Abstract
Urine represents a noninvasive source in which proteins and nucleic acids can be assessed. Such analytes may function as biomarkers to monitor kidney graft pathology at every desired frequency, thereby providing a time window to prevent graft damage by therapeutic intervention. Recently, several proteins have been measured in urine as markers of graft injury. However, the specificity is limited, and measuring urinary proteins generally lacks the potential to predict early kidney graft damage. Currently, urinary mRNA and microRNA are being investigated to evaluate the prognostic value of changes in gene expression during the initial stages of graft damage. At such time point, a change in treatment regimen and dosage is expected to have maximum potency to minimize future decline in graft function. Both mRNA and microRNAs have shown promising results in both detection and prediction of graft injury. An advantage of microRNAs compared to mRNA molecules is their stability, a characteristic that is beneficial when working with urine samples. In this review, we provide the current state of urinary biomarkers in renal transplantation, with a focus on urinary microRNA. In addition, we discuss the methods used to study urinary microRNA expression.
Keywords: basic (laboratory) research/science, clinical research/practice, immunobiology, kidney transplantation/nephrology, genetics, biomarker, kidney (allograft) function/dysfunction, genetics, graft survival
Short abstract
Urinary microRNA is a promising new biomarker in renal transplantation.
Abbreviations
- IF/TA
interstitial fibrosis and tubular atrophy
- IRI
ischemia–reperfusion injury
- qPCR
quantitative real‐time polymerase chain reaction
- uEV
urinary extracellular vesicles
Introduction
Kidney transplantation is the treatment of choice for patients with end‐stage renal disease. After transplantation, the graft is subject to injury by several pathological processes, such as ischemia–reperfusion injury, acute and chronic rejection, infections, and recurrence of original kidney disease. In addition, the use of the calcineurin inhibitors tacrolimus and cyclosporine, which are part of the standard immunosuppressive treatment, can lead to renal damage 1.
Two biomarkers commonly used to monitor renal graft status are serum creatinine and proteinuria. Serum creatinine is used as a measure of the glomerular filtration rate, and proteinuria indicates structural damage to the glomerular filtration barrier. Although practical as global indicators of the condition of the graft, increases in serum creatinine and proteinuria are generally not seen before injury to the transplant already has progressed considerably. Furthermore, these parameters do not allow for discrimination between different types of underlying pathology. The traditional “gold standard” to evaluate the severity and cause of kidney graft injury is examination of a renal biopsy. However, taking a kidney graft biopsy is an invasive procedure that has a ≈3% complication risk 2. In addition, the focal nature of several types of kidney graft pathologies is a limiting factor for the diagnostic value of a renal biopsy. Because of these aforementioned limitations of the renal biopsy, it is important to identify novel biomarkers in blood or urine that can provide information on various types of graft pathology at an early stage in a noninvasive manner. This review addresses currently available urinary biomarkers that may be used to evaluate the type and severity of different forms of graft injury. After a brief summary of the use of urinary messenger RNA (mRNA), we will in particular discuss recent data on the potential use of urinary microRNA as biomarker.
mRNAs in Urine as Biomarkers of Graft Performance
After centrifugation, mRNA can be isolated from cells that are present in the urine. An important advantage of urinary mRNA derived from cells as compared with urinary proteins is that proteinuria and tubular reabsorption have less effect on mRNA measurements. In a landmark paper, the measurement of mRNA encoding the cytotoxic proteins perforin and granzyme B was shown to enable the diagnosis of acute renal allograft rejection in a noninvasive way 3. In addition, increased expression of urinary mRNA encoding CD103, a marker of alloreactive CD8+ cytotoxic T cells, could predict the simultaneous presence of renal allograft rejection with a sensitivity of 59% and a specificity of 75% in a cohort of 79 patients 4. Other urinary mRNAs that could serve a similar goal are interferon‐γ and TIM‐3 mRNA 5. Urinary granulysin mRNA was the first described mRNA that can diagnose renal allograft rejection at an early stage (i.e. before elevation of serum creatinine) 6. Also, urinary CXCL‐10 mRNA has been shown to be elevated up to 6 to 7 days prior to an acute rejection episode 7. In a recent prospective multicenter study, urinary mRNA of CXCL‐9 (as well as urinary CXCL‐9 protein) was shown to be predictive for the presence of acute rejection 8.
