Abstract
Glycomics has become an increasingly important field of research since glycans play critical roles in biology processes ranging from molecular recognition and signaling to cellular communication. Glycans often conjugate with other biomolecules, such as proteins and lipids, and alter their properties and functions, so glycan characterization is essential for understanding the effects they have on cellular systems. However, the analysis of glycans is extremely difficult due to their complexity and structural diversity (i.e., the number and identity of monomer units, and configuration of their glycosidic linkages and connectivities). In this work, we coupled ion mobility spectrometry with mass spectrometry (IMS-MS) to characterize glycan standards and biologically important isomers of synthetic αGal-containing O-glycans including glycotopes of the protozoan parasite Trypanosoma cruzi, which is the causative agent of Chagas disease. IMS-MS results showed significant differences for the glycan structural isomers when analyzed in positive and negative polarity and complexed with different metal cations. These results suggest that specific metal ions or ion polarities could be used to target and baseline separate glycan isomers of interest with IMS-MS.
Keywords: Ion mobility spectrometry, Mass spectrometry, Glycans, O-Glycans, Isomers
Introduction
Free glycans have multiple biological functions. When glycans covalently link with other biomolecules, such as proteins and lipids to form glycoconjugates, this modification often alters their properties and functions. Various glycoconjugates are found inside the cell and on cellular surfaces with important roles in biological processes such as protein folding, cell signaling, neural development, and hormone activity [1–9]. Specific cellular interactions are thought to be controlled by glycans, and recent work has suggested that subtle glycan structural changes can result in vastly different cellular exchanges, causing severe morphogenic and metabolic defects and diseases [10, 11]. In addition, cell surface glycans often act as specific binding targets for microbes and microbial toxins, allowing them to play a key role in host-pathogen recognition [10]. A human host may also have an immune response against glycans that exist on the cell surfaces of pathogens. For example, patients with acute or chronic Chagas disease produce specific and protective (trypanolytic) anti-α-galactopyranosyl (anti-α-Gal) antibodies that target O-glycans with terminal α-Gal moieties present on cell surface glycoproteins of the parasite Trypanosoma cruzi, the causative agent of Chagas disease [12–16]. These complex biological roles make understanding how glycan structures correlate with their functions and dysfunctions crucial for unraveling their role in human diseases and determining therapeutic strategies for mammalian-host infection.
To date, elucidating detailed information about glycans and glycoconjugates has been limited due to their structural complexity [1, 17]. Furthermore, challenges such as the heterogeneity of glycans isolated from biological materials, difficulties in purifying these glycans, low availability of glycan standards and suitable analytical methods for unequivocal identification have hampered progress in understanding glycan functions [18, 19]. Mass spectrometry (MS) alone or combined with other structural characterization methods such as NMR [20] is often used to characterize glycans and their conjugates [21, 22]. A number of separation methods, including gas chromatography, liquid chromatography, and capillary electrophoresis, have also been interfaced with MS to assist in the separation and characterization of glycans [23]. However, optimizing the conditions for conventional methods is often very time consuming and costly. Ion mobility spectrometry coupled with MS (IMS-MS) [24, 25] is a technique capable of separating molecules that have the same mass-to-charge (m/z) ratio but different shapes or sizes and has become an appealing tool for characterizing the structures of biomolecules [26–28], including glycans [29–35]. Most IMS-MS studies of glycans have mainly focused on positive ion mode characterization due to its higher ionization efficiency. Here we use an IMS-QTOF-MS platform to characterize standard glycans in both positive and negative polarities and also explore the effects of metal ions on enhancing glycan isomer separations. We next applied the optimized IMS-MS method to study synthetic α-Gal-containing O-glycans, some of which contain an immunodominant T. cruzi glycotope that is a potential biomarker for the diagnosis of Chagas disease [12, 36, 37].
Methods
Materials and sample preparation
Standards and reagents were either purchased from Sigma-Aldrich, V-labs Inc., Megazyme (Wicklow, Ireland), or United States Biological Corp. (Swampscott, MA). The O-glycans α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-β-d-Glc-(CH2)3SH (1)[36], α-d-Gal-(1 → 6)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (2) [36], α-d-Gal-(1 → 3)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (3) [Schocker, N.S., Almeida, I.C., Michael, K., unpublished data], β-d-Galf-(1 → 4)- [β-d-Gal-(1 →6)]-α-d-GlcNAc-(CH2)3SH (4) [Schocker, N.S., 2016, Ph.D. Dissertation, University of Texas at El Paso], α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-α-d-GlcNAc-(CH2)3SH (5)[37], α-l-Rha-(1 → 2)-α-l-Fuc-(CH2)3SH (6), and α-l-Rha-(1 → 3)-α-l-Fuc-(CH2)3SH (7) [Khamsi, J., Michael, K., unpublished data] were synthesized in the Michael lab. The glycans were dissolved in a MeOH/H2O/formic acid solution (49.45:49.45:0.1, v/v/v) to a final concentration of ∼5 μM or lower.
