Abstract
Necroptosis is a cellular mechanism of necrotic cell death induced by apoptotic stimuli in the form of death domain receptor engagement by their respective ligands under conditions where apoptotic execution is prevented. Although it occurs under regulated conditions, necroptotic cell death is characterized by the same morphological features as unregulated necrotic death. Here we report that necrostatin-1, a previously identified small-molecule inhibitor of necroptosis, is a selective allosteric inhibitor of the death domain receptor–associated adaptor kinase RIP1 in vitro. We show that RIP1 is the primary cellular target responsible for the antinecroptosis activity of necrostatin-1. In addition, we show that two other necrostatins, necrostatin-3 and necrostatin-5, also target the RIP1 kinase step in the necroptosis pathway, but through mechanisms distinct from that of necrostatin-1. Overall, our data establish necrostatins as the first-in-class inhibitors of RIP1 kinase, the key upstream kinase involved in the activation of necroptosis.
Apoptosis, a genetically encoded and highly regulated pathway of cell death, plays an important role in the normal development of multi-cellular eukaryotes and in the maintenance of adult organism homeostasis1. However, it has also become increasingly clear that cells have alternative means to trigger regulated cell death, thereby allowing elimination of damaged or inappropriately stimulated cells even in the absence of functional apoptosis. Mutant mice that are deficient in key apoptotic factors (for example, members of the caspase or Bcl-2 families), while exhibiting certain developmental abnormalities, are still able to survive to adulthood2,3. Furthermore, certain well-characterized developmental cell death events (for example, interdigital tissue elimination4 or motor neuron cell death5) occur normally under these apoptosis-deficient conditions with a distinct non-apoptotic morphology. Importantly, activation of non-apoptotic cell death has been observed in diverse animal models of human pathologies, such as neonatal brain hypoxia6, acute pancreatitis7 and multiple organ failure8, further establishing the importance of determining molecular mechanisms of non-apoptotic cell death. Multiple types of non-apoptotic cell death have been described in the literature, including autophagic cell death, necroptosis, aponecrosis and paraptosis; however, the underlying molecular mechanisms of these processes are just beginning to emerge.
Necroptosis is a notable example of non-apoptotic cell death. It is triggered by the same stimuli that normally activate apoptosis, which underscores the notion that it is a regulated process of cell death. However, necroptosis is clearly distinct from apoptosis, as it does not involve key apoptosis regulators such as caspases and Bcl-2 family members, or cytochrome c release from mitochondria9. Furthermore, the cell morphology of necroptotic demise, including early loss of plasma membrane integrity, lack of nuclear fragmentation, mitochondrial dysfunction and oxidative stress, is very similar to that of necrosis. Notably, cell death with necrotic features is prevalent in an array of acute human pathologies, including myocardial infarction, cerebral ischemia and acute organ failure. However, inhibition of necrosis is not currently perceived as a viable therapeutic strategy because necrosis is believed to be an unregulated process caused by overwhelming external stress. As “apoptotic” stimuli, now found to also trigger programmed necrosis, are well established as contributing to many instances of pathologic injury1, the discovery of necroptosis offers the possibility that a subset of pathologic necrotic cell death is regulated by a distinct cellular mechanism, and therefore is amenable to therapeutic intervention.
Important insights into the signaling mechanism of regulated necrosis have been recently reported. Serine/threonine kinase activity of the death domain receptor (DR)-associated molecule RIP1 was found to be essential for Fas ligand–induced and tumor necrosis factor-α (TNFα)-induced necrosis in Jurkat cells10. However, the mechanism that is responsible for the execution of necroptosis is not yet clear. We have previously reported the identification of a potent and selective inhibitor of necroptosis, necrostatin-1 (Nec-1, 1), which efficiently suppresses necroptotic cell death triggered by an array of stimuli in a variety of cell types9. Furthermore, we and others have found that Nec-1–inhibitable non-apoptotic death is an important contributor to pathologic injury in mouse models of cerebral ischemia9 and myocardial infarction11. The target of Nec-1 therefore represents a critical mediator in the necroptosis pathway that is relevant to regulation of pathologic necrosis.
In the present study, we explored the mechanism of action of Nec-1 and other necrostatins. We report that three structurally distinct necrostatins—necrostatin-1, necrostatin-3 (Nec-3, 2) and necrostatin-5 (Nec-5, 3)—all inhibit RIP1 kinase activity, thereby highlighting the role of this molecule as a key step in the necroptosis pathway. Furthermore, we present initial analysis of the mechanism of action of three necrostatins that suggests distinct modes of RIP1 inhibition. Overall, these data establish RIP1 kinase as a new target for therapeutic drug development for human diseases involving necrotic tissue injury, and they establish necrostatins as first-in-class potent and selective inhibitors of RIP1 kinase.
Results
Nec-1 is a RIP1 kinase inhibitor in vitro
Our previous analysis9 showed that 1 is capable of inhibiting necroptosis triggered by dimerization of the RIP1 kinase domain, which suggests that Nec-1 exerts its activity at or downstream from the step of RIP1 kinase activation. Furthermore, because indoles similar to 1 (Fig. 1a) are a common structural feature in kinase inhibitors (for example, the CDK2 inhibitor SU9516 (4)12), we considered the possibility that 1 may directly inhibit RIP1 kinase activity. Though no direct RIP1 kinase substrate has been thus far identified, previous studies have suggested13 that RIP1 can autophosphorylate based on incorporation of radioactive phosphate in an in vitro kinase reaction using overexpressed and immunoprecipitated RIP1. We first sought to confirm that phosphorylation of RIP1 in this assay results from its own kinase activity and is not due to other associated kinases. Indeed, we observed robust phosphorylation of overexpressed wild type, but not the kinase-inactive K45M mutant of RIP1 (Fig. 1b).
Figure 1.
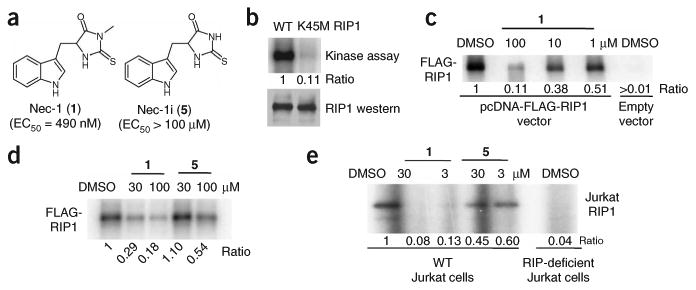
Nec-1 (1) is an inhibitor of RIP1 kinase. (a) Structures of 1 and 5. EC50 values for inhibition of cellular necrosis in TNFα-treated FADD-deficient Jurkat cells were determined as described in the Methods and were previously reported14. (b) Phosphorylation of RIP1 requires its kinase activity. Expression constructs of FLAG-tagged wild-type (WT) or a kinase-inactive point mutant of RIP1 (K45M) were transfected into 293T cells and RIP1 kinase assay was performed as described in the Methods in the presence of [γ-32P]ATP for 30 min at 30 °C. Samples were subjected to SDS-PAGE and RIP1 band was visualized by autoradiography. Relative intensities of radioactive bands were quantified and are shown (ratio) in this and all other autoradiographs. In parallel to kinase reactions, a sample of beads was subjected to western blot analysis using anti-RIP1 antibody to ensure equal protein amounts in kinase reactions. (c) 1 inhibits the autophosphorylation of overexpressed RIP1 in vitro in a dose-dependent fashion. Assay was performed as in b, except different amounts of 1 were added 15 min before ATP. (d) Inactive analog of 1 (5) displays substantially reduced activity against RIP1 kinase in vitro. Assay was performed as in c with indicated concentrations of 1 and 5. (e) 1 inhibits the autophosphorylation of endogenous RIP1 in vitro. RIP1 was immunoprecipitated from lysates of WT and RIP1-deficient Jurkat cells using agarose-conjugated anti-RIP1 antibody, and kinase reactions were performed as in c in the presence of the indicated amounts of 1 or 5. Assays were performed at least two or three times, and similar results were obtained each time. The representative images are shown.