Besides the studies described above showing a correlation between urinary expression of a single mRNA and acute cellular rejection, the group of Suthanthiran has shown in a large cohort of 471 patients that acute cellular rejection can be separated from other causes of graft dysfunction by combined measuring of the expression of CD3ε, CXCL10, and 18S rRNA 9. In another study, a five‐gene signature consisting of mRNAs for CD3ε, CD105, CD14, CD46, and 18S rRNA was also able to distinguish cellular rejection from antibody‐mediated rejection 10. Taken together, measurement of urinary mRNA expression can certainly be helpful in the noninvasive diagnosis of graft damage. Most studies have focused on acute rejection and not on other kinds of graft damage such as virus‐induced pathologies 5, 6. The studies showing that a signature of multiple urinary mRNA transcripts can predict graft injury are quite promising. Further research should focus on implementation of measuring mRNA profiles in complex patient groups in which multiple conditions such as cellular rejection, antibody‐mediated rejection, drug‐induced nephrotoxicity, and viral infection can be present simultaneously.
MicroRNAs in Renal Physiology and Pathology
MicroRNAs are 19–23‐nucleotide‐long noncoding single‐stranded RNA molecules that act as regulators of gene expression 11. Precursor microRNAs, known as pri‐microRNAs, are generated by RNA polymerase II and have as hallmark 5′ and 3′ poly‐A tails. These pri‐microRNAs undergo a series of alterations in both the nucleus and the cytoplasm before they become mature microRNAs, which cause breakdown of the mRNA transcript or inhibition of translation of the mRNA to protein (Figure 1).
Figure 1.
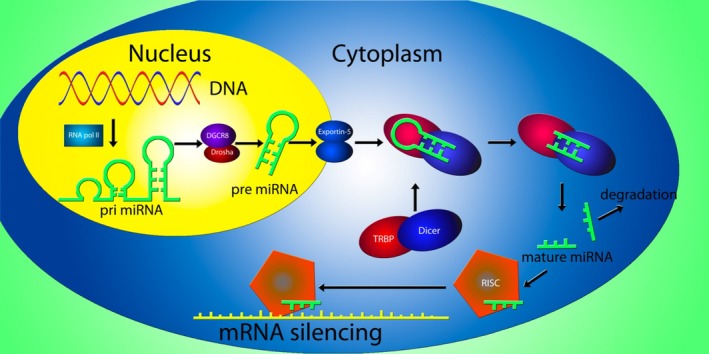
MicroRNA generation and mRNA silencing in humans. Following transcription of a microRNA gene by RNA polymerase II (RNA pol II), a pri‐microRNA stemloop structure is generated. The pri‐microRNA is consequently processed by the Drosha/DGCR8 complex into a pre‐microRNA, which is transported to the cellular cytoplasm by the exportin‐5 protein. In the cytoplasm, the Dicer/TRBP complex removes the stemloop and microRNA duplexes are formed. After removal of the passenger strand, the mature microRNA is incorporated into the RNA‐induced silencing complex and guides the functional protein to a complementary mRNA strand, inducing mRNA silencing by either translational repression or mRNA breakdown, depending on the complementarity of the RNA strands.
Many microRNAs display tissue‐specific expression patterns and are involved in the development and maintenance of organ function. Studies in conditional podocyte‐specific Dicer knockout mice have revealed roles for miRNAs in the development and physiology of the kidney, as renal failure quickly developed and the mice died within 6–8 weeks after birth 12.
In human chronic kidney disease, microRNAs are implicated in transforming growth factor β1–mediated fibrosis 13. Because of their important role in kidney physiology and pathology, microRNAs have potential to convey information about the graft status in kidney transplant patients.