RapidFire SPE-IMS-QTOF MS method
Standard glycans and synthetic O-glycans were analyzed with both a home-built IMS-QTOF MS [38–41] and an Agilent 6560 ion mobility QTOF MS platform [28]. A RapidFire solid-phase extraction (SPE) system (Agilent Technologies, Santa Clara, CA) was coupled to the 6560 IMS-QTOF-MS platform, enabling fast sample handling, clean-up, and delivery to the IMS-QTOF MS platform. A detailed description of RapidFire can be found elsewhere [Zhang et al. (2016) A Metabolomics Platform for Sub-minute Comprehensive Disease and Exposure Surveillance, submitted to Clinical Mass Spectrometry]. A graphitic carbon cartridge was chosen for glycans after optimization based on maximum binding efficiency and minimum carry-over. Briefly the samples were loaded using water containing 0.1 % formic acid (v/v) with a flow rate of 1.5 mL/min, washed by 100 % methanol with 1.25 mL/min flow rate, and eluted using water/acetonitrile/ace-tone (2: 1: 1, v/v/v) containing 0.1% formic acid at 0.6 mL/min. The times for aspiration, load/wash, elution, and re-equilibration were 0.6, 3, 6, and 1 s, respectively, and the total cycle time was ∼10 s. A jet stream orthogonal electrospray ionization source was used to connect the RapidFire system with the IMS-QTOF MS platform. After ionization, ions were passed through the inlet glass capillary, focused by a high-pressure ion funnel, and accumulated in a lower-pressure ion funnel trap (IFT). Ions were then pulsed into the 89 cm long IMS drift tube filled with ∼4 Torr of nitrogen gas, where they travel under a uniform electric field. Ions exiting the drift tube were refocused by a rear ion funnel prior to QTOF MS detection. See [28] for additional instrumentation details.
Density functional theory calculations
Molecular geometries were predicted for d-(+)-xylose, d-(–)-arabinose, d-(–)-fructose, and d-(+)-glucose using protocols similar to that outlined in Graham et al. and Ma et al. [in press]. Briefly, 2D structure files (.mol) were obtained using ChemSpider [42], and analyzed using the Marvin pKa plugin (Marvin 15.9.14, 2015, ChemAxon) for adduct site prediction [43]. Initial geometry relaxation was performed using the Merck molecular force field (MMFF94) [44] implemented in Avogadro (v1.1.1) [45]. Final geometry optimization was completed using density functional theory (DFT) calculations with NWChem (v6.6) [46]. The B3LYP exchange-correlation functional was used for all calculations [47–50], and a single-step optimization was performed with Pople basis sets at the 6-31G* level (a double-zeta valence potential basis set having a single polarization function) [51–53]. Finally, collision cross sections were calculated using the IMPACT method [54].
Results and discussion
Glycan standards analyses in positive and negative polarities
To understand how IMS aids in glycan isomer separation, commercially available glycan standards and synthetic glycans were studied in both positive and negative polarities. Interestingly, the IMS profiles for glycans showed significant differences between the two ion modes (Fig. 1). For example, the monosaccharides d-(+)-xylose and d-(−)-arabinose showed similar drift time profiles in negative mode and were not separable (Fig. 1a). However, in positive ion mode, the sodiated form of xylose displayed a much shorter drift time than sodiated arabinose, providing baseline separation and suggesting differences in the glycan conformations. The monosaccharides d-(–)-fructose and d-(+)-glucose, which are important in human carbohydrate metabolism (Fig. 1b) [55], also showed a similar pattern with no separation in negative mode, but distinction in positive mode with sodium addition. To understand this effect, theoretical modeling was performed on d-(+)-xylose, d-(–)-arabinose, d-(–)-fructose, and d-(+)-glucose, and low-energy density functional theory (DFT) structures were analyzed. The lowest energy deprotonated structures of xylose and arabinose in their furanose form revealed very similar conformations; however, in the positive ion mode, the sodium ion binds to different locations on the ring causing different structural sizes. Due to the preferential binding to oxygen for xylose, the sodium preferred to sit on top of the furanose ring, but for arabinose the sodium ion complexed to the side of the ring causing it to be larger and likely explaining the baseline separation observed upon sodium addition. A similar trend was also observed for cyclic versions of fructose and glucose in their pyranose form, where the deprotonated structures look very similar, but the sodium ion binds to different positions on the ring for fructose and glucose. We realize that these glycans can have many different structural forms in solution (linear, furanose, pyranose, and α and β), but these initial calculations provide a small understanding of why the structures may be separating by IMS.
Fig. 1.