Using this assay, we next tested whether RIP1 kinase activity might be inhibited in the presence of 1. Indeed, 1 efficiently inhibited kinase activity of overexpressed protein (Fig. 1c) in a dose-dependent manner. To establish the specificity of 1, we took advantage of the very specific structure-activity relationship (SAR) of 1 established in the cellular necroptosis assays9. For example, we previously found that elimination of a single methyl group (5, Fig. 1a) results in only marginal antinecroptotic activity9. Compound 5 was similarly much less active in the in vitro RIP1 kinase assay (Fig. 1d). Next, we determined that endogenous RIP1, immunoprecipitated from Jurkat cells, also displays activity in the in vitro kinase assay (Fig. 1e). Similar to the results with overexpressed protein, 1 (but not 5) efficiently suppressed endogenous RIP1 kinase activity (Fig. 1e). These results show that 1 can inhibit RIP1 kinase in vitro, and this activity correlates with 1's ability to inhibit necroptotic cell death.
To further verify this conclusion, we again took advantage of the extensive SAR for 1 from the cellular assay (Fig. 2a and ref. 14). We selected three additional analogs (6–8) differing from 1 by the addition of a single methyl group at various positions. All three compounds showed substantially reduced antinecroptotic activity and were likewise ineffective at inhibiting RIP1 kinase activity in vitro (Fig. 2b). At the same time, we previously determined that of all the positions on 1, only replacement of the sulfur of the thiohydantoin with oxygen (as in 9) or changes to the 7 position of the indole ring (for example, addition of chlorine as in 10) retain cellular necroptotic inhibitory activity14. Both of these derivatives were also active in the in vitro kinase assay (Fig. 2c). Finally, we previously reported that the (R)-7-chloromethylhydantoin-Trp ((R)-7-Cl-MH-Trp or (R)-7-Cl-O-Nec-1, 11, Supplementary Fig. 1 online) analog of 1 is ∼10-fold more active in the cell-based necroptosis assay than the original molecule14. Similarly, this molecule was also substantially more active in the in vitro kinase assay (effector concentration for half-maximum response (EC50) = 182 nM for 1 against endogenous RIP1 from Jurkat cells versus EC50 = 18 nM for 11, Supplementary Fig. 1). Overall, these data show that the Nec-1 SAR perfectly translates from the cellular activity to the in vitro inhibition of RIP1 kinase activity, thereby further supporting the notion that inhibition of RIP1 kinase may be the key to inhibition of necroptosis in cells by 1.
Figure 2.
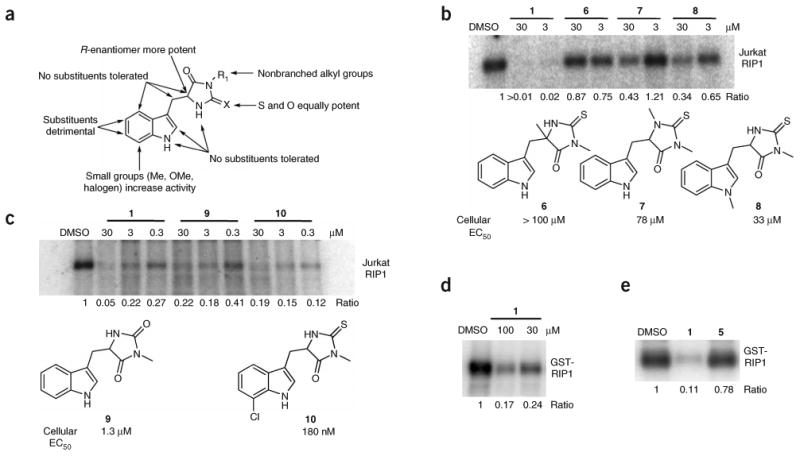
SAR analysis of RIP1 inhibition by necrostatins. (a) Summary of cellular SAR of 1 based on ref. 14. (b,c) In vitro activity of select inactive (b) and active (c) 1 analogs. Endogenous RIP1 kinase assays were performed as in Figure 1e in the presence of indicated amounts of 1 analogs. Structures of the derivatives are shown. EC50 values for inhibition of cellular necrosis in TNFα-treated FADD-deficient Jurkat cells by necrostatins were determined as described in the Methods and were previously reported14. (d,e) 1 inhibits kinase activity of recombinant RIP1 expressed in Sf9 cells. Recombinant RIP1, expressed in Sf9 cells, was subjected to in vitro kinase assay in the presence of indicated amounts (d) or 100 μM (e) of 1 (d,e) or 5 (e). Assays were performed at least two or three times, and similar results were obtained each time. The representative images are shown.
To characterize the mechanism of RIP1 kinase inhibition, we first determined whether 1 acts through an ATP-competitive mechanism. We performed RIP1 kinase assay in the presence of increasing concentrations of nonradioactive ATP. Indeed, increasing amounts of cold ATP resulted in decreased incorporation of the radioactive phosphate and suppression of the inhibitory effect by 1, which is consistent with an ATP-competitive mechanism of inhibition by 1 (Supplementary Fig. 2a online).
To establish that 1 exerts its inhibitory effect directly on RIP1, we purified recombinant baculovirally expressed human RIP1 from Sf9 cells (Supplementary Fig. 2b). Compound 1 efficiently inhibited the activity of recombinant RIP1 in the in vitro kinase reaction (Fig. 2d), which suggests a direct inhibitory effect of 1 on RIP1, whereas 5 again showed no effect (Fig. 2e).
Finally, to verify the specificity of 1 activity, we analyzed whether 1 might inhibit the activity of RIP2 (also known as RICK), the closest homolog of RIP1 (33% sequence identity in the kinase domain, determined using the TCoffee alignment algorithm from http://www.tcoffee.org), using the same in vitro autophosphorylation assay. Compound 1 had no effect on RIP2 autophosphorylation (Supplementary Fig. 2c), even though it was expressed at a level comparable to that of RIP1. At the same time, autophosphorylation of RIP2 was inhibited by PD169316 (12, Supplementary Fig. 2d) as previously reported15. In a similar experiment, we also determined that 1 does not affect the activity of RIP3 (Supplementary Fig. 2e), another closely related RIP1 homolog (33% identity in the kinase domain), which suggests that 1 specifically inhibits RIP1.