In a kidney transplantation setting, Anglicheau et al have found that acute T cell–mediated rejection could be predicted by measuring levels of miR‐142‐5p, ‐155, and ‐223 in graft tissue 14. Soltaninejad et al have found that high expression of miR‐142‐3p and miR‐223 in both peripheral blood mononuclear cells and biopsy material could discriminate between acute rejection and normal graft function 15. Ischemia–reperfusion‐induced kidney injury (IRI) is the first threat to graft function after renal transplantation. Mice lacking Dicer expression in their proximal tubular epithelial cells were shown to be resistant to IRI, demonstrating the importance of microRNAs in the pathogenesis of IRI 16. The important role for microRNAs in various kidney transplant pathologies has been nicely summarized in several recent review papers 17, 18, 19. Although the use of urinary miRNA as biomarker is highlighted in these aforementioned reviews, the methodological aspects are only briefly addressed and will therefore be discussed more extensively in this minireview.
The Potential of Urinary MicroRNAs as Biomarkers of Graft Injury
Urinary microRNA can be present either in cells originating from the kidney or in cells that infiltrated the renal tissue and thereafter were shed in the urine. Moreover, free microRNAs either bound to proteins or included in extracellular vesicles may be released from kidney cells or enter the urine from the circulation by glomerular filtration. MicroRNAs are relatively stable in urine, with around 50% of urinary microRNA remaining after 5 days of storage at 4°C, tested by quantitative real‐time polymerase chain reaction (qPCR) analysis of miR‐16 and miR‐21 20. The stability of mature microRNAs is explained by the fact that they are encapsulated in Argonaut proteins, which are part of the RISC complex. This prevents breakdown by RNAses, and cells lacking the Argonaut proteins showed reduced half lives for multiple microRNAs, an effect that can be reversed by rescuing the Argonaut expression 21. Because of their stability and accessible nature, urinary microRNAs have high potential as noninvasive biomarkers for graft injury. First, we will provide an overview of the methodology that has been used to measure microRNAs in urine. Next, we will summarize the currently available data on the use of urinary microRNA as marker of acute rejection, interstitial fibrosis, and tubular atrophy, and viral infection of the graft in renal transplant patients.
Methodology for Measuring MicroRNAs in Urine
Over the last few years, urinary microRNAs have been analyzed in a number of studies concerning different conditions. The methods used throughout the studies are far from consistent with respect to the source of the microRNA, the method of quantification, and the way data were normalized.
An important first question is whether microRNAs should be measured in whole urine, the cell pellet, or the supernatant. After centrifugation of a urine sample, higher concentrations of microRNA are measured in the extracted pellet than in the cell‐free supernatant or in noncentrifuged urine 20, 22. In a study comparing patients with autosomal‐dominant polycystic kidney disease with patients with other kidney diseases, both cell pellets and cell‐free supernatant were used. With both sources, nonoverlapping results were obtained with regard to differential microRNA expression between the patient groups 22. In pediatric lupus patients, a relationship between the level of several urinary microRNAs and lupus nephritis activity was found when cell‐free supernatant was used, but not with the cell pellet 23. These studies indicate that using several sources of urinary microRNA can have added value. Until now, most studies investigating urinary microRNA levels have used urinary cell pellets as their source of microRNAs, although the way that the cell pellet was obtained is not consistent between studies or is often not described in detail. Since cells from the lower urinary tract may contaminate the urinary cell pellet, we recommend using midstream urine specimens as source when using microRNA as biomarker for kidney pathology.
Of interest is the presence of microRNAs in urinary extracellular vesicles (uEV). The concentration of microRNA in uEV was shown to be higher than in cell‐free urine, but lower than in the cell pellet 24. Various methods of uEV isolation (e.g. precipitation vs. ultracentrifugation) and microRNA extraction have been compared, but a “gold standard” for microRNA profiling in uEV has not been identified yet 25.
Quantification Methods
qPCR is the hallmark method for microRNA profiling, but newer high‐throughput technologies are being investigated. Next‐generation sequencing has provided good results in both solid tissue and biofluids 26, especially in samples with high‐quality microRNA 27. Comparative studies on urinary microRNA, which is usually less abundant and of lower quality, are scarce. One group has compared qPCR‐based profiling and RNA sequencing in urine from rats with tubular injury and found limited agreement comparing these two methods, which they attributed to a higher sensitivity of qPCR 28. The authors suggest that microRNA sequencing might have higher specificity, but is unable to detect low abundant microRNAs, whereas qPCR profiling could not detect microRNA isoforms but was able to pick up lower abundant sequences in urine samples. In a study in which 12 commercially available microRNA expression analysis platforms were compared, the finding that qPCR has a higher sensitivity than RNA sequencing was confirmed in human serum samples 29. In addition, qPCR‐based expression analysis was shown to be more reproducible and accurate.
