Abstract
We have mapped protein conformational space from two to seven residue lengths by employing multidimensional scaling on a data matrix composed of pair-wise angular distances for multiple ϕ-Ψ values collected from high-resolution protein structures. The resulting global maps show clustering of peptide conformations that reveals a dramatic reduction of conformational space as sampled by experimentally observed peptides. Each map can be viewed as a higher order ϕ-Ψ plot defining regions of space that are conformationally allowed.
Keywords: global peptide conformational mapping, multidimensional scaling, Ramachandran map, global mapping
The Ramachandran map was conceived as a theoretical means of predicting the allowed conformational space of a single amino acid in a peptide by means of a hard sphere model that allowed for the steric coupling effects of both ϕ and Ψ angles (1). This work showed that protein conformations are substantially restricted, due to steric hindrances from what one might expect without considering the coupling of ϕ and Ψ angles. Conformations of experimental structures can be plotted into this ϕ-Ψ space. If this plot is constructed from a database of protein structures that are well resolved, the 2D plot discriminates ϕ-Ψ space into “allowed” and “disallowed” regions by outlining the most populated regions. The experimentally observed conformations from well resolved structures basically correspond to those regions of ϕ-Ψ space initially predicted by Ramachandran. New structures can be analyzed for the fraction of residues within allowable regions. This type of analysis is implemented in commonly used validation tools for protein structure, such as procheck (2). In this way, the ϕ-Ψ plot has proven itself as an unequalled tool in understanding the conformational space available for proteins and in the refinement and analysis of newly determined protein structures.
It is also possible to validate protein structures by means of longer fragment lengths. Protein substructures or building blocks have been used for modeling earlier by Unger et al. (3) and Alwyn Jones and Thirup (4). Most recently, Micheletti et al. (5) have also demonstrated that the conformational spaces for peptides are restricted, and almost any known protein structure can be reconstructed within 1 Å rms deviation by using a representative set of polypeptide units of four to seven residues in length with between 28 and 2,500 representative conformations, respectively (5), suggesting that it is possible to define an allowable space for polypeptides longer than three residues. The conformation of a dipeptide fragment, that is, two complete residues in length (with attached C- and N-terminal peptide bonds), can be described by four torsion angles (two pairs of ϕ-Ψ values) around two central Cα atoms. We refer to polypeptide units of a given length by the number of ϕ-Ψ pairs: (ϕ, Ψ)1, which is equivalent to a Ramachandran map, (ϕ, Ψ)2, (ϕ, Ψ)3, and so forth. Unfortunately, the 4D space of the (ϕ, Ψ)2 unit (and the subsequent higher dimensional spaces of longer units) cannot be readily visualized in two or even three dimensions. However, the multidimensional scaling (MDS) method often allows one to reduce the number of dimensions and view the conformational space in a reduced (e.g., three) dimensional representation. A family of statistical methods exists that can be used for dimensional reduction, of which we have used classical MDS in interpreting the conformational space of each polypeptide length. The technique of mapping by means of dimensional reduction has been applied successfully in nucleic acid conformational space as well as protein fold space (6–8). We have implemented MDS for extending conformational space analysis to peptide fragments of longer length beyond (ϕ,Ψ)1, the conventional ϕ-Ψ map.
In this work, we cluster the high-resolution peptide conformations of two to five ϕ-Ψ pairs long in 3D space, where clusters of recurring peptide conformations can be visualized. We observe that the number of conformational clusters is drastically smaller than the values predicted from theoretical consideration, suggesting that the conformational space sampled by a growing peptide is considerably smaller than generally assumed.
Materials and Methods
Data. A reference database of high-resolution structures (≤1.0 Å) was created from a nonredundant PDB structure collection from PDBSELECT (April, 2003) with <25% sequence identity (see Table 2, which is published as supporting information on the PNAS web site, for structures) (9). This set contains 51 structures in 44 Structural Classification of Proteins (SCOP) folds, providing 10,976 ϕ-Ψ pairs (10). Structures determined at ≤1.0-Å resolution are of near atomic resolution, excluding the possibility of any model fitting bias of conformational restraints incorporated into the refinement procedures of structures at lower resolutions. Torsion angles (ranging from 0° to 360°), ϕ and Ψ, were calculated for the reference structures by using dssp (11). Each structure was segmented into (ϕ, Ψ)n length units by a sliding window of length n, for values of n = 1–5. Every (ϕ, Ψ)n unit is represented by a vector of 2n torsion angles. Next, 6,000 fragments were randomly chosen from the set of each unit length. This random sampling was necessary because of matrix size restrictions in the MDS algorithm (which is discussed further below).