Identification of RIP1 autophosphorylation sites
To characterize the mechanism of RIP1 autophosphorylation, we determined residues phosphorylated during the kinase assay. We expressed FLAG-tagged human RIP1 in 293T cells and immunoprecipitated it using anti-FLAG M2 antibody beads. Before and after the kinase reaction, we subjected the samples to SDS-PAGE. We performed phosphorylation analysis by in-gel digestion of RIP1, followed by enrichment on immobilized metal affinity chromatography (IMAC) resin16,17 and mass spectrometry analysis of the resulting phosphopeptides. To maximize the coverage of RIP1, we digested RIP1 protein band extracted from the gel independently using three proteases (trypsin, chymotrypsin and Glu-C), providing almost complete (90.2%) coverage of the RIP1 molecule. This analysis identified a number of phosphorylated residues. We found some of these sites, notably Ser14/15 (it should be noted that phosphorylation of adjacent sites cannot be reliably distinguished by mass spectrometry), Ser20, Ser161 and Ser166 (Supplementary Fig. 3a online), phosphorylated only in the samples subjected to the kinase reaction. Thus, they may represent RIP1 autophosphorylation sites. On the other hand, we found several other residues (Ser6, Ser25, Ser303, Ser320, Ser330/331 and Ser333) to be phosphorylated even in the absence of the kinase reaction. Importantly, we found that the kinase-inactive K45M RIP1 mutant was unable to phosphorylate a subset of the above-described phosphorylation sites in an in vitro kinase assay (Ser14/15, Ser20, Ser161 and Ser166), while retaining other phosphorylation events. These data suggest that Ser14/15, Ser20, Ser161 and Ser166 represent autophosphorylation sites in vitro, detected in the RIP1 kinase assay (Fig. 1), whereas Ser6, Ser25, Ser303, Ser320, Ser330/331 and Ser333 are likely to have been phosphorylated in the cells before immunoprecipitation, independent of RIP1 kinase activity. Notably, while RIP1 (671 amino acids) contains 86 evenly distributed Ser/Thr residues, all of the detected autophosphorylation sites are located in the N-terminal kinase domain (Supplementary Fig. 3b), which suggests that phosphorylation may regulate the kinase activity.
RIP1 T-loop controls inhibition by Nec-1
Analysis of RIP1 against the global human kinase alignment (http://cellsignaling.lanl.gov/structure/kinase/kinase.html0) suggests that the activation segment (T-loop) of RIP1 is very similar to that of B-RAF (ref. 18) (Fig. 3a). In particular, the RIP1 Ser161 autophosphorylation site corresponds to the Thr598 autophosphorylation site of B-RAF, which has been shown to be important for the regulation of the catalytic activity of B-RAF (ref. 19). The activation segment is an important regulatory element, controlling kinase activity by occluding the catalytic cleft of the kinase (the “closed form”). This inhibition can be relieved by destabilization of the “closed” T-loop conformation through a number of mechanisms, including (i) phosphorylation within the activation segment, for example, autophosphorylation of Thr598 in B-RAF (ref. 19) or phosphorylation of Thr160 in CDK2 by CAK (ref. 20); (ii) mutations within the activation segment, for example, oncogenic mutation V599E in B-RAF (ref. 18); and (iii) allosteric regulation affecting the conformation of the T-loop, for example, induced by cyclin A binding to CDK2 (ref. 21). Homology modeling (Fig. 3b) showed that RIP1 can assume a conformation very similar to that of B-RAF, which suggests a potential role of Ser161 phosphorylation in the regulation of RIP1 kinase activity. Indeed, the S161A RIP1 mutant was substantially less active in the in vitro autophosphorylation assay compared with wild-type kinase (Fig. 3c). Because Ser161 also serves as one of the acceptors of radioactive phosphate in autophosphorylation, we must consider the possibility that the reduction of autophosphorylation in the S161A mutant might be due to the loss of a phosphorylation site, rather than the kinase activity per se. However, because the S161E mutant did not show a significant decrease in autophosphorylation (Fig. 3d), we interpret these data to suggest that the S161A mutation indeed decreases RIP1 kinase activity and that Ser161 autophosphorylation positively contributes to RIP1 kinase activity. Consistent with this conclusion, the decrease in the kinase activity of the S161A mutant translated into 40% attenuation in necroptosis induction in Jurkat cells (Fig. 3e). However, mutation of Ser161 did not result in a complete loss of activity, which suggests that other important inputs contribute to RIP1 kinase activation during necroptosis in the cells, promoting either RIP1 kinase activity or substrate availability.
Figure 3.
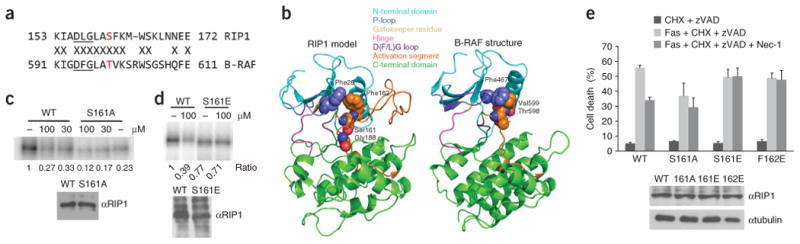
Effect of Ser161 and Phe162 mutations on necroptosis and inhibition by 1. (a) Sequence alignment of the magnesium binding and activation segments of B-RAF and RIP1. Thr598 of B-RAF and Ser161 of RIP1 are shown in red. Kinase-conserved DFG motif is underlined. (b) Homology model of RIP1. (c,d) S161A (c) and S161E (d) mutations attenuate RIP1 kinase sensitivity to 1 in vitro. Kinase assays of RIP1 mutants overexpressed in 293T cells were performed as described in Figure 1. “–” indicates DMSO. In parallel to kinase reactions, a sample of beads was subjected to western blot analysis using anti-RIP1 antibody to ensure equal protein amounts in kinase reactions. Assays were performed at least two or three times, and similar results were obtained each time. The representative images are shown. (e) Mutations of Ser161 and Phe162 attenuate inhibition of necroptosis by 1. Corresponding pcDNA-FLAG-RIP1 vectors were transiently electroporated into RIP1-deficient Jurkat cells along with pEGFP. Cells were allowed to recover for 48 h, treated with anti-FAS antibody, cycloheximide and zVAD-fmk to induce necroptosis and 30 μM 1, followed by analysis by FACS as described in the Methods. The data were obtained in a single experiment performed in triplicate and represent mean values ± s.d. Experiment was repeated multiple times, and similar results were obtained each time. Equal expression of RIP1 mutants was confirmed by western blot using anti-RIP1 and anti-tubulin antibodies.
Curiously, the S161A mutation also reduced the degree of RIP1 kinase inhibition by 1 in vitro and in cells (Fig. 3d,e), which suggests that the inhibition demonstrated by 1 may be related to the structure and/or function of the T-loop. To further explore this hypothesis, we tested mutations that are predicted to destabilize the “closed” conformation of the T-loop (that is, S161E and F162E), thereby promoting an “open” conformation. These mutations are based on structurally equivalent activating mutants of B-RAF (T598D and V599E)18,19. Consistent with our hypothesis, the S161E mutant was fully active in vitro, unlike the S161A mutant, and furthermore, it was no longer sensitive to inhibition by 1 (Fig. 3d). RIP1-deficient Jurkat cells are completely insensitive to necroptotic death triggered by Fas, cycloheximide and benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (zVAD-fmk, 13) (ref. 10 and Supplementary Fig. 4a online). Introducing wild-type RIP1 kinase into RIP1-deficient Jurkat cells restores necroptotic cell death, inhibitable by 1 (Fig. 3e). Introduction of the S161E mutant into these cells also fully restored induction of necroptosis, which is consistent with its full kinase activity in vitro, and, importantly, 1 was no longer capable of inhibiting necroptotic cell death mediated by this mutant, which is consistent with its resistance to inhibition by 1 in vitro (Fig. 3e). We obtained the same results with the second T-loop mutant, F162E, which was also no longer inhibited by 1 in vitro or in cells (Fig. 3e and Supplementary Fig. 4b). Loss of inhibition by 1 of necroptotic cell death mediated by 1-insensitive RIP1 mutants provides an important functional confirmation that RIP1 kinase is a primary cellular target of Nec-1. Notably, none of these mutants induced necroptosis in Jurkat cells in the absence of Fas antibody, which suggests that additional inputs from the receptor are required.