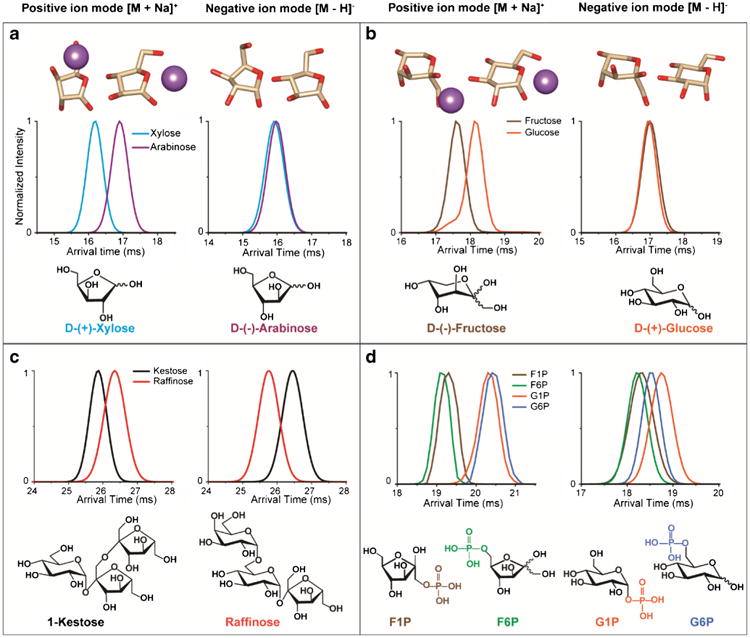
IMS profiles in both positive and negative ion mode for the a) pentoses, d-(+)-xylose and d-(–)-arabinose, b) hexoses, d-(–)-fructose and d-(+)-glucose, c) isomeric trisaccharides, 1-kestose and raffinose, and d) monosaccharide phospho-derivatives, (d)-fructose-1-phosphate (F1P), d-fructose 6-phosphate (F6P), α-d-glucose-1-phosphate (G1P), and d-(+)-glucose 6-phosphate (G6P). The lowest energy density functional theory structures with and without sodium are shown above d-(+)-xylose, d-(–)-arabinose, d-(–)-fructose, and d-(+)-glucose illustrating their deprotonated structural similarities (in negative mode) and differences upon sodium addition (in positive mode)
We also investigated other glycan isomers and found completely different scenarios where the glycans could be separated in negative mode but not positive mode (such as Fig. 5b which will be discussed later) and cases where the isomers inverted position in the different modes. For instance, the trisaccharides, 1-kestose (β-d-Fruf-(2 → 1)-β-d-Fruf-(2 → 1)-α-d-Glcp) and raffinose (α-d-Galp-(1 → 6)-α-d-Glcp-(1 → 2)-β-d-Fruf) showed very different IMS profiles in the different polarities (Fig. 1c). In the positive ion mode, raffinose traveled slower than kestose while the opposite scenario was observed in the negative ion mode. This trend was also observed for the four isomeric monosaccharide phospho-derivatives, fructose-1-phosphate (F1P), fructose-6-phosphate (F6P), glucose-1-phosphate (G1P), and glucose-6-phosphate (G6P), which are all important in glycolysis (Fig. 1d). In the positive ion mode, sodiated F1P and F6P had earlier arrival times than sodiated G1P and G6P, and the isomers appeared to group together by the size of their carbon ring (pentose or hexose). However, in the negative ion mode, F1P and F6P remained overlapping while G1P and G6P switched places and had better separation from each other, but less separation from F1P and F6P. These observations all indicate that glycan conformations can be significantly different in the two polarities and upon cation addition.
Fig. 5.
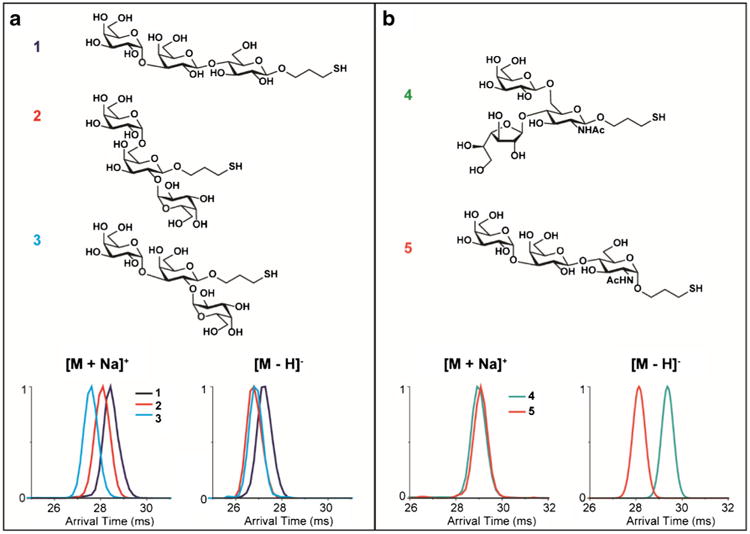
IMS separation of synthetic O-glycan isomers in both positive and negative ion modes for a) α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-β-d-Glc-(CH2)3SH (1), α-d-Gal-(1 → 6)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (2), and α-d-Gal-(1 → 3)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (3); and b) β-d-Galf-(1 → 4)-[β-d-Gal-(1 → 6)]-α-d-GlcNAc-(CH2)3SH (4) and α-d-Gal-(1→ 3)-β-d-Gal-(1 → 4)-α-d-GlcNAc-(CH2)3SH (5)
IMS trend lines for standard glycans
Since IMS profiles for glycans might change dramatically from positive to negative ion mode and use of both polarities could improve glycan isomer separation, we characterized 43 glycan standards in both modes by drift times and m/z (Fig. 2). A list of the glycan standards studied and their corresponding drift times from monosaccharides to hexasaccharides is included in Table S1 in the Electronic Supplementary Material (ESM). Overall, we were able to ionize standard glycans in both ion modes; however, it is noted that monosaccharides did not ionize as well in the negative mode. When the trendlines in the positive ion mode were analyzed (Fig. 2a), the sodiated forms of smaller saccharide isomers displayed better separation than their negative counterparts (Fig. 2b), suggesting the sodium ion had a significant effect on the small glycans' conformations. However, the higher order oligo-saccharides displayed improved separation in negative ion mode possibly due to the charge repulsion. In positive ion mode, sodium complexation caused the oligosaccharides to adopt more compact structures due to the presence of all the available oxygens throughout the structure conjugating with the sodium and folding the molecules. For example, the lactose-based hexasaccharides, lacto-N-hexaose (LNH, β-d-Gal-(1 → 3)-β-d-GlcNAc-(1 → 3)- [β-d-Gal-(1 → 4)-β-d-GlcNAc-(1 → 6)]-β-d-Gal-(1 → 4)-d-Glc) and lacto-N-neohexaose (LNnH, β-d-Gal-(1 → 4)-β-d-GlcNAc-(1 → 3)-[β-d-Gal-(1 → 4)-β-d-GlcNAc-(1 → 6)]-β-d-Gal-(1 → 4)-d-Glc), which are the largest m/z isomers on the plots, clearly illustrated this trend with baseline separation in negative mode, but much less distinction in positive mode.
