS-48. This work further establishes important features of the KAHA ligation: its utility for the synthesis of highly hydrophobic proteins by forming depsipeptide ligation products and operating under acidic conditions in the presence of organic solvents – in this case HFIP/CH3CO2H – that excel at dissolving even difficult sequences. It is notable that the cyclization proceeded well on the denatured, linear protein, in contrast to enzymatic cyclizations that require a prefolded structure. The ease of preparing long linear peptides by KAHA ligation of segments bearing photoprotected α-ketoacids and hydroxylamines by Fmoc–SPPS will make this approach ideal for preparing other types of cyclic proteins.
Acknowledgments
This work was supported by the Swiss National Science Foundation (150073, 169451). Prof. Mercedes Maqueda (University of Granada) is gratefully acknowledged for the generous gift of an authentic sample of AS-48. We thank the MS service of the Laboratorium für Organische Chemie at ETH Zürich for analyses, and Prof. Peter Kast, Thibault Harmand and Dr Vijaya Pattabiraman for helpful discussions.
Footnotes
References
- Sánchez-Hidalgo M., Montalbán-López M., Cebrián R., Valdivia E., Martínez-Bueno M., Maqueda M. Cell. Mol. Life Sci. 2011;68:2845–2857. doi: 10.1007/s00018-011-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande Burgos M. J., Pulido R. P., del Carmen López Aguayo M., Gálvez A., Lucas R. Int. J. Mol. Sci. 2014;15:22706–22727. doi: 10.3390/ijms151222706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrián R., Martínez-Bueno M., Valdivia E., Albert A., Maqueda M., Sánchez-Barrena M. J. J. Struct. Biol. 2015;190:162–172. doi: 10.1016/j.jsb.2015.03.006. [DOI] [PubMed] [Google Scholar]
- Samyn B., Martinez-Bueno M., Devreese B., Maqueda M., Gálvez A., Valdivia E., Coyette J., Van Beeumen J. FEBS Lett. 1994;352:87–90. doi: 10.1016/0014-5793(94)00925-2. [DOI] [PubMed] [Google Scholar]
- Sánchez-Barrena M. J., Martínez-Ripoll M., Gálvez A., Valdivia E., Maqueda M., Cruz V., Albert A. J. Mol. Biol. 2003;334:541–549. doi: 10.1016/j.jmb.2003.09.060. [DOI] [PubMed] [Google Scholar]
- González C., Langdon G. M., Bruix M., Gálvez A., Valdivia E., Maqueda M., Rico M. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11221–11226. doi: 10.1073/pnas.210301097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- Sánchez-Hidalgo M., Fernández-Escamilla A. M., Martínez-Bueno M., Valdivia E., Serrano L., Maqueda M. Protein Pept. Lett. 2010;17:708–714. doi: 10.2174/092986610791190390. [DOI] [PubMed] [Google Scholar]
- Cobos E. S., Filimonov V. V., Gálvez A., Maqueda M., Valdívia E., Martínez J. C., Mateo P. L. FEBS Lett. 2001;505:379–382. doi: 10.1016/s0014-5793(01)02841-1. [DOI] [PubMed] [Google Scholar]
- Cebrián R., Arévalo S., Ananou S., Arias-Santiago S., Riazzo C., Dolores Rojo M., Bermudez-Ruiz M. P., Valdivia E., Martinez-Bueno M., Maqueda M. PeerJ Prepr. 2016 doi: 10.7287/peerj.preprints.2107v1. [DOI] [Google Scholar]
- Bondalapati S., Jbara M., Brik A. Nat. Chem. 2016;8:407–418. doi: 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]
- Harmand T. J., Murar C. E., Bode J. W. Curr. Opin. Chem. Biol. 2014;22:115–121. doi: 10.1016/j.cbpa.2014.09.032. [DOI] [PubMed] [Google Scholar]
- Kleineweischede R., Hackenberger C. P. R. Angew. Chem., Int. Ed. 2008;47:5984–5988. doi: 10.1002/anie.200801514. [DOI] [PubMed] [Google Scholar]
- Saxon E., Armstrong J. I., Bertozzi C. R. Org. Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- Nilsson B. L., Kiessling L. L., Raines R. T. Org. Lett. 2000;2:1939–1941. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu C., Lam H. Y., Lee C. L., Li X. Proc. Natl. Acad. Sci. U. S. A. 2013:201221012. [Google Scholar]
- Wang J.-X., Fang G.-M., He Y., Qu D.-L., Yu M., Hong Z.-Y., Liu L. Angew. Chem., Int. Ed. 2015;54:2194–2198. doi: 10.1002/anie.201408078. [DOI] [PubMed] [Google Scholar]
- Boll E., Ebran J.-P., Drobecq H., El-Mahdi O., Raibaut L., Ollivier N., Melnyk O. Org. Lett. 2015;17:130–133. doi: 10.1021/ol503359w. [DOI] [PubMed] [Google Scholar]
- Moyal T., Bavikar S. N., Karthikeyan S. V., Hemantha H. P., Brik A. J. Am. Chem. Soc. 2012;134:16085–16092. doi: 10.1021/ja3078736. [DOI] [PubMed] [Google Scholar]
- Camarero J. A., Pavel J., Muir T. W. Angew. Chem., Int. Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Hemu X., Qiu Y., Nguyen G. K. T., Tam J. P. J. Am. Chem. Soc. 2016;138:6968–6971. doi: 10.1021/jacs.6b04310. [DOI] [PubMed] [Google Scholar]
