Abstract
Baculoviral inhibitor of apoptosis repeat-containing (Birc)6 gene/BIRC6 (Bruce/APOLLON) encodes an inhibitor of apoptosis and a chimeric E2/E3 ubiquitin ligase in mammals. The physiological role of Bruce in antiapoptosis is unknown. Here, we show that deletion of the C-terminal half of Bruce, including the UBC domain, causes activation of caspases and apoptosis in the placenta and yolk sac, leading to embryonic lethality. This apoptosis is associated with up-regulation and nuclear localization of the tumor suppressor p53 and activation of mitochondrial apoptosis, which includes up-regulation of Bax, Bak, and Pidd, translocation of Bax and caspase-2 onto mitochondria, release of cytochrome c and apoptosis-inducing factor, and activation of caspase-9 and caspase-3. Mutant mouse embryonic fibroblasts are sensitive to multiple mitochondrial death stimuli but resistant to TNF. In addition, eliminating p53 by RNA interference rescues cell viability induced by Bruce ablation in human cell line H460. This viability preservation results from reduced expression of proapoptotic factors Bax, Bak, and Pidd and from prevention of activation of caspase-2, -9, and -3. The amount of second mitochondrial-derived activator of caspase and Omi does not change. We conclude that p53 is a downstream effector of Bruce, and, in response to loss of Bruce function, p53 activates Pidd/caspase-2 and Bax/Bak, leading to mitochondrial apoptosis.
Programmed cell death or apoptosis is critical for development and homeostasis in metazoans (1). The inhibitor of apoptosis (IAP) proteins antagonize cell death by suppressing active caspases. This inhibition can be reversed by IAP antagonists Reaper, Grim, and Hid in flies and the second mitochondrial-derived activator of caspase (Smac)/Diablo family of proteins in vertebrates (2–5). Despite their well known anti-death function in vitro, IAPs have not been shown to be required for apoptosis inhibition in vertebrate development. Gene ablation of baculoviral IAP repeat-containing (Birc)4 (XIAP), the most potent caspase inhibitory IAP in cultured cells, does not perturb mouse development (6). In addition, Birc1a (Naip)-deficient mice do not manifest developmental abnormalities or spinal muscular atrophy, even though the deletion of BIRC1 (NAIP) correlates with the severity of this disease in a significant proportion of spinal muscular atrophy patients (7, 8). Although tissue-specific ablation of Birc5 (survivin) in mice has demonstrated that Birc5 is required for thymocyte development (9) and the resistance of keratinocytes to UVB-induced apoptosis (10), Birc5-null embryos die primarily because of deficient mitotic division instead of apoptosis (11, 12). Taken together, the physiological function of mammalian IAP family proteins in antagonizing apoptosis remains elusive in vivo.
Birc6 (Bruce) is a large mouse IAP with a molecular mass of 528 kDa (13). Similar to other IAPs, Bruce promotes cell survival. It inhibits apoptosis by binding to caspases through its BIR domain, and its caspase inhibitory activity also requires the C-terminal UBC domain (14). Likewise, APOLLON, the human Bruce ortholog, shares 92% identity with Bruce and contributes to the chemotherapy resistance in certain cancer cells (15). In addition, Drosophila Bruce suppresses apoptosis induced by Reaper and Grim but not Hid, and as a result, Drosophila Bruce mutants are viable but the males are infertile (16). Moreover, Drosophila Bruce is also required to protect the sperm nucleus against hypercondensation and degeneration during sperm differentiation (17). A recent report (18) shows that gene-targeted knockout of Birc6 in mice results in defective development of placenta, leading to embryonic lethality. In contrast to the antiapoptosis activity of Bruce in cells (14), no apoptosis was found in the mutant placenta and mouse embryonic fibroblasts (MEFs). Because Bruce is a chimeric E2/E3 ubiquitin ligase and can ubiquitylate Smac/Diablo and active caspase-9 in vitro (14, 19), Bruce is thought to preserve cell survival by antagonizing apoptosis induced by spontaneously released Smac (19).
Here, we report the analysis of mutant mice lacking the C-terminal half of Bruce, including the UBC domain, identified in a screen of gene-trap insertions that cause recessive lethal phenotypes (20). We show that embryos homozygous for the gene-trap insertion in Birc6 died in utero due to extensive apoptotic cell death in the placenta and yolk sac, and the mutation of Bruce led to up-regulation of the tumor suppressor p53, leading to mitochondrial apoptosis.