Fig. 2.

IMS trend lines for standard glycans ranging from monosaccharides to hexasaccharides observed as a) sodiated in positive ion mode and b) deprotonated in negative ion mode. Both pentoses and hexoses are included. For the tetra-, penta-, and hexasaccharides, the lower m/z components correspond to standard glycans, while the higher molecular weight species are lactose-based O-glycans. The name and connectivity information of each glycan analyzed is illustrated in Table S1 in the ESM
IMS separation of glycan isomers with subtle structural differences
To further examine IMS separations of glycans, isomer pairs with subtle structural differences were studied with both polarities (Fig. 3). As shown in Fig. 3a, two pentasac-charides, maltopentaose [α-d-Glc-(1 → 4)]4-d-Glc and cellopentaose [β-d-Glc-(1 → 4)]4-d-Glc, with five d-glucose units linked by either an α- or β-linkage caused distinct IMS profiles. The arrival time for cellopentaose was observed to be shorter than that for maltopentaose in both positive and negative ion modes, allowing IMS separation. Oligosaccharides with the same type of linkage but at different positions were also distinguishable by IMS. For instance, the tetrasaccharides, lacto-N-tetraose (LNT, β-d-Gal-(1 → 3)-β-d-GlcNAc-(1 → 3)-β-d-Gal-(1 → 4)-d-Glc) and lacto-N-neotetraose (LNnT, β-d-Gal-(1 → 4)-β-d-GlcNAc-(1 → 3)-β-d-Gal-(1 → 4)-d-Glc), which both have β-glycosidic linkages but in LNT the non-reducing galactose unit has a 1,3-glycosidic linkage to N-acetyl-glucosamine, while LNnT has a 1,4-glycosidic linkage. As shown in Fig. 3b, LNT and LNnT have very different drift time profiles, where LNT travels much faster than LNnT in both positive and negative ion modes, illustrating a more compact structure. Note that these two tetrasaccharides showed almost baseline separations, although LNnT displayed a broader IMS peak in the negative ion mode, indicating LNnT may adopt multiple conformations. Similarly, oligosaccharides with linear and branched connectivities were also examined. When comparing linear mannopentaose (L_Man5, [β-d-Man-(1 → 4)]4-d-Man) with branched 3α,6α-mannopentaose (B_Man5, α-d-Man-(1 → 3)[α-d-Man-(1 → 6)]-α-d-Man-(1 → 6)[α-d-Man-(1 → 3)]-d-Man), L_Man5 displayed a shorter drift time than B_Man5 in both positive and negative ion modes indicating it was more compact than its branched counterpart (Fig. 3c). These observations indicate that IMS is able to reveal subtle structural differences both in positive and negative ion modes for standard glycans, including those with different linkages, connectivities, and configurations.
Fig. 3.
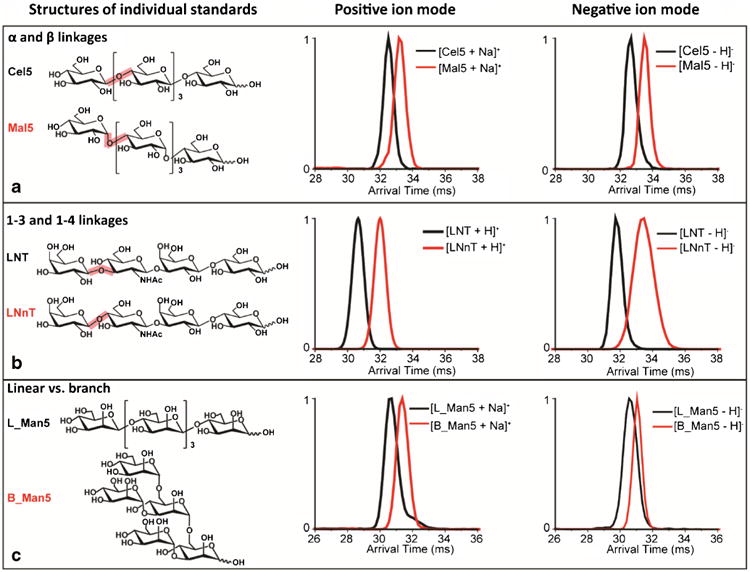
IMS separation of standard glycan isomers with subtle structure differences such as a) α- versus β-linkages: maltopentaose (Mal5) and cellopentaose (Cel5); b) 1–3 versus 1–4 connectivity: lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT); and c) linear versus branched oligosaccharides: linear and branched mannopentaose (L_Man5 and B_ Man5). The structures for each standard are shown in the left panel, and the IMS spectra in positive and negative ion modes are shown in the middle and right panels
Metal ions enable better separations of glycan isomers
While most glycan isomers were distinguishable in either the positive or negative ion mode, they were not baseline resolved in many cases due to their similar sizes and structures. For instance, although the protonated tetrasaccharides LNT and LNnT were almost baseline resolved (Fig. 3b), their sodiated forms displayed very similar IMS profiles and were inseparable (Fig. 4, top left panel). This again indicates that the addition of metal ions can dramatically change the conformations of glycans (and potentially their configuration in case of hemiacetals) as indicated by previous studies [35, 56–58]. Since many isomers have very similar structures, we further explored this mechanism for obtaining better separations. As shown in Fig. 4, the addition of sodium to LNT and LNnT resulted in similar IMS profiles. In contrast, the addition of potassium provided partial separation (Fig. 4, top left panel), indicating the potassium ion drives the two saccharides toward different conformations. Moreover, the addition of different divalent cations, e.g., Mn2+, Cu2+, or Zn2+, showed even better separation of the isomers. For instance, the addition of a Zn2+ ion significantly increased the structural separation of LNT and LNnT (Fig. 4, bottom left panel). Interestingly, LNT and LNnT with two sodium ions attached were baseline resolved (Fig. 4, bottom right panel), but in a different order than the K+ and Zn2+ separations. However, not all glycans bind two sodium ions, especially the smaller ones, so this will not work in all cases. Thus, these results illustrate that the addition of metal ions can significantly change the IMS profiles of glycan standards and can be used as an effective tool for separating isomers that have high structural similarity.