The results of the RIP1 mutagenesis studies, which suggest the involvement of the activation segment of RIP1 in the inhibition by 1, are reminiscent of imatinib's (STI-571, 14) mode of Abl kinase inhibition. The crystal structure of the imatinib–Abl complex revealed that the inhibitor targets the closed inactive conformation of the kinase22. Similar to our data, mutations destabilizing the inactive conformation, such as H396P in the activation loop, have been found to eliminate sensitivity of Bcr-Abl to imatinib23. Therefore, 1 may belong to a class of allosteric inhibitors selectively recognizing the adaptive pocket on RIP1, which is present in the inactive conformation of the RIP1 kinase. Furthermore, based on the attenuation of 1's inhibition in the case of the S161A mutant, we hypothesize that Ser161 may participate in direct (structurally equivalent to the interaction between the P-loop Ser466 and the T-loop Ala587 in the published B-RAF structure (Protein Data Bank code 1UWH), but in reverse direction) or indirect (through a water-mediated hydrogen bond) interactions that partially contribute to stabilization of the closed conformation of RIP1. Consistent with this proposal, our molecular model predicts that Ser161 forms a hydrogen bond with the backbone of Gly188 (Fig. 3b), which may explain the less efficient inhibition of the S161A mutant by 1.
Molecular modeling of the RIP1–Nec-1 complex
To dissect the interaction of 1 with RIP1 kinase further, we established a binding mode hypothesis of the most potent analog of 1, compound 11 (Supplementary Fig. 1), with RIP1 kinase using molecular modeling techniques. Based on the binding mode of the structurally related CDK2 inhibitor SU9516 (4) and on the finding that RIP1 kinase inhibition by 1 was competitive with ATP (Supplementary Fig. 2a), we hypothesized that 11 binds into the ATP pocket. Structural ordering of the DFG motif, which forms a part of the magnesium-binding pocket (“DFG-in” state)24, is one of the important conformational transitions leading to kinase activation, along with structural changes of the adjacent activation loop. The key difference between SU9516 and 11 is that SU9516 binds to the active DFG-in state of CDK2, whereas our data indicate that 11 may target the inactive state of RIP1. Notably, RIP1 has a DLG (Fig. 3a) rather than a DFG motif (as in CDK2) immediately preceding the activation segment, and it has been shown that smaller amino acids in the second position of this motif promote formation of the inactive “DLG-out” conformation of kinases24. Therefore, we concentrated on modeling 11 into the closed DLG-out state of RIP1 kinase and verified that the SAR of 11 is consistent with this conformation of RIP1. Inhibitor 11 forms two classes of interactions with RIP1 (Fig. 4): on the left, the heteroatoms of the hydantoin and indole rings form five hydrogen bonds with hinge residues Glu93, Tyr94, Glu96, Lys97 and Asn99, while the hydrophobic indole ring is located in the deep pocket formed on the top by Leu23 of the P-loop, on the bottom by Leu143 of the C-terminal domain, in the back by the gatekeeper residue Met92 and on the right by Leu157 of the DLG motif (Fig. 4). Importantly, our model correctly predicts the preference for the R enantiomer over the S enantiomer, which would clash with the hinge of RIP1 (ref. 14). Our model also provides a good fit for the available SAR in this series of inhibitors (Fig. 2a)14. In particular, the amino groups of the indole ring and the hydantoin moiety are predicted to form multiple hydrogen bonds, which explains the loss of activity upon methylation of these moieties. The amide carbonyl of the hydantoin ring is postulated to form a hydrogen bond with Asn99, which is consistent with the complete loss of activity when it is replaced with a methylene (15, Supplementary Fig. 5 online)14. The N-methyl on the hydantoin is highly favored in the hydrophobic environment of the Leu23 residue, which explains the decreased activity of 5, which lacks this group. According to our model, small substituents in the 7 position of the indole ring make contacts with Met92, which is consistent with the increased activity displayed by the 7-Me, 7-Cl and 7-Br analogs14. At the same time, larger substituents cannot be accommodated due to steric hindrance, which is indeed observed in the case of the 7-OBn derivative14. Substituents on positions 4, 5 and 6 of the indole ring result in the loss of necroptosis activity14, which is consistent with their proximity to Leu157 of the DLG motif in the inactive, DLG-out conformation (Fig. 4a shows the difference in DLG position between DLG-out and DLG-in states).
Figure 4.
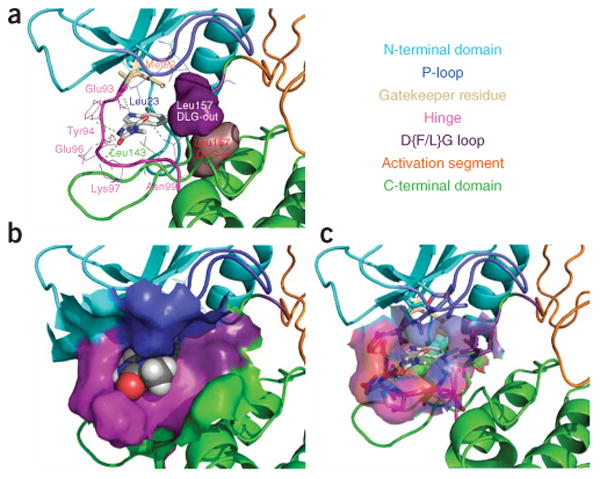
Molecular model of the RIP1–11 complex. (a) RIP1 is shown in ribbon representation with the binding site amino acids shown in wireframe representation, and with the gatekeeper residue and 11 shown in stick representation. Solid surfaces of DLG leucine in its predicted DFG-in and DFG-out conformations are highlighted in purple and brown, respectively. (b) RIP1 is shown in ribbon representation with the binding site solid surface displayed, and 11 is shown in spacefill representation. (c) RIP1 is shown in ribbon representation with the binding site transparent surface displayed, and binding site amino acids and 11 are shown in stick representation. Different structural features of RIP1 kinase are shown in different colors and are annotated on the included text panel.