MDS. To visualize (ϕ, Ψ)n conformations, these higher dimensional spaces must be embedded into a lower dimensional space by employing, for example, classical (metric) MDS (12). We use MDS as implemented in the cmdscale function from the multivariate analysis package of r (13). MDS transforms a matrix of pair-wise distances into a reduced dimensional space, e.g., 3D Cartesian space, where Euclidean distances in the reduced space are approximately proportional to the original higher-dimensional distances.
A distance matrix, DE, can be formed with Euclidean distances, where each matrix element is calculated,
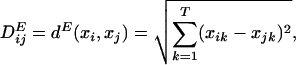 |
[1] |
where T = 2n is the number of torsion angles in each (ϕ-Ψ)n unit, xi and xj are the ϕ-Ψ angle vectors for each peptide fragment that range over the entire reference database.
However, because angles are circular quantities, the angular distance matrix, DA, can also be constructed using modified forms of Euclidean distances that account for angular circularity by calculating the minimum angular distances ≤180°. The angular distance measure is the physically meaningful way to calculate distances between angles (14). The form of the distance matrix we use is calculated by means of Eq. 2.
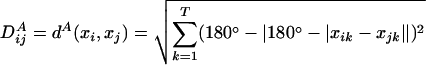 |
[2] |
We had tried using other distance metrics to construct the distance matrix, for example, rms deviation. However, the clustering we obtained was not as well defined as the results we present here using angular distances.
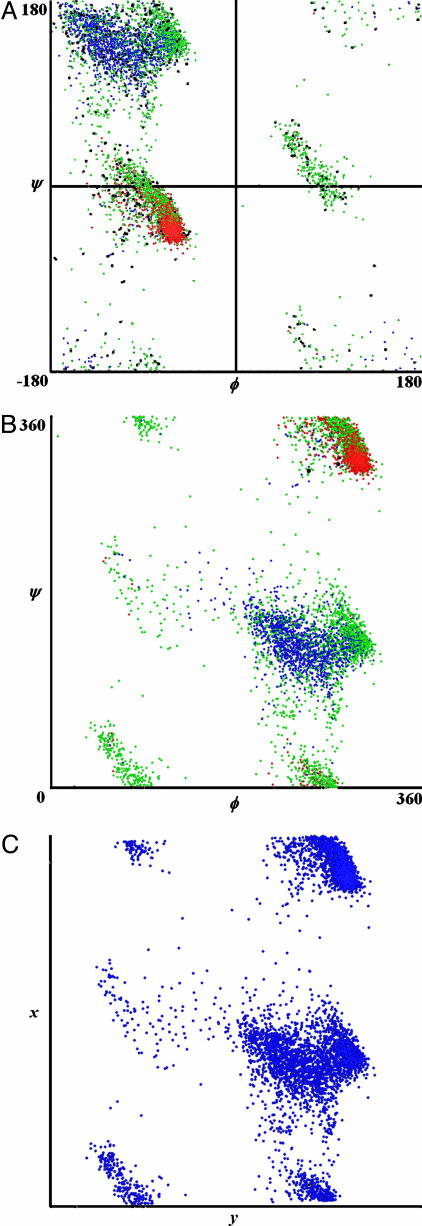
MDS Equivalence of the ϕ-Ψ Map. To test the equivalence of our method to the 2D ϕ-Ψ map (Fig. 1 A and B) we constructed DE using Eq. 1 for (ϕ, Ψ)1 units. Next, the distance information in DE was scaled with MDS, which revealed only two eigenvalues. The mapping of the results in two dimension (Fig. 1C) reproduced exactly the familiar ϕ-Ψ space, thus, validating the MDS method for the conformational clustering analysis. When DA is constructed by using Eq. 2 for (ϕ, Ψ)1 units, MDS returned three major eigenvalues and, the conformational space in 3 dimensions appears as a toroid corresponding to the folding of the 2D surface of Fig. 1 due to the angular identity of 0 and 360°.
Fig. 1.
Equivalence of ϕ-Ψ map and 2D MDS map. (A) A ϕ-Ψ map of high-resolution protein structures (1.0 Å or better) from a PDB collection of nonredundant protein structures. Colors indicate dssp defined secondary structures: helix (red), sheet (blue), and coil or any other type (green). (B) The ϕ-Ψ map where all angles are transformed to a 0°–360° scale, to compare with our MDS map of the same data. (C)A(ϕ,Ψ)1 MDS map constructed from normal Euclidean distances (Eq. 1) yields conformational clusters almost identical to those of the conventional ϕ-Ψ map. The first and second principal components are indicated by x and y.