Fig. 4.
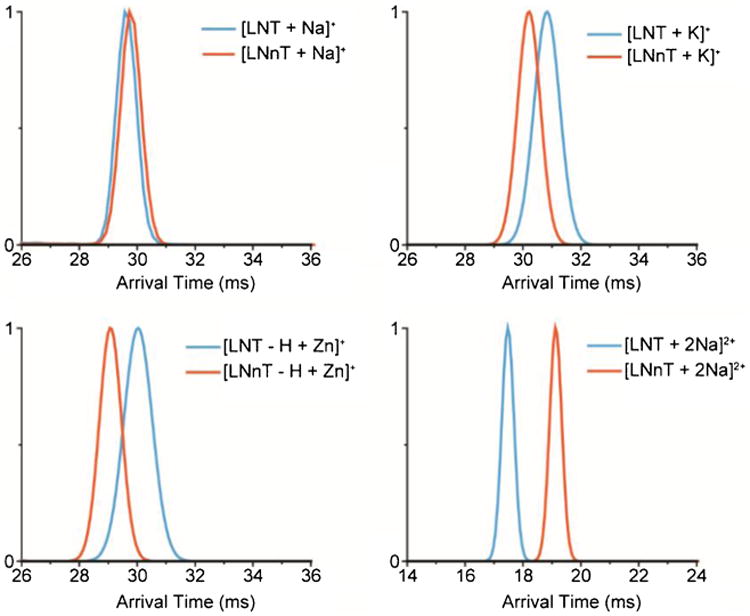
Metal ions were found to improve the separation of glycan isomers. The IMS spectra of the tetrasaccharides LNT and LNnT with one Na+, K+, Zn2+, and two Na+ ions are illustrated
Separating Chagas disease-related synthetic α-Gal-containing O-glycans
The glycan standard results demonstrated that positive and negative mode structural profiles and the addition of metal ions can lead to significant changes in glycan conformations and enable better isomer separations. Using this knowledge, the IMS-MS platform was applied to biologically relevant synthetic α-Gal-containing O-glycans that are important targets for protective anti-α-Gal antibodies against the parasite T. cruzi, which causes Chagas disease. The surface of the protozoan parasite T. cruzi is coated by glycoproteins, which contain highly immunogenic O-glycans. Among these glycotopes, the trisaccharide α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-α-d-GlcNAc is highly expressed on the O-glycosylated glycosylphosphatidylinositol-anchored mucins (GPI-mucins) in the T. cruzi trypomastigote stage but not in human cells [12]. Thus, it is highly immunogenic to humans and, moreover, triggers high levels of protective anti-α-Gal-antibodies (Abs) in infected individuals [12–14, 16, 59]. Although O-glycans of GPI-mucins from the insect-derived parasite forms have been well characterized in several parasite strains and genotypes, the exact structural information of most O-glycans for the mammal-dwelling tGPI-mucins remains unknown. Partial structural analysis and immunoassays have revealed that many of these trypomastigote-derived GPI-mucin (tGPI-mucin) O-glycans contain a terminal α-Gal residue, which is non-reducing and conserved on tGPI-mucins from at least four major Chagas disease causing T. cruzi genotypes [15, 60, 61]. These tGPI-mucin glycans are predominantly branched, and highly heterogeneous with different connectivities of the terminal α-Gal moiety to another sugar unit [12, 62]. Understanding these O-glycan structures is of importance for the identification of potential T. cruzi α-Gal-containing biomarkers for the diagnosis of Chagas disease, follow-up of chemotherapy, and the development of preventative and therapeutic vaccines for Chagas disease. The synthetic glycans analyzed were mercaptopropyl glycosides, where mercaptopropyl groups were installed to allow conjugation of these glycans to maleimide-activated bovine serum albumin needed for the generation of a glycan array [36, 37].