In conclusion, qPCR‐based profiling and RNA sequencing are reported to give comparable results for high‐quality microRNA with sufficient amounts of material 28, but when microRNA yield is limited, one should be aware of possible differences in outcomes with alternative quantification methods.
Normalization
After microRNA sequencing or qPCR, several statistical methods are used for normalization in order to correct for technical variation. Data normalization following microRNA sequencing was evaluated using seven different strategies (global normalization, Lowess normalization, trimmed mean method, quantile normalization, scaling normalization, variance stabilization, and invariant method) and it was concluded that Lowess normalization, correcting for the intensity‐dependent variation in dye bias, and quantile normalization, used to equalize the statistical properties, were the best‐performing methods 30.
For qPCR, the expression of one or more housekeeping genes is usually included for correction of the data. Although an accepted housekeeping gene for urinary microRNA is not available, RNU6 and RNU48 are commonly used for this purpose. The downside of using small nucleolar RNAs as reference genes is that they are longer than microRNAs and are therefore not necessarily degraded at the same rate. Alternatively, nonhuman microRNA, such as Caenorhabditis elegans miR‐39, can be spiked into the sample to allow correction for variability during purification. Using endogenous household genes requires less handling of the samples than spiking nonhuman microRNA, reducing the chance of contamination of samples, which is important when samples are limited in availability or quality. Using spiked‐in nonhuman microRNA at specific concentrations has the advantage of eliminating the problem of differences in expression in endogenous housekeeping genes, making this method preferable in situations where microRNA is abundant and of high quality. The disadvantage of using spiked‐in nonhuman microRNAs is that they do not correct for the actual amount of cellular input, making them vulnerable for interfering factors that occur during handling procedures.
Urinary MicroRNAs as Biomarkers of Graft Injury
Rejection
A single study has addressed the association between urinary microRNA and acute T cell–mediated rejection 31. In this study, the urinary microRNA transcriptome profile was first examined in pooled urine samples of five patients with acute rejection and five patients with stable graft function. miR‐10a levels were significantly higher in the urine of rejection patients, whereas miR‐10b and miR‐210 levels were downregulated in the urine of rejection patients compared to the controls (Table 1). In a validation cohort of 62 patients with acute rejection (subclinical in 55 cases), 19 control patients, and 13 stable transplant patients with urinary tract infection, miR‐210 levels were decreased in patients with acute rejection. Moreover, miR‐210 levels were significantly lower during acute rejection as compared to before rejection (n = 12 patients) or after treatment of the rejection (n = 7 patients). Finally, low miR‐201 levels were associated with a stronger decline in GFR at 1 year after transplantation. Due to the large overlap in urinary miR‐210 levels between patients with acute rejection and controls, which might partly be explained by additional biopsy findings such as chronic changes and acute tubular necrosis in both groups, the diagnostic value of urinary miR‐210A was limited. A detailed time course of miR‐210 levels might provide more insight into the functional relevance of miR‐210 in the development of acute rejection.
Table 1.