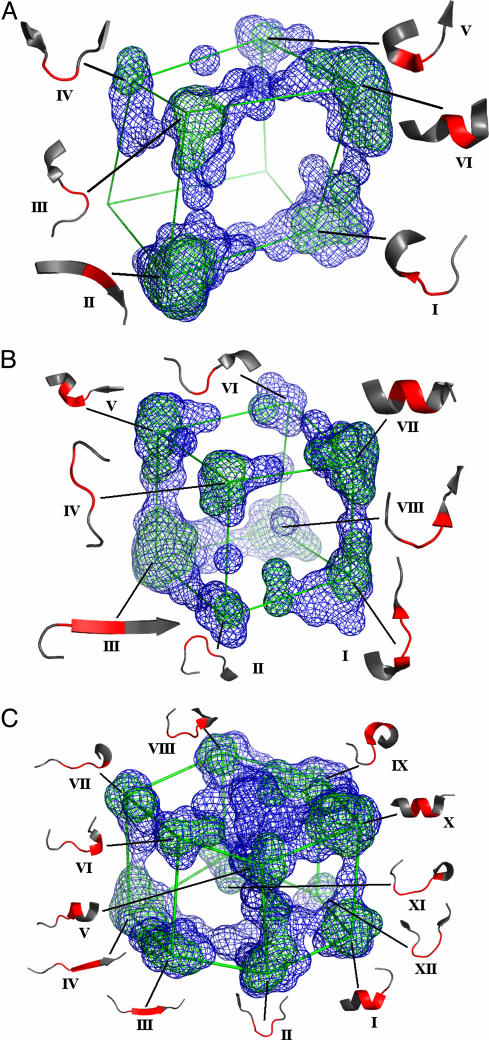
Global Mapping of Protein Conformations. Conformational space maps were constructed from the high-resolution reference set by using minimum angular distances (Eq. 2) for units of size (ϕ, Ψ)2 to (ϕ, Ψ)5. First, eigenvalues from the MDS were examined to assess the validity of using the first three components to approximately represent the conformational space of higher order ϕ-Ψ descriptions of the peptide fragments. In all cases, the three largest eigenvalues were significantly greater than the rest. The distribution of Cartesian coordinates representing all sampled peptide units of a given length obtained from the MDS procedure was converted to a density contour by means of the following procedure. Each of the three dimensions x, y, and z was divided into 70 bins, creating 3.43 × 105 cubes. Each cube was assigned the frequency of peptide units occurring within that bin. The average frequency of all cubes, n, and standard deviation, σ, were calculated, and the space was contoured at the n + 2σ and the n + 3σ levels. Fig. 2 depicts the 3D projections of each of these conformational spaces. If vertices are placed in the highest densities of the map and connected, the resulting objects represent familiar polyhedral shapes (cube and hexagonal prism) with varying structural occupancy at each of the vertices.
Fig. 2.
Higher order ϕ-Ψ maps and representative conformations. The MDS projections are represented as wire mesh surfaces at two colored contour (σ) levels (blue, 2σ; green, 3σ). Wire frame polyhedrons connect cluster centers to indicate the distance relationships of the conformational clusters at each vertex. Each conformation (in red) is indicated in the context of the local structure in which the sample conformation was found. (A) For (ϕ, Ψ)2, clusters are arranged as a cube with six of eight vertices densely occupied. For each cluster, some annotated conformations are listed (residue positions in fragment are indicated where appropriate): I, turn-VIII; II, β-extended; III, turn-III, turn-Via; IV, turn-II; V, helical C-cap; VI, α-helix, 310, and π, turn-I. (B) For (ϕ, Ψ)3, the figure is also cubic in shape, but with all vertices densely occupied (blue, 2σ; green, 3σ). Annotated conformations are as follows: I, turn II (1–2); II, turn II- (1–2); III, β-extended; IV, turn-I (3–4); V, helical N-cap; VI, turn-II (2–3); VII, helix; VIII, turn-I (2–3). (C) For (ϕ, Ψ)4, the figure is hexagonal in shape (blue, 3σ; green, 4σ). Annotated conformations are as follows: I, helix C-cap; II, turn-I (3–4); III, turn-II (3–4); IV, β-extended; V, helix N-cap; VI, turn-I (4–5); VII, turn-I (5–6); VIII, turn-II (1–2); IX, turn-I (residues 2–3); X, helix; XI, turn-II (1–2); XII, turn-II (2–3). See Rose et al. (16) for structural definitions. The ray traced figures were drawn by using pymol (17).