Materials and Methods
ES Cells, Mice, and MEF Cells. The gene trap ES cell line (Ex54, available from Baygenomics, which can be accessed at http://baygenomics.ucsf.edu) was of the 129/Ola strain, and germ-line chimeras were initially crossed to C57BL/6 strain. Backcrosses to C57BL/6 were carried out for eight generations before analysis. MEF cells were prepared from embryonic day (E)13.5–E14.5 embryos by using standard procedures (21) and were genotyped by PCR as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Antibodies. Bruce antibodies: Anti-BIR and Anti-UBC IgGs were obtained from rabbits immunized with purified recombinant His6-tagged protein of the respective domains by Rockland Immunochemicals. P53 antibodies: clone 1C12 was purchased from Cell Signaling Technology (Beverly, MA), clone PAb 122 was from Pharmingen, and clone DO-1 was from Santa Cruz Biotechnology. Antibodies against caspases, Bax and Bak, were purchased from Cell Signaling Technology, and β-gal antibody was from Promega. Anti-Pidd serum was obtained from rabbits immunized with the C-terminal half of human Pidd protein from Rockland Immunochemicals.
In Situ Hybridization. Mouse embryos from E7.5–E11.5 fixed in 4% paraformaldehyde were used for in situ hybridization to determine Bruce mRNA expression by using digoxigenin-labeled RNA probes. The mRNA was detected with anti-digoxigenin-Ap (Roche Molecular Biochemicals, Indianapolis) and developed with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution.
Hematoxylin/Eosin Staining. Mouse placentas, yolk sacs, and embryos were fixed for 24–48 h in 4% paraformaldehyde at 4°C, then embedded in paraffin, sectioned at 3–4 μm, and stained with hematoxylin/eosin.
Immunohistochemistry. The paraffin-embedded tissue sections were deparaffinized and rehydrated. The antigens were unmasked by heating, and endogenous peroxidase was eliminated by treatment with 0.3–1% hydrogen peroxide in methanol. Immunohistochemical staining of the sections was performed by using antibodies directly against the respective proteins. Secondary antibodies were horseradish peroxidase-conjugated and developed with 3-amino-9-ethylcarbazole reagent. The sections were counterstained with hematoxylin/eosin.
Immunocytochemistry. Cells cultured in eight-well slide chambers were fixed with 4% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 for 10 min on ice. The cells were incubated in 3% BSA (in PBS) for 30 min, followed by incubation with the respective primary antibody at 4°C for overnight. After washing with PBS, the cells were incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies for 1 h at room temperature. Nuclei were counterstained with DAPI. To label the mitochondria, MitoTracker Red CMXRos (100–500 nm) was added to the culture media for 30–45 min before fixation. Photos of the stained cells were taken by using the Axioplan2 imaging system.
TUNEL Staining of Placentas and Yolk Sacs. Apoptotic cells in paraffin-embedded tissue sections were detected by the TUNEL method (Roche Molecular Biochemicals).
Results
Bruce Mutant Mice by Gene-Trap Insertion Are Embryonic Lethal and Defective in Placentas and Yolk Sacs. An insertional mutation in the murine Birc6 gene was isolated in a gene-trap screen by using the pGT1.8TM vector in mouse ES cells (20). The sequence tag associated with the Ex54 gene trap ES cells (available from Baygenomics) matches exons 29 and 30 of Birc6. We performed inverse PCR and confirmed that the insertion in Birc6 occurred in intron 30 (Fig. 1). We verified at the mRNA and protein level that this insertion truncates the Birc6 at amino acid 2146, fusing the N-terminal half of Birc6 to the βgeo reporter gene of the gene-trap vector and eliminating the C-terminal half of the protein, including the UBC domain (Fig. 6, which is published as supporting information on the PNAS web site). Hereafter, we use Birc6GT to denote this insertional mutation. In situ hybridization of mouse embryos displayed high expression of Birc6 mRNA in the extraembryonic and embryonic tissues (Fig. 7A, which is published as supporting information on the PNAS web site). Compared with wild type, heterozygous mutant embryos are normal; however, among 271 embryos that we analyzed, all homozygous Birc6GT embryos died during E11.5–E16.5. The defects are as follows: (i) the early development of homozygous Birc6GT embryos delayed 1–2 days as manifested by the delayed body turning; (ii) severe dilation, disruption, and hemorrhage of vasculature structure in various tissues of the mutant embryos including skin, yolk sac, and placenta; and (iii) the mutant placenta was smaller and structurally disorganized with a much thinner layer of spongiotrophoblasts (Fig. 7 B–E). These defects are similar but more severe than those generated by gene-targeted knockout of Birc6 (18).
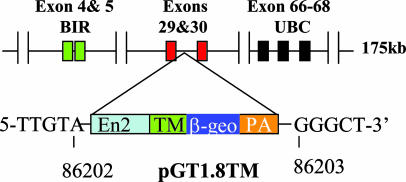
Fig. 1.