Urinary microRNA levels in kidney transplant pathologies
| MicroRNA | Transplant pathology | Increased/decreased levelsa | Ref(s) | Putative mechanistic pathwayb |
|---|---|---|---|---|
| miR‐210 | Acute rejection | Decreased | 31 | Prevents cell division via Mnt 39 |
| miR‐142‐3p | CAD‐IF/TA | Increased | 32, 33 | Decreases activity of regulatory T cells via adenyl cyclase‐9 and cyclic AMP 40 |
| miR‐204 | CAD‐IF/TA | Decreased | 32, 33 | Inhibits apoptosis 41 |
| miR‐211 | CAD‐IF/TA | Decreased | 32, 33 | Inhibits proapoptotic chop/gadd153 42 |
| miR‐125b | CAD‐IF/TA | Decreased | 33 | Inhibits TNF‐α, decreasing inflammation 43 |
| miR‐203 | CAD‐IF/TA | Decreased | 33 | Inhibits cell proliferation 44 |
| miR‐200b | CAD‐IF/TA | Decreased/increased | 33, 34 | Inhibits epithelial to mesenchymal transition 45 |
| miR‐21 | CAD‐IF/TA | Increased | 34 | Promotes renal fibrosis 46 |
CAD‐IF/TA, chronic allograft dysfunction with interstitial fibrosis and tubular atrophy; TNF‐α, tumor necrosis factor‐α; AMP, adenosyl monophosphate.
As compared to normal graft function.
Example of mechanistic pathway known to be related to the particular microRNA. It should be noted that each microRNA may be involved in many pathophysiologic processes and it is unknown whether the given pathways are involved in the respective transplant pathologies.
Interstitial fibrosis and tubular atrophy
Interstitial fibrosis and tubular atrophy (IF/TA) is a common hallmark of chronic kidney graft damage. After identifying five microRNAs that were differentially expressed between normal allograft tissue and tissue with IF/TA, Scian et al measured the levels of these five microRNAs in the urinary cell pellets of seven kidney transplant patients with biopsy‐confirmed IF/TA and seven patients with normal allograft tissue 32. They observed a significant upregulation of miR‐142‐3p and downregulation of both miR‐211 and miR‐204 in the urine of patients with IF/TA (Table 1). In addition, when the levels of these microRNAs were analyzed in the urine of a separate cohort of 36 kidney transplant recipients undergoing protocol biopsy at 3, 9, and 12 months posttransplantation, hierarchical clustering separated two clusters of samples: one with a microRNA profile similar to that observed in patients with established IF/TA and one with a microRNA characteristic of patients with normal allograft histology. Importantly, at the time of urine collection, estimated GFR did not differ between the two groups. The predictive value of the urinary microRNA profile for graft outcome was not evaluated in this study.
In a follow‐up study by the same group, the urine microRNA content of patients who developed histologically proven IF/TA or retained stable graft function was not only compared at the time of established injury, thereby confirming and extending earlier findings, but also at an earlier time point to identify molecular markers that are predictive of future events 33. Based on microRNA levels in tissue and urine of patients with established IF/TA versus normal histology, and on microRNA levels in urine of patients with good versus poor graft function at 24 months after transplantation, a set of 12 microRNAs was longitudinally tested in a cohort of 66 patients. From this selected panel of microRNAs, miR‐99a, miR‐140‐3p, miR‐200b, and miR‐200* were differentially expressed in the urinary cell pellet at 3.7 ± 1.3 as well as at 20 ± 4 months posttransplantation between patients with good graft function (n = 41) and those with unfavorable outcome during a 2‐year follow‐up period (n = 25). Validation of the predictive value of these particular markers and correlation with the exact cause of poor graft function is still required. Together, these studies on IF/TA show that as for mRNAs, a panel of microRNAs rather than a single microRNA will be more useful for graft monitoring.
A recent, relatively small study reported the ability of urinary miR‐21 and miR‐200b to discriminate between renal transplant patients with IF/TA and stable graft function 34. Remarkably, in this study with living donors, IF/TA was associated with higher miR‐200b levels as compared to stable graft function, while the study of Maluf et al, which included recipients of deceased donors, found decreased levels of miR200b in patients with IF/TA.
Virally Encoded MicroRNAs in Kidney Transplantation
Members of certain virus families are known to encode their own microRNAs. Reactivation of BK virus is a frequent phenomenon after kidney transplantation, with BK virus copies identified in blood and urine of 30–50% of patients during the first year after transplantation 35. BK virus encodes a single precursor microRNA, bkv‐miR‐B1, which generates a mature ‐3p and ‐5p variant 36. These microRNAs have been demonstrated to target viral protein T antigen, thereby increasing DNA replication of the viral genome 37. In addition, BK virus microRNA can interfere with the production of ULBP3, which is recognized by NK cells, potentially helping BK virus–infected cells to evade the immune system 38. A strong correlation was found between BK virus–associated nephropathy (BKVAN) and the presence of BKV‐encoded microRNA in both blood and urine, but the study population was too small (31 BK‐positive patients compared to 15 controls) to determine whether BK microRNA detection has a higher predictive value than BK viral load for the diagnosis of BKVAN 36. The involvement of other virally encoded microRNAs in the pathophysiology of transplanted kidneys has not yet been elucidated.