Results
Global Mapping of Peptide Conformations. Our method demonstrates equivalence between MDS clustering and the traditional ϕ-Ψ plot at the level of (ϕ, Ψ)1 as shown in Fig. 1 A and B. As mentioned earlier, for (ϕ, Ψ)2 through (ϕ, Ψ)5, the MDS method using DA matrices reveals that in all cases the three largest eigenvalues are significantly larger than all of the remaining eigenvalues, thus justifying the dimensional reduction to 3D as a reasonable approximation, which provides an intuitively understandable representation (Table 1). Each 3D representation produces occupancy distributions of the higher order peptide conformational space that resemble geometric shapes (Fig. 2). The vertices of each polyhedra roughly correspond to the centers of conformational clusters. See Table 3, which is published as supporting information on the PNAS web site, for angle statistics for each cluster. In the (ϕ, Ψ)2 map, only six conformational clusters are observed, and they occupy six of eight vertices of a virtual cube (Fig. 2 A). In the (ϕ, Ψ)3 map, only eight conformational clusters are observed, and they occupy all eight vertices of the cube. Curiously, not a great amount of conformational variation is introduced by lengthening (ϕ, Ψ)2 unit size to (ϕ, Ψ)3, suggesting that steric hindrances play a greater role in restricting the conformational space available as the peptide grows longer. The (ϕ, Ψ)4 map resembles a hexagonal prism representing an increase in cluster number. Adding a further degree of complexity, (ϕ, Ψ)5 appears as a three-layer hexagonal prism. Beyond (ϕ, Ψ)5, the structure of the MDS plots becomes much more dispersed. In all of the maps, the first component (the most significant eigenvalue) can roughly discriminate between helical-like and extended-like peptide fragments.
Table 1. Normalized eigenvalues from MDS.
| Eigenvalue index (dimension)
|
|||||
|---|---|---|---|---|---|
| (ϕ-ψ)n | 1 | 2 | 3 | 4 | 5 |
| 1 | 1 | 0.25 | 0.01 | 0 | 0 |
| 2 | 1 | 0.30 | 0.16 | 0.16 | 0.13 |
| 3 | 1 | 0.31 | 0.21 | 0.13 | 0.12 |
| 4 | 1 | 0.32 | 0.21 | 0.17 | 0.11 |
| 5 | 1 | 0.33 | 0.21 | 0.16 | 0.14 |
Discussion
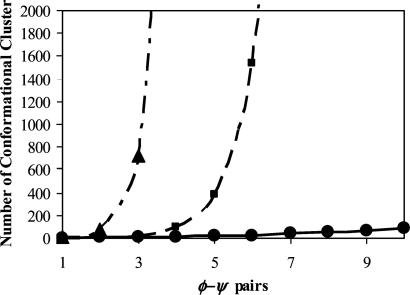
The MDS method for representing protein conformations of peptide units longer than three residues is a valuable tool for conceptualizing conformational spaces that would otherwise be difficult to visualize and interpret. In these maps, we were able to capture the simplicity of the original Ramachandran concept in terms of allowed/disallowed space for longer peptides from experimentally determined protein structures. The geometrical nature of each of the maps is also a fascinating result, and not purely an artifact of the scaling procedure. Each vertex of the polyhedral shapes represents a conformational cluster. We have made the angular statistics of each cluster available in Table 3. The number of clusters in each map for every ϕ-Ψ unit length may be tallied by counting the occupied vertices of each polyhedron. In this case, a relationship between peptide length and number of conformational clusters is obtained that contrasts with Levinthal's paradox and the combinatorial extension of the Ramachandran plot (Fig. 3) (15). Levinthal suggested that each amino acid residue adopts its conformation independent of each other, and that the configurations of each residue are independent of the preceding and following residues. If we allow each ϕ-Ψ pair to be constructed only with Levinthal angles (60°, 180°, 300°), this results in 3 × 3 = 9 conformations per ϕ-Ψ pair. The extrapolated result is the steepest exponential curve, represented by 9N, where N is the number of ϕ-Ψ pairs. Allowing four conformational clusters per ϕ-Ψ pair based upon the Ramachandran map (Fig. 1 A) indicates a less steep curve of 4N. However, the curve plotted by our method of conformational cluster analysis is not as steeply exponential (≈1.6N). For example, one hundred ϕ-Ψ pairs will have 5.95 × 1015, 1.61 × 1060, or 2.66 × 1095 conformational clusters using MDS, Ramachandran, or Levinthal models, respectively. Performing conformational searches through this reduced allowed conformational space may not be as impossible as the extreme cases implied by the Levinthal and Ramachandran models.