Generation of Birc6GT mutant mice by pGT1.8TM vector. The diagram shows the gene trap vector pGT1.8TM and the insertion site within the Bruce gene in ES clone Ex54. The BIR domain is encoded by exons 4 and 5 and the UBC domain is encoded by exons 66–68. The insertion site was determined by RACE and confirmed by inverse PCR. In the Ex54 ES clone, pGT1.8TM was inserted in intron 30 of Birc6 between nucleotides 86202 and 86203; this insertion truncates the Bruce protein (4,845 aa total) after the amino acid residue 2146, deleting the C-terminal half of Bruce, including the UBC domain.
Birc6GT Mutation Causes Apoptotic Cell Death in Placentas and Yolk Sacs. Given the fact that Drosophila Bruce inhibits apoptosis (16), we speculated that the deteriorating structures in Birc6GT mutant conceptuses might be the consequence of apoptotic cell death. Examination of the placentas revealed apoptotic nuclei in all three layers of mutant placentas by TUNEL staining (Fig. 2A) and activation of caspase-3 by immunohistochemical staining with an antibody that only recognizes the cleaved caspase-3 (Fig. 2B). In addition, caspase-7 was also activated (data not shown). Likewise, TUNEL staining was positive in Birc6GT mutant yolk sacs, and especially in the endoderm cells (Fig. 2C), and the amount of active caspase-3 in yolk sacs was much higher in the mutant than in the wild type (Fig. 2D). Together, these results demonstrate that eliminating the C-terminal half of Bruce in BruceGT mutants leads to spontaneous cell death in the placentas and yolk sacs.
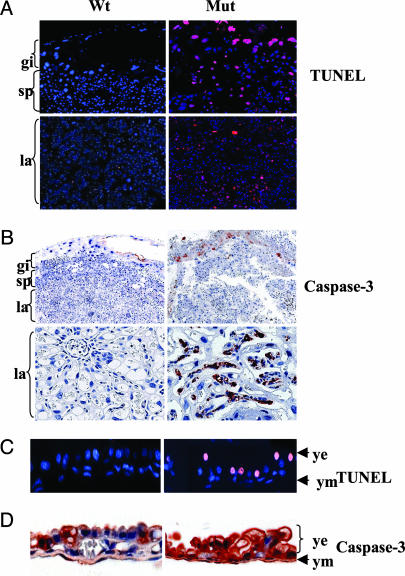
Fig. 2.
Apoptotic cell death in Birc6GT mutant placentas and yolk sacs. (A and C) TUNEL staining (red) of wild-type (Wt) and Birc6GT mutant (Mut) placentas and yolk sacs with the nuclei counterstained by DAPI (blue). (B and D) Immunohistochemical staining of Wt and Birc6GT Mut placentas and yolk sacs with the cleaved caspase-3 antibody (Asp-175 and Asp-5A1).
Ablation of Bruce Activates the Mitochondrial Pathway of Apoptosis. Bruce is reported to ubiquitylate Smac/Diablo by which Bruce scavenges spontaneously released Smac and preserves cell survival (19). We crossed Smac-null mice (22) with Bruce heterozygotes, and no BruceGT newborns with a Smac-null background were found alive among 90 newborns (data not shown), indicating that the predominant survival function of Bruce in vivo is not targeting Smac for degradation. In addition, Bruce does not appear to be involved in the TNF/cell death receptor pathway of apoptosis because Birc6GT MEFs are not sensitive to TNF treatment compared with wild type (Fig. 3D). Moreover, Bruce does not seem to be involved in PI-3/Akt survival pathway because phosphorylated Akt increased and Bad was phosphorylated in Bruce mutant placentas (data not shown).
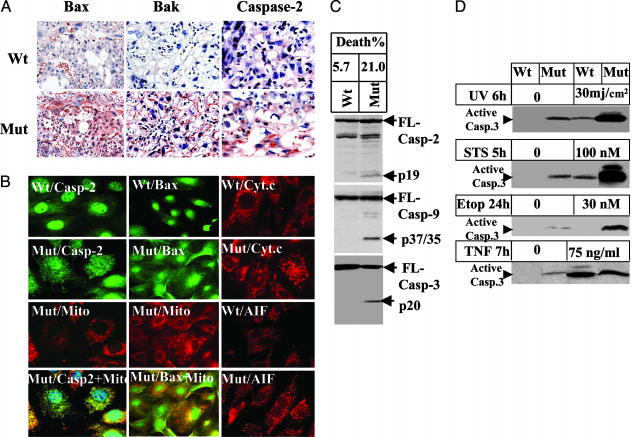
Fig. 3.