Conclusion and Future Prospects
There is an urgent need for noninvasive biomarkers that can convey information about the status of the transplanted kidney before extensive irreversible damage has occurred through rejection, drug toxicity, or other pathological events. Urinary proteins have clear advantages over serum creatinine because they reflect earlier and more specific markers of renal damage. However, they do currently lack specificity at an early stage of the disease, which is required to come up with timely therapeutic intervention and prevent progression of tissue damage that otherwise is irreversible. As an alternative, mRNA expression in urine has successfully been used to identify and predict graft pathology, but the downside of urinary mRNA is its instability.
Measurement of microRNAs in urine for the discovery of new biomarkers is still in a pioneering stage. The studies described in this review show clear potential for the use of urinary microRNAs as biomarkers in the field of renal transplantation. In addition, the stability of microRNAs in urine compared to other biomarkers is a favorable characteristic.
When designing a path from discovery of urinary microRNA as biomarkers to clinical application, three stages can be distinguished. The first stage is the identification of potential biomarkers in an unbiased approach by using sequencing techniques or microarrays with known microRNAs. The patient cohorts used for this discovery phase should be quite homogeneous and similar, only differing for the condition of interest (e.g. presence or absence of acute cellular rejection). Items for which the interindividual variation should be minimized in kidney transplant recipients include the donor source (living vs. deceased), donor age, interval after transplantation, type of immunosuppressive medication, and viral infections (especially cytomegalovirus and BKV). Because urinary cell pellets, cell‐free urine, and urinary exosomes can yield different outcomes 22, 23, consideration should be given to evaluate these urinary microRNA sources in parallel because they can all be obtained from the same urinary specimen. Uniform protocols should be applied for the isolation of microRNA and the measurement of its concentration. In many cases, multiple microRNAs are found to be correlated with the condition of interest. Selection of the most promising biomarkers can be based on the strength of the correlation and on putative functional relevance for which algorithms for target prediction in terms of regulating mRNA expression and intracellular pathways can provide useful insight. Importantly, selected microRNAs should be validated in an independent patient cohort, which resembles as much as possible the discovery cohort. Because multiple microRNAs are usually included in this validation step, a statistical method to correct for false discovery rate should be applied. Typically, the size of the cohorts used for these discovery and validation stages amounts to some dozens of patients per condition. Finally, the crucial third stage of biomarker development concerns the evaluation of its diagnostic and prognostic potential in daily clinical practice. While in this phase complex patient populations are included with sometimes multiple conditions being present simultaneously, appropriate interpretation of the results requires detailed documentation of relevant donor and patient characteristics as outlined above. As demonstrated for mRNA as well as for microRNA, a panel of biomarkers forming a signature can be expected to be best suited for graft monitoring 9, 10, 32, 33, reflecting the fact that different molecular pathways are usually involved in the pathophysiology of any type of graft injury. Moreover, biomarkers should be measured at different time points during follow‐up, capturing the period preceding the presence of the pathological condition as well as the recovery phase, if applicable, to assess the prognostic potential of microRNAs and their value in monitoring response to therapy. This final stage of biomarker development requires large multicenter studies with hundreds of patients.