Three trisaccharide O-glycan isomers with different connectivities were initially studied: α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-β-d-Glc-(CH2)3SH (1), α-d-Gal-(1 → 6)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (2), and α-d-Gal-(1 → 3)-[α-d-Gal-(1 → 2)]-β-d-Gal-(CH2)3SH (3). Each isomer displayed a distinct IMS profile (Fig. 5a) with the arrival time for 1 being longer than 2 or 3 in both positive and negative ion modes, suggesting 1 had a more extended structure than the others. The trisaccharides 2 and 3 on the other hand had very similar negative mode IMS profiles and were hardly separated. However, in the positive ion mode, they could be distinguished, suggesting the addition of a sodium ion changed the conformation of one or both of the glycans. In Fig. 5b, another set of isomeric trisaccharide O-glycans, β-d-Galf-(1 → 4)-[β-d-Gal-(1 → 6)]-α-d-GlcNAc-(CH2)3SH (4) and α-d-Gal-(1 → 3)-β-d-Gal-(1 → 4)-α-d-GlcNAc-(CH2)3SH (5), illustrated very similar positive arrival time distributions for their sodiated forms. However, they were almost baseline resolved in the negative ion mode, which was consistent with the glycan standard results for Fig. 1 showing that different polarities may affect glycan structures distinctly. These glycans acted similarly to the linear and branched mannose-containing pentasaccharides L_Man5 and B_Man5 (Fig. 3c), where in negative ion mode the branched trisaccharide 4 traveled slower than the linear isomer 5 (Fig. 5b, bottom right panel). Furthermore, when 4 and 5 were mixed, they showed one broad peak in the positive ion mode while in the negative ion mode baseline separation was achieved (see ESM Fig. S1).
Because conformational changes caused by sodium addition have been observed in both O-glycan isomer groups, we applied different metal ions (K+, Rb+, Cu2+, and Zn2+) to those that did not separate well in either polarity, i.e., the disaccha-ride O-glycan isomers α-l-Rha-(1 → 2)-α-l-Fuc-(CH2)3SH (6) and α-l-Rha-(1 → 3)-α-l-Fuc-(CH2)3SH (7) (Fig. 6, top panel). For these two isomers, the addition of a potassium or rubidium monovalent ion showed conformational effects similar to that of sodium, which did not significantly improve isomer separation. However, divalent cations like copper and zinc showed great enhancement in the isomer separation, with zinc having the most effect. These results are consistent with the glycan standards shown in Fig. 4 and further indicate that the use of different metal ions allows better separations for distinct glycan isomers. Thus, specific metal ions could be used to target and baseline separate certain isomers in later focused studies.
Fig. 6.

Metal ions enable better separations for the disaccharide O-glycans α-l-Rha-(1 → 2)-α-l-Fuc-(CH2)3SH (6) and α-l-Rha-(1 → 3)-α-l-Fuc-(CH2)3SH (7)
Conclusions
In this work, we have applied a new rapid SPE-IMS-QTOF-MS platform to characterize glycan standards with subtle structural differences caused by different configurations of glycosidic linkages (α or β) and different linear or branched connectivities. We found that the IMS profiles of glycans can change dramatically from positive to negative ion mode, and a specific polarity might achieve better separations or in some cases both are needed for confident identifications. Moreover, we have shown that the addition of metal ions can lead to significant changes in glycan conformations (and potentially configurations in case of reducing end hemiacetals) and enable baseline isomer separations through metal complexation. Using all of these capabilities, biologically relevant synthetic αGal-containing O-glycans, which are important targets for the identification of immunodominant T. cruzi glycotopes, were also separated illustrating the potential for IMS-MS in biological and structural studies. Our results strongly suggest that IMS-MS is a powerful tool for identifying glycans of biological origin and distinguishing them from their isomers. Technical advances are also in progress to allow even better IMS glycan separations [63]; however, at present standards will initially be needed for confident identification of each glycan isomer.
Supplementary Material
Acknowledgments
Portions of this research were supported by grants from the National Institute of Environmental Health Sciences of the NIH (R01ES022190) (ESB), NIH grant R21CA199744 (KT), NIH grants R21AI079618 and R21AI115451 (ICA and KM), Robert J. Kleberg Jr. and Helen C. Kleberg Foundation grant (ICA and KM), Bridges to the Doctorate scholarship (NSF grants HRD-1139929) (NSS), Biomolecule Analysis Core Facility at UTEP, NIHMD grant G12MD007592, National Institute of General Medical Sciences grants P41 GM103493 (RDS) and P41 GM104603 (CEC), the Laboratory Directed Research and Development Program, and the Microbes in Transition (MinT) Initiative at Pacific Northwest National Laboratory. This research utilized capabilities developed by the Pan-omics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program). This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL01830.
Footnotes
Electronic supplementary material: The online version of this article (doi:10.1007/s00216-016-9866-4) contains supplementary material, which is available to authorized users.