Ablation of Bruce activates the mitochondrial pathway of apoptosis. (A) Immunohistochemical staining of placentas with antibodies against Bax, Bak, and caspase-2. (B) Subcellular localization of proapoptotic factors in Birc6GT MEFs. Mitochondria were stained with Mito Tracker red. Caspase-2, Bax, and cytochrome c were stained by their primary antibody followed by Alexa Fluor 488-conjugated secondary antibody (green) for caspase-2 and Bax, and by Texas red for cytochrome c and AIF (red). The yellow fluorescence in the overlay of caspase-2 or Bax with Mito Tracker indicates mitochondrial colocalization of the two molecules. Smear staining of cytochrome c in the mutant MEFs indicates the release of cytochrome c. Nuclear staining of AIF indicates translocation of AIF to the nucleus in mutant MEFs. (C) Apoptosis in MEFs. Fifty micrograms of protein extract was assayed by Western blot for cleavage of caspase-2 (Top), caspase-9 (Middle), and caspase-3 (Bottom). The spontaneous death rate of Birc6GT mutant MEFs was 21%, as determined by propidium iodine staining and scored for subdiploid DNA contents by flow cytometry analysis. (D) Sensitivity of MEFs to apoptotic stimuli. MEFs were subjected to UV, staurosporin, etoposide, or TNF plus 0.5 μg/ml cycloheximide. Cell death was scored by Western blot for the activation of caspase-3.
Multiple death signals affect mitochondria in apoptosis. We examined whether the mitochondrial (intrinsic) pathway contributed to the apoptosis induced by the loss of Bruce function. Because activation of the proapoptotic Bcl-2 members Bax and Bak and the apical caspase-2 is essential for mitochondrial permeabilization required for apoptosis (23, 24), we assessed the expression level of these proteins in mouse placenta by immunohistochemical staining. The expression level of all three molecules was found to be up-regulated in mutant placentas compared with their basal level of expression in wild type (Fig. 3A). Permeabilization of mitochondria by Bax and caspase-2 releases cytochrome c, an essential proapoptotic factor for the activation of caspase-9 in the mitochondrial apoptosis pathway (25, 26). We assessed whether this enhanced expression of Bax and caspase-2 had caused the permeabilization of mitochondria by immunocytochemical staining of Bruce MEFs. In mutant MEFs, Bax and caspase-2 were found to have translocated onto mitochondria, cytochrome c was released to the cytoplasm, and another mitochondrial proapoptotic factor, apoptosis-inducing factor (AIF), was released and entered into the nucleus (Fig. 3B). In addition, ≈20% of the BruceGT MEF population spontaneously died in the absence of apoptotic stimuli, which coincided with autonomous activation of apical caspase-2 and caspase-9 and effector caspase-3 (Fig. 3C). Moreover, BruceGT MEFs displayed increased sensitivity to apoptosis induced by the mitochondrial pathway stimuli, such as UV irradiation, staurosporin, and etoposide. In contrast, the mutant MEFs were weakly sensitive to apoptosis induced by extrinsic pathway stimulus TNF and, to our surprise, the mutant MEFs were resistant to TNF compared with the wild-type MEFs (Fig. 3D). Together, these results indicate that eliminating the C-terminal half of Bruce leads to the activation of the intrinsic mitochondrial pathway of apoptosis.
Inactivation of Bruce Leads to Up-Regulation of Tumor Suppressor p53 in Placentas and MEFs. p53 is a short-lived protein whose abundance and transcriptional activity are kept at low levels in unstressed cells; however, upon genotoxic stress, p53 is up-regulated by stabilization and/or transcriptional activation and accumulates in the nucleus to activate gene expression (27). Bax is a p53-responsive gene (28), and its up-regulation in BruceGT mutant placentas and MEFs may reflect transcriptional activation of Bax by p53. Likewise, enhanced caspase-2 also suggests p53 activation because caspase-2 is activated by Pidd, another p53-responsive gene (29, 30). Immunohistochemical staining for p53 displayed an expected low level in wild-type placentas, but, in Birc6GT mutant placentas, p53 was enhanced in all three layers with the highest expression in the spongiotrophoblasts (Fig. 4A). In wild-type MEFs, the expression level of p53 was undetectable (Fig. 4B, lane 1) and only weakly visible in cells (Fig. 4C Upper), whereas it became stabilized after the MEFs were challenged with the genotoxic stress-inducer etoposide (Fig. 4B, lane 3). In contrast, in Birc6GT MEF p53 was readily detected in the absence of apoptotic treatment (Fig. 4B, lane 2) and accumulated in the nucleus (Fig. 4C Lower). The up-regulation of p53 reflects protein stabilization, rather than gene transcription, because Northern blot analysis displayed no increase of p53 mRNA in mutant MEFs (Fig. 4D).