Other necrostatins also inhibit RIP1 kinase
We have previously reported the identification and SAR analysis of two additional potent necrostatins: necrostatin-3 (Nec-3, 2) and necrostatin-5 (Nec-5, 3) (Fig. 5a)25,26. Using RIP1 kinase assays (described in Fig. 1), we investigated whether these two distinct necrostatins also target RIP1 kinase activity. To our surprise, considering that 2 and 3 are structurally very different from 1, both molecules potently inhibited RIP1 kinase, immunoprecipitated from Jurkat cells (Fig. 5b). Furthermore, their effect was specific, as closely related inactive analogs (16 and 17, respectively) lacked activity. Compound 2 also potently inhibited recombinant RIP1, expressed in Sf9 cells, whereas (surprisingly) 3 was essentially inactive (Fig. 5c). Inhibition of recombinant RIP1 by 2 again correlated with its cellular activity, since 16 was much less active (Fig. 5d). Based on these results, we conclude that 2 inhibits RIP1 kinase directly, similar to 1. On the other hand, it is likely that 3 acts on RIP1 indirectly, through interactions with another component present in the endogenous RIP1 complex, but absent in the recombinant RIP1 sample.
Figure 5.
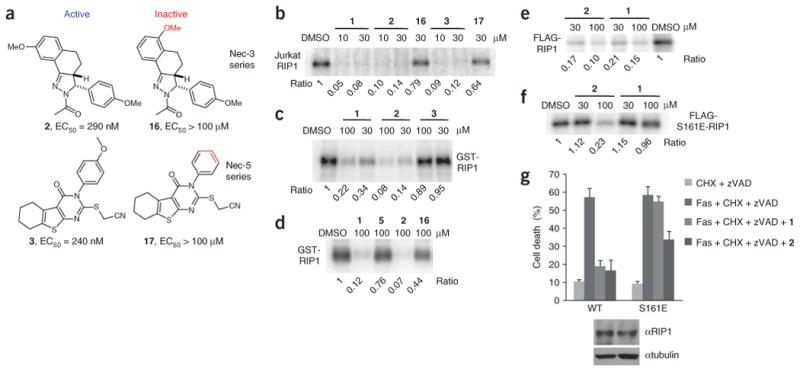
Necrostatin-3 and necrostatin-5 inhibit RIP1 kinase activity. (a) Structures of necrostatin-3 and necrostatin-5 analogs. Cellular EC50 in TNFα-treated FADD-deficient Jurkat cells are shown. (b) Additional Necs inhibit activity of Jurkat cell RIP1. In vitro kinase reaction in the presence of the indicated concentrations of necrostatins was performed as in Figure 1e. (c,d) 2, but not 16, specifically inhibits recombinant RIP1 kinase, expressed in Sf9 cells. In vitro kinase reactions in the presence of the indicated concentrations of necrostatins were performed as in Figure 2d. (e,f) 2 still inhibits activity of S161E mutant of RIP1 in vitro. WT FLAG-RIP1 (e) and S161E FLAG-RIP1 (f) were expressed in 293T cells and subjected to in vitro kinase assay in the presence of the indicated concentrations of necrostatins as in Figure 1c. Assays were performed at least two or three times, and similar results were obtained each time. The representative images are shown. (g) 2 attenuates necroptosis mediated by S161E mutant of RIP1. Recapitulation of WT and S161E mutants in RIP-deficient Jurkat cells and necroptosis viability assay were performed as described in Figure 3e. Compounds were used at 30 μM. The data were obtained in a single experiment performed in triplicate and represent mean values ± s.d. Experiment was repeated multiple times, and similar results were obtained each time. Equal expression of RIP1 mutants was confirmed by western blot using anti-RIP1 and anti-tubulin antibodies.
Because inhibitor 2 is structurally different from 1, we explored whether these two molecules inhibit RIP1 differently. Indeed, unlike 1, which completely lacks activity against the S161E mutant of RIP1, 2 still inhibited the autophosphorylation of this mutant in vitro, albeit less effectively than in the case of wild-type RIP1 (Fig. 5e,f). Furthermore, consistent with the ability of 2 to inhibit the autophosphorylation of the S161E RIP1 mutant and in contrast to that of 1, 2 was capable of inhibiting cellular necroptosis mediated by S161E RIP1 (Fig. 5g), though it did so less effectively than in cells expressing wild-type RIP1. We are currently pursuing further structural analyses of the interactions of 1 and 2 with RIP1 to determine the mechanistic basis for their differential mode of RIP1 inhibition.
Necrostatin inhibition of RIP1 is independent of Hsp90
RIP1 is a client protein of Hsp90 (ref. 27), and geldanamycin (18, a specific ATP-competitive inhibitor of Hsp90) has been previously reported to inhibit RIP1 kinase-dependent necroptosis10. To further investigate the mechanistic basis of RIP1 kinase inhibition by necrostatins, we explored whether small molecules, in particular 3 acting on RIP1 kinase indirectly, might affect RIP1 through the regulation of Hsp90 chaperone (see Supplementary Methods online for assay details). However, using the kinase assay lysis buffer conditions, we were unable to detect the presence of Hsp90 in either the samples of endogenous Jurkat cell RIP1 or human RIP1 expressed in Sf9 cells (Supplementary Fig. 6a online). These results suggest that the Hsp90– RIP1 complex is labile and may not tolerate immunoprecipitation conditions used for RIP1 kinase assays. Thus, it is unlikely that Hsp90 is critical for RIP1 kinase activity in vitro, and therefore Hsp90 is unlikely to be the target of necrostatins. Consistent with this proposal, geldanamycin did not significantly affect the activity of either the endogenous RIP1 or the protein expressed in Sf9 cells (Supplementary Fig. 6b). Furthermore, using an established fluorescence polarization assay28, we found that none of the Necs are direct ATP-competitive inhibitors of Hsp90 in Jurkat cell lysates, as they failed to disrupt FITC-geldanamycin binding to Hsp90 (Supplementary Fig. 6c). In addition, the cellular activity of the Necs is distinguishable from the effect of inhibiting Hsp90, as none of the Necs reduced protein levels of RIP1 kinase as is seen with geldanamycin27 (Supplementary Fig. 6d). Finally, we found that none of the necrostatins significantly inhibit RIP1-Hsp90 interaction (Supplementary Fig. 6e), which was detectable using hypotonic cell lysis conditions. Based on these data we conclude that the activity of necrostatins is not related to the regulation of Hsp90 activity or its association with RIP1. This conclusion is also consistent with the lack of necrostatin effect on NFkB activation (Supplementary Fig. 7 online), which is prominently inhibited by geldanamycin27.
Discussion
In summary, we show that all three classes of potent necrostatins identified in a random unbiased cell-based screen target RIP1 kinase activity through distinct mechanisms—T-loop–dependent inhibition of RIP1 kinase by 1, partially T-loop–independent inhibition of RIP1 by 2 and indirect inhibition of RIP1 by 3—thus highlighting the critical role of RIP1 kinase activity for necroptotic cell death. Our discovery that multiple unrelated necrostatins isolated in a cell-based assay all target RIP1 kinase is unexpected. Our results demonstrate that a key step in the necroptosis pathway (RIP1 kinase activity) is highly amenable to small-molecule inhibition. Considering the emerging importance of necroptosis in multiple paradigms of pathologic injury in vivo9,11, our results suggest that RIP1 kinase may represent a promising new target for drug discovery. Furthermore, high potency and the well-explored SAR of identified necrostatins may provide a strong basis for their further therapeutic development. Finally, the ability of an unbiased screen to identify potent and specific small-molecule inhibitors of a key regulator in a cellular pathway demonstrates the power of cell-based screening for small-molecule discovery research.