Compliance with ethical standards: Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Bertozzi CR, Rabuka D, Varki A. Essentials of Glycobiology. 2009 [PubMed] [Google Scholar]
- 2.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat Methods. 2005;2(11):817–24. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 3.Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004;3(10):863–73. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 4.Kleene R, Schachner M. Glycans and neural cell interactions. Nat Rev Neurosci. 2004;5(3):195–208. doi: 10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- 5.Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73(1):491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- 6.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43R–56. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 7.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3(2):97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3(6):313–20. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 9.Dwek RA. Glycobiology: toward understanding the function of sugars. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 10.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7(7):537–51. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 12.Almeida IC, Ferguson MA, Schenkman S, Travassos LR. Lytic anti-α-galactosyl antibodies from patients with chronic Chagas' disease recognize novel O-linked oligosaccharides on mucin-like glycosyl-phosphatidylinositol-anchored glycoproteins of Trypanosoma cruzi. Biochem J. 1994;304(3):793–802. doi: 10.1042/bj3040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazzinelli RT, Pereira ME, Romanha A, Gazzinelli G, Brener Z. Direct lysis of Trypanosoma cruzi: a novel effector mechanism of protection mediated by human anti-gal antibodies. Parasite Immunol. 1991;13(4):345–56. doi: 10.1111/j.1365-3024.1991.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 14.Almeida IC, Milani SR, Gorin PA, Travassos LR. Complement-mediated lysis of Trypanosoma cruzi trypomastigotes by human anti-alpha-galactosyl antibodies. J Immunol. 1991;146(7):2394–400. [PubMed] [Google Scholar]
- 15.Almeida IC, Krautz GM, Krettli AU, Travassos LR. Glycoconjugates of Trypanosoma cruzi: a 74 kD antigen of trypomastigotes specifically reacts with lytic anti-alpha-galactosyl antibodies from patients with chronic Chagas disease. J Clin Lab Anal. 1993;7:307–16. doi: 10.1002/jcla.1860070603. [DOI] [PubMed] [Google Scholar]
- 16.Milani SR, Travassos LR. Anti-alpha-galactosyl antibodies in chagasic patients. Possible biological significance. Braz J Med Biol Res. 1988;21(6):1275–86. [PubMed] [Google Scholar]
- 17.Boltje TJ, Buskas T, Boons GJ. Opportunities and challenges in synthetic oligosaccharide and glycoconjugate research. Nat Chem. 2009;1(8):611–22. doi: 10.1038/nchem.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings Richard D, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21(1):1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasnova L, Wong CH. Understanding the chemistry and biology of glycosylation with glycan synthesis. Annu Rev Biochem. 2016;85:599–630. doi: 10.1146/annurev-biochem-060614-034420. [DOI] [PubMed] [Google Scholar]
- 20.Duus JO, Gotfredsen CH, Bock K. Carbohydrate structural determination by NMR spectroscopy: modern methods and limitations. Chem Rev. 2000;100:4589–614. doi: 10.1021/cr990302n. [DOI] [PubMed] [Google Scholar]
- 21.Venetz D, Hess C, Lin CW, Aebi M, Neri D. Glycosylation profiles determine extravasation and disease-targeting properties of armed antibodies. Proc Natl Acad Sci. 2015;112(7):2000–5. doi: 10.1073/pnas.1416694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–6. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 23.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6(10):713–23. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 24.Wyttenbach T, Bowers M. Gas-Phase Conformations: The Ion Mobility/Ion Chromatography Method. In: Schalley C, editor. Modern Mass Spectrometry Topics in Current Chemistry. Springer; Berlin Heidelberg: 2003. pp. 207–32. [Google Scholar]
- 25.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6(4):281–94. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 26.Baker ES, Burnum-Johnson KE, Ibrahim YM, Orton DJ, Monroe ME, Kelly RT, et al. Enhancing bottom-up and top-down proteomic measurements with ion mobility separations. Proteomics. 2015;15(16):2766–76. doi: 10.1002/pmic.201500048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–55. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 28.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, et al. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86(4):2107–16. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray CJ, Thomas B, Upton R, Migas LG, Eyers CE, Barran PE, et al. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochimica et Biophysica Acta (BBA) - General Subjects. 2006;1860(8):1688–709. doi: 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhu M, Bendiak B, Clowers B, Hill HH., Jr Ion mobility-mass spectrometry analysis of isomeric carbohydrate precursor ions. Anal Bioanal Chem. 2009;394:1853–67. doi: 10.1007/s00216-009-2865-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenn LS, McLean JA. Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys. 2011;13:2196–205. doi: 10.1039/c0cp01414a. [DOI] [PubMed] [Google Scholar]
- 32.Both P, Green AP, Gray CJ, Šardzík R, Voglmeir J, Fontana C, et al. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat Chem. 2014;6(1):65–74. doi: 10.1038/nchem.1817. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann J, Hahm HS, Seeberger PH, Pagel K. Identification of carbohydrate anomers using ion mobility-mass spectrometry. Nature. 2015;526(7572):241–4. doi: 10.1038/nature15388. [DOI] [PubMed] [Google Scholar]