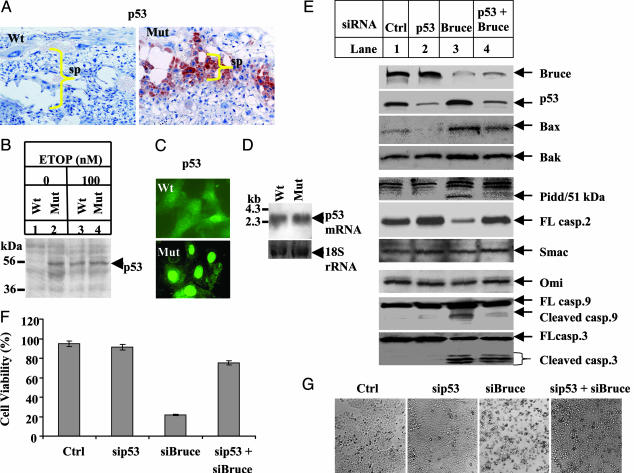
Fig. 4.
p53 is required for initiating mitochondrial apoptosis in Bruce mutants. (A) Immunohistochemical staining of placenta with p53 antibody (1C12). (B) p53 Western blot in MEFs. Cell extracts were prepared from Wt and Birc6GT MEFs treated with 100 nM etoposide (ETOP) for 27 h or left untreated. Fifty micrograms of this protein extract was assayed with a p53 antibody (PAb122). (C) Subcellular localization of p53 in MEFs. Wild-type (Wt) and mutant (Mut) MEFs were fixed and immunostained with a p53 antibody (1C12) followed by staining with a secondary antibody conjugated with Alexa Fluor 488 (green). (D) p53 Northern blot in MEFs. Blots containing total RNA isolated from Wt and Mut MEFs were hybridized with a digoxigenin-labeled probe specific for p53 gene. The 18S rRNA shows equal RNA loading. (E) siRNA experiments in H460 cells. A total of 2 × 105 cells per well were plated for 24 h and transfected with siRNA against Bruce and p53 (Supporting Materials and Methods) for 36 h by GeneSilencer reagent. siRNA (20 nM) was transfected into cells in the single knock-down samples and 20 nM sip53 plus 80 nM siBruce in the double silencing. The control siRNA was the nontarget from Dharmacon. Protein extract (20 μg) was assayed for protein expression. (F) Cell viability. Cells after siRNA treatment were trypsinized for Trypan blue staining, and viable cells were counted. (G) Cell morphology. Phase-contrast photos of H460 cells after siRNA treatment.
Silencing p53 Expression Down-Regulates the Mitochondrial Apoptosis Induced by Bruce Inactivation and Rescues Cell Viability. To verify that both the apoptotic phenotypes and the p53 up-regulation in Birc6GT mutants are general consequences of a deficiency in Bruce protein, we silenced Bruce expression by small interfering RNA (siRNA) in the human lung cancer cell line H460 (p53+/+). Approximately 80% of the cell population was apoptotic (Fig. 4F) and this change occurred with p53 up-regulation and the activation of the mitochondrial pathway of apoptosis, including up-regulation of Bax, Bak, and Pidd, as well as the activation of caspase-2, -9, and -3 (Fig. 4E, lane 3). It is proposed that Bruce ubiquitylates and eliminates Smac and inhibits Smac-mediated apoptosis (19). However, we did not observe an increase in the total amount of Smac or Omi upon Bruce silencing (Fig. 4E, lane 3). To examine whether p53 was responsible for this mitochondrial apoptosis, Bruce and p53 expression was simultaneously silenced in H460 cells, and the results showed a rescue of cell viability to 75%, compared with 20% viability induced by silencing Bruce alone (Fig. 4 F and G). As shown in Fig. 4E, lane 4, this ablation of p53 reduced the expression of the p53-responsive genes Bax and Pidd, and a p53-inducible gene, Bak (31). In addition, the reduced expression of Pidd, an activator of caspase-2, correlated with the restoration of caspase-2 zymogen. The amount of Smac and Omi, in contrast, did not change. In response to this dampened activation of proapoptotic factors, mitochondrial activation of the downstream caspase-9 and caspase-3 was reduced. It is noted that activation of caspase-3 was only partially decreased, indicating that caspase-3 is also activated by a p53-independent signal. Together, these results indicate that silencing Bruce activates p53. p53 up-regulation is required for the activation of Bax, Bak, and of Pidd/caspase-2, which permeabilize mitochondria and release cytochrome c, leading to caspase activation and apoptosis.