Our previous analysis suggested that the signaling step, targeted by 1, plays a universally important role in multiple cellular paradigms of necroptosis triggered by DR ligands (that is, TNFα, Fas ligand and tumor necrosis factor–related apoptosis-inducing ligand (TRAIL, also known as Apo2L)) in various primary cells and cell lines of epithelial, fibroblast and hematopoietic origin9. These results contrast with the emerging data regarding the cell type specificity for the mechanisms of necroptosis execution. For instance, while reactive oxygen species (ROS) generation through the formation of the RIP1–Rac1– NADPH oxidase complex has been recently proposed to be a key mediator of DR-induced necrotic death in mouse fibrosarcoma L929 cells29, neither oxidative stress nor NADPH oxidase activity appear to play a role in other paradigms of necroptosis (for example, in Jurkat cells; ref. 9 and Supplementary Fig. 7a,b). Similarly, autophagy has been proposed to represent a key execution mode of necrosis in some cell lines, for example, L929 and human monocytic U-937 cells9,30, but it was found to play only a secondary role in other cases, for example, mouse embryonic fibroblasts and Jurkat cells9. Therefore, our data identify RIP1 kinase as a key universal upstream step in the necroptosis pathway that is potentially followed by divergent downstream execution steps, which are dependent on the cellular context. Furthermore, our findings identify RIP1 as a specific protein target for various paradigms of pathologic cell death, for which inhibition by 1 has been demonstrated both in vitro and in vivo, including in animal models of brain ischemia9 and myocardial infarction11, and in death triggered by glutamate31, plant sterols32 and killing of apoptosis-resistant cancer cells by the chemotherapeutic agent shikonin33.
RIP1 protein consists of three major domains, which have been linked to distinct activities. The C-terminal death domain is important for homologous interaction with death domain receptors34 and/or the DR-associated adaptor TRADD (ref. 35). The central region of RIP1 has been implicated in NFκB activation through polyubiquitination of Lys377 (ref. 36), nucleating the formation of NFκB-inducing signaling complex I (refs. 37,38). While RIP1 was originally reported not to contribute to apoptosis induction in Jurkat cells13, recent evidence suggests that it is an intrinsic part of the DR-induced apoptosis signaling complex II (DISC)39–41, although the domains required for this activity have not been identified. The N-terminal Ser/Thr kinase domain of RIP1 is required for necroptosis10 but is dispensable for NFkB activation42 and apoptosis in Jurkat cells10. At the same time, the kinase activity was found to be dispensable for NFκB activation42 and apoptosis in Jurkat cells10. Consistent with these results, necrostatins showed no effect on NFκB activation (Supplementary Fig. 7c,d); we have also not observed attenuation of necroptosis in Jurkat cells reconstituted with the K377R mutant of RIP1, which is defective in NFκB signaling36 (Supplementary Fig. 4a). Similarly, we have not observed any inhibition of apoptosis by 1 in multiple cellular systems in which 1 was found to efficiently inhibit necroptosis9. These data are consistent with the selective role of RIP1 kinase in necroptotic cell death. Considering the potency and selectivity of 1 for inhibiting RIP1 kinase and the complexity of RIP1 signaling, 1 and its active analogs represent a unique and very useful class of probes for RIP1 kinase activity in vitro and in vivo.
Finally, our molecular modeling data presents a feasible model of RIP1 inhibition by Nec-1, which will be further validated once the structure of the RIP1 kinase domain becomes available. According to our model, the predicted position of Leu157 in its active DLG-in conformation is approximately 10 Å away from the predicted binding pose of 11 (Fig. 4a) and therefore cannot account for the sharp SAR (Fig. 2a)14 on the 4, 5 and 6 positions of the indole ring. This observation further supports the notion that the Nec-1 series of inhibitors may target the inactive DLG-out state of RIP1 kinase. The plausible explanation for such selectivity, based on our model, may be that the hydrophobic 4, 5 and 6 positions of the indole ring would be exposed to solvent in the DLG-in conformation, destabilizing 11 binding to RIP1. At the same time, the DLG-in conformation would also promote ATP binding into RIP1's catalytic center by removing Leu157 of the DLG motif from the ATP path, thus promoting competition with 11. It should be also noted that RIP1 appears to have substantial activity even in the basal state when isolated from unstimulated Jurkat cells (Fig. 1e), which is consistent with only partial attenuation of its activity and necroptosis activation by S161A mutation (Fig. 3c,e). Based on these data, the modeling results and the inability of 1 to inhibit T-loop mutants, we propose a plausible model that RIP1 kinase exists in equilibrium between inactive DLG-out and active DLG-in conformations, and 1 preferentially binds to and stabilizes the inactive conformation, thereby shifting the equilibrium toward the inactive state. Conversely, T-loop–mediated activation will shift the equilibrium in the opposite direction, potentially functioning to modulate the kinetics of necroptosis initiation and/or to render RIP1 kinase activation irreversible. While our current study focused primarily on the identification of the cellular target of necrostatins, our molecular modeling provides a framework for further analysis of the molecular mechanism of RIP1 kinase activation and inhibition, and for future rational design of small-molecule inhibitors of RIP1 kinase activity.
Methods
Chemical compounds
All small-molecule inhibitors used in the paper have been previously described. 1 was from Sigma. zVAD-fmk (13) was purchased from Axxora. NADPH oxidase inhibitors apocynin (19) and diphenyleneiodonium chloride (DPI, 20) were obtained from Cayman Chemical. Geldanamycin (18) was purchased from Invivogen. FITC-geldanamycin was from Biomol. The synthesis of all necrostatins used in this study has previously been described: compounds 5–11 and 15 (refs. 14,43), compounds 2 and 16 (ref. 25) and compounds 3 and 17 (ref. 26).
Reagents
We purchased human TNFα from Cell Sciences. Mouse RIP1 antibody was from BD Biosciences. Rabbit RIP1 agarose-conjugated beads were obtained from Santa Cruz. Protein A magnetic beads were purchased from Dynal and covalently coupled to mouse RIP1 antibody using dimethyl pimelimidate (DMP) reagent (Pierce) according to the standard protocol from New England Biolabs. Rat Hsp90 and mouse β-tubulin antibody were obtained from Stressgene. We purchased FLAG M2 agarose beads from Sigma and glutathione-sepharose from Amersham. Other common reagents and chemicals were purchased from Sigma.
Cell lines
Fas-associated via death domain (FADD)-deficient and RIP1-deficient human Jurkat T cells, Spodoptera frugiperda Sf9 cells and SV40-transformed human kidney embryonic epithelial HEK 293T cells were purchased from ATCC. Jurkat and 293T cells were maintained in RPMI1640 or DMEM (Invitrogen), respectively, supplemented with 10% fetal bovine serum (Sigma) and antimycotic-antibiotic mix (Invitrogen). Sf9 cells were maintained in SF-900 II SFM medium (Invitrogen).
DNA plasmids
Human pcDNA3-FLAG-RIP1 expression vector was a generous gift of J. Tschopp (University of Lausanne). RIP1 point mutations were introduced into pcDNA3-FLAG-RIP1 vector using QuikChange mutagenesis kit (Stratagene). pcDNA3-RIP2-Myc expression vector was a generous gift of G. Nunez (University of Michigan). RIP3 complementary DNA was amplified by PCR using MGC:75066 EST clone (ATCC) along with FLAG sequence, introduced into 5′ primer, and cloned into pcDNA3.1 vector (Invitrogen). RIP1 baculoviral expression vector was generated by PCR amplification of residues 1–375 of human RIP1, which were subcloned between the EcoRI and PstI sites of the pAcGHLT vector, containing GST tag (BD Biosciences).