The promising data obtained until now should encourage researchers and clinicians to design additional studies along this path to identify patients at risk for kidney graft loss at a time when medical intervention might still affect outcome.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Acknowledgments
This work was supported by the Dutch Kidney Foundation (program grant CP09.04).
van de Vrie M, Deegens JK, Eikmans M, van der Vlag J & Hilbrands LB. Urinary MicroRNA as Biomarker in Renal Transplantation. Am J Transplant 2017; 17: 1160–1166
References
- 1. Pascual J, Perez‐Saez MJ, Mir M, Crespo M. Chronic renal allograft injury: Early detection, accurate diagnosis and management. Transplant Rev (Orlando) 2012; 26: 280–290. [DOI] [PubMed] [Google Scholar]
- 2. Korbet SM. Nephrology and the percutaneous renal biopsy: A procedure in jeopardy of being lost along the way. Clin J Am Soc Nephrol 2012; 7: 1545–1547. [DOI] [PubMed] [Google Scholar]
- 3. Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal‐allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med 2001; 344: 947–954. [DOI] [PubMed] [Google Scholar]
- 4. Ding R, Li B, Muthukumar T, et al. CD103 mRNA levels in urinary cells predict acute rejection of renal allografts. Transplantation 2003; 75: 1307–1312. [DOI] [PubMed] [Google Scholar]
- 5. Renesto P, Ponciano V, Cenedeze M, Câmara N, Pacheco‐Silva A. High expression of Tim‐3 mRNA in urinary cells from kidney transplant recipients with acute rejection. Am J Transplant 2007; 7: 1661–1665. [DOI] [PubMed] [Google Scholar]
- 6. Kotsch K, Mashreghi MF, Bold G, et al. Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation 2004; 77: 1866–1875. [DOI] [PubMed] [Google Scholar]
- 7. Matz M, Beyer J, Wunsch D, et al. Early post‐transplant urinary IP‐10 expression after kidney transplantation is predictive of short‐ and long‐term graft function. Kidney Int 2006; 69: 1683–1690. [DOI] [PubMed] [Google Scholar]
- 8. Hricik DE, Nickerson P, Formica R, et al. Multicenter validation of urinary CXCL9 as a risk‐stratifying biomarker for kidney transplant injury. Am J Transplant 2013; 13: 2634–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suthanthiran M, Schwartz JE, Ding R, et al. Urinary‐cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med 2013; 369: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matignon M, Ding R, Dadhania DM, et al. Urinary cell mRNA profiles and differential diagnosis of acute kidney graft dysfunction. J Am Soc Nephrol 2014; 25: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lorenzen JM, Thum T. Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol 2012; 7: 1528–1533. [DOI] [PubMed] [Google Scholar]
- 12. Shi S, Yu L, Chiu C, et al. Podocyte‐selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 2008; 19: 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 2015; 11: 23–33. [DOI] [PubMed] [Google Scholar]
- 14. Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci USA 2009; 106: 5330–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soltaninejad E, Nicknam MH, Nafar M, et al. Differential expression of microRNAs in renal transplant patients with acute T‐cell mediated rejection. Transpl Immunol 2015; 33: 1–6. [DOI] [PubMed] [Google Scholar]
- 16. Wei Q, Bhatt K, He H‐Z, Mi Q‐S, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia‐reperfusion injury. J Am Soc Nephrol 2010; 21: 756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as biomarkers in solid organ transplantation. Am J Transplant 2013; 13: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Akker EK, Dor FJ, IJzermans JN, de Bruin RW. MicroRNAs in kidney transplantation: Living up to their expectations? J Transplant 2015; 2015: 354826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilflingseder J, Reindl‐Schwaighofer R, Sunzenauer J, et al. MicroRNAs in kidney transplantation. Nephrol Dial Transplant 2015; 30: 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mall C, Rocke DM, Durbin‐Johnson B, Weiss RH. Stability of miRNA in human urine supports its biomarker potential. Biomark Med 2013; 7: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 2011; 8: 1149–1157. [DOI] [PubMed] [Google Scholar]
- 22. Ben‐Dov IZ, Tan Y‐C, Morozov P, et al. Urine microRNA as potential biomarkers of autosomal dominant polycystic kidney disease progression: Description of miRNA profiles at baseline. PLoS ONE 2014; 9: e86856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abulaban KM, Fall N, Nunna R, et al. Relationship of cell‐free urine MicroRNA with lupus nephritis in children. Pediatr Rheumatol Online J 2016; 14: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non‐exosomal miRNA in human urine. Kidney Int 2014; 86: 433–444. [DOI] [PubMed] [Google Scholar]