Discussion
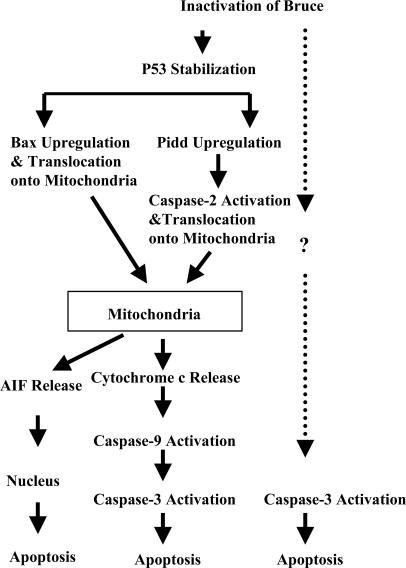
The Primary Function of Bruce Is to Regulate p53 and the Mitochondrial Pathway of Apoptosis. The study by Hao et al. (19) reported that Bruce is an E2/E3 ligase for ubiquitylating Smac and active caspase-9 in vitro, and it thus has an essential role in preventing apoptosis and preserving cell survival (19). Our study of Birc6GT mutant mice indicates Bruce mechanistically acts at a step upstream of mitochondria. We reason that in a healthy nonapoptotic cell, Smac and Bruce are unable to react as enzyme and substrate because of their different subcellular localization: Smac stays inside mitochondria, whereas Bruce localizes in the cytoplasm as well as in the Golgi apparatus (2, 13). Therefore, Bruce can bind to Smac to catalyze Smac ubiquitylation only after Smac release postmitochondrial damage. Similarly, caspase-9 is cleaved and activated by apoptosome only after mitochondrial damage (26). Because spontaneous Smac release and caspase-9 activation seldom occur, Bruce's primary survival function cannot be to scavenge these apoptotic molecules. As illustrated in Fig. 5, our studies on BruceGT mutant mice indicate that Bruce's primary survival function resides upstream of mitochondria. Loss of Bruce function triggers the up-regulation of p53. The transcriptional activity of p53 is responsible for the activation of Pidd, Bax, and Bak, which in turn activate mitochondria, leading to apoptosis.
Fig. 5.
Diagram of Bruce activation of apoptosis. Stabilization of p53 induced by Bruce inactivation turns on two parallel apoptotic signals. One signal is Bax/Bak, and the other signal is Pidd/caspase-2. Bax, Bak, and caspase-2 change the permeability of mitochondria and release cytochrome c and AIF. Cytochrome c, in turn, activates caspase-9 and caspase-3, whereas AIF kills cells in a caspase-independent manner. The dotted arrow indicates p53-independent activation of caspase-3, because ablation of p53 cannot eliminate caspase-3 activation induced by Bruce ablation (Fig. 4E).
p53 Is a Key Downstream Effector of Bruce. The tumor suppressor p53 integrates diverse cellular stress signals to induce cell-cycle arrest, apoptosis, or senescence (27). In this study, deletion of the C-terminal half of Bruce leads to stabilization of p53 and apoptosis, thereby making p53 a downstream effector of Bruce. How Bruce causes p53 stabilization is not known, but, from this study, we found that Bruce does not regulate the transcription of p53 (Fig. 4D). Considering that Bruce itself is an E2/E3 ubiquitin ligase, Bruce could act similarly to Mdm2, Pirh2, or COP1 to directly catalyze p53 ubiquitylation and proteasome degradation (32, 33). Alternatively, it could indirectly affect certain regulators of p53. Because p53 is activated by multiple stress signals, whether Bruce is functionally related to any stress, and if so, what kind of stress Bruce senses and responds to, will be interesting to address. It is of great significance to decipher how Bruce regulates p53 because this information will provide the molecular basis for clinical intervention. The identification of Bruce as an upstream regulator of p53 raises the possibility that therapeutic inactivation of Bruce activity could keep p53 levels high, and thus sensitize p53-mediated apoptosis in p53 wild-type tumors.
Why Does Apoptosis Appear in the Birc6GT Mutants but Not in the Gene-Targeted Bruce Knockout Mice? We observed apoptosis in our gene-trapped Bruce mutant mice and MEFs, but no apoptosis was found in the gene-targeted Bruce mutant mice by Lotz et al. (18). This phenotypic discrepancy raises the possibility that the fusion protein of the N-terminal half of Bruce with the βgeo reporter in Birc6GT mutant mice may exert a dominant-negative activity in vivo and accounts for the apoptosis. However, mice heterozygous for the insertional mutation are phenotypically normal. In addition, the apoptosis phenotypes in Birc6GT mutant mice are consistent with the antiapoptosis function of Drosophila Bruce in flies (16). Moreuover, down-regulation of the endogenous Bruce mRNA by siRNA produced spontaneous cell death in various human cell lines by different laboratories, including our own (15, 34), and this result recapitulated what we observed in MEFs expressing truncated Bruce protein. Finally, the UBC domain is essential for Bruce to repress apoptosis (14). Taken together, these results argue against the possibility that the apoptosis observed in our gene-trapped mice is the result of a nonphysiologic activity of the Bruce-β-gal fusion protein.