- Nguyen G. K. T., Kam A., Loo S., Jansson A. E., Pan L. X., Tam J. P. J. Am. Chem. Soc. 2015;137:15398–15401. doi: 10.1021/jacs.5b11014. [DOI] [PubMed] [Google Scholar]
- Nguyen G. K. T., Hemu X., Quek J.-P., Tam J. P. Angew. Chem., Int. Ed. 2016;55:12802–12806. doi: 10.1002/anie.201607188. [DOI] [PubMed] [Google Scholar]
- Nguyen G. K. T., Qiu Y., Cao Y., Hemu X., Liu C.-F., Tam J. P. Nat. Protoc. 2016;11:1977–1988. doi: 10.1038/nprot.2016.118. [DOI] [PubMed] [Google Scholar]
- At the present time, active butelase cannot be produced by recombinant expression. Approximately 1 mg is obtained by extraction of 1 kg of pods of Clitoria ternatea See: Nguyen G. K. T., Wang S., Qiu Y., Hemu X., Lian Y., Tam J. P., Nat. Chem. Biol., 2014, 10 , 732 –738 . [DOI] [PubMed] [Google Scholar]
- Bode J. W., Fox R. M., Baucom K. D. Angew. Chem., Int. Ed. 2006;45:1248–1252. doi: 10.1002/anie.200503991. [DOI] [PubMed] [Google Scholar]
- Rohrbacher F., Wucherpfennig T. G. and Bode J. W., in Protein Ligation and Total Synthesis II, ed. L. Liu, Springer International Publishing, 2014, pp. 1–31. [Google Scholar]
- Pattabiraman V. R., Ogunkoya A. O., Bode J. W. Angew. Chem., Int. Ed. 2012;51:5114–5118. doi: 10.1002/anie.201200907. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig T. G., Rohrbacher F., Pattabiraman V. R., Bode J. W. Angew. Chem., Int. Ed. 2014;53:12244–12247. doi: 10.1002/anie.201406097. [DOI] [PubMed] [Google Scholar]
- Pusterla I., Bode J. W. Nat. Chem. 2015;7:668–672. doi: 10.1038/nchem.2282. [DOI] [PubMed] [Google Scholar]
- Harmand T. J., Murar C. E., Bode J. W. Nat. Protoc. 2016;11:1130–1147. doi: 10.1038/nprot.2016.052. [DOI] [PubMed] [Google Scholar]
- Thuaud F., Rohrbacher F., Zwicky A., Bode J. W. Org. Lett. 2016;18:3670–3673. doi: 10.1021/acs.orglett.6b01692. [DOI] [PubMed] [Google Scholar]
- Zheng J.-S., Yu M., Qi Y.-K., Tang S., Shen F., Wang Z.-P., Xiao L., Zhang L., Tian C.-L., Liu L. J. Am. Chem. Soc. 2014;136:3695–3704. doi: 10.1021/ja500222u. [DOI] [PubMed] [Google Scholar]
- Thuaud F., Rohrbacher F., Zwicky A., Bode J. W. Helv. Chim. Acta. 2016;99:868–894. [Google Scholar]
- Hasan A., Stengele K.-P., Giegrich H., Cornwell P., Isham K. R., Sachleben R. A., Pfleiderer W., Foote R. S. Tetrahedron. 1997;53:4247–4264. [Google Scholar]
- Hirota N., Goto Y., Mizuno K. Protein Sci. 1997;6:416–421. doi: 10.1002/pro.5560060218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Tang S., Liu L. Sci. China: Chem. 2011;54:110–116. [Google Scholar]
- During the cyclization, a small amount of amide product (S7; see the ESI) at the cyclization site is formed. 4 rearranges quickly to S7 and more slowly to 5. Based on these observations, we assume that the rearrangement at E49T§50 proceeds quickly, whereas the rearrangement at V25T§26 is slower
- Two days after the first antibacterial assay, the experiment was repeated with unchanged activity
- Sánchez-Hidalgo M., Martínez-Bueno M., Fernández-Escamilla A. M., Valdivia E., Serrano L., Maqueda M. J. Antimicrob. Chemother. 2008;61:1256–1265. doi: 10.1093/jac/dkn126. [DOI] [PubMed] [Google Scholar]
- Cebrián R., Maqueda M., Neira J. L., Valdivia E., Martínez-Bueno M., Montalbán-López M. Appl. Environ. Microbiol. 2010;76:7268–7276. doi: 10.1128/AEM.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz V. L., Ramos J., Melo M. N., Martinez-Salazar J. Biochim. Biophys. Acta, Biomembr. 2013;1828:2524–2531. doi: 10.1016/j.bbamem.2013.05.036. [DOI] [PubMed] [Google Scholar]
- Gálvez A., Maqueda M., Martínez-Bueno M., Valdivia E. J. Bacteriol. 1991;173:886–892. doi: 10.1128/jb.173.2.886-892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley S. D., Francis M. B. J.Am. Chem. Soc. 2006;128:1080–1081. doi: 10.1021/ja057106k. [DOI] [PubMed] [Google Scholar]
- In preliminary experiments using 80 µM of wildtype AS-48 we had observed precipitation after incubation of MLVs with 80 µM AS-48 at pH 8 without SDS. In a control experiment, MLVs were incubated with pH 8 buffer without AS-48. No precipitation was observed. This indicated that – similar to the assumed biological mode of action – AS-48 was able to lyse MLVs. A low concentration of SDS (7 mM) proved to be effective to prevent lysis of the MLVs
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.