In vitro kinase assay
The assay was performed essentially as described13. 293T cells were transfected with pcDNA3-FLAG-RIP1 vector, vectors encoding RIP1 mutant proteins or pcDNA3-RIP2-Myc and pcDNA3-FLAG-RIP3 vectors using standard Ca3(PO4)2 precipitation procedure. Culture medium was replaced 6 h after the transfection and cells were lysed 48 h later in the TL buffer consisting of 1% Triton X-100, 150 mM NaCI, 20 mM HEPES, pH 7.3, 5 mM EDTA, 5 mM NaF, 0.2 mM NaVO3 (ortho) and complete protease inhibitor cocktail (Roche). Immunoprecipitation was carried out for 16 h at 4 °C using anti-FLAG M2 agarose beads (Sigma), followed by three washes with TL buffer and two washes with 20 mM HEPES, pH 7.3. Beads were incubated in 15 μl of the reaction buffer containing 20 mM HEPES, pH 7.3, 10 mM MnCl2 and 10 mM MgCl2 for 15 min at 23–25 °C in the presence of different concentrations of necrostatins. For these assays, compound stocks (in DMSO) were diluted to appropriate concentrations in DMSO before the addition to the reactions to maintain final concentration of DMSO for all samples at 3%. Kinase reaction was initiated by addition of 10 μM cold ATP and 1 mCi of [γ-32P] ATP, and reactions were carried out for 30 min at 30 °C. Reactions were stopped by boiling in SDS-PAGE sample buffer and subjected to 8% SDS-PAGE. RIP1 band was visualized by analysis in a Storm 8200 Phosphorimager (Molecular Dynamics). Similar protocol was used for endogenous RIP1 kinase reactions, except mouse monoclonal RIP1 antibody (BD Biosciences) and protein magnetic beads (Dynal) or rabbit RIP1 antibody-coupled agarose beads (Santa Cruz) were used. For recombinant baculovirally expressed RIP1, protein was expressed in Sf9 cells according to manufacturer's instructions (BaculoGold system, BD Biosciences) and purified using glutathione-sepharose beads (Amersham). Protein was eluted in 50 mM Tris-HCl, pH 8.0 supplemented with 10 mM reduced glutathione, and eluted protein was used in the kinase reactions, supplemented with 5 × kinase reaction buffer (100 mM HEPES, pH 7.3, 50 mM MnCl2, 50 mM MgCl2, 50 μM cold ATP and 5 μCi of [γ-32P]ATP).
RIP1 reconstitution in RIP1-deficient Jurkat cells
For analysis of RIP1 mutants, 5 × 106 RIP1-deficient Jurkat cells were electroporated with 2.7 μg of pcDNA3-FLAG-RIP1 vectors and 0.3 μg pEGFP vector (Clontech), serving to identify transfected cells, using a Nucleofector II device with reagent V (Amaxa). 48 h after electroporation, live cells were isolated by centrifugation (1,400 r.p.m., 40 min) over the layer of Ficoll-PAQUE (Amersham). Cells were treated with 1 mg ml cycloheximide, 100 mM zVAD-fmk and 200 ng ml−1 agonistic Fas antibody (clone 7C11, Beckman Coulter) for 24 h and stained with 2 μg ml−1 propidium iodide (Roche). Percentages of green fluorescent protein (GFP)-positive and propidium iodide–negative cells were determined using FACS (FACSCalibur, BD Biosciences) at Tufts Flow Cytometry Core Facility.
Cellular EC50 determination of necrostatins
Determination of EC50 was performed in FADD-deficient Jurkat cells treated with human TNFα as previously described14. Briefly, cells were seeded into 96-well plates and treated with a range of necrostatin concentrations (30 nM to 100 μM, 11 dose points) in the presence and absence of 10 ng ml–1 human TNFα for 24 h. For these and all other cellular assays, compound stocks (in DMSO) were diluted to appropriate concentrations in DMSO before addition to the cells to maintain final concentration of DMSO for all samples at 0.5%. Cell viability was determined using CellTiter-Glo luminescent cell viability assay (Promega). Ratio of luminescence in compound and TNF-treated wells to compound-treated, TNF-untreated wells was calculated (viability, %) and used to calculate EC50 by nonlinear regression in GraphPad Prizm.
Mass spectrometry analysis of RIP1 autophosphorylation sites
HEK 293T cells (two 15 cm2 plates) were transfected with 20 mg of pcDNA3-FALG-RIP1 vector as described above. 48 h after transfection, cells were lysed and immunoprecipitated using anti-FLAG M2 antibody beads as described above. Samples were eluted using modified SDS sample buffer (70 mM Tris-HCl, pH 8.1, 2% SDS, 5 mM DTT, bromphenol blue) by boiling for 5 min at 95 °C. Free thiols were alkylated using 13 mM iodoacetamide (30 min in the dark). Excess of iodoacetamide was quenched by addition of 5 mM DTT for 10 min. Samples were separated by 8% SDS-PAGE, and RIP1 band was visualized by staining with GelCode Blue reagent (Pierce). In-gel digestion of RIP1 band, IMAC purification, LC-MS/MS and data analysis were performed as previously described44.
Molecular modeling of RIP1 and RIP1–11 complex
Homology model of RIP1 kinase domain was built with MODELER45 using the structure of B-RAF (ref. 18) as template. The homology model was minimized in CHARMm46, and the conformation of the side chains was predicted by the SCWRL3 algorithm (http://dunbrack.fccc.edu/SCWRL3.php). For modeling of the RIP1–11 complex, the indole ring 11 was initially placed in the area occupied by the indole ring of SU9516 in CDK2. The subsequent extensive refinement of this complex was carried out in CHARMm46. The models were analyzed using PyMOL visualization software (http://pymol.sourceforge.net).
Supplementary Material
Acknowledgments
The authors thank J. Tschopp (University of Lausanne), I. Verma (Salk Institute) and G. Nunez (University of Michigan) for the generous gifts of the plasmids. This work was supported in part by grants from the National Institute of General Medicine (R01 GM64703), the National Institute on Aging (R37 AG012859) and the National Institute of Neurological Disorders and Stroke (UO1 NS050560) to J.Y., and by funding from the Harvard NeuroDiscovery Center to X.T. and G.D.C. A.D. is a recipient of a National Institute on Aging Mentored Research Scientist Career Development Award and a Massachusetts Medical Foundation Smith Family New Investigator Award.
Footnotes
Accession codes. Protein Data Bank: the structure of B-RAF was deposited as part of a previous study under PDB code 1UWH.
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
Author Contributions: A.D., J.H., I.L.C., O.K. and S.M.H. analyzed inhibition of RIP1 kinase and Hsp90 by necrostatins. A.D., M.G. and O.K. performed RIP1 mutagenesis and characterized cellular and in vitro kinase activity of the mutants. A.D. and J.H. generated samples for mass spectrometry analysis, which was carried out by S.A.G. A.L. and G.W. performed molecular modeling of RIP1. X.T., G.D.C. and C.Y. designed and synthesized the necrostatin analogs. D.A. prepared the baculoviral RIP1 expression vector and optimized expression conditions. The project was designed and coordinated by A.D. and J.Y. A.D. and J.Y. wrote the manuscript. All authors discussed the results and contributed to discussion of relevant experimental results in the manuscript.