In the case of gene-targeted disruption of the Bruce gene, the knockout strategy used by Lotz et al. (18) eliminates essential portions of the BIR domain. The authors explain the absent apoptosis in their mice as the consequence of possible overlapping IAP activities. They also proposed that although the level of other IAPs is not elevated in their mutant mice and MEFs, other IAPs are likely to compensate the phenotype because Bruce is 10-fold weaker than XIAP. Our explanation for this phenotypic discrepancy is that the BIR domain maintained in our Birc6GT mutant mice may have been interpreted by the genome as an “active” IAP molecule and thus prevents functional contribution from other IAPs. However, this truncated Bruce fails to inhibit apoptosis because of the elimination of UBC domain, which is essential for Bruce's antiapoptosis activity (14). Therefore, the Birc6GT mutant mice manifest apoptosis. Future investigation from conditional knockout of the Bruce gene in mice and mapping other functional domains on Bruce will help to address this phenotypic discrepancy.
Supplementary Material
Acknowledgments
We thank Dr. Robb Krumlauf for critical comments on the manuscript; Michelle L. Newton, Kyle D. Stanosheck, Carolyn Foster, and Steve Punke for discussions and technical support; Debra Grant and Jean-Philippe Rey for their tremendous support and contribution of histologic techniques and analysis; Sach Jayasinghe and Jeff Haug for flow cytometry; Eric Schuenemann and Heather Newkirk for preparing in situ hybridization probes; and Donna di Natale for assistance with editing the manuscript.
Author contributions: J.R., W.C.S., and C.D. designed research; J.R., M.S., R.L., T.J., and C.D. performed research; W.C.S. contributed new reagents/analytic tools; J.R., T.J., and C.D. analyzed data; and W.C.S. and C.D. wrote the paper.
Abbreviations: IAP, inhibitor of apoptosis; Birc, baculoviral IAP repeat-containing; AIF, apoptosis-inducing factor; MEF, mouse embryonic fibroblast; Smac, second mitochondrial-derived activator of caspase; En, embryonic day n; siRNA, small interfering RNA.
References
- 1.Vaux, D. L. & Korsmeyer, S. J. (1999) Cell 96, 245-254. [DOI] [PubMed] [Google Scholar]
- 2.Du, C., Fang, M., Li, Y., Li, L. & Wang, X. (2000) Cell 102, 33-42. [DOI] [PubMed] [Google Scholar]
- 3.Verhagen, A. M., Ekert, P. G., Pakusch, M., Silke, J., Connolly, L. M., Reid, G. E., Moritz, R. L., Simpson, R. J. & Vaux, D. L. (2000) Cell 102, 43-53. [DOI] [PubMed] [Google Scholar]
- 4.Goyal, L., McCall, K., Agapite, J., Hartwieg, E. & Steller, H. (2000) EMBO J. 19, 589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, S. L., Hawkins, C. J., Yoo, S. J., Muller, H. A. & Hay, B. A. (1999) Cell 98, 453-463. [DOI] [PubMed] [Google Scholar]
- 6.Harlin, H., Reffey, S. B., Duckett, C. S., Lindsten, T. & Thompson, C. B. (2001) Mol. Cell. Biol. 21, 3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy, N., Mahadevan, M. S., McLean, M., Shutler, G., Yaraghi, Z., Farahani, R., Baird, S., Besner-Johnston, A., Lefebvre, C., Kang, X., et al. (1995) Cell 80, 167-178. [DOI] [PubMed] [Google Scholar]
- 8.Holcik, M., Thompson, C. S., Yaraghi, Z., Lefebvre, C. A., MacKenzie, A. E. & Korneluk, R. G. (2000) Proc. Natl. Acad. Sci. USA 97, 2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada, H., Bakal, C., Shahinian, A., Elia, A., Wakeham, A., Suh, W. K., Duncan, G. S., Ciofani, M., Rottapel, R., Zuniga-Pflucker, J. C. & Mak, T. W. (2004) J. Exp. Med. 199, 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman, D., Kim, P. J., Blanc-Brude, O. P., Brash, D. E., Tognin, S., Marchisio, P. C. & Altieri, D. C. (2001) J. Clin. Invest. 108, 991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uren, A. G., Wong, L., Pakusch, M., Fowler, K. J., Burrows, F. J., Vaux, D. L. & Choo, K. H. (2000) Curr. Biol. 10, 1319-1328. [DOI] [PubMed] [Google Scholar]