- 25. Lv L‐L, Cao Y, Liu D, et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci 2013; 9: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meng F, Hackenberg M, Li Z, Yan J, Chen T. Discovery of novel microRNAs in rat kidney using next generation sequencing and microarray validation. PLoS ONE 2012; 7: e34394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tam S, de Borja R, Tsao MS, McPherson JD. Robust global microRNA expression profiling using next‐generation sequencing technologies. Lab Invest 2014; 94: 350–358. [DOI] [PubMed] [Google Scholar]
- 28. Nassirpour R, Mathur S, Gosink MM, et al. Identification of tubular injury microRNA biomarkers in urine: Comparison of next‐generation sequencing and qPCR‐based profiling platforms. BMC Genom 2014; 15: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 2014; 11: 809–815. [DOI] [PubMed] [Google Scholar]
- 30. Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA‐Seq data. RNA 2012; 18: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lorenzen J, Volkmann I, Fiedler J, et al. Urinary miR‐210 as a mediator of acute T‐cell mediated rejection in renal allograft recipients. Am J Transplant 2011; 11: 2221–2227. [DOI] [PubMed] [Google Scholar]
- 32. Scian M, Maluf D, David K, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant 2011; 11: 2110–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maluf DG, Dumur CI, Suh JL, et al. The urine microRNA profile may help monitor post‐transplant renal graft function. Kidney Int 2014; 85: 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zununi Vahed S, Omidi Y, Ardalan M, Samadi N. Dysregulation of urinary miR‐21 and miR‐200b associated with interstitial fibrosis and tubular atrophy (IFTA) in renal transplant recipients. Clin Biochem 2016. doi: 10.1016/j.clinbiochem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 35. Bressollette‐Bodin C, Coste‐Burel M, Hourmant M, Sebille V, Andre‐Garnier E, Imbert‐Marcille BM. A prospective longitudinal study of BK virus infection in 104 renal transplant recipients. Am J Transplant 2005; 5: 1926–1933. [DOI] [PubMed] [Google Scholar]
- 36. Li JY, McNicholas K, Yong TY, et al. BK virus encoded microRNAs are present in blood of renal transplant recipients with BK viral nephropathy. Am J Transplant 2014; 14: 1183–1190. [DOI] [PubMed] [Google Scholar]
- 37. Fanning E, Zhao K. SV40 DNA replication: From the A gene to a nanomachine. Virology 2009; 384: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bauman Y, Mandelboim O. MicroRNA based immunoevasion mechanism of human polyomaviruses. RNA Biol 2011; 8: 591–594. [DOI] [PubMed] [Google Scholar]
- 39. Zhang Z, Sun H, Dai H, et al. MicroRNA miR‐210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009; 8: 2756–2768. [DOI] [PubMed] [Google Scholar]
- 40. Huang B, Zhao J, Lei Z, et al. miR‐142‐3p restricts cAMP production in CD4 + CD25− T cells and CD4 + CD25 + TREG cells by targeting AC9 mRNA. EMBO Rep 2009; 10: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scian MJ, Maluf DG, Archer KJ, et al. Gene expression changes are associated with loss of kidney graft function and interstitial fibrosis and tubular atrophy: Diagnosis versus prediction. Transplantation 2011; 91: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chitnis NS, Pytel D, Bobrovnikova‐Marjon E, et al. miR‐211 is a prosurvival microRNA that regulates chop expression in a PERK‐dependent manner. Mol Cell 2012; 48: 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tili E, Michaille J‐J, Cimino A, et al. Modulation of miR‐155 and miR‐125b levels following lipopolysaccharide/TNF‐α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007; 179: 5082–5089. [DOI] [PubMed] [Google Scholar]
- 44. Viticchiè G, Lena AM, Latina A, et al. MiR‐203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 2011; 10: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 45. Xiong M, Jiang L, Zhou Y, et al. The miR‐200 family regulates TGF‐β1‐induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol 2012; 302: F369–F379. [DOI] [PubMed] [Google Scholar]
- 46. Chau BN, Xin C, Hartner J, et al. MicroRNA‐21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 2012; 4: 121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]