Competing Interests Statement: The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturechemicalbiology/.
References
- 1.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 2.Lindsten T, Thompson CB. Cell death in the absence of Bax and Bak. Cell Death Differ. 2006;13:1272–1276. doi: 10.1038/sj.cdd.4401953. [DOI] [PubMed] [Google Scholar]
- 3.Los M, Wesselborg S, Schulze-Osthoff K. The role of caspases in development, immunity, and apoptotic signal transduction: lessons from knockout mice. Immunity. 1999;10:629–639. doi: 10.1016/s1074-7613(00)80062-x. [DOI] [PubMed] [Google Scholar]
- 4.Chautan M, Chazal G, Cecconi F, Gruss P, Golstein P. Interdigital cell death can occur through a necrotic and caspase-independent pathway. Curr Biol. 1999;9:967–970. doi: 10.1016/s0960-9822(99)80425-4. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheim RW, et al. Programmed cell death of developing mammalian neurons after genetic deletion of caspases. J Neurosci. 2001;21:4752–4760. doi: 10.1523/JNEUROSCI.21-13-04752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West T, Atzeva M, Holtzman DM. Caspase-3 deficiency during development increases vulnerability to hypoxic-ischemic injury through caspase-3-independent pathways. Neurobiol Dis. 2006;22:523–537. doi: 10.1016/j.nbd.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Mareninova OA, et al. Cell death in pancreatitis: caspases protect from necrotizing pancreatitis. J Biol Chem. 2006;281:3370–3381. doi: 10.1074/jbc.M511276200. [DOI] [PubMed] [Google Scholar]
- 8.Cauwels A, Janssen B, Waeytens A, Cuvelier C, Brouckaert P. Caspase inhibition causes hyperacute tumor necrosis factor-induced shock via oxidative stress and phospholipase A2. Nat Immunol. 2003;4:387–393. doi: 10.1038/ni914. [DOI] [PubMed] [Google Scholar]
- 9.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 10.Holler N, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 11.Smith CC, et al. Necrostatin: a potentially novel cardioprotective agent? Cardiovasc Drugs Ther. 2007;21:227–233. doi: 10.1007/s10557-007-6035-1. [DOI] [PubMed] [Google Scholar]
- 12.Moshinsky DJ, et al. SU9516: biochemical analysis of cdk inhibition and crystal structure in complex with cdk2. Biochem Biophys Res Commun. 2003;310:1026–1031. doi: 10.1016/j.bbrc.2003.09.114. [DOI] [PubMed] [Google Scholar]
- 13.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 14.Teng X, et al. Structure-activity relationship study of novel necroptosis inhibitors. Bioorg Med Chem Lett. 2005;15:5039–5044. doi: 10.1016/j.bmcl.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 15.Argast GM, Fausto N, Campbell JS. Inhibition of RIP2/RIck/CARDIAK activity by pyridinyl imidazole inhibitors of p38 MAPK. Mol Cell Biochem. 2005;268:129–140. doi: 10.1007/s11010-005-3701-0. [DOI] [PubMed] [Google Scholar]
- 16.Ficarro SB, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 17.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 18.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 19.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buck V, Russell P, Millar JB. Identification of a cdk-activating kinase in fission yeast. EMBO J. 1995;14:6173–6183. doi: 10.1002/j.1460-2075.1995.tb00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 22.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 23.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 24.Bukhtiyarova M, Karpusas M, Northrop K, Namboodiri HV, Springman EB. Mutagenesis of p38alpha MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-OUTstate. Biochemistry. 2007;46:5687–5696. doi: 10.1021/bi0622221. [DOI] [PubMed] [Google Scholar]
- 25.Jagtap P, et al. Structure activity relationship study of tricyclic necroptosis inhibitors. J Med Chem. 2007;50:1886–1895. doi: 10.1021/jm061016o. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, et al. Structure-activity relationship analysis of a novel necroptosis inhibitor, necrostatin-5. Bioorg Med Chem Lett. 2007;17:1455–1465. doi: 10.1016/j.bmcl.2006.11.056. [DOI] [PubMed] [Google Scholar]
- 27.Lewis J, et al. Disruption of hsp90 function results in degradation of the death domain kinase, receptor-interacting protein (RIP), and blockage of tumor necrosis factor-induced nuclear factor-kappaB activation. J Biol Chem. 2000;275:10519–10526. doi: 10.1074/jbc.275.14.10519. [DOI] [PubMed] [Google Scholar]
- 28.Gooljarsingh LT, et al. A biochemical rationale for the anticancer effects of Hsp90 inhibitors: slow, tight binding inhibition by geldanamycin and its analogues. Proc Natl Acad Sci USA. 2006;103:7625–7630. doi: 10.1073/pnas.0602650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YS, Morgan MJ, Choksi S, Liu ZG. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol Cell. 2007;26:675–687. doi: 10.1016/j.molcel.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Yu L, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, et al. Necrostatin-1 protects against glutamate-induced glutathione depletion and caspase-independent cell death in HT-22 cells. J Neurochem. 2007;103:2004–2014. doi: 10.1111/j.1471-4159.2007.04884.x. [DOI] [PubMed] [Google Scholar]
- 32.Bao L, Li Y, Deng SX, Landry D, Tabas I. Sitosterol-containing lipoproteins trigger free sterol-induced caspase-independent death in ACAT-competent macrophages. J Biol Chem. 2006;281:33635–33649. doi: 10.1074/jbc.M606339200. [DOI] [PubMed] [Google Scholar]
- 33.Hong Q, et al. Zfra affects TNF-mediated cell death by interacting with death domain protein TRADD and negatively regulates the activation of NF-kappaB, JNK1, p53 and WOX1 during stress response. BMC Mol Biol. 2007;8:50. doi: 10.1186/1471-2199-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 35.Thakar J, Schleinkofer K, Borner C, Dandekar T. RIP death domain structural interactions implicated in TNF-mediated proliferation and survival. Proteins. 2006;63:413–423. doi: 10.1002/prot.20895. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for TNFalpha-induced NF-kappa B activation. J Biol Chem. 2006;281:13636–13643. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 37.Muppidi JR, Tschopp J, Siegel RM. Life and death decisions: secondary complexes and lipid rafts in TNF receptor family signal transduction. Immunity. 2004;21:461–465. doi: 10.1016/j.immuni.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 39.Jin Z, El-Deiry WS. Distinct signaling pathways in TRAIL- versus tumor necrosis factor-induced apoptosis. Mol Cell Biol. 2006;26:8136–8148. doi: 10.1128/MCB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Donnell MA, Legarda-Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr Biol. 2007;17:418–424. doi: 10.1016/j.cub.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi R, et al. Focal adhesion kinase determines the fate of death or survival of cells in response to TNFalpha in the presence of actinomycin D. Biochim Biophys Acta. 2007;1770:518–526. doi: 10.1016/j.bbagen.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Lee TH, Shank J, Cusson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185–33191. doi: 10.1074/jbc.M404206200. [DOI] [PubMed] [Google Scholar]
- 43.Cuny GD, Yuan J, Jagtap P, Degterev A. Inhibitors of cellular necrosis. United States patent US 20050119260. 2005 [Google Scholar]
- 44.Li X, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 45.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 46.Brooks BR, et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