- 34.Pagel K, Harvey DJ. Ion mobility-mass spectrometry of complex carbohydrates: collision cross sections of sodiated N-linked glycans. Anal Chem. 2013;85(10):5138–45. doi: 10.1021/ac400403d. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Dodds ED. Discrimination of isomeric carbohydrates as the electron transfer products of Group II Cation Adducts by ion mobility spectrometry and tandem mass spectrometry. Anal Chem. 2015;87(11):5664–8. doi: 10.1021/acs.analchem.5b00759. [DOI] [PubMed] [Google Scholar]
- 36.Ashmus RA, Schocker NS, Cordero-Mendoza Y, Marques AF, Monroy EY, Pardo A, et al. Potential use of synthetic [small alpha]-galactosyl-containing glycotopes of the parasite Trypanosoma cruzi as diagnostic antigens for Chagas disease. Org Biomol Chem. 2013;11(34):5579–83. doi: 10.1039/c3ob40887f. [DOI] [PubMed] [Google Scholar]
- 37.Schocker NS, Portillo S, Brito CRN, Marques AF, Almeida IC, Michael K. Synthesis of Galα(1,3)Galβ(1,4)GlcNAcα-, Galβ(1, 4)GlcNAcα- and GlcNAc-containing neoglycoproteins and their immunological evaluation in the context of Chagas disease. Glycobiology. 2016;26(1):39–50. doi: 10.1093/glycob/cwv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang K, Shvartsburg AA, Lee HN, Prior DC, Buschbach MA, Li F, et al. High-sensitivity ion mobility spectrometry/mass spectrometry using electrodynamic ion funnel interfaces. Anal Chem. 2005;77(10):3330–9. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibrahim Y, Tang K, Tolmachev AV, Shvartsburg AA, Smith RD. Improving mass spectrometer sensitivity using a high-pressure electrodynamic ion funnel interface. J Am Soc Mass Spectrom. 2006;17(9):1299–305. doi: 10.1016/j.jasms.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belov ME, Clowers BH, Prior DC, Danielson WF, III, Liyu AV, Petritis BO, et al. Dynamically multiplexed ion mobility time-of-flight mass spectrometry. Anal Chem. 2008;80(15):5873–83. doi: 10.1021/ac8003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clowers BH, Ibrahim YM, Prior DC, Danielson WF, Belov ME, Smith RD. Enhanced ion utilization efficiency using an electrodynamic ion funnel trap as an injection mechanism for ion mobility spectrometry. Anal Chem. 2008;80(3):612–23. doi: 10.1021/ac701648p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pence HE, Williams A. ChemSpider: an online chemical information resource. J Chem Educ. 2010;87(11):1123–4. [Google Scholar]
- 43.Csizmadia JSaF., editor. A method for calculating the pKa values of small and large molecules. American Chemical Society Spring meeting; March 25-29th, 2007. [Google Scholar]
- 44.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17(5–6):490–519. [Google Scholar]
- 45.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform. 2012;4(1):1–17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, Van Dam HJJ, et al. NWChem: a comprehensive and scalable open-source solution for large scale molecular simulations. Comput Phys Commun. 2010;181(9):1477–89. [Google Scholar]
- 47.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98(7):5648–52. [Google Scholar]
- 48.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37(2):785–9. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 49.Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: a critical analysis. Can J Phys. 1980;58(8):1200–11. [Google Scholar]
- 50.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J Phys Chem. 1994;98(45):11623–7. [Google Scholar]
- 51.Francl MM, Pietro WJ, Hehre WJ, Binkley JS, Gordon MS, DeFrees DJ, et al. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for second-row elements. J Chem Phys. 1982;77(7):3654–65. [Google Scholar]
- 52.Hariharan PC, Pople JA. The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chim Acta. 1973;28(3):213–22. [Google Scholar]
- 53.Rassolov VA, Ratner MA, Pople JA, Redfern PC, Curtiss LA. 6-31G* basis set for third-row atoms. J Comput Chem. 2001;22(9):976–84. [Google Scholar]
- 54.Marklund Erik G, Degiacomi Matteo T, Robinson Carol V, Baldwin Andrew J, Benesch Justin LP. Collision cross sections for structural proteomics. Structure. 2015;23(4):791–9. doi: 10.1016/j.str.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 55.Feinman RD, Fine EJ. Fructose in perspective. Nutr Metab. 2013;10(1):1–12. doi: 10.1186/1743-7075-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y, Dodds ED. Ion mobility studies of carbohydrates as group I adducts: isomer specific collisional cross section dependence on metal ion radius. Anal Chem. 2013;85(20):9728–35. doi: 10.1021/ac402133f. [DOI] [PubMed] [Google Scholar]
- 57.Huang Y, Dodds ED. Ion-neutral collisional cross sections of carbohydrate isomers as divalent cation adducts and their electron transfer products. Analyst. 2015;140(20):6912–21. doi: 10.1039/c5an01093d. [DOI] [PubMed] [Google Scholar]
- 58.Gaye MM, Nagy G, Clemmer DE, Pohl NLB. Multidimensional analysis of 16 glucose isomers by ion mobility spectrometry. Anal Chem. 2016;88(4):2335–44. doi: 10.1021/acs.analchem.5b04280. [DOI] [PubMed] [Google Scholar]
- 59.Avila JL, Rojas M, Galili U. Immunogenic Gal alpha 1–3Gal carbohydrate epitopes are present on pathogenic American Trypanosoma and Leishmania. J Immunol. 1989;142(8):2828–34. [PubMed] [Google Scholar]
- 60.Soares RP, Torrecilhas AC, Assis RR, Rocha MN, Castro FAM, Freitas GF, et al. Intraspecies variation in Trypanosoma cruzi GPI-mucins: biological activities and differential expression of α-galactosyl residues. Am J Trop Med Hyg. 2012;87(1):87–96. doi: 10.4269/ajtmh.2012.12-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Izquierdo L, Marques AF, Gallego M, Sanz S, Tebar S, Riera C, et al. Evaluation of a chemiluminescent enzyme-linked immunosorbent assay for the diagnosis of Trypanosoma cruzi infection in a nonendemic setting. Mem Inst Oswaldo Cruz. 2013;108:928–31. doi: 10.1590/0074-0276130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buscaglia CA, Campo VA, Frasch ACC, Di Noia JM. Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nat Rev Microbiol. 2006;4(3):229–36. doi: 10.1038/nrmicro1351. [DOI] [PubMed] [Google Scholar]
- 63.Deng L, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Zhang X, et al. Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. ChemistrySelect. 2016;1(10):2396–9. doi: 10.1002/slct.201600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