- 12.Conway, E. M., Pollefeyt, S., Steiner-Mosonyi, M., Luo, W., Devriese, A., Lupu, F., Bono, F., Leducq, N., Dol, F., Schaeffer, P., Collen, D. & Herbert, J. M. (2002) Gastroenterology 123, 619-631. [DOI] [PubMed] [Google Scholar]
- 13.Hauser, H. P., Bardroff, M., Pyrowolakis, G. & Jentsch, S. (1998) J. Cell. Biol. 141, 1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartke, T., Pohl, C., Pyrowolakis, G. & Jentsch, S. (2004) Mol. Cell 14, 801-811. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Z., Naito, M., Hori, S., Mashima, T., Yamori, T. & Tsuruo, T. (1999) Biochem. Biophys. Res. Commun. 264, 847-854. [DOI] [PubMed] [Google Scholar]
- 16.Vernooy, S. Y., Chow, V., Su, J., Verbrugghe, K., Yang, J., Cole, S., Olson, M. R. & Hay, B. A. (2002) Curr. Biol. 12, 1164-1168. [DOI] [PubMed] [Google Scholar]
- 17.Arama, E., Agapite, J. & Steller, H. (2003) Dev. Cell 4, 687-697. [DOI] [PubMed] [Google Scholar]
- 18.Lotz, K., Pyrowolakis, G. & Jentsch, S. (2004) Mol. Cell. Biol. 24, 9339-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao, Y., Sekine, K., Kawabata, A., Nakamura, H., Ishioka, T., Ohata, H., Katayama, R., Hashimoto, C., Zhang, X., Noda, T., et al. (2004) Nat. Cell. Biol. 6, 849-860. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell, K. J., Pinson, K. I., Kelly, O. G., Brennan, J., Zupicich, J., Scherz, P., Leighton, P. A., Goodrich, L. V., Lu, X., Avery, B. J., et al. (2001) Nat. Genet. 28, 241-249. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, B., Beddington, R. S., Constantini, F. & Lacy, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 22.Okada, H., Suh, W. K., Jin, J., Woo, M., Du, C., Elia, A., Duncan, G. S., Wakeham, A., Itie, A., Lowe, S. W., et al. (2002) Mol. Cell. Biol. 22, 3509-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lassus, P., Opitz-Araya, X. & Lazebnik, Y. (2002) Science 297, 1352-1354. [DOI] [PubMed] [Google Scholar]
- 24.Wei, M. C., Zong, W. X., Cheng, E. H., Lindsten, T., Panoutsakopoulou, V., Ross, A. J., Roth, K. A., MacGregor, G. R., Thompson, C. B. & Korsmeyer, S. J. (2001) Science 292, 727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. (1996) Cell 86, 147-157. [DOI] [PubMed] [Google Scholar]
- 26.Li, P., Nijhawan, D., Budihardjo, I., Srinivasula, S. M., Ahmad, M., Alnemri, E. S. & Wang, X. (1997) Cell 91, 479-489. [DOI] [PubMed] [Google Scholar]
- 27.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita, T. & Reed, J. C. (1995) Cell 80, 293-299. [DOI] [PubMed] [Google Scholar]
- 29.Lin, Y., Ma, W. & Benchimol, S. (2000) Nat. Genet. 26, 122-127. [DOI] [PubMed] [Google Scholar]
- 30.Tinel, A. & Tschopp, J. (2004) Science 304, 843-846. [DOI] [PubMed] [Google Scholar]
- 31.Kannan, K., Amariglio, N., Rechavi, G., Jakob-Hirsch, J., Kela, I., Kaminski, N., Getz, G., Domany, E. & Givol, D. (2001) Oncogene 20, 2225-2234. [DOI] [PubMed] [Google Scholar]
- 32.Leng, R. P., Lin, Y., Ma, W., Wu, H., Lemmers, B., Chung, S., Parant, J. M., Lozano, G., Hakem, R. & Benchimol, S. (2003) Cell 112, 779-791. [DOI] [PubMed] [Google Scholar]
- 33.Dornan, D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G. D., Dowd, P., O'Rourke, K., Koeppen, H. & Dixit, V. M. (2004) Nature 429, 86-92. [DOI] [PubMed] [Google Scholar]
- 34.Qiu, X. B., Markant, S. L., Yuan, J. & Goldberg, A. L. (2004) EMBO J. 23, 800-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.