Fig. 3.
Extrapolating the number of conformational clusters. Exponential curves compare the number of clusters for the nth (ϕ, Ψ) unit. Shown are combinatorial extension of four conformational clusters per ϕ-Ψ pair in Ramachandran (dot-dash), combinatorial extension of nine clusters per ϕ-Ψ pair based on Levinthal assumption (dashed), and our estimate of the number of peptide conformational clusters from MDS (solid).
Conclusion
Our maps of the experimental peptide conformational space from two to five ϕ-Ψ pairs show conformational clusters, the number of which is drastically smaller than those predicted from theoretical consideration, and each map can be interpreted as a higher order ϕ-Ψ plot. Protein conformational space has been thoroughly studied for single ϕ-Ψ pairs, but less for higher order ϕ-Ψ pairs, which are expected to be highly restricted due to steric hindrance. However, it was not known to what extend the space would be restricted. As shown in Fig. 3, the restriction of the conformational space is dramatic.
Another possible utility for the clustering of higher order conformations is to evaluate the likelihood of protein structure models, predicted or determined experimentally by using low-resolution data. Just as the Ramachandran map is commonly used to assess the fraction of protein structures that are in allowed regions of single ϕ-Ψ pairs, our results can provide a similar assessment of higher order ϕ-Ψ pairs. In higher order spaces, there are defined regions that are allowed, and we hope that this restricted conformational space can reduce the conformational search space in theoretical protein folding studies, and ab initio model builders can use this conformational space as a tool to gauge the quality of their predicted structural models, or even employ the space in the model building process.
Supplementary Material
Acknowledgments
We thank our colleagues Chao Zhang, Jingtong Hou, Se-Ran Jun, Jaimyoung Kwon, and Jennifer Sims for their help, advice, and discussions throughout the course of this work. This work was supported by National Science Foundation Grant DBI-0114707, by National Institutes of Health Grants GM-62412 (to S.-H.K.) and GM-0829515/HG-0004705 (to G.E.S.), and by the National Energy Research Scientific Computing Center at Lawrence Berkeley National Laboratory, which is supported by the Department of Energy.
Author contributions: G.E.S., I.-G.C., and S.-H.K. designed research, performed research, analyzed data, and wrote the paper.
Abbreviation: MDS, multidimensional scaling.
References
- 1.Ramachandran, G. N. & Sasisekharan, V. (1963) J. Mol. Biol. 7, 95-99. [DOI] [PubMed] [Google Scholar]
- 2.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 3.Unger, R., Harel, D., Wherland, S. & Sussman, J. L. (1989) Proteins 5, 355-377. [DOI] [PubMed] [Google Scholar]
- 4.Alwyn Jones, T. & Thirup, S. (1986) EMBO J. 5, 819-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micheletti, C., Seno, F. & Maritan, A. (2000) Proteins Struct. Funct. Genet. 40, 662-674. [DOI] [PubMed] [Google Scholar]
- 6.Sims, G. E. & Kim, S.-H. (2003) Nucleic Acids Res. 31, 5607-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou, J., Sims, G. E., Zhang, C. & Kim, S.-H. (2003) Proc. Natl. Acad. Sci. USA 100, 2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm, L. & Sander, C. (1999) Nucleic Acids Res. 27, 244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobohm, U. & Sander, C. (1994) Protein Sci. 3, 522-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536-540. [DOI] [PubMed] [Google Scholar]
- 11.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577-2637. [DOI] [PubMed] [Google Scholar]
- 12.Torgerson, W. S. (1952) Psychometrika 17, 401-419. [Google Scholar]
- 13.Ihaka, R. & Gentleman, R. (1996) J. Comput. Graph. Stat. 5, 299-314. [Google Scholar]
- 14.Reijmers, T. H., Wehrens, R. & Buydens, L. M. C. (2001) Chemometr. Intell. Lab. 56, 61-71. [Google Scholar]
- 15.Levinthal, C. (1968) J. Chim. Phys. PCB 65, 44-45. [Google Scholar]
- 16.Rose, G. D., Gierasch, L. M. & Smith, J. A. (1985) Adv. Protein Chem. 37, 1-109. [DOI] [PubMed] [Google Scholar]
- 17.Delano, W. L. (2002) The PyMol Molecular Graphics System (DeLano Scientific, